Abstract
p53 is essential for the cellular responses to DNA damage that help to maintain genomic stability. However, the great majority of human cancers undergo disruption of the p53-network. Identification and characterization of molecular components important in both p53-dependent and -independent apoptosis might be useful in developing novel therapies for cancers. In the complete absence of p53, cells treated with N-(phosphonacetyl)-L-aspartate (PALA) continue to synthesize DNA slowly and eventually progress through S phase, suffering severe DNA damage that in turn triggers apoptosis, whereas cells with functional p53 undergo growth arrest. In the present study, we investigated apoptotic signaling in response to PALA and the role of p53 expression in this pathway. We found that treatment of cells lacking p53 with PALA induced TAp73, Noxa, and Bim and inactivation of these proteins with dominant negative plasmids or siRNAs significantly inhibited apoptosis, suggesting that PALA-induced apoptosis was mediated via TAp73-dependent expression of Noxa and Bim. However, PALA treatment inhibited the expression of ΔNp73 only in cells lacking p53 but not in cells expressing p53. In addition, PALA treatment inhibited Bcl-2, and overexpression of Bcl-2 significantly inhibited PALA-induced apoptosis. Moreover, expression of p53 in these cells protected them from PALA-induced apoptosis by activating p21, sustaining the expression of ΔNp73 and inhibiting the induction of Noxa and Bim. Taken together, our study identifies novel but opposing roles for the p53 and TAp73 in the induction of Noxa and Bim and regulation of apoptosis. Our data will help to develop strategies to eliminate cancer cells lacking p53 while protecting normal cells with wild-type p53.
Keywords: DNA damage, apoptosis, signal transduction, aspartate transcarbamylase inhibitor, p53 family, replicative stress
Introduction
p53 plays a pivotal role in the pathways that prevent the development of cancer by inducing apoptosis, DNA repair and cell-cycle arrest in response to different types of cellular stress (1–3). p53 is also critical for the action of many chemotherapy and chemopreventive drugs (4–7). Unfortunately, the majority of tumors harbor mutations affecting the p53 gene, and those tumors that have wild-type p53 protein most probably lack a functional p53 response as a result of mutations affecting other genes that function in the same pathways as p53 (8, 9). In addition to p53, mammalian cells contain two closely related proteins, p63 and p73, which share overlapping functions with p53 and regulate cell cycle arrest and apoptosis (10–14). However, some of the genes transcribed by p73 are specific for p73 (15). Because of the presence of an alternative promoter in the third intron and due to alternative splicing at the C-terminal end, p73 protein has multiple isoforms with distinct functions (14). Usage of the most upstream P1 promoter generates the transactivating (TA) isoforms, whereas that of the downstream P2 promoter yields amino-terminal truncated proteins (ΔNp73) that lack the TA domain. At least 7 different transcripts (α, β, γ, ζ, δ, ε, η) are known to be generated due to alternative splicing at the C-terminus. Another group of ΔNp73 isoforms, known as ΔTAp73, were generated from alternative splicing targeting the N-terminus of the transcript generated from the P1 promoter (14). Studies suggest that p73 plays essential roles in determining cellular sensitivity to many anticancer drugs, particularly in tumors lacking functional p53 (12, 13, 16–18). However, TAp73 and ΔNp73 have opposing roles. Although TAp73 induces cell cycle arrest and apoptosis, ΔNp73 functions as a dominant negative for TAp73 and promotes survival and has an oncogenic function.
Unlike p53, mutations in p73 are extremely rare in human cancers (14, 16, 19–21). However, the expression of p73 is frequently dysregulated in human tumors (14). Overexpression of p73 mRNA and/or protein was observed in breast (22, 23), ovarian (24), hepatocellular (25), neuroblastoma (26), bladder (27), prostate (28) and colorectal (29) cancers. On the other hand, frequent loss of p73 was observed in pancreatic adenocarcinoma (30). Isoform-specific differences in the expression of p73 were also observed between normal and malignant tissues; α and β are the predominant isoforms in normal tissues whereas other C-terminal splice variants were expressed in tumor tissues. Silencing of p73 due to the loss of allele 1 at 1p36 and promoter methylation or mutation was found in neuroblastoma, acute lymphoblastic leukemia, and Burkitt’s and non-Hodgkin lymphomas. In most thyroid carcinomas (31) and B-cell chronic leukemias (32), both TA and δN isoforms are overexpressed. In contrast, expression of γ, δ, ε, and θ isoforms is also strongly upregulated in acute myeloid leukemia and chronic myeloid leukemia (33). Expression of ΔNp73 isoforms were also increased in head and neck cancers (34).
N-(phosphonacetyl)-L-aspartate (PALA) is a potent and reversible inhibitor of pyrimidine nucleotide synthesis in cells (35). Upon treatment with PALA, cells containing wild-type p53 undergo p53-dependent protective arrest either at G1 and G2 checkpoints or in S-phase, depending on the levels of p53 in the cells (36–39). In contrast, cells lacking p53 continue through the cell cycle and incur severe DNA damage, followed by apoptosis (40). Since most cancer cells lack p53, PALA holds strong promise as a chemotherapeutic agent targeting cancer cells lacking p53 while protecting normal cells with wild-type p53. Previous studies from our laboratory have shown that restoration of wild-type p53 in cells lacking it restored the normal response to PALA (36, 37, 39). However, the signaling pathways that execute cell death in response to PALA remain to be elucidated.
Activation of caspases is a prerequisite for most apoptosis. Caspases can be activated by either of two major pathways: the extrinsic pathway, triggered by death receptors such as Fas, DR5, DR4, and the intrinsic pathway, triggered by cellular stresses (cytokine deprivation, DNA damage) (41, 42). The Bcl2 family of proteins, which can be subdivided into two major classes, plays a critical role in the intrinsic pathway of apoptosis (43–46). The first group consists of the anti-apoptotic members (Bcl2, Bcl-xL, Bcl-w and Mcl-1), overexpression of which can prevent apoptosis (43). The second group, or the proapoptotic Bcl2 members, is divided into effector proteins and the BH3-only proteins. The effector group consists of the pro-apoptotic Bcl2 family members, Bcl2-associated X protein (Bax) and Bcl2 antagonist/killer (Bak), deletion of which renders cells highly resistant to apoptosis. The BH3-only pro-apoptotic subgroup includes Bim, Bad, Puma and Noxa, all of which contain a BH3 domain (45, 46). Members of this group promote apoptosis when overexpressed, by inhibiting anti-apoptotic members and/or directly activating Bax/Bak (45, 46). Bim is one of the most potent proapoptotic BH3-only proteins and binds to all prosurvival Bcl2 family members with high affinity (47). Bim has three isoforms generated through alternative splicing. All three Bim isoforms namely BimEL, BimL and BimS induce apoptosis. However, the shorter isoforms are more potent inducers of apoptosis than the longer isoforms (48) and the basal expression of different Bim isoforms varies among cancer cell types (49). It was reported previously that Bim is a direct transcriptional target of p73 (50, 51) although p53 negatively regulates the expression of Bim (52, 53).
In the current study, we showed for the first time that the p53 family member p73 plays an essential role in PALA-induced apoptosis by transcriptionally upregulating the proapoptotic BH3-only proteins Bim and Noxa and downregulating Bcl-2 in cells lacking p53. Rescue of p53 functions in these cells negatively regulates the expression of Bim and Noxa and has a cytoprotective effect.
Results
p53 and TAp73 play opposing roles in PALA-induced apoptosis
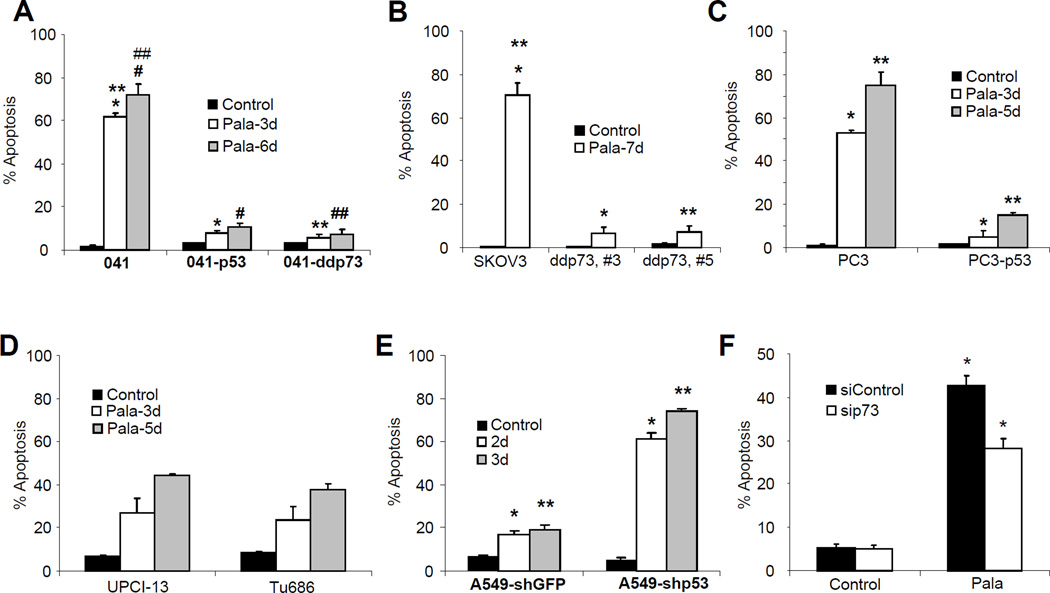
It was previously established that in the complete absence of p53, PALA-treated cells continue to synthesize DNA slowly and eventually progress through S phase, suffering severe DNA damage that in turn triggers apoptosis (40). In contrast, PALA-treated cells expressing p53 underwent growth arrest without apoptosis (36–39). In order to study the signaling pathways that trigger apoptosis in response to imbalance in DNA precursors, we have employed several cell lines expressing wild-type p53, mutant p53 or no p53 and their isogenic pairs with reconstituted p53 or expressing dominant negative p73 (Materials and Methods). Expression of dominant negative p73 in 041 and SKOV3 cells was confirmed by Western blotting using T7 antibody (Supplementary Fig. 1A and B). All the cells were treated with 250 µM PALA for different time periods, fixed in 70% ethanol, and labeled with fluorescein-tagged bromodeoxy-uridine triphosphate (Br-dUTP) and propidium iodide for apoptosis assay (TUNEL). As expected, all cells lacking p53 underwent time-dependent massive apoptosis in response to PALA and those with wild-type p53 were protected (Fig. 1, A–C). Interestingly, cells expressing dominant negative p73 were also protected significantly (p values <0.05) from PALA-induced apoptosis (Fig. 1, A and B). Cells expressing mutant p53 were also sensitive to PALA-induced apoptosis, but to a lesser extent as compared to cells lacking p53 (Fig. 1D), most probably due to inhibition of TAp73 by mutant p53 (54). To further confirm the protective role of p53 in PALA-induced apoptosis, we knocked down p53 in A549 cells which express wild-type p53 using lentivirus-mediated shRNA (55). A549 cells transduced with control and p53 shRNA were treated with PALA and apoptosis was measured (Fig. 1E). A549 cells transduced with shGFP were almost resistant to PALA, whereas ablation of p53 drastically increased apoptosis, further confirming the protective role of p53. We also confirmed the proapoptotic role of TAp73 by ablating TAp73 expression in SKOV3 cells using siRNA (supplementary Fig. S1C). Ablation of p73 significantly inhibited apoptosis (Fig. 1F). Taken together, these results suggest that the family members p53 and TAp73 play opposing roles in response to PALA; p53 protects cells, whereas TAp73 mediates cells death. Since cells expressing dominant negative p73 do not undergo apoptosis, we examined whether PALA treatment induces growth inhibition or cell cycle arrest of these cells. Although cells expressing dominant negative p73 initially accumulated at G1, this was transient and cells started to grow again (supplementary Fig. S2). However, the growth rate is little slower than the parental cells.
Figure 1.
p53 and TAp73 play opposing roles in PALA-induced apoptosis. (A) 041 and its derivatives expressing either wild-type p53 (041-p53) or dominant negative p73 (041-ddp73) were treated with 250 µM/L of PALA for 3 and 6 days and apoptosis was measured by TUNEL staining. p values for *, **, # and ## are 0.0008, 0,0011, 0.0018 and 0.0013, respectively. (B) Ovarian cancer cell line SKOV3 and two clones expressing dominant negative p73 (ddp73, #3 and ddp73, #5) were treated with 250 µM/L of PALA for 7 days and apoptosis was measured by TUNEL staining. p values for * and ** are 0.0011 and 0.0035, respectively. (C) Prostate cancer cell line PC3 and isogenic cells with wild-type p53 (PC3-p53) were treated with 250 µM/L of PALA for 3 and 5 days and the apoptotic population was measured by TUNEL staining. p values for * and ** are 0.0005 and 0.003, respectively. (D) PCI-13 and Tu686 cells expressing mutant p53 were treated with 250 µM/L of PALA for 3 and 5 days and apoptosis was measured. (E) A549 and its derivative with ablated p53 were treated with 250 µM/L of PALA for 2 and 3 days and apoptosis was measured. p values for * and ** are 0.0039 and 0.0001, respectively. (F) SKOV3 cells transfected with control siRNA and siRNA targeting TAp73 were treated with 250 µM/L of PALA for 3 days and apoptosis was measured. *=0.003. For A–F, average results from three independent experiments with standard deviations as error bars are shown. p values were determined by Student’s T-test and values less than 0.05 were considered statistically significant.
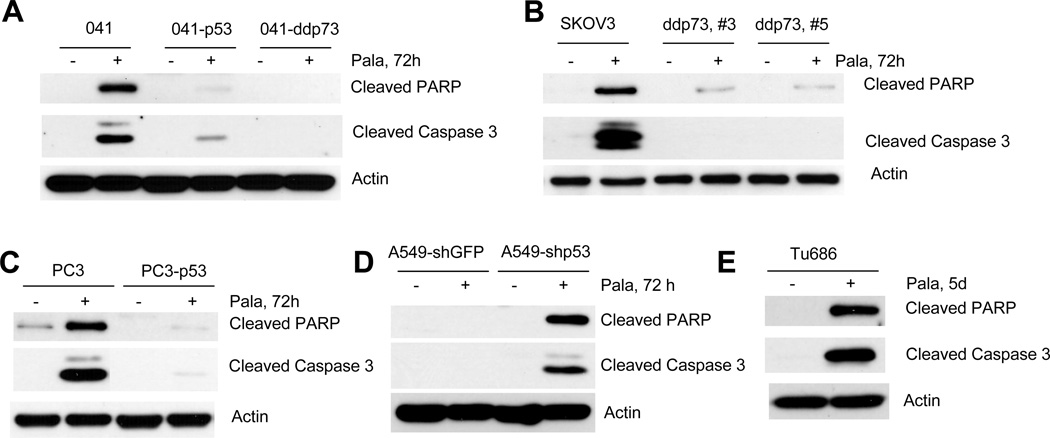
In order to further confirm the flow cytometry results, we treated cells with PALA and total cell lysates were immunoblotted with anti-PARP and anti-caspase-3 using antibodies specific for their cleaved forms. As shown in Fig. 2A–2E, PALA induced PARP and caspase-3 degradation in cells lacking p53 or expressing mutant p53, and this process was strongly inhibited in cells expressing dominant negative p73 or wild-type p53. Moreover, ablation of p53 in cells expressing wild-type p53 rescued PALA-induced PARP and caspase-3 cleavage (Fig. 2D). These results also suggest that PALA-induced apoptosis is mediated by TAp73 and that p53 protects cells from PALA-induced apoptosis.
Figure 2.
Cleavage of PARP and caspase-3 by PALA: (A) 041 and its derivatives 041-p53 and 041-ddp73; (B) SKOV3 and its derivatives ddp73, #3 and ddp73, #5; (C) PC3 and PC3-p53; (D) A549-shGFP and A549-shp53 and (E) Tu686 cells were treated with 250 µM/L of PALA for the indicated times and total cell lysates were immunoblotted with PARP and caspase-3 antibodies that detect only the cleaved form of PARP and caspase-3. All experiments were repeated three times for reproducibility and representative data are shown.
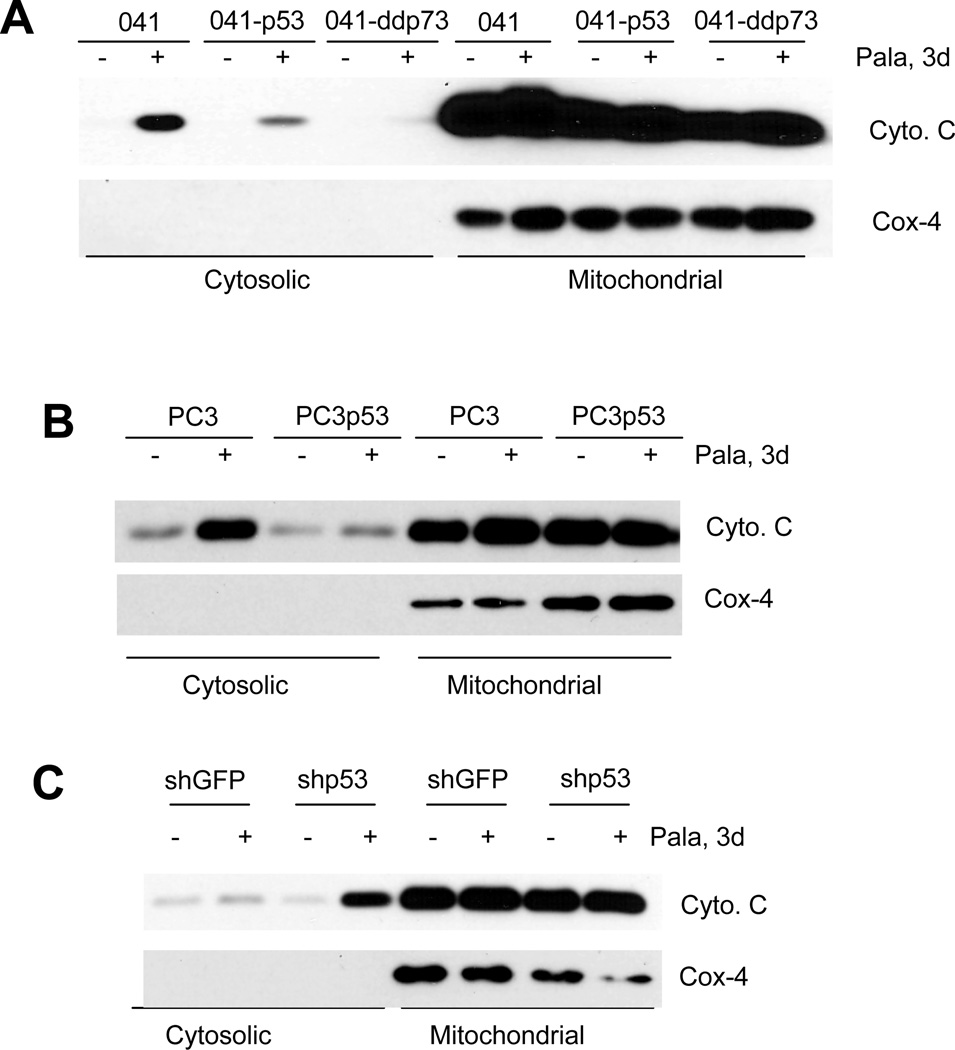
We next examined whether PALA induced mitochondria-dependent or -independent apoptosis by checking the release of cytochrome c from mitochondria to the cytoplasm. Mitochondrial and cytoplasmic fractions were separated as described previously (55). As shown in Fig. 3A–C, treatment with PALA efficiently released cytochrome c into the cytoplasm in cells lacking p53. In contrast, the release of cytochrome c into the cytoplasm was inhibited in cells expressing dominant negative p73 or wild-type p53. The efficiency of fractionation was confirmed by the lack of expression of COX4, a mitochondrial protein, in the cytoplasmic fractions. This result suggests that PALA-induced apoptosis is mediated via mitochondrial membrane depolarization and further confirms that p53 and TAp73 play opposing roles in PALA-induced apoptosis: TAp73-mediates apoptosis, whereas p53 protects from apoptosis.
Figure 3.
Release of cytochrome C by PALA treatment: (A) 041 and its derivatives 041-p53 and 041-ddp73; (B) PC3 and pC3-p53; and (C) A549-shGFP and A549-shp53 cells were treated with 250 µM/L of PALA for 72 h and expression of cytochrome c and Cox4 in the cytosolic and mitochondrial fractions were examined after cytoplasmic and mitochondrial fractionation. Representative results from three independent experiments are shown.
Activation of p53 and TAp73 by PALA
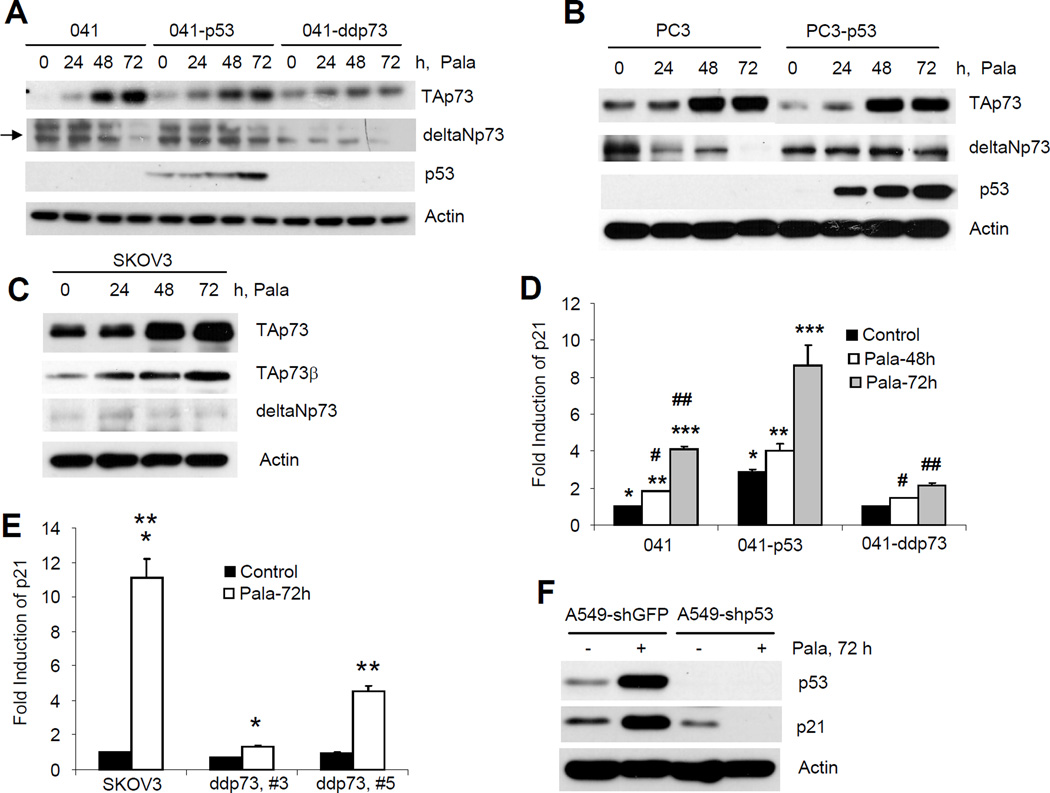
Since cells expressing dominant negative p73 or wild-type p53 are resistant to PALA-induced apoptosis, we next examined the expression of TAp73 using an antibody that specifically detects all TA isoforms but not the ΔN isoforms (Materials and Methods) in 041, PC3, SKOV3 and their derivative cell lines. As shown in Fig. 4A–C, treatment with PALA efficiently induced the expression of TAp73 protein. Expression of p53 had little effect on TAp73 expression. In 041 and PC3 cells, PALA increased the TAp73 protein, which has a molecular weight ~42kDa (corresponds to the molecular weight of TAp73δ). In SKOV3 cells, the molecular weight of TAp73 induced by PALA was ~68kDa (corresponds to the molecular weight of β-isoform). We next confirmed that the isoform induced in SKOV3 cells was indeed TAp73β using an antibody that specifically detects the β-isoform (Fig. 4C). We also examined the expression of ΔNp73 using an antibody that does not cross-react with TAp73 isoforms. All cells expressed ΔNp73, which was decreased after PALA treatment in cells lacking p53; however, in cells expressing wild-type p53, the expression of ΔNp73 was sustained (Fig. 4A–C). Moreover, PALA treatment efficiently induced the expression of p53 (Fig. 4A, B and F). To confirm transcriptional activation of TAp73 and p53, we measured p21 mRNA, a known transcriptional target of p53 family proteins by real-time PCR in 041, SKOV3 cells and their derivatives expressing p53 and dominant negative p73. As shown in Fig. 4D, expression of p53 significantly increased the basal and PALA-induced level of p21. In contrast, expression of dominant negative p73 had an insignificant effect on the basal level of p21. PALA treatment significantly increased the expression of p21 in parental cells which was significantly inhibited after inactivation of p73 (Fig. 4D). TAp73-dependent p21 expression was also confirmed in SKOV3 cells (Fig. 4E). We also measured p21 expression in A549 cells transduced with shGFP and shp53 which suggested that PALA induced p53-dependent expression of p21. Taken together, the results presented in Fig. 4 thus suggest that PALA treatment activates both p53 and TAp73.
Figure 4.
Activation of p53 and TAp73 by PALA treatment. (A) 041 and derivatives; (B) PC3 and PC3-p53 and (C) SKOV3 cells were treated with 250 µM/L of PALA for the indicated times and expression of TAp73, ΔNp73 and p53 were examined by Western blotting. Expression of TAp73β was also examined in SKOV3 cells (C). Representative data from three independent experiments are presented. (D) 041 and derivatives expressing p53 and dominant negative p73; and (E) SKOV3 and its derivatives expressing dominant negative p73 were treated with 250 µM/L PALA for the indicated times and expression levels of p21 mRNA were examined by real time PCR. The experiments were done in triplicate and average results with standard deviations as error bars are shown. p values <0.05 were considered statistically significant. For (D), p values for *, **, ***, #, and ## are 0.003, 0.0077, 0.02, 0.005 and 0.0001, respectively. For (E) p values for * and ** are 0.004 and 0.008, respectively. (F) A549-shGFP and A549-shp53 cells were treated with 250 µM/L of PALA for 72 h and expression of p53 and p21 were examined by Western blotting.
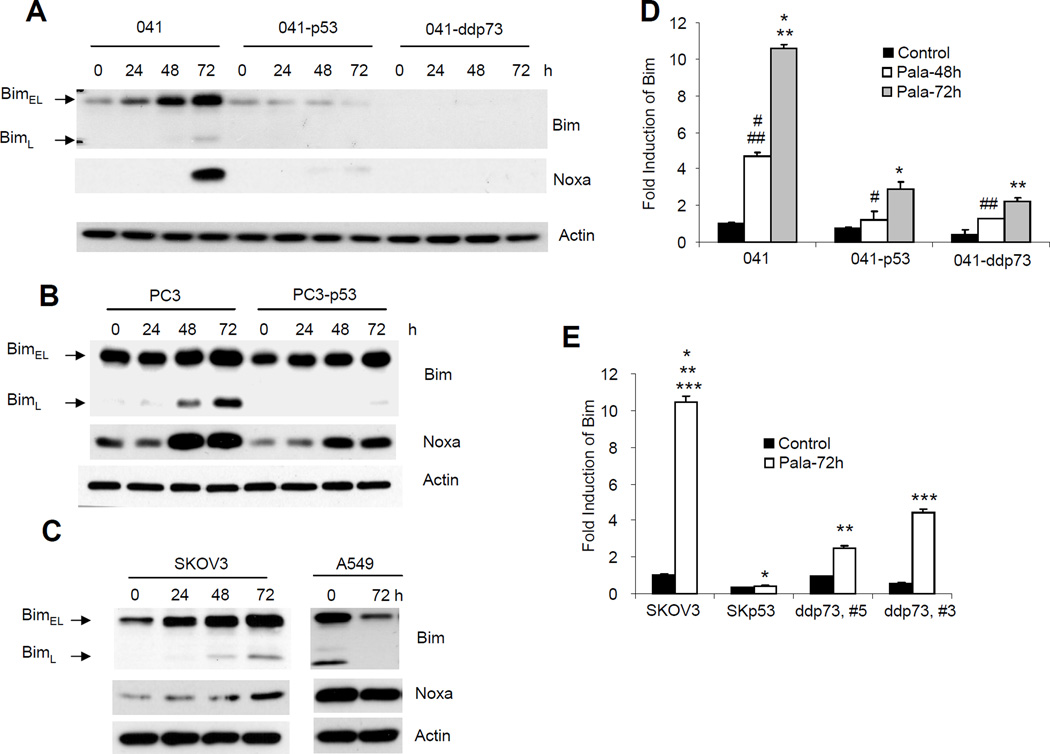
Differential regulation of PALA-induced Bim and Noxa expression by p53 and TAp73
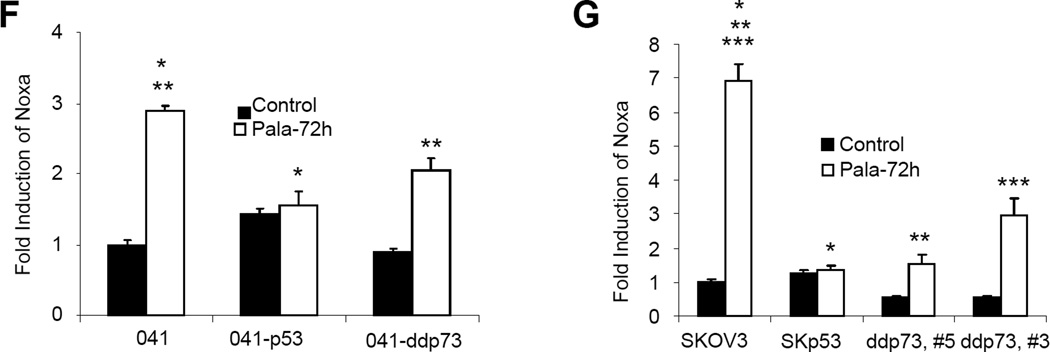
The pro-apoptotic members of Bcl2 family that contain a BH3 domain play essential roles in the regulation of mitochondria-mediated apoptosis in response to various forms of stress. We have previously reported that Concanavalin A induced Bim in a p73-dependent manner and that this process was inhibited after rescue of p53 function (12, 52). In order to investigate the downstream target of TAp73 responsible for mediating PALA-induced mitochondria-mediated apoptosis, we next examined the expression of Bim and Noxa after PALA treatment. As shown in Fig. 5A–C, PALA treatment efficiently induced the expression of Bim and Noxa in all three cell lines lacking p53 and undergoing apoptosis. In 041 and SKOV3 cell lines, induction of the BimEL (23 kDa) isoform by PALA was more pronounced than that of the BimL isoform. On the other hand, PC3 cells expressed a high basal level of the BimEL isoform, which was slightly increased after PALA treatment (Fig. 5C). Increase in expression of the BimL (19 kDa) isoform by PALA was more predominant in this cell line. The expression of BimS was very low in these cell lines and a long exposure was required to detect it (data not shown). This result is consistent with a previous report that the basal expression of different Bim isoforms varies with cell lines and that induction of the BimL isoform is more predominant when the basal level of BimEL is high (49). Moreover, it was previously shown that the BimL isoform is more potent in inducing apoptosis, suggesting that induction of BimL in PC3 cells might induce apoptosis in this cell line (48). Interestingly, expression of p53 in all three cell lines drastically inhibited the expression of Bim and Noxa, suggesting a p53-dependent suppression of PALA-induced Bim and Noxa expression. PALA also failed to increase Bim and Noxa in A549 cells which is p53 wild-type (Figure 5C). Moreover, inactivation of TAp73 also inhibited the expression of Bim and Noxa. To investigate whether the induction of Bim and Noxa by PALA occurred at the transcriptional level, we measured the expression of Bim and Noxa mRNA after PALA treatment in 041 and SKOV3 derivative cell lines by real time PCR (Fig. 5D–G). The primer set used for Bim can detect all three isoforms of Bim. Treatment with PALA strongly increased the level of Bim and Noxa mRNA in parental cells (more than 10-fold at 72 h). In contrast, expression of p53 or inhibition of TAp73 significantly inhibited the induction of Bim transcripts (Fig. 5D and E). PALA treatment also increased the level of Noxa transcripts in parental cells, which was significantly inhibited by inactivation of TAp73 or expression of p53 (Fig. 5F and G). These results suggest that, as for apoptosis, p53 and TAp73 differentially regulate PALA-induced Bim and Noxa expression.
Figure 5.
p53 and TAp73 play opposing role in PALA-induced expression of Bim and Noxa: (A) 041 and derivatives; (B) PC3 and PC3-p53 and (C) SKOV3 cells were treated with 250 µM/L of PALA for the indicated times and total cell lysates were immunoblotted with anti-Bim and anti-Noxa. (D) O41 and its derivative cell lines and (E) SKOV3 and its derivatives expressing p53 and dominant negative p73 were treated with 250 µM/L of PALA for 48 or 72 h. Total RNA was used for the expression of Bim mRNA by real time PCR. For (D), p values for #, ##, * and ** are 0.0002, 0.0009, 0.0002 and 0.00002, respectively. For (E), p values for *, ** and *** are 0.0003, 0.0004 and 0.0008, respectively. (F, G) Cells were treated with PALA and expression levels of Noxa mRNA were examined by real time PCR. For (F), p values for * and ** are 0.004 and 0.012, respectively. For (G), p values for *, ** and *** are 0.001, 0.0004 and 0.007, respectively.
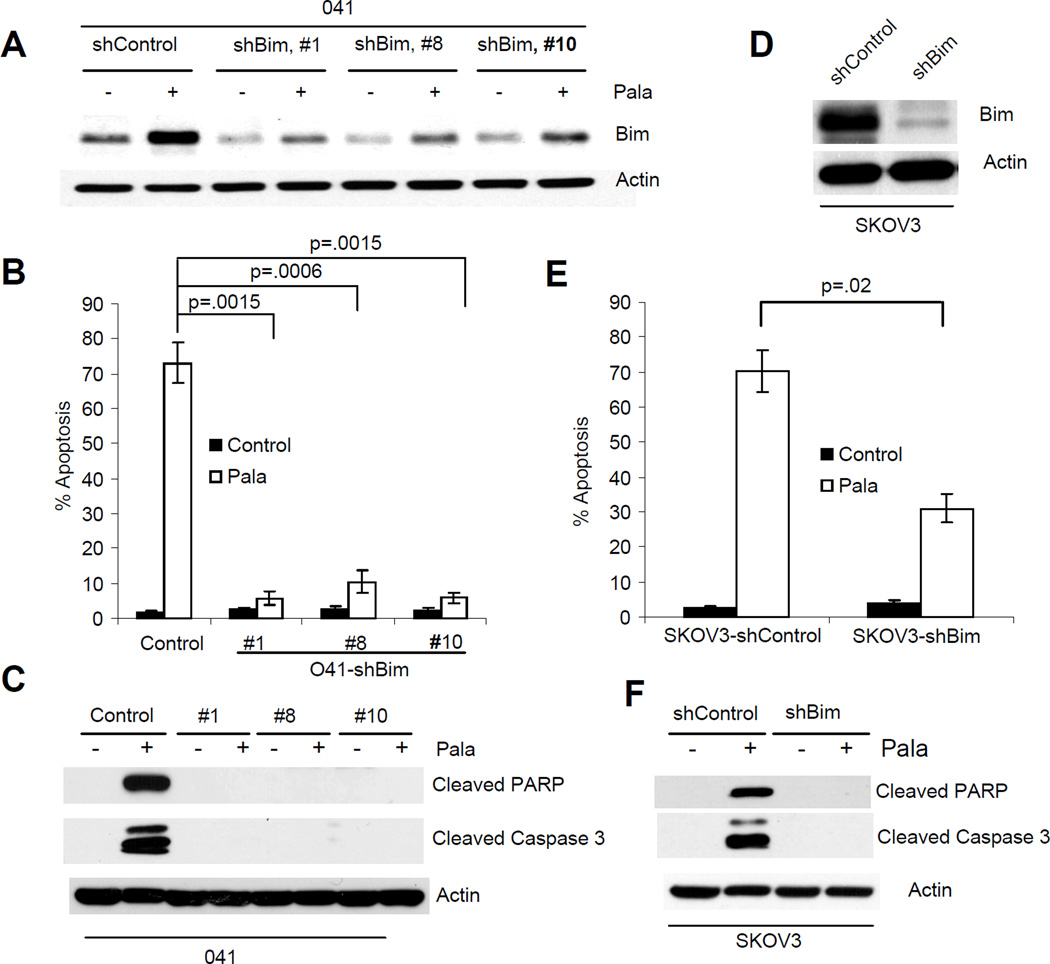
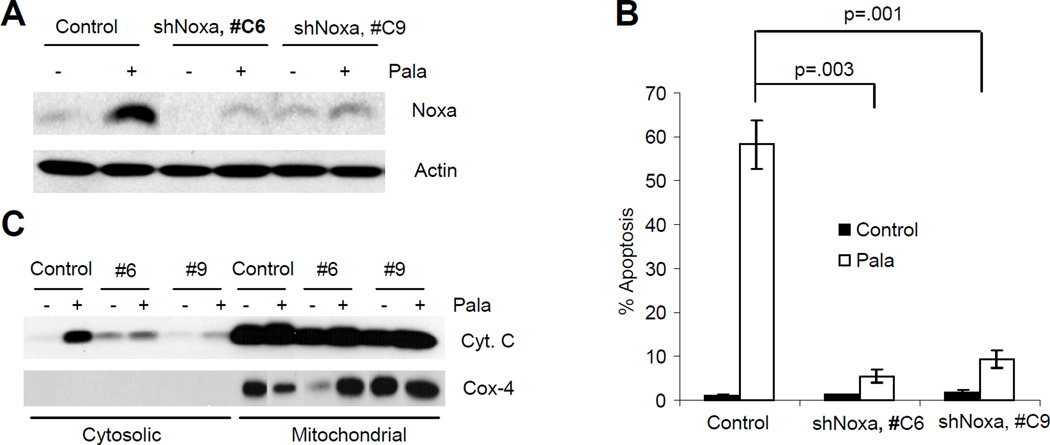
Role of Bim and Noxa in PALA-induced apoptosis
To investigate the role of Bim in PALA-induced apoptosis, we used shRNA to suppress Bim expression in 041 and SKOV3 cells (Fig. 6A and D) and measured apoptosis in these cells. Knockdown of Bim expression significantly inhibited PALA-induced apoptosis in both the cell lines (Fig. 6B and E). Inhibition of PALA-induced apoptosis after ablation of Bim was also supported by inhibition of PARP and caspase-3 cleavage (Fig. 6C and F). We also knocked down the expression of Noxa in SKOV3 cells using an shNoxa plasmid (SuperArray Bioscience Corporation, Frederick, MD) and two stable clones with ablated Noxa expression were established by puromycin selection (Fig. 7A). Cells expressing control siRNA or shNoxa were treated with PALA and apoptosis and release of cytochrome c were examined. Treatment with PALA induced efficient apoptosis in cells transfected with control siRNA, which was significantly decreased after knock down of Noxa expression (Fig. 7B). Ablation of Noxa also inhibited the release of cytochrome c (Fig. 7C). These results suggest that Noxa is also required for PALA-induced apoptosis.
Figure 6.
PALA-induced apoptosis is mediated via Bim. (A) 041 cells were transduced with either scrambled siRNA (shControl) or shRNA against Bim (shBim) and clones were established by puromycin selection. Total cell lysates from PALA-treated and untreated cells were immunoblotted with anti-Bim. (B) Cells were treated with 250 µM/L of PALA for 6 days and apoptosis was measured by TUNEL. (C) Cells were treated with PALA for 72 h and cleavage of PARP and caspase-3 were examined by Western blotting. (D) Expression of Bim was ablated in SKOV3 cells. (E) SKOV3 cells with ablated Bim were treated with 250 µM/L of PALA for 7 days and apoptosis was measured by TUNEL staining. (F) Cells were treated with PALA for 72 h and cleavage of PARP and caspase-3 were examined by Western blotting. For B and E, average apoptosis from three independent experiments with standard deviation as error bars are shown. p values were determined by Student’s T-test and values <0.05 are considered significant.
Figure 7.
(A) Expression of Noxa was ablated in SKOV3 cells. Total cell lysates from PALA-treated and untreated cells expressing scrambled siRNA or Noxa shRNA were immunoblotted with Noxa. (B) SKOV3 cells transfected with shNOXA or control siRNA were treated with 250 µM/L of PALA for 6 days and apoptosis was measured by TUNEL assay. Experiments were repeated three times and average values for apoptosis with standard deviation as error bars are shown. p values were determined by Student’s T-test and values less than 0.05 are considered significant. (C) SKOV3 cells transfected with shNOXA or control siRNA were treated with 250 µM/L of PALA and release of cytochrome C into the cytosol was examined.
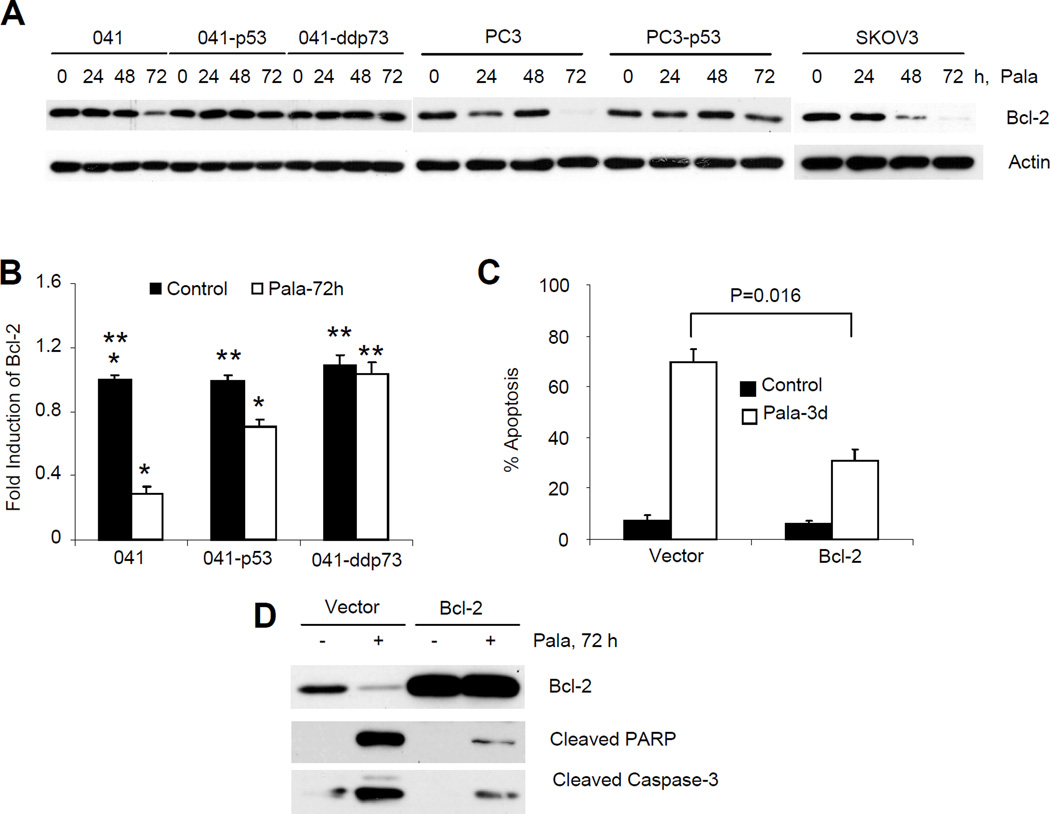
Role of inhibition of Bcl-2 in PALA-induced apoptosis
Since it is the ratio of pro- and anti-apoptotic Bcl-2 that is critical for apoptosis, we examined the expression of anti-apoptotic Bcl-2 protein and mRNA and found that PALA also inhibited the expression of Bcl-2 protein and transcripts in the parental cells lacking p53 (Fig. 8A and B). The inhibition of Bcl-2 was less prevalent in cells expressing p53 or dominant negative p73. To assess the role of inhibition of Bcl-2 in PALA-induced apoptosis, we overexpressed Bcl-2 in 041 cells, and measured apoptosis after treatment with PALA. As shown in Fig. 8C, overexpression of Bcl-2 significantly inhibited PALA-induced apoptosis. Cleavage of PARP and caspase-3 also demonstrated that overexpression of Bcl-2 inhibited PALA-induced apoptosis (Fig. 8D). Taken together, our findings from Figs. 6–8 thus suggest that PALA induces apoptosis of cells lacking functional p53 by modulating multiple pro- and anti-apoptotic Bcl-2 proteins.
Figure 8.
Role of inhibition of Bcl-2 in PALA-induced apoptosis: (A) Cells were treated with 250 µM/L of PALA for the indicated times and expression of Bcl-2 was examined by Western blotting. (B) 041 and derivative cell lines were treated with 250 µM/L PALA for 72 h and expression of Bcl-2 mRNA was examined by real time PCR. Experiments were done in triplicate. Average results with standard deviations as error bars are presented. p values were determined by Student’s t-test and values <0.05 were considered significant. * indicates significant and ** not significant. (C) Bcl-2 was overexpressed in 041 cells from a retroviral plasmid and apoptosis was measured after treatment with 250 µM/L of PALA. Average results from three independent experiments are presented. (D) 041 and 041-Bcl2 cells were treated with 250 µM PALA for 72 h and expression of Bcl-2 and cleavage of PARP and caspase-3 were examined by Western blotting.
Discussion
PALA, a potent and reversible inhibitor of aspartate transcarbamylase, blocks de novo pyrimidine nucleotide biosynthesis, induces growth arrest of cells with p53, but kills cells lacking p53 (35–40). Treatment of cells with PALA depletes the pyrimidine nucleotide pool and thus exerts replicative stress, which rapidly activates p53, leading to reversible cell-cycle arrest (56). In contrast, in the absence of p53, PALA treatment leads to DNA damage (40) without cell-cycle arrest, cells continue DNA synthesis and are very likely to incur more DNA damage. In cells where the degree of DNA damage is beyond the capacity for repair, a signal (not known yet) is generated that leads to apoptosis. The properties of PALA are ideal for an anti-tumor drug, because virtually all cancer cells have some disruption of p53 function. Studies in cell culture and animal models revealed PALA to be highly promising as an anti-tumor drug, and it has been tested in many clinical trials (57–59). However, several important questions still remain unanswered: How is a halt in replication linked to mitochondria-mediated apoptosis? What are the initiators of mitochondrial depolarization? What happens to this signaling pathway in the presence of p53? The findings presented in this manuscript have addressed these questions.
For the first time, we have shown that PALA-induced apoptosis is mediated by TAp73-dependent expression of Noxa and Bim. We found that treatment of cells lacking functional p53 with PALA activates TAp73, as evidenced by an increase in TAp73 expression, and inhibition of PALA-induced p21 expression after inactivation of TAp73 using a dominant negative construct. Moreover, inactivation of TAp73 significantly inhibited PALA-induced apoptosis, suggesting that activation of TAp73 is required for PALA-induced apoptosis. We also found that treatment of cancer cells lacking p53 with PALA increased the expression of several pro-apoptotic Bcl-2 proteins, such as Bim and Noxa, and decreased the anti-apoptotic protein Bcl-2. We also established that the expression of Bim and Noxa is dependent on TAp73. The ratio of pro-apoptotic to anti-apoptotic Bcl-2 proteins is highly critical for mitochondria-mediated apoptosis. Our results also showed that PALA induced mitochondria-mediated apoptosis. Moreover, ablation of the expression of Bim and Noxa or overexpression of Bcl-2 significantly inhibited PALA-induced apoptosis. Taken together, these data suggest that PALA-induced apoptosis is mediated by TAp73-dependent expression of Bim and Noxa and involves the mitochondria-mediated pathway. Although most cancers lack functional p53, they still respond to DNA damaging agents or chemotherapy that induces DNA damage. An increasing body of evidence suggests that activation of TAp73 plays a critical role in regulating apoptosis in these circumstances (60–63). Moreover several studies suggest that Bim and Noxa cooperate to induce apoptosis (64–66). Our findings are consistent with these observations that TAp73 mediates PALA-induced apoptosis by upregulating Bim and Noxa.
Interestingly, expression of p53 in cells lacking p53 inhibited the expression of both Noxa and Bim, although the expression of p21 was increased. It was previously established that the expression of p21 in these cells is dependent on p53 and is responsible for G1 cell cycle arrest (36–39). However, in the absence of p21, PALA-induced p53-dependent S-phase arrest without significant apoptosis (36, 37, 67). We have also reported that macrophage inhibitory cytokine 1 (MIC-1) mediates p53-dependent S phase arrest in response to PALA (37). Although it was not clear how p53 inhibited the expression of Noxa and Bim after PALA treatment, this might be due to rapid activation of p21 or other cell cycle regulatory protein such as (MIC-1) and growth arrest in these cells. Since treatment of cells with PALA induced limited DNA damage (68), sufficient to activate p53-dependent growth arrest, the synthesis of other DNA is halted. On the other hand, since cells lacking p53 do not undergo arrest, DNA damage accumulates through cell cycle progression, which later activates TAp73 as reported previously for other DNA damaging agents (69, 70). Although it is not clear from our study how PALA-induced p73 expression, it might be via a similar mechanism (c-abl-dependent posttranslational modification) like other DNA-damaging agents since PALA also induced DNA-damage (40, 68). It was also reported that transcriptional repression of certain genes by p53 can also occur indirectly, through the ability of p53 to transactivate p21 (71–73). Thus, p53-dependent p21 expression may also be responsible for inhibition of Noxa and Bim in cells expressing p53. Studies also suggest that upon DNA damage, ΔNp73 is rapidly degraded which facilitates apoptosis (74). Knock out of ΔNp73 also increased the expression of p53 target genes and increased DNA damage-induced apoptosis (75). Since PALA induced severe DNA damage and degraded ΔNp73 in cells lacking p53, but only minimal DNA damage in the presence of p53 where the expression of ΔNp73 was also sustained, these events might also be responsible for the inhibition of apoptosis and expression of Bim and Noxa in the presence of p53. However, further studies are warranted to confirm how p53 suppresses the expression of Bim and Noxa. In conclusion, our results demonstrate a novel mechanism by which PALA induces the apoptosis of cancer cells lacking functional p53, which may be valuable for the further development of PALA for cancer treatment.
Materials and Methods
Cell culture and cell treatment
Characteristics of the cell lines used in the study are presented in Table 1. MDAH041 (041) is a Li-Fraumeni Syndrome cell line with no p53 (76). 041-p53 (C11) was derived from 041, which expresses wild-type p53 and is arrested in S-phase upon PALA-treatment (36). 041-ddp73 (C17) from 041, expresses T7-tagged dominant negative p73 (12). Expression of T7 was confirmed (supplementary Fig. S1A). PC3 (from ATCC; Bethesda, MD) is a prostate cancer cell line lacking expression of p53. PC3-p53 was derived from PC3 expressing wild-type p53 (77). SKOV3 (from ATCC; Bethesda, MD), which lacks p53, is an ovarian cancer cell line. SKp53 which express wild-type p53 were derived from SKOV3 and undergo S-phase arrest (37). C3 and C5 are two independent clones derived from SKOV3 expressing dominant negative p73. SKOV3 cells were transfected with dominant negative p73 plasmid which inactivates all the TAp73 isoforms (78). Expression of exogenous plasmid was confirmed by immunoblotting with T7 (supplementary Fig. S1B). PCI-13, a head and neck cancer cell line which expresses mutant p53 (E286K) (79) was obtained from Dr. Ferris laboratory (University of Pittsburgh Cancer Institute). All these cell lines were maintained in DMEM containing 10% fetal bovine serum (FBS) in a CO2 incubator. Tu686 is also a head and neck cancer cell line expressing mutant p53 (55) and maintained in DMEM/F12 (1:1) supplemented with 10% (FBS). A549 is a non small cell lung cancer cell line expressing wild-type p53 and is maintained in RPMI medium supplemented with 10% FBS. Stable knock down of p53 in the A549 cell line was described elsewhere (55). For PALA treatment, either 2 × 105 (041 and derivatives), 3 × 105 (SKOV3 and PC3 derivatives) or 5 × 105 (Tu686, A549 and PCI-13) cells were plated in a 10-cm culture dish. After overnight incubation, the media was replaced with fresh media containing 10% (vol/vol) dialyzed FCS and the cells were treated with 250 µM/L of PALA for the indicated times.
Table 1.
Characteristics of the cells used in the study.
| Cell Name | Origin | p53 status | p73 status | Reference |
|---|---|---|---|---|
| MDAH041 (041) | Li-Fraumeni Syndrome | Negative | Endogenous | Bischoff et al 1991 |
| 041-p53 (C11) | 041 | Wild-type | Endogenous | Agarwal et al 1998a |
| 041-ddp73 (C17) | 041 | Negative | Inactive by dominant negative | Amin et al 2007a |
| shBim, #1 | 041 | Negative | Endogenous | Amin et al 2010a |
| shBim, #8 | 041 | Negative | Endogenous | New |
| shBim, #10 | 041 | Negative | Endogenous | Amin et al 2010a |
| SKOV3 | Ovarian cancer | Negative | Endogenous | Agarwal et al 2006 |
| SK-p53 | SKOV3 | Wild-type | Endogenous | Agarwal et al 2006 |
| SKOV3-ddp73, #C3 | SKOV3 | Negative | Inactive (dominant negative) | New |
| SKOV3-ddp73, #C5 | SKOV3 | Negative | Inactive by dominant negative | New |
| SKOV3-shNoxa, #6 | SKOV3 | Negative | Endogenous | New |
| SKOV3-shNoxa, #9 | SKOV3 | Negative | Endogenous | New |
| SKOV3-shBim, #6 | SKOV3 | Negative | Endogenous | Amin et al 2010a |
| PC3 | Prostate cancer | Negative | Endogenous | Hastak et al 2005 |
| PC3-p53 | PC3 | Wild-type | Endogenous | Hastak et al 2005 |
| A549 | Lung cancer | Wild-type | Endogenous | Amin et al 2010b |
| PCI-13 | Head and Neck cancer | Mutant | Endogenous | Heo et al 1989 |
| Tu686 | Head and Neck cancer | Mutant | Endogenous | Amin et al 2010b |
Materials and chemicals
PALA (NSC224131) was obtained from the Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, National Cancer Institute (Bethesda, MD). Anti-p53 (DO-1), anti-p21 and anti-Bcl-2 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), anti-p73 (catalog #A300-126A, specific for all the TA isoforms) from Bethyl Laboratories (Montgomery, TX), anti-Bim (which detects all Bim isoforms), anti-p73β (ab33131) and anti-ΔNp73 (ab11997) from Abcam (Cambridge, MA), and anti-Noxa from Sigma-Aldrich (St. Louis, MO). Antibodies that recognize cleaved PARP (Asp214, catalog #9541) and caspase-3 (Asp175, catalog #9661) were purchased from Cell Signaling Technology (Danvers, MA). p73 siRNA was purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and transfected into SKOV3 cells using Lipofectamine 2000 transfection reagent.
Terminal deoxynucleotidyl transferase–mediated dUTP nick end labeling (TUNEL) assay
For the apoptosis assay, cells were treated with PALA for the indicated times, fixed in 70% ethanol and labeled with fluorescein-tagged bromo-dUTP and propidium iodide per the manufacturer's protocol using the apoptosis kit (Phoenix Flow Systems, San Diego, CA).
RNA isolation and real-time PCR
Total RNA was extracted from the cells using a Qiagen RNeasy mini kit according to the manufacturer's protocol. Amplification of the corresponding gene was performed using primer sets supplied by Applied Biosystems, Inc. The data were analyzed for fold induction of each gene compared with the untreated sample after normalization with β-actin expression.
Western blotting analyses
SDS-PAGE and immunoblotting were performed as described elsewhere (12). Briefly, total cellular proteins were isolated by lysing the cells in 20 mmol/L Tris-HCl (pH 7.5), 2% (w/v) SDS, 2 mmol/L benzamidine, and 0.2 mmol/L phenylmethylsulfonyl fluoride. Protein concentrations were determined by the Bradford method. Proteins were resolved on SDS-10–12% polyacrylamide gels and then transferred to polyvinylidene difluoride membranes. The membranes were blocked in 5% nonfat skimmed milk and incubated with the respective antibody, followed by incubation with a secondary antibody. Immunostained protein bands were detected with an enhanced chemiluminescence kit (Thermo Scientific).
Supplementary Material
Acknowledgements
The authors are thankful to Dr. George R Stark (Cleveland Clinic Foundation) for his continuous encouragement and criticism throughout the course of the work and critically reading and editing the manuscript, and to Dr. Anthea Hammond (Emory University) for editorial assistance. We also thank Dr. Rajib K Paul for screening SKp53 cells. This work was supported by National Institutes of Health Grants R01 CA98916 to MLA and P50 CA128613 to DMS and ARA. ARA is a recipient of Career Development Award.
Footnotes
Conflict of Interest: None
References
- 1.Farnebo M, Bykov VJ, Wiman KG. The p53 tumor suppressor: a master regulator of diverse cellular processes and therapeutic target in cancer. Biochem Biophys Res Commun. 2010;396:85–89. doi: 10.1016/j.bbrc.2010.02.152. [DOI] [PubMed] [Google Scholar]
- 2.Gottlieb E, Vousden KH. p53 regulation of metabolic pathways. Cold Spring Harb Perspect Biol. 2010;2:a001040. doi: 10.1101/cshperspect.a001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zilfou JT, Lowe SW. Tumor suppressive functions of p53. Cold Spring Harb Perspect Biol. 2009;1:a001883. doi: 10.1101/cshperspect.a001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joerger AC, Fersht AR. The tumor suppressor p53: from structures to drug discovery. Cold Spring Harb Perspect Biol. 2010;2:a000919. doi: 10.1101/cshperspect.a000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sedletska Y, Giraud-Panis MJ, Malinge JM. Cisplatin is a DNA-damaging antitumour compound triggering multifactorial biochemical responses in cancer cells: importance of apoptotic pathways. Curr Med Chem Anticancer Agents. 2005;5:251–265. doi: 10.2174/1568011053765967. [DOI] [PubMed] [Google Scholar]
- 6.Chung J, Irwin MS. Targeting the p53-family in cancer and chemosensitivity: triple threat. Curr Drug Targets. 2010;11:667–681. doi: 10.2174/138945010791170833. [DOI] [PubMed] [Google Scholar]
- 7.Amin AR, Kucuk O, Khuri FR, Shin DM. Perspectives for cancer prevention with natural compounds. J Clin Oncol. 2009;27:2712–2725. doi: 10.1200/JCO.2008.20.6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiman KG. Pharmacological reactivation of mutant p53: from protein structure to the cancer patient. Oncogene. 2010;29:4245–4252. doi: 10.1038/onc.2010.188. [DOI] [PubMed] [Google Scholar]
- 9.Agarwal ML, Taylor WR, Chernov MV, Chernova OB, Stark GR. The p53 network. J Biol Chem. 1998;273:1–4. doi: 10.1074/jbc.273.1.1. [DOI] [PubMed] [Google Scholar]
- 10.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 11.Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 12.Amin AR, Paul RK, Thakur VS, Agarwal ML. A novel role for p73 in the regulation of Akt-Foxo1a-Bim signaling and apoptosis induced by the plant lectin, Concanavalin A. Cancer Res. 2007;67:5617–5621. doi: 10.1158/0008-5472.CAN-07-0655. [DOI] [PubMed] [Google Scholar]
- 13.Amin AR, Thakur VS, Paul RK, Feng GS, Qu CK, Mukhtar H, et al. SHP-2 tyrosine phosphatase inhibits p73-dependent apoptosis and expression of a subset of p53 target genes induced by EGCG. Proc Natl Acad Sci U S A. 2007;104:5419–5424. doi: 10.1073/pnas.0700642104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rufini A, Agostini M, Grespi F, Tomasini R, Sayan BS, Niklison-Chirou MV, et al. p73 in Cancer. Genes Cancer. 2011;2:491–502. doi: 10.1177/1947601911408890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.John K, Alla V, Meier C, Putzer BM. GRAMD4 mimics p53 and mediates the apoptotic function of p73 at mitochondria. Cell Death Differ. 2011;18:874–886. doi: 10.1038/cdd.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 17.Melino G, Lu X, Gasco M, Crook T, Knight RA. Functional regulation of p73 and p63: development and cancer. Trends Biochem Sci. 2003;28:663–670. doi: 10.1016/j.tibs.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Lunghi P, Costanzo A, Mazzera L, Rizzoli V, Levrero M, Bonati A. The p53 family protein p73 provides new insights into cancer chemosensitivity and targeting. Clin Cancer Res. 2009;15:6495–6502. doi: 10.1158/1078-0432.CCR-09-1229. [DOI] [PubMed] [Google Scholar]
- 19.Dotsch V, Bernassola F, Coutandin D, Candi E, Melino G. p63 and p73, the ancestors of p53. Cold Spring Harb Perspect Biol. 2010;2:a004887. doi: 10.1101/cshperspect.a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ichimiya S, Nimura Y, Kageyama H, Takada N, Sunahara M, Shishikura T, et al. p73 at chromosome 1p36.3 is lost in advanced stage neuroblastoma but its mutation is infrequent. Oncogene. 1999;18:1061–1066. doi: 10.1038/sj.onc.1202390. [DOI] [PubMed] [Google Scholar]
- 21.Douc-Rasy S, Barrois M, Echeynne M, Kaghad M, Blanc E, Raguenez G, et al. DeltaN-p73alpha accumulates in human neuroblastic tumors. Am J Pathol. 2002;160:631–639. doi: 10.1016/s0002-9440(10)64883-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dominguez G, Garcia JM, Pena C, Silva J, Garcia V, Martinez L, et al. DeltaTAp73 upregulation correlates with poor prognosis in human tumors: putative in vivo network involving p73 isoforms, p53, and E2F-1. J Clin Oncol. 2006;24:805–815. doi: 10.1200/JCO.2005.02.2350. [DOI] [PubMed] [Google Scholar]
- 23.Zaika AI, Kovalev S, Marchenko ND, Moll UM. Overexpression of the wild type p73 gene in breast cancer tissues and cell lines. Cancer Res. 1999;59:3257–3263. [PubMed] [Google Scholar]
- 24.Ng SW, Yiu GK, Liu Y, Huang LW, Palnati M, Jun SH, et al. Analysis of p73 in human borderline and invasive ovarian tumor. Oncogene. 2000;19:1885–1890. doi: 10.1038/sj.onc.1203512. [DOI] [PubMed] [Google Scholar]
- 25.Tannapfel A, Wasner M, Krause K, Geissler F, Katalinic A, Hauss J, et al. Expression of p73 and its relation to histopathology and prognosis in hepatocellular carcinoma. J Natl Cancer Inst. 1999;91:1154–1158. doi: 10.1093/jnci/91.13.1154. [DOI] [PubMed] [Google Scholar]
- 26.Kovalev S, Marchenko N, Swendeman S, LaQuaglia M, Moll UM. Expression level, allelic origin, and mutation analysis of the p73 gene in neuroblastoma tumors and cell lines. Cell Growth Differ. 1998;9:897–903. [PubMed] [Google Scholar]
- 27.Yokomizo A, Mai M, Tindall DJ, Cheng L, Bostwick DG, Naito S, et al. Overexpression of the wild type p73 gene in human bladder cancer. Oncogene. 1999;18:1629–1633. doi: 10.1038/sj.onc.1202474. [DOI] [PubMed] [Google Scholar]
- 28.Yokomizo A, Mai M, Bostwick DG, Tindall DJ, Qian J, Cheng L, et al. Mutation and expression analysis of the p73 gene in prostate cancer. Prostate. 1999;39:94–100. doi: 10.1002/(sici)1097-0045(19990501)39:2<94::aid-pros3>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 29.Sunahara M, Ichimiya S, Nimura Y, Takada N, Sakiyama S, Sato Y, et al. Mutational analysis of the p73 gene localized at chromosome 1p36.3 in colorectal carcinomas. International journal of oncology. 1998;13:319–323. [PubMed] [Google Scholar]
- 30.Loukopoulos P, Shibata T, Katoh H, Kokubu A, Sakamoto M, Yamazaki K, et al. Genome-wide array-based comparative genomic hybridization analysis of pancreatic adenocarcinoma: identification of genetic indicators that predict patient outcome. Cancer Sci. 2007;98:392–400. doi: 10.1111/j.1349-7006.2007.00395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malaguarnera R, Vella V, Vigneri R, Frasca F. p53 family proteins in thyroid cancer. Endocr Relat Cancer. 2007;14:43–60. doi: 10.1677/erc.1.01223. [DOI] [PubMed] [Google Scholar]
- 32.Leupin N, Luthi A, Novak U, Grob TJ, Hugli B, Graber H, et al. P73 status in Bcell chronic lymphocytic leukaemia. Leuk Lymphoma. 2004;45:1205–1207. doi: 10.1080/10298190310001623829. [DOI] [PubMed] [Google Scholar]
- 33.Tschan MP, Grob TJ, Peters UR, Laurenzi VD, Huegli B, Kreuzer KA, et al. Enhanced p73 expression during differentiation and complex p73 isoforms in myeloid leukemia. Biochem Biophys Res Commun. 2000;277:62–65. doi: 10.1006/bbrc.2000.3627. [DOI] [PubMed] [Google Scholar]
- 34.Faridoni-Laurens L, Tourpin S, Alsafadi S, Barrois M, Temam S, Janot F, et al. Involvement of N-terminally truncated variants of p73, deltaTAp73, in head and neck squamous cell cancer: a comparison with p53 mutations. Cell Cycle. 2008;7:1587–1596. doi: 10.4161/cc.7.11.5894. [DOI] [PubMed] [Google Scholar]
- 35.Swyryd EA, Seaver SS, Stark GR. N-(phosphonacetyl)-L-aspartate, a potent transition state analog inhibitor of aspartate transcarbamylase, blocks proliferation of mammalian cells in culture. J Biol Chem. 1974;249:6945–6950. [PubMed] [Google Scholar]
- 36.Agarwal ML, Agarwal A, Taylor WR, Chernova O, Sharma Y, Stark GR. A p53-dependent S-phase checkpoint helps to protect cells from DNA damage in response to starvation for pyrimidine nucleotides. Proc Natl Acad Sci U S A. 1998;95:14775–14780. doi: 10.1073/pnas.95.25.14775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agarwal MK, Hastak K, Jackson MW, Breit SN, Stark GR, Agarwal ML. Macrophage inhibitory cytokine 1 mediates a p53-dependent protective arrest in S phase in response to starvation for DNA precursors. Proc Natl Acad Sci U S A. 2006;103:16278–16283. doi: 10.1073/pnas.0607210103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin Y, Tainsky MA, Bischoff FZ, Strong LC, Wahl GM. Wild-type p53 restores cell cycle control and inhibits gene amplification in cells with mutant p53 alleles. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]
- 39.Agarwal ML, Agarwal A, Taylor WR, Stark GR. p53 controls both the G2/M and the G1 cell cycle checkpoints and mediates reversible growth arrest in human fibroblasts. Proc Natl Acad Sci U S A. 1995;92:8493–8497. doi: 10.1073/pnas.92.18.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paulson TG, Almasan A, Brody LL, Wahl GM. Gene amplification in a p53- deficient cell line requires cell cycle progression under conditions that generate DNA breakage. Mol Cell Biol. 1998;18:3089–3100. doi: 10.1128/mcb.18.5.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moffitt KL, Martin SL, Walker B. From sentencing to execution--the processes of apoptosis. J Pharm Pharmacol. 2010;62:547–562. doi: 10.1211/jpp.62.05.0001. [DOI] [PubMed] [Google Scholar]
- 42.Brenner D, Mak TW. Mitochondrial cell death effectors. Curr Opin Cell Biol. 2009;21:871–877. doi: 10.1016/j.ceb.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 43.Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leibowitz B, Yu J. Mitochondrial signaling in cell death via the Bcl-2 family. Cancer Biol Ther. 2010;9:417–422. doi: 10.4161/cbt.9.6.11392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghiotto F, Fais F, Bruno S. BH3-only proteins: the death-puppeteer's wires. Cytometry A. 2010;77:11–21. doi: 10.1002/cyto.a.20819. [DOI] [PubMed] [Google Scholar]
- 46.Lomonosova E, Chinnadurai G. BH3-only proteins in apoptosis and beyond: an overview. Oncogene. 2008;27(Suppl 1):S2–S19. doi: 10.1038/onc.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 48.O'Connor L, Strasser A, O'Reilly LA, Hausmann G, Adams JM, Cory S, et al. Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J. 1998;17:384–395. doi: 10.1093/emboj/17.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li R, Moudgil T, Ross HJ, Hu HM. Apoptosis of non-small-cell lung cancer cell lines after paclitaxel treatment involves the BH3-only proapoptotic protein Bim. Cell Death Differ. 2005;12:292–303. doi: 10.1038/sj.cdd.4401554. [DOI] [PubMed] [Google Scholar]
- 50.Toh WH, Nam SY, Sabapathy K. An essential role for p73 in regulating mitotic cell death. Cell Death Differ. 2010;17:787–800. doi: 10.1038/cdd.2009.181. [DOI] [PubMed] [Google Scholar]
- 51.Busuttil V, Droin N, McCormick L, Bernassola F, Candi E, Melino G, et al. NF-kappaB inhibits T-cell activation-induced, p73-dependent cell death by induction of MDM2. Proc Natl Acad Sci U S A. 2010;107:18061–18066. doi: 10.1073/pnas.1006163107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Amin AR, Thakur VS, Gupta K, Jackson MW, Harada H, Agarwal MK, et al. Restoration of p53 functions protects cells from concanavalin A-induced apoptosis. Mol Cancer Ther. 2010;9:471–479. doi: 10.1158/1535-7163.MCT-09-0732. [DOI] [PubMed] [Google Scholar]
- 53.Miyaguchi Y, Tsuchiya K, Sakamoto K. P53 negatively regulates the transcriptional activity of FOXO3a under oxidative stress. Cell Biol Int. 2009;33:853–860. doi: 10.1016/j.cellbi.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 54.Di Como CJ, Gaiddon C, Prives C. p73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol Cell Biol. 1999;19:1438–1449. doi: 10.1128/mcb.19.2.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Amin AR, Wang D, Zhang H, Peng S, Shin HJ, Brandes JC, et al. Enhanced antitumor activity by the combination of the natural compounds (-)-epigallocatechin-3-gallate and luteolin: potential role of p53. J Biol Chem. 2010;285:34557–34565. doi: 10.1074/jbc.M110.141135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Linke SP, Clarkin KC, Di Leonardo A, Tsou A, Wahl GM. A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev. 1996;10:934–947. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- 57.Wadler S, Gleissner B, Hilgenfeld RU, Thiel E, Haynes H, Kaleya R, et al. Phase II trial of N-(phosphonacetyl)-L-aspartate (PALA), 5-fluorouracil and recombinant interferon-alpha-2b in patients with advanced gastric carcinoma. Eur J Cancer. 1996;32A:1254–1256. doi: 10.1016/0959-8049(96)00035-4. [DOI] [PubMed] [Google Scholar]
- 58.Redei I, Green F, Hoffman JP, Weiner LM, Scher R, O'Dwyer PJ. Phase II trial of PALA and 6-methylmercaptopurine riboside (MMPR) in combination with 5-fluorouracil in advanced pancreatic cancer. Invest New Drugs. 1994;12:319–321. doi: 10.1007/BF00873047. [DOI] [PubMed] [Google Scholar]
- 59.Grem JL, McAtee N, Steinberg SM, Hamilton JM, Murphy RF, Drake J, et al. A phase I study of continuous infusion 5-fluorouracil plus calcium leucovorin in combination with N-(phosphonacetyl)-L-aspartate in metastatic gastrointestinal adenocarcinoma. Cancer Res. 1993;53:4828–4836. [PubMed] [Google Scholar]
- 60.Ibrahim N, He L, Leong CO, Xing D, Karlan BY, Swisher EM, et al. BRCA1-associated epigenetic regulation of p73 mediates an effector pathway for chemosensitivity in ovarian carcinoma. Cancer Res. 2010;70:7155–7165. doi: 10.1158/0008-5472.CAN-10-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chakraborty J, Banerjee S, Ray P, Hossain DM, Bhattacharyya S, Adhikary A, et al. Gain of cellular adaptation due to prolonged p53 impairment leads to functional switchover from p53 to p73 during DNA damage in acute myeloid leukemia cells. J Biol Chem. 2010;285:33104–33112. doi: 10.1074/jbc.M110.122705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vilgelm A, El-Rifai W, Zaika A. Therapeutic prospects for p73 and p63: rising from the shadow of p53. Drug Resist Updat. 2008;11:152–163. doi: 10.1016/j.drup.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ozaki T, Nakagawara A. p73, a sophisticated p53 family member in the cancer world. Cancer Sci. 2005;96:729–737. doi: 10.1111/j.1349-7006.2005.00116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kirschnek S, Vier J, Gautam S, Frankenberg T, Rangelova S, Eitz-Ferrer P, et al. Molecular analysis of neutrophil spontaneous apoptosis reveals a strong role for the proapoptotic BH3-only protein Noxa. Cell Death Differ. 2011 doi: 10.1038/cdd.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Santidrian AF, Gonzalez-Girones DM, Iglesias-Serret D, Coll-Mulet L, Cosialls AM, de Frias M, et al. AICAR induces apoptosis independently of AMPK and p53 through up-regulation of the BH3-only proteins BIM and NOXA in chronic lymphocytic leukemia cells. Blood. 2010;116:3023–3032. doi: 10.1182/blood-2010-05-283960. [DOI] [PubMed] [Google Scholar]
- 66.Morales AA, Gutman D, Lee KP, Boise LH. BH3-only proteins Noxa, Bmf, and Bim are necessary for arsenic trioxide-induced cell death in myeloma. Blood. 2008;111:5152–5162. doi: 10.1182/blood-2007-10-116889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 68.Hastak K, Paul RK, Agarwal MK, Thakur VS, Amin AR, Agrawal S, et al. DNA synthesis from unbalanced nucleotide pools causes limited DNA damage that triggers ATR-CHK1-dependent p53 activation. Proc Natl Acad Sci U S A. 2008;105:6314–6319. doi: 10.1073/pnas.0802080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gong JG, Costanzo A, Yang HQ, Melino G, Kaelin WG, Jr, Levrero M, et al. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399:806–809. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 70.Yuan ZM, Shioya H, Ishiko T, Sun X, Gu J, Huang YY, et al. p73 is regulated by tyrosine kinase c-Abl in the apoptotic response to DNA damage. Nature. 1999;399:814–817. doi: 10.1038/21704. [DOI] [PubMed] [Google Scholar]
- 71.Gottifredi V, Karni-Schmidt O, Shieh SS, Prives C. p53 down-regulates CHK1 through p21 and the retinoblastoma protein. Mol Cell Biol. 2001;21:1066–1076. doi: 10.1128/MCB.21.4.1066-1076.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lohr K, Moritz C, Contente A, Dobbelstein M. p21/CDKN1A mediates negative regulation of transcription by p53. J Biol Chem. 2003;278:32507–32516. doi: 10.1074/jbc.M212517200. [DOI] [PubMed] [Google Scholar]
- 73.Shats I, Milyavsky M, Tang X, Stambolsky P, Erez N, Brosh R, et al. p53-dependent down-regulation of telomerase is mediated by p21waf1. J Biol Chem. 2004;279:50976–50985. doi: 10.1074/jbc.M402502200. [DOI] [PubMed] [Google Scholar]
- 74.Maisse C, Munarriz E, Barcaroli D, Melino G, De Laurenzi V. DNA damage induces the rapid and selective degradation of the DeltaNp73 isoform, allowing apoptosis to occur. Cell Death Differ. 2004;11:685–687. doi: 10.1038/sj.cdd.4401376. [DOI] [PubMed] [Google Scholar]
- 75.Wilhelm MT, Rufini A, Wetzel MK, Tsuchihara K, Inoue S, Tomasini R, et al. Isoform-specific p73 knockout mice reveal a novel role for delta Np73 in the DNA damage response pathway. Genes Dev. 2010;24:549–560. doi: 10.1101/gad.1873910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bischoff FZ, Strong LC, Yim SO, Pratt DR, Siciliano MJ, Giovanella BC, et al. Tumorigenic transformation of spontaneously immortalized fibroblasts from patients with a familial cancer syndrome. Oncogene. 1991;6:183–186. [PubMed] [Google Scholar]
- 77.Hastak K, Agarwal MK, Mukhtar H, Agarwal ML. Ablation of either p21 or Bax prevents p53-dependent apoptosis induced by green tea polyphenol epigallocatechin-3-gallate. FASEB J. 2005;19:789–791. doi: 10.1096/fj.04-2226fje. [DOI] [PubMed] [Google Scholar]
- 78.Irwin M, Marin MC, Phillips AC, Seelan RS, Smith DI, Liu W, et al. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature. 2000;407:645–648. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- 79.Heo DS, Snyderman C, Gollin SM, Pan S, Walker E, Deka R, et al. Biology, cytogenetics, and sensitivity to immunological effector cells of new head and neck squamous cell carcinoma lines. Cancer Res. 1989;49:5167–5175. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.