Abstract
The smooth muscle isoform of myosin light chain kinase (MLCK) is a Ca2+-calmodulin-activated kinase that is found in many tissues. It is particularly important for regulating smooth muscle contraction by phosphorylation of myosin. This review summarizes selected aspects of recent biochemical work on MLCK that pertains to its function in smooth muscle. In general, the focus of the review is on new findings, unresolved issues, and areas with the potential for high physiological significance that need further study. The review includes a concise summary of the structure, substrates, and enzyme activity, followed by a discussion of the factors that may limit the effective activity of MLCK in the muscle. The interactions of each of the many domains of MLCK with the proteins of the contractile apparatus, and the multi-domain interactions of MLCK that may control its behaviors in the cell are summarized. Finally, new in vitro approaches to studying the mechanism of phosphorylation of myosin are introduced.
Introduction
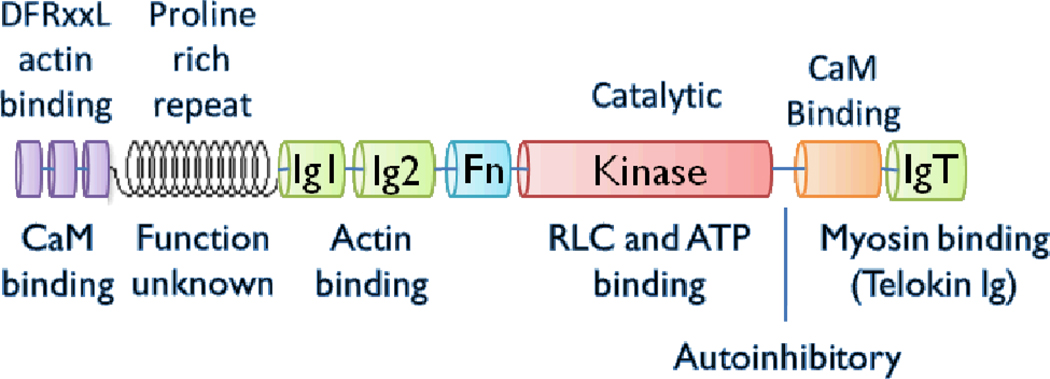
Myosin light chain kinase (MLCK; EC 2.7.11.18) is a ubiquitous Ca2+/CaM-activated kinase found in smooth, cardiac and skeletal muscle as well as in mammalian non-muscle cells. The focus of this review is the so-called smooth muscle MLCK (called MLCK here), which in humans is encoded by the single copy MYLK1 gene. This smooth muscle isoform is not restricted to smooth muscle tissues. Regulation of expression of MYLK1 in smooth muscle and other tissues has been recently reviewed (1). MYLK2 encodes the skeletal MLCK and MYLK3 encodes a cardiac-specific MLCK ((2); GenBank accession number EU403565). The nomenclature can be confusing because MYLK1 gene encodes both the so-called nonmuscle and the smooth muscle isoforms of MLCK in addition to telokin (also called kinase related protein or KRP) by alternative initiation sites (3). The nonmuscle isoforms are longer than the smooth muscle isoforms due to an N-terminal ~900 amino acid extension. Therefore, nonmuscle MLCK is often referred to as the long isoform and smooth muscle the short isoform. There are several nonmuscle variants in humans that are generated by alternative splicing (Q15746-1,2,3a,3b,4,5,6), but the smooth muscle isoform (Q15746-7) and telokin (Q15746-8) have no alternative spliced variants. Therefore, the nonmuscle and smooth muscle isoforms share identical sequences in the region shown in Figure 1 and telokin contains only the IgT portion of MLCK.
Figure 1.
Domain structure of short or smooth muscle isoform of mammalian MLCK.
The avian gene is worth mentioning here because many studies have been done with the easily-purified MLCK from the abundant chicken or turkey gizzard tissue (4–9). The avian MLCK gene (10) encodes three proteins by alternative initiation, the long (P11799-2 or MLCK-210) and short (P11799-1 or MLCK-108) isoforms are known as the nonmuscle and smooth muscle, respectively, and telokin (P11799-1). Avian MLCK is unique in that it lacks a portion of the proline rich repeat region found in the mammalian MLCKs. The chicken gizzard smooth muscle MLCK is composed of 972 amino acid residues with a calculated Mr value of 107,534.
Structure of MLCK
MLCK contains conserved domains in a conserved order
The conserved domains of the short or smooth muscle isoform of MLCK is shown in Figure 1. The shorter isoform found in avian muscle lack the proline rich repeats. MLCK has an actin-binding domain, composed of three DFRxxL motifs at the N-terminus (11–14), which binds weakly to purified F-actin (15). Ca2+-CaM also binds the DFRXXL region resulting in weakened actin binding (16), but the physiological significance of this is not known. There is some evidence that Ig1 and/or Ig2 also bind to actin (14). The IgT domain binds to smooth muscle myosin (SMM) (17, 18) and the kinase domain binds to ATP and to the N-terminal phosphorylation domain of the myosin regulatory light chain (RLC) to phosphorylate S19. MLCK binds to SMM filaments (15, 19–21) and SMM monomers (21, 22) at the junction between the two SMM heads and the tail (17, 19) through the IgT domain (17, 18), although other MLCK-SMM interaction sites have been proposed (15, 23). The functions of the fibronectin domain (Fn; Type 3) and proline rich repeat regions are not known. In general, the order of the conserved domains in MLCK is also highly conserved, strongly suggesting that specific interactions between domains, potentially involving the linker regions, are important for function.
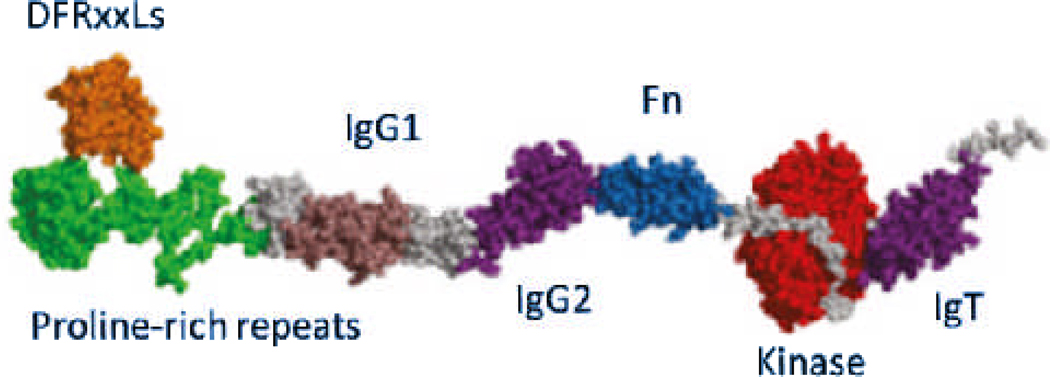
MLCK is an elongated and potentially flexible molecule
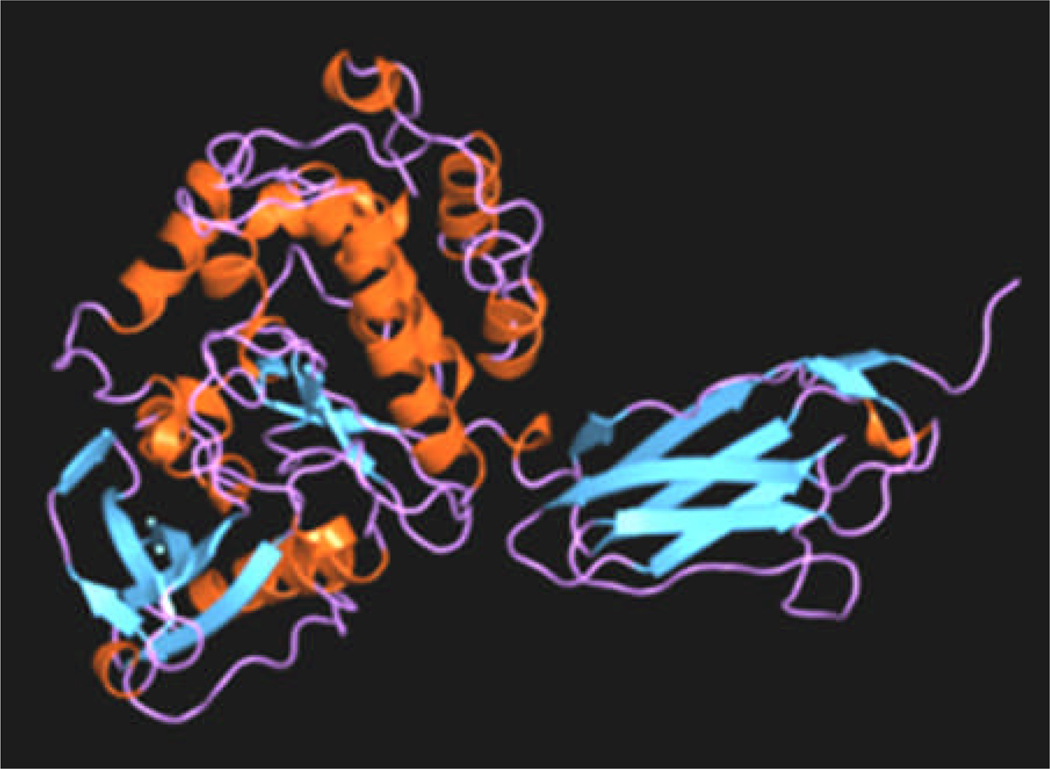
MLCK is an elongated and potentially flexible molecule (19, 24, 25). There are no crystal structures of the full length kinase, but several structures in the database are representative of various domains (25). Figure 2 shows a proposed molecular model of the rabbit kinase based upon known structures from homologous proteins and structural predictions based upon sequence (25). Visualization of the full length molecule by electron microscopy suggests that it can adopt many conformations from elongated, to compact (19, 25). This suggests that the molecule is highly flexible. The extended length of MLCK is sufficient to span between the thick and thin filaments in smooth muscle, and the length could be longer if the proline rich repeat segment is modeled as an extensible linker (25). The distance between the thick and thin filaments at rest is ~ 26 nm (26). Figure 3 shows the crystal structure of a fragment of twitchin kinase, which is likely to be structurally homologous to MLCK, although it binds S100 instead of Ca2+-CaM. Note the close juxtaposition of the IgT domain (right) with the kinase domain (left) creating a large interacting surface. If MLCK adopts a similar conformation, this interface would be disrupted by CaM binding.
Figure 2.
Molecular model of predicted rabbit MLCK structure. Length = ~35 nm. Reproduced from (25).
Figure 3.
Crystal structure of a fragment of twitchin kinase from C. Elegans (PDF entry 1KOA; reproduced from (23)). Kinase domain (left) and the Ig domain (right) are similar to expected structure of the MLCK kinase and IgT domains, respectively.
Substrates of MLCK
Relative kinetics of SMM versus NMM phosphorylation by MLCK
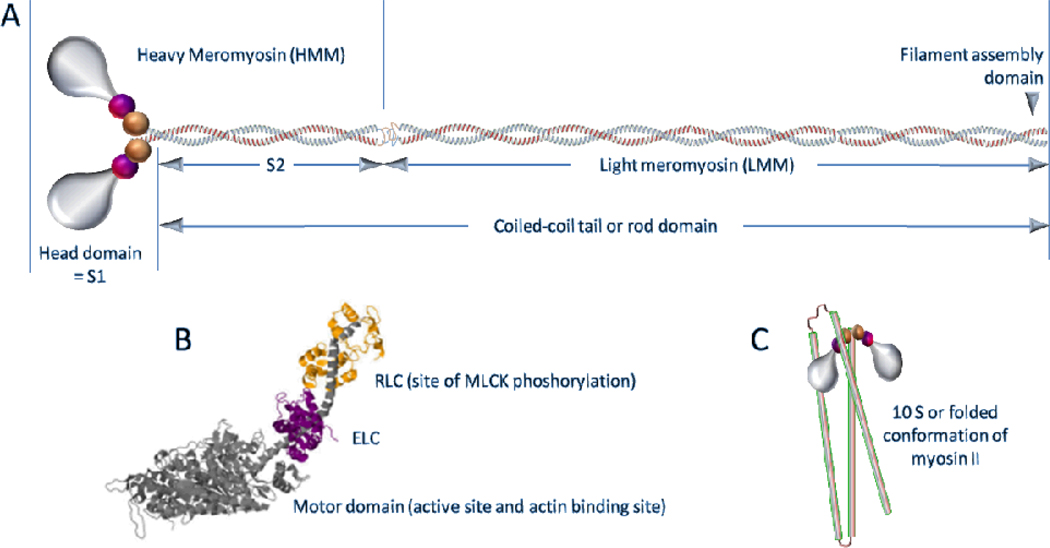
NMM (nonmuscle myosin) and SMM (smooth muscle myosin) are the only known substrates of MLCK in vivo (27, 28). Figure 4 shows a schematic of the structure of myosin II, a class that includes both SMM and NMM. Phosphorylation of myosin on the regulatory light chains (RLC; also called LC20) activates the actin-activated myosin ATPase activity, which is necessary and sufficient for muscle contraction. MLCK-mediated phosphorylation in tracheal, bronchial (29), and gastrointestinal (30) smooth muscle cannot be rescued by other kinases in knockout mice, demonstrating its pivotal position in signaling pathways that regulate force generation.
Figure 4.
A. Cartoon of molecular structure of myosin II. Smooth and nonmuscle myosins have the same subunit composition and fall into the myosin II class. Myosin II is a hexamer composed of 2 heavy chains, 2 RLC and 2 ELC. The names of the sub-fragments (labeled S1, S2, LMM, and HMM) are derived from pioneering proteolysis studies and are commonly used today. Constructs with similar compositions are often expressed. B. Atomic model of the head domain showing the heavy chain in grey with the two light chains binding to the long heavy chain α-helix of the lever arm domain. C. Cartoon of the folded or 10S conformation of myosin. The heads are as in A, but the tail domain (show as tubes) is now folded into thirds. The region of the rod near the upper hinge interacts with the RLC of one or both heads (97, 123, 124).
All smooth muscles express two forms of myosin II, SMM (most abundant; includes 4 alternatively-spliced variants derived from a single smooth muscle myosin heavy chain gene: 1A, 1B, 2A and 2B) and NMM (IIA, IIB major, IIC minor). A number of groups have provided biochemical (31, 32) as well as mechanical (33–35) evidence that NMM participates in a physiologically relevant pathway for force maintenance in smooth muscle. Remarkably, it has only been recently shown directly that NMM is phosphorylated by MLCK in smooth muscle (28). There are no published kinetic studies of the interaction of MLCK with NMM. Our preliminary data using expressed nonmuscle RLC or HMM IIB and smooth muscle RLC or HMM shows that even though the sequence identity of these isoforms is 90%, the nonmuscle proteins are not as good substrates for MLCK as are the smooth muscle proteins (lower Vmax; W. Pearce collaboration, unpublished). During the rapid phase of contraction, it might be that MLCK phosphorylates SMM more rapidly than NMM. This would prevent a situation in which phosphorylated NMM IIB, with a slower ADP release rate than SMM, from serving as a brake on contraction, even though the assisting load from the faster SMM would provide some level of enhancement of ADP release from the acto-NMM complex (31). During the force maintenance phase the more slowly phosphorylated NMM will have attained a significant level of phosphorylation to maintain force, most likely in the ADP state. Further studies are required to test this idea.
Nucleotide substrates
Of course, ATP is the other substrate of MLCK, which is hydrolyzed to ADP at the active site in the kinase domain. Typical Kms for ATP are around 50–150 µM. ATP can bind to the catalytic core regardless of whether the autoinhibitory sequence occupies the surface of the large lobe of the core. MLCK can also use ATPγS as a substrate, which is useful to essentially irreversibly phosphorylate myosin in the presence of phosphatase activity, which only very slowly dethiophosphorylates myosin (36).
MLCK enzyme activity
MLCK is sensitive to proteases
Like many contractile proteins, MLCK is susceptible to proteolytic degradation during purification from the tissue (37). Even highly purified preparations are susceptible to slow proteolysis in our hands (21). The enzyme activity is not particularly stable, we find that inclusion of reducing agents in buffers helps to some extent, but freezing and thawing are not recommended. Rabbit MLCK has been expressed in insect cells with full activity (38) (25, 39), where proteases are less of a problem, but in modest yields. There is one report of high yield expression in E. coli (16).
Proteolytic digestion studies have revealed regions of flexibility in MLCK and were useful to determine the sequence of domains that were later confirmed by direct sequence analysis. Digestion of chicken gizzard MLCK with α-chymotrypsin in the absence of Ca2+-CaM generates a ~95 kDa fragment that lacks the telokin domain and the N-terminal 63 residues but retains intact autoinhibitory and calmodulin-binding domains (19, 40). Similar digestion with chymotrypsin but in the presence of Ca2+-CaM yields a Ca2+-CaM-independent kinase of ~80,000 (41). Digestion with trypsin initially generates a 64 kDa fragment that lacks Ca2+-CaM-sensitive activity because CaM binding site has been removed but the inhibitory segment is retained. Further digestion releases the inhibitory segment to generate constitutively fragment of 61 kDa (42). This 61 kDa fragment does not bind or binds weakly to actin and has therefore lost an undetermined portion of the N-terminal actin binding domain.
Ca2+-CaM binding is the most important regulator of MLCK enzyme activity
The residues of MLCK that are required for catalytic activity have been well-characterized (reviewed in (26)). The most important regulator of MLCK activity is Ca2+-CaM. MLCK is catalytically inactive unless Ca2+-CaM binds to a region (Figs. 1 & 2) between the kinase and IgT domains. Thus, Ca2+-CaM activates MLCK by reversal of an auto-inhibited state. This induces a conformational change, which involves displacement of the auto-inhibitory sequence (Figure 1) from the surface of the catalytic core, thereby allowing substrate (RLC) access (23) and a global rearrangement of the interactions between the kinase domain and CaM (27). The specific amino acids in CaM, the kinase catalytic core, and CaM-binding region, and the partially overlapping auto-inhibitory segment that are involved in this mechanism have been characterized (see (27) for review).
MLCK activity assay and kinetic constants
MLCK specifically phosphorylates S19 on the RLC, whereas other kinases known to phosphorylate myosin in smooth muscle can in addition phosphorylate other sites, most importantly T18 (43, 44). In vitro, the steady-state kinase activity of MLCK is typically measured in low ionic strength buffers at pH 7–7.8 containing 5–10 mM MgCl2, 0.1–1 mM CaCl2, various CaM concentrations, and 1 mM γ[P32]-ATP at low ratios of enzyme to substrate. Intrinsic MLCK activity with RLC as the substrate is decreased about 10–20 fold by increasing the KCL concentration 10 fold from 0.05 to 0.5 M (45). A common substrate is the purified RLC from chicken gizzards. The essential light chain of myosin is not a substrate, nor is the heavy chain (but see (46)). The most common assay method is to apply aliquots of reaction mixtures, incubated for various times after addition of ATP, to paper discs which bind strongly to anionic proteins. After quenching the reaction on the discs in acid, extensive washing steps remove free ATP to allow the phosphorylated substrate (RLC or myosin) to be quantified by scintillation counting of the discs. A compilation of kinetic constants for MLCK can be found in the BRENDA enzyme database (http://www.brenda-enzymes.org). Typical specific activities of MLCK, calculated from initial velocities, using chicken or turkey gizzard RLC as a substrate are 5–20 µmol/min-mg under Vmax conditions. Apparent Kms for RLC and CaM are typically ~5–10 µM and 1–2 nM, respectively.
Steady-state mechanism of the myosin phosphorylation reaction
The steady-state mechanism of the myosin phosphorylation reaction catalyzed by the smooth muscle isoform of Ca2+-CaM-MLCK has not been fully characterized. For the skeletal isoform interacting with the RLC (47) kinetic studies are consistent with a rapid-equilibrium random bi-bi reaction model. This means that MLCK can bind either RLC or ATP first to form the MLCK · ATP · RLC complex before conversion to the MLCK·ADP·PRLC complex. The binding of ATP to MLCK and MLCK·RLC are similar, and similarly, the binding of RLC to MLCK and MLCK·ATP are similar. The skeletal isoform can form a dead end MLCK·ADP·RLC complex. It is not clear whether this is true for the smooth muscle isoform, which may not be significantly inhibited by ADP (47). This needs to be clarified because free ADP concentrations can be rather high in some smooth muscles. For example ADPfree in an unstimulated carotid artery can be from 50–150 µM (48). Both the skeletal and smooth (20, 47) isoforms show strong product inhibition, meaning that the PRLC and the RLC bind with similar affinity. An integrated study is in order because important differences likely exist between the isoforms and substrates cannot always be directly compared, i.e. RLC versus SMM. However, SMM is a difficult substrate to study because it has two active sites and forms filaments in a phosphorylation-dependent manner. An alternative substrate is the double-headed HMM, which does not contain the filament-assembly domain at the C-terminus of the tail or rod domain.
Factors limiting the effective activity of MLCK in the muscle
Factors that likely limit the effective MLCK activity and thus the rate of contraction in the muscle are: 1) MLCK is a low abundance enzyme relative to myosin, 2) MLCK is tightly bound to the contractile apparatus (5, 21, 49), 3) MLCK can be phosphorylated by other kinases at a serine in the CaM binding region, which weakens Ca2+-CaM binding (50–53), phosphorylation rates can be modulated by telokin (see below), 4) the accessible pool of CaM is often not sufficient for MLCK saturation (53–56), and 5) other unknown factors limit the fractional activation of MLCK (57). Even with these limitations and constraints, MLCK is able to rapidly phosphorylate ~80–90%% of the myosin upon agonist stimulation (see below). Typically however, maximal phosphorylation levels observed in intact muscles are usually below 50%, with basal levels commonly at ~10–30%
Abundance of MLCK in smooth muscles
Several studies have investigated the concentration of MLCK in muscle tissue. Typical values range from 1 to 8 µM (53, 57–59). In comparison, the concentration of myosin is much higher, for example it is ~50–80 µM in gizzard tissue and 100 µM in ovine carotid arteries (57). Actin concentrations are higher yet, ranging from 0.8–1.6 mM (59–61). The exact ratios of the proteins most likely varies greatly from muscle to muscle. For example, the ratio of actin to myosin in common carotid arteries is 30:1 (62).
Fractional activation of MLCK in muscle
It has been shown by several groups that the in situ specific activity of MLCK in smooth muscle is far below the values expected for maximal activation. Recent careful quantification of this effect in intact ovine carotid arteries shows that the fractional activation of MLCK may be developmentally regulated (57). By comparing the in situ specific MLCK activity and the specific activity of muscle homogenates saturated with added Ca2+-CaM (maximal), it was found that the fractional activation of MLCK was only 1.7% in adult and 9% in the fetal arteries. These values are reasonably consistent with other similar measurements (54). Age-related shifts in MLCK isoforms, or age-related differences in rates of change in cytosolic Ca2+ concentration did not appear to be the reason for the decrease in MLCK specific activity during postnatal maturation. Furthermore, in this case the in situ CaM concentrations were actually greater in adult versus fetal tissue, and were probably sufficient to saturate MLCK. This is not always true in all smooth muscles (53, 54). It is possible that the MLCK is post-translationally-modified but rates of MLCK phosphorylation are typically inhibited if MLCK is bound to CaM. Other potential reasons include the presence of an MLCK population that is not co-localized near myosin that does not produce force or phosphorylation of myosin.
Mechanism of MLCK access to SMM in the muscle
An interesting question is how MLCK phosphorylates smooth and nonmuscle myosins in the 3D environment of the muscle. It is not understood how MLCK catalyzes the phosphorylation of the majority of myosin in smooth muscle if it remains tightly bound to the contractile apparatus. MLCK is an elongated and potentially flexible molecule (19, 24, 25) with dimensions that allow it to simultaneously bind actin and myosin. The actin and myosin binding domains described above, although each with relatively weak affinity, could explain the tight interaction with the contractile apparatus. It is possible that the putative extensible or flexible characteristics of MLCK are sufficient to allow it to access a sufficient amount of myosin to initiate contraction, especially if only one head of myosin must be phosphorylated for mechanical activation (63). Calculations considering molecular dimensions suggest that at most 8 local myosin heads could be phosphorylated by one MLCK. Further phosphorylation could then proceed as contraction brings the actin-tethered (see below) MLCK proximal to the remaining myosin. Alternatively, the known large conformational changes in myosin upon phosphorylation (64) may play a role, where phosphorylation itself could enhance access of the MLCK to other myosin heads. Any model requiring contraction is weakened by the fact that ~90% of all the myosin in permeabilized chicken gizzard tissue can be thiophosphorylated in the absence of any contraction (65). This suggests that relative sliding of the thick and thin filaments are not required for MLCK to access most of the myosin. This experiment is possible because myosin hydrolyzes ATPγS 500 times more slowly than ATP (66, 67) and does not support motion in an in vitro motility assay. It is interesting that MLCK can access and thiophosphorylate ATPγS-myosin heads effectively, because this nucleotide traps myosin in the weak actin-binding state resembling the pre-hydrolysis ATP-myosin intermediate, which is thought to be a poor substrate for MLCK (see below). Also under this condition, no tension is generated and most of the myosin heads would not be expected to be bound to actin, which might limit the access of MLCK to SMM.
Does the affinity of MLCK for the contractile apparatus weaken upon contraction?
A possible explanation for “apparent mobility” of MLCK on the contractile apparatus is that it might somehow be more mobile or more weakly bound in activated muscle, thus explaining its apparent accessibility to the myosin. Studies in cultured A7r5 cells suggests that this is not the case (38). However, in tracheal smooth muscle, while permeabilization of strips with 1% Triton-X-100 caused no loss in MLCK, ~25% of it was lost from the muscle after one contraction cycle. Interestingly, there was also a 25% decrease in total protein after one contraction cycle, consistent with other studies (68). Therefore it is not clear whether the loss of MLCK was a specific effect, or due to a more general damage to the tissue upon contraction. Further quantitative studies like these are needed to address this important point.
Proposed myosin-assisted movement of MLCK on the contractile apparatus
It has also been suggested that MLCK moves on myosin filaments in a myosin-assisted manner (69, 70). This idea arose from kinetic studies of the rate of phosphorylation of SMM in solution with known low amounts of added MLCK-CaM. Monomeric myosin was compared with filamentous myosin. All the monomeric myosin could be eventually phosphorylated but phosphorylation of filamentous myosin could only reach sub-stoichiometric levels. This fit with a model in which the MLCK remained tightly bound to filaments and was unable to diffuse between filaments, whereas SMM monomers could be phosphorylated by freely diffusing MLCK allowing for complete phosphorylation. Neither the actin-binding nor the myosin-binding domains of MLCK are required for this behavior, because the catalytic core of MLCK also behaved in the same manner. This is unexpected because the catalytic core of MLCK binds more weakly to myosin filaments than full length MLCK because it lacks the IgT domain. The authors have further proposed that the mechanism is “vectorial” or directional, where the MLCK translocates along the myosin filaments during the phosphorylation process, but this has never been directly demonstrated. The mechanism is proposed to be myosin-assisted because single-headed myosin did not exhibit the vectorial-like kinetics. However, a more simple explanation is that the MLCK simply does not bind as tightly to single-headed versus double-headed myosin.
Increased MLCK expression and activity have been linked to many disease states
Both the nonmuscle and smooth muscle isoforms of MLCK have been directly linked to many chronic and acute human diseases. Selective over-expression of MLCK in asthmatic airway smooth muscle contributes to airway hyperresponsiveness observed in asthma (71, 72) and variants in the enzyme are associated with severe asthma (73) and increased susceptibility to sepsis (74). Over-expression of MLCK in the vascular endothelium of transgenic mice leads to susceptibility to inflammatory lung disease (75). The hyper-contractility of sensitized venous smooth muscle is correlated to an increase in MLCK and is likely to be relevant to the known in vivo hyper-reactivity in anaphylactic shock (72, 76). Lessons from studies of the nonmuscle isoforms of MLCK, suggest that even very small increases in MLCK expression can lead to serious dysfunction. Increased nonmuscle MLCK activity has been shown to be necessary and sufficient for several barrier dysfunctions (lung and intestinal epithelium (77–79), and microvasculature (80). These dysfunctions have been linked to human diseases and conditions such as colitis (81), Crohn’s disease, inflammatory bowel disease (82, 83), diarrhea (84) and increased vascular permeability due to burn-induced edema (85) and sepsis (74). Membrane-permeant MLCK inhibitors have been shown to restore intestinal barrier dysfunction (86). Although increased MLCK expression has been linked to most of these diseases, it remains unclear why the relatively small increases in MLCK content (~2-fold) have such profound consequences.
MLCK domain interactions with contractile proteins
Interactions of MLCK with myosin
The catalytic core binds to myosin
There are two sites on MLCK that interact with myosin. The structure of myosin is summarized in Figure 4. First, the catalytic core of MLCK must interact with the highly positively charged and flexible N-terminal domains of the two RLC, which contain the phosphorylated S19s to allow for the phosphorylation reaction. Specifically, R16, Q20, V21, F22, and undefined structural determinants in the N-lobe of the RLC are important for substrate recognition by MLCK (27).
The IgT domain binds to myosin
The other site of interaction of MLCK with myosin is through the IgT domain. Deletion of the IgT domain from MLCK increases the Km for SMM by about a factor 3, but has little effect on the Km for RLC (19). This suggests that the role of the IgT domain of MLCK is to improve catalytic activity by binding to structural determinants near the RLC. Much has been learned about the interaction of the IgT domain of MLCK with myosin by studying telokin (87), which is an independently expressed protein that is identical in sequence to the IgT domain of MLCK. Telokin competes with MLCK for binding to myosin (18, 21), and telokin binding increases the Km but not the Vmax of phosphorylation of HMM by MLCK (17, 88) or SMM, but not the RLC (19). HMM is a truncated SMM fragment that lacks the C-terminal 2/3 of the tail or rod domain, but retains an intact head-tail junction and a full light chain complement (Figure 4). The apparent inhibition constant (Ki) of telokin is 20 µM (19) to 40 µM (18). Telokin interacts with unphosphorylated myosin with 1:1 stoichiometry with a Kd of ~5–10 µM (17). The affinity to phosphorylated myosin is very weak (15). The affinity of tissue-purified chicken MLCK for unphosphorylated myosin is approximately 10 fold stronger, Kd is 0.8 µM (15).
MLCK binds to the SMM head-tail junction
There are several lines of evidence that the IgT domain of MLCK binds to the SMM head-tail junction. The head-tail junction of SMM is where the rod or tail domain merges with the heads or S1 domains (Figure 4). The RLC subunits reside in this region of the head, placing the N-terminal lobe and thus the phosphorylated S19s of the two RLC proximal to the head-tail junction. First, telokin binding inhibits the rate of proteolysis at the head-tail junction of both the RLC and the heavy chain of SMM (17). Myosin fragments that lack an intact head-tail junction bind weakly. For example the isolated head or S1 domain and the isolated rod domain do not interact with as high affinity as HMM, and soluble fragments of SMM that lack the motor domains but contain the S2 and RLC binding regions of the heavy chain together can compete with SMM for telokin binding (17). S2, the region of the rod or tail that is proximal to the myosin heads, can also compete for binding but not as efficiently (Kd not determined). Although weaker, the interaction of telokin with the S2 region may be interesting. The SMM rod contains a motif (1739LEARIAQLEEELDEEHS1756) that is homologous to a skeletal myosin rod sequence that binds to titin’s myosin-binding Ig domain (89) and might similarly bind MLCK’s IgT domain. This binding site may be exposed during cycling of the myosin heads with actin, when it is thought that the S2 region detaches from the filament backbone (90–92).
Crosslinking studies have shown that treatment of a mixture of telokin and myosin with EDC, a cross-linker that couples D and E to K side chains, results in crosslinking to the RLC and the SMM heavy chain (17, 93). Consistently, proteolytic studies show that telokin protects the RLC and the heavy chain from digestion (17). It would be of interest to determine the sites of crosslinking in this complex, as it might shed light on the potential dual binding modes of telokin to myosin.
Visualization of the MLCK-SMM interactions
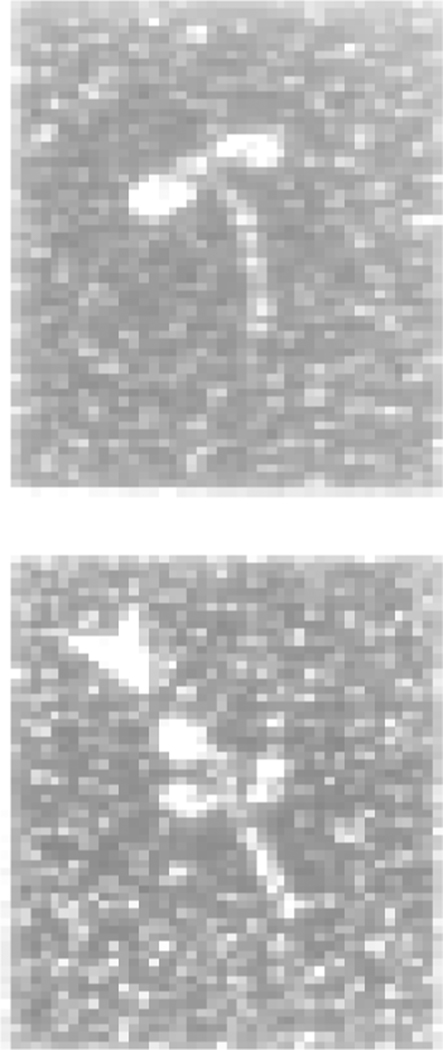
The interaction of both MLCK and telokin with the SMM head-tail junction has been visualized by EM. The relatively weak (low µM range) Kds prevent visualization of the unmodified complexes, but they were evident after EDC cross-linking. Figure 5 shows electron micrographs of full length MLCK cross-linked to SMM (19). Similar complexes of telokin-SMM have been visualized after rotary shadowing (93). A clear interaction of MLCK and telokin with the head-tail junction of unphosphorylated myosin is apparent, consistent with the biochemical studies summarized above.
Figure 5.
Molecules of HMM (top) and EDC cross-linked HMM-MLCK complexes (bottom) observed with the electron microscope after rotary-shadowing with platinum. Arrows indicate the position of MLCK. Data reproduced from (19).
The C-terminus of the IgT domain is important for MLCK binding to myosin
The crystal structure of turkey gizzard telokin has been solved at 2.8Å resolution ((94);PDB 1TLK) with a refinement to 2.0Å (PDB 1FHG). Of the 157 amino acids, 103 were visible in the electron density map. The amino-terminal 35 residues and the carboxyl-terminal 19 residues were unresolved in the crystal structure. The structural core is a seven-stranded C2 immunoglobulin-like beta barrel. Eight amino acids on the N-terminal side of the barrel were visible in an extended structure but amino acids 1–35 were not ordered. This N-terminal region contains several phosphorylation sites. Neither phosphorylation nor deletion of this N-terminal region affected the interaction with myosin (17). Telokin is an acidic protein with a pI value of 4.5, largely due to the disordered C-terminal region, which is highly acidic (residues 136–157; Figure 6). It is this region that is most important for telokin-myosin interaction and myosin filament stabilization properties ((17); see below). In vitro assays with bacterially-expressed telokin truncation mutants indicated that sequence G138–E150 is the primary determinant of myosin binding. The C-terminal sequence is AMISG… 139GK(EG)4EEDEEEEEE157. Removing the residues 151 to 157 had little effect, but further truncation from the C-terminus weakened binding and diminished filament stabilizing effects. The minimal binding locus was found to be GK(EG)4EE Sequence alignment (Figure 6) shows that the first EGEGE is highly conserved and all forms of telokin and MLCK contain between 5 and 10 additional C-terminal repeating E residues. We have found that Sf9 cell-expressed rabbit MLCK binds to myosin about 3–4 times more weakly than tissue-purified chicken MLCK (data not shown), consistent with a prior study (18). Interestingly, the two avian isoforms seen in Figure 6 are different in the C-terminal region from the mammalian isoforms, possibly explaining the differences in binding affinity. The β-barrel motif binds only weakly to myosin (17). The fact that telokin interacts with myosin largely through a potentially disordered acidic domain suggests that it interacts with a basic region on myosin, which is a property of the RLC N-terminus and the myosin heavy chain region of the head-tail junction.
Figure 6.
Amino acid sequence alignment of the C-terminal region of MLCKs from various sources showing, the differences in the glutamyl residues at the C-terminus. “*” indicates conserved residues and “:” indicates amino acid similarity. Underlined region is described in the text.
MLCK and telokin in stomach and uterus are substrates for the protein-deglutamylating enzyme CCP1
It has recently been shown that both MLCK and telokin found in the stomach and the uterus are substrates for the protein-deglutamylating enzyme CCP1, which removes the last 7 E residues from the C-terminus of both proteins (95). The functional significance of this deletion is not known. Interestingly, a similar enzyme activity may be present in avian gizzard because telokin isolated from this tissue is heterogeneous at the C-terminus. Six different C-terminal peptides corresponding to the removal of one to six C-terminal glutamyl residues were characterized by mass spectrometry (96). It is likely that MLCK also shows similar heterogeneity, but this has not been demonstrated, nor have similar C-terminal deletion studies been done with MLCK as described above for telokin. In light of the potential for physiological significance, and the relationship to myosin binding affinity, caution should be paid to MLCK heterogeneity from tissue-purified preparations, and potentially also expressed recombinant preparations.
Telokin-SMM interaction stabilizes SMM filaments
Telokin binding to SMM promotes filament assembly in vitro (18). If telokin is cross-linked to SMM, the SMM can no longer adopt the self-inhibited 10S intramolecularly folded conformation (64) that is normally stable in the presence of ATP (93)(Figure 4). Also, MLCK binds weakly to 10S myosin (22) but more strongly to 6S myosin (extended). This explains why telokin stabilizes filaments, which are normally in rapid equilibrium with 10S in the presence of ATP. The simplest explanation is that telokin and the portion of the tail that interacts with the heads in 10S myosin compete for the same binding site at the head-tail junction. We suggest that the telokin domain on MLCK may not serve only to target the MLCK to myosin but more specifically to target MLCK to the contractile apparatus versus the cytosolic 10S monomers (97). This would place the MLCK on the filaments, which is the force generating form of myosin. If this is true, it might impact the effective concentration of MLCK on the filaments.
The inhibitory effect of telokin may not be restricted to MLCK-SMM interactions
Recently, Shcherbakova et al (88) confirmed that telokin can inhibit the rate of phosphorylation of purified HMM. Interestingly, their studies also show that telokin strongly inhibits the rate of phosphorylation of HMM by a 61kD fragment of chicken gizzard MLCK that lacks the IgT domain. This suggests that the inhibitory properties of IgT are not solely due to direct competition of IgT for MLCK binding, but also might include masking the phosphorylation site itself. C-terminally truncated telokin, lacking the known SMM binding region (discussed above), no longer inhibited the activity of the 61 kD fragment of MLCK. Furthermore, telokin also inhibits contraction and RLC phosphorylation in Triton-skinned taenia coli fibers elicited by non-canonical RLC kinases such as such as zipper-interacting kinase and/or integrin-linked kinase (88). This raises the important possibility that the inhibitory effect of telokin in vivo is broader than originally envisioned and not restricted to inhibition of MLCK interactions with SMM.
The CaM-MLCK complex co-purifies with SMM from tissue
Although MLCK binds to SMM filaments in vitro, (15, 19–21) through the IgT domain and the kinase domain, it has not been established that this interaction is important to stabilizing MLCK on the contractile apparatus in vivo. However, many studies (20–22, 98) report that MLCK and CaM co-purify with SMM using various large scale preparations of SMM from chicken gizzards. Indeed, it is usually sufficient to add ATP and Ca2+ to purified SMM preparations to effect phosphorylation by the co-purifying MLCK-CaM. Hong et al (21) quantified the co-purifying MLCK and found that the MLCK:SMM ratio in the purified preparations was very high, about 23–37% of the ratio found in the tissue. Two groups have reported that the co-purifying MLCK may be heterogeneous with regard to myosin binding affinity, with one fraction binding with much tighter affinity, 30 nM (20) or 200 nM (21), and the other with weaker affinity, 1.3 µM or 10 µM, respectively. Binding heterogeneity may be related to C-terminal deletions of E residues as discussed above, but this remains to be determined. Both groups report that phosphorylation of the myosin weakened the affinity of MLCK but not to the extent reported for the interaction of purified MLCK with SMM.
Since the prevailing evidence suggests that actin-MLCK interactions are the most important for high affinity MLCK binding to myofilaments (see below), many reasoned that the co-purification of MLCK with SMM might simply be due to MLCK co-purifying with contaminating actin in the SMM preparations. However, recently it has been shown that this is not the case, and that the MLCK is binding directly to SMM (21). Furthermore the co-purifying MLCK has normal enzyme activity and the SMM that it phosphorylates functions normally in an in vitro motility assay. This suggests that the MLCK-SMM complex is functional.
Ca2+-CaM weakens MLCK-myosin interactions
It is interesting that Ca2+-CaM weakens the binding of MLCK to SMM by about 3 fold (15). The IgT domain of myosin has no Ca2+-CaM binding affinity. Therefore, Ca2+-CaM binding to its binding domain, which is adjacent to the IgT domain, may alter the IgT conformation in an allosteric manner, which may lead to weakened interactions with SMM. Alternatively, the effect may be unrelated to the IgT domain but rather affecting the interactions of N-terminus of the RLC directly. Further work is needed to address the mechanism for this effect.
Interactions of MLCK with actin
Three DFRxxL motifs at the N-terminus of MLCK interact with actin
It is clearly established that MLCK binds to F-actin. All studies have used actin that was purified from mammalian skeletal muscle (15, 59). When tropomyosin is complexed with F-actin the affinity of MLCK for actin increases 2–3 fold (15). As can be seen in Table 1, the measured binding affinities to purified F-actin vary considerably, even though buffer conditions are similar. The binding of F-actin to MLCK is weakened at increasing ionic strength, although reports differ significantly concerning the magnitude of this effect. Dabrowska reports a minor weakening of the affinity by about 20% from 0.1 to 0.2 M NaCl (59), but Sellers and Pato (15) report a much larger effect where the Kd changes from 1 to 50 µM between 50 and 150 mM NaCl concentration. It may be that the small differences in ionic strength or buffers contribute to the variances seen in Table 1. This effect should be further clarified due to the potential for physiological significance.
Table 1.
Comparison of binding affinities of different MLCK constructs to smooth muscle myofilaments and to F-actin purified from skeletal muscle.
| Filament(1) | Kd(2) (µM) |
MLCK construct | Buffer conditions | Reference |
|---|---|---|---|---|
| Chicken gizzard myofilaments (detergenht-washed myofibrils) | 15 | Rabbit (recombinant and purified from tissue) | 50 mM MOPS, 0.5 mM EGTA, 10% glycerol, 1 mM DTT, pH 7.0 | (38) |
| Turkey gizzard myofilaments (detergent-washed, MgCl2 pre-treated myofibrils) | 1.2 | Rabbit MLCK (recombinant) | 10 mM MOPS pH 7, 50 mM NaCl, 2 mM DTT, 1 mg/ml BSA, 1 mM MgCl2, 0.1 mM EGTA | (39) |
| Chicken gizzard myofilaments (detergent-washed myofibrils) | 0.1 | Rabbit GST-1-75 Phosphorylated at S73 | 10 mM imidazole, pH 7.2, 50 mM KCl, 1 mM MgCl2, 1 mM DTT, 0.1 mM EGTA, 10% glycerol and 0.2 mg/ml BSA | (102) |
| Purified F-actin | >35 25(3) | Rabbit MLCK (Recombinant) | 10 mM MOPS pH 7, 50 mM NaCl, 2 mM DTT, 1 mg/ml BSA, 1 mM MgCl2, 0.1 mM EGTA | (39) |
| Purified F-actin | 0.8 | Rabbit GST-1-75 Phosphorylated at S73 | 10 mM imidazole pH 7.2, 50 mM KCl, 1 mM MgCl2, I mM DTT, 0.1mM EGTA, 10% glycerol, 0.2 mg/ml BSA | (102) |
| Purified F-actin | 13.3 | Peptide 1–114 MLCK Both from chicken | 50 mM KC1, 25 mM Tris-HC1 (pH 7.6), 0°C | (11) |
| Purified F-actin | 4 14(4) | Tissue-purified chicken MLCK | 50 mM NaC1, 10 mM Mops (pH 7.0), 1 mM MgCl2, 1 mM dithiothreitol, 0.1 mM EGTA, 0.5 mg/ml BSA, 25 °C | (15) |
| Purified F-actin | 2.2 | Tissue-purified chicken MLCK | 50 mM KCl, 20mM Tris-HCl (pH 7.5), 5 mM MgCl2, and 0.1 mM EGTA | (14) |
| Purified F-actin | 2.7 | Bovine Peptide 1–337 | 50 mM KCl, 20mM Tris-HCl (pH 7.5), 5 mM MgCl2, and 0.1 mM EGTA | (14) |
| Purified F-actin | 5.5 | Bovine peptide 319–721(5) | 50 mM KCl, 20mM Tris-HCl (pH 7.5), 5 mM MgCl2, and 0.1 mM EGTA | (14) |
| Purified F-actin with bound tropomyosin | 1 2(4) | Tissue-purified chicken MLCK | 50 mM NaC1, 10 mM MOPS (pH 7.0), 1 mM MgCl2, 1 mM dithiothreitol, 0.1 mM EGTA, 0.5 mg/ml BSA, 25 °C | (15) |
All myofilament values are expressed relative to the amount of actin in the myofilaments. Therefore, values are not necessarily actual binding constants to actin because myofilaments also contain a large amount of myosin.
Apparent equilibrium dissociation constants.
Quoted by the authors.
In the presence of saturating Ca2+-CaM.
Not sensitive to Ca2+-CaM.
A portion of the MLCK-actin binding interaction is due to three DFRxxL motifs at the N-terminus of MLCK (11–14). Negative staining and helical 3-D reconstruction (2.5–3.0 nm resolution) of F-actin after decoration with a peptide containing the NH2-terminal 147 residues of MLCK showed that. one side of MLCK-147 is close to COOH-terminal residues Arg374, Val372, Glu366, and Tyr364 on subdomain-1 of one actin monomer and that the opposite side of the peptide closely approached an α-helix (actin residues 228–232) protruding from subdomain-4 of the neighboring monomer (99). This latter site of interaction on F-actin does not appear to overlap with any other actin-binding protein interactions (100). However, the interaction near the C-terminus of actin may be within a hydrophobic cleft between actin subdomains 1 and 3 that appears to be a ‘hot spot’ for actin-binding proteins (101). If this is true, it suggests that MLCK-actin interactions may be regulated by other actin-binding proteins.
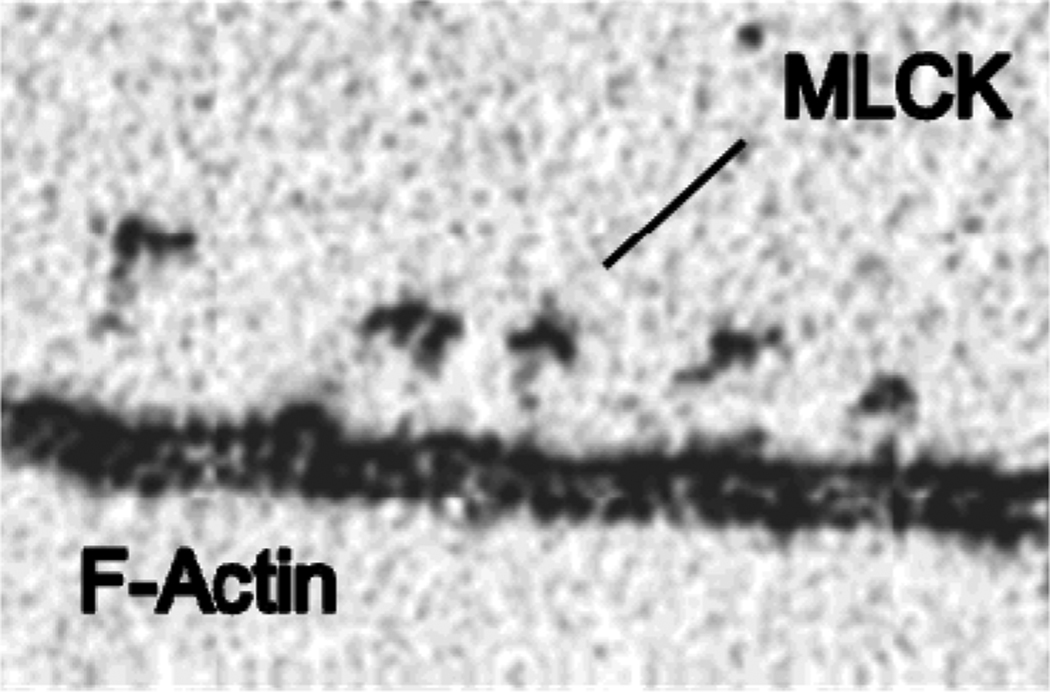
Figure 7 shows an electron micrograph of rabbit MLCK molecules cross-linked to F-actin, presumably through the DFRxxL domains. Smith and Stull (102) found that a peptide constituting the first 75 residues of MLCK bound to actin with a stoichiometry of 3 actin monomers/peptide. They proposed a model in which each of the 3 DRFxxL motifs binds on adjacent F-actin monomers, based upon unpublished findings that the peptide showed no secondary structure by circular dichroism. So far however, there is no direct structural evidence for this interesting model.
Figure 7.
Molecules of MLCK that were cross-linked to F-actin through the DRFxxL domains observed with the electron microscope after rotary-shadowing with platinum. Data from (25).
Ca2+-CaM weakens the binding of MLCK to actin by binding to the N-terminus
Ye et al (14) have shown that the 3rd DFRxxL motif (Pro26-Pro41 of bovine MLCK) is a Ca2+-CaM binding site with homology to other known Ca2+-CaM-binding proteins such as myosin V and neuromodulin. This explains the ability of Ca2+-CaM to weaken actin binding, a phenomenon observed by many investigators. Sellers and Pato showed that the presence of Ca2+-CaM weakens MLCK binding to actin from to 4 to 14 µM (15)(Table 1). Although more data is needed, it appears that Ca2+-CaM binds to this region of MLCK with a weak affinity of ~5 µM (14). This suggests that it would be at least partially occupied with CaM in the muscle, assuming the freely diffusible pool of CaM is ~10% of the total or about 3 µM (53). With regard to kinetic properties, data from the Stull lab showed that a construct of rabbit MLCK lacking the first 655 residues has a very weak KCaM of 28 µM versus the full length MLCK of 0.7 µM (39). Therefore, the full length MLCK can be enzymatically activated at a much lower Ca2+-CaM concentration, suggesting that Ca2+-CaM binding to the N-terminus somehow affects the activation of the kinase activity. This is an area that needs further study due to the potential for high physiological significance.
MLCK contains an additional actin binding region that is not sensitive to Ca2+-CaM
Ye et al (14) have shown that there is a Ca2+-CaM-insensitive binding site for F-actin between residues Gly338 and Val721 of bovine MLCK. This region contains IgG1 and IgG2 and the Fn domain (Figure 1), each of which are ~100 residues. The binding affinity of this fragment (5 µM) is very similar to that of the N-terminus (residues 1–117). Further studies are required to determine which domains within this region are most important for actin binding. The IgG domains are likely culprits based upon homology with other cytoplasmic IgG domains (103) that bind to actin such as palladin and the giant muscle proteins that include titin and twitchin. These structural motifs are conserved and therefore are predicted to have functional significance. Analogous to twitchin (104), myosin interactions with actin may compete with the binding of the MLCK IgGs to actin in activated muscle.
MLCK interactions with the thin filament
Several groups report that isolated thin filaments do not contain MLCK or contain very little MLCK (105, 106). However, this needs further investigation because the ratio of MLCK to actin is very low in smooth muscle (~1/100) and quantitative studies have not been pursued. Typical protocols for preparation of thin filaments from muscle (106–109) have no obvious steps that might promote weak interactions of MLCK with actin, such as high ionic strength or high MgCl2. Washed myofibrils are simply treated with ATP to dissociate actin and myosin, residual acto-myosin filaments are removed, and the clarified supernatants are centrifuged at high g-force to pellet the thin filaments and leave the 10S myosin in the supernatant. There could be several reasons for the lack of MLCK in thin filaments, such as proteolysis and weak binding of tropomyosin and other actin binding proteins at 4°C (105). Thin filaments can also be reconstituted (110). Our preliminary studies (data not shown; collaboration with Mike Walsh) suggest that MLCK binds with similar affinity to reconstituted actin-Tm and actin-Tm-caldesmon, but further work is needed.
Multi-domain interactions of MLCK in the cell and with macromolecular structures
Difficulty in measuring MLCK interactions
The published binding constants for MLCK to actin and myosin vary by large factors between various studies and simple binding isotherms often do not fit the data well. For myosin, the main differences appear when comparing affinities of exogenously added purified MLCK versus MLCK that co-purifies with myosin. With regard to actin binding, the two structurally separated binding sites may play a role, with different conditions affecting each differently. A number of factors can complicate the normal sedimentation approach used for actin and myosin interactions. MLCK-induced actin bundling and MLCK polymerization or aggregation may be complicating factors. It has been shown that EDC and other cross-linkers generates dimers and other oligomers of MLCK (111, 112) which are more prominent at the lower ionic strengths commonly used for sedimentation studies (<100 mM added salts).
MLCK weakly stimulates myosin ATPase activity without phosphorylating the myosin light chain
The Kohama group has several reports of MLCK and MLCK constructs that stimulate the myosin ATPase activity without phosphorylating the myosin light chain (16, 113–116). The maximal observed stimulation is about 4-fold. Therefore, the activated myosin does not behave like phosphorylated myosin, which is activated ~500–1000 fold by phosphorylation. Furthermore, it is not clear whether the effect has physiological significance because very high concentrations of MLCK are required to observe the effect. Progress has been made on understanding the region of MLCK required for this effect (114). Interesting studies show that this nonkinase activity of MLCK may play a role in elongated filopodia formation and chemotaxis of vascular smooth muscle cells toward sphingosylphosphorylcholine (117).
MLCK interactions in the cell
The prevailing evidence suggests that the most important interaction of the MLCK in smooth muscle cells is binding to actin through 3 actin-binding motifs in the N-terminal actin-binding domain (11, 102). The N-terminal actin-binding domain of MLCK is necessary and sufficient for binding to myofilaments. (12) However, a potential caveat is that these studies were done with myofilaments that were extracted with 50 mM MgCl2 to remove the endogenous kinase. It is not known why high MgCl2 concentrations effectively extract MLCK. However, one interaction that is known to be weakened by MgCl2 is between MLCK and purified actin (13). It is possible that proteins critical for tight binding of MLCK to SMM filaments were also extracted from the myofilaments under these conditions. For example, myosin phosphatase can also be extracted under these conditions (118). Consistent with this idea is that the actin-binding domain of MLCK (GST-N1-75MLCK), even at concentrations much higher than the measured Kd, displaces only about 50% of the endogenous MLCK from gizzard myofilaments that have not been treated with 50 mM MgCl2 (12, 21)). However, if the endogenous kinase is extracted with MgCl2 first, followed by incubation of the myofilaments with an excess of the first 75 residues of MLCK (GST-N1-75MLCK), binding of exogenously added MLCK is completely prevented (12). Other important observations of localization of fluorescently-labeled MLCK in transfected A7r5 cells strongly suggest that MLCK binds to cellular stress fibers only through the actin-binding domain (38). However, in these experiments GFP was fused to the C-terminus of MLCK, which could potentially affect SMM filament binding. In addition, some experiments were performed under conditions that strongly promote depolymerization of SMM to the 10S conformation, which can diffuse out of permeabilized cells (119) and which does not bind to MLCK (22, 45). Other experiments with Cy3-labeled MLCK in Swiss 3T3 cells or in primary bovine tracheal smooth muscle cells in culture (39) do not exclude the possibility that some of the MLCK co-localizes with SMM. It is known, for example, that the expression of SMM is down-regulated in such cells (120). Also, conclusions here were based upon a construct that has a 10-fold higher apparent Km for the RLC suggesting it does not bind as strongly as wild type. Other studies have shown that the 130 kDa isoform of MLCK co-localized with myosin IIA but not with myosin IIB or F-actin in bovine pulmonary artery endothelial cells.(120) Similarly, antibodies to smooth MLCK co-localize with the periodic distribution of myosin along the stress fibers of gerbil fibroma nonmuscle cells.
MLCK interactions with the myofilament
Several studies have examined the in vitro interaction of MLCK in myofilaments purified from smooth muscle (Table 1). Myofilaments contain many proteins, principally actin, myosin, caldesmon, calponin, tropomyosin, CaM, and MLCK. Typically, the muscle is minced, homogenized and washed in a low ionic strength buffer containing Triton-X-100 to release any soluble proteins, leaving the contractile proteins in the form of washed myofibrils. Myofibrils are incubated with MLCK and the amount of MLCK in the supernatant versus the pellet is determined after sedimentation of the myofibrils. Values are relative to the amount of actin in the myofibrils. More data are needed but it appears that pre-treating the myofibrils with MgCl2 (which extracts endogenous MLCK from the filaments) significantly increases their affinity for rabbit MLCK. It is not clear though why the peptide 1–75 binds very tightly even without pre-treating the myofibrils with MgCl2. This is not consistent with other studies from the same laboratory showing that the peptide 1–75 competes with endogenous MLCK on myofibrils (not MgCl2 extracted), but with very weak apparent affinity (~80 µM), unless there is a kinetic effect that limits the rate of dissociation under the experimental protocol. This is inconsistent with the very high affinity for direct binding of the peptide 1–75 to myofibrils (Table 1). A similar protocol in which a tagged MLCK competes with the peptide 1–75 for binding to MgCl2-extracted myofibrils gave an apparent binding affinity of the peptide of 10 µM with complete inhibition of binding of the tagged MLCK. Therefore further studies are needed to clarify these points. It is not clear whether or not the myofilament affords a higher affinity binding environment for MLCK than the purified F-actin or purified myosin filaments. It also is not clear whether the MLCK state found in endogenous unextracted myofibrils can be achieved by adding purified MLCK to myofibrils.
The direct in vitro binding studies summarized in Table 1 concerning myofilaments cannot explain why endogenous MLCK has such a tight association with the contractile apparatus in the muscle. None of the individual domains appear to bind with high enough affinity to account for this. Perhaps the combination of the many weak interactions is sufficient to keep it from dissociating, or other proteins such as the phosphatase targeting subunit, or caldesmon, may be involved. There is evidence that at least some of the MLCK that is bound to SMM during purification is perhaps more tightly associated than purified MLCK. It is possible that it is in a different conformational state.
In vitro assays of MLCK interactions with actin and myosin
MLCK interactions with actin and myosin using an in vitro motility assay
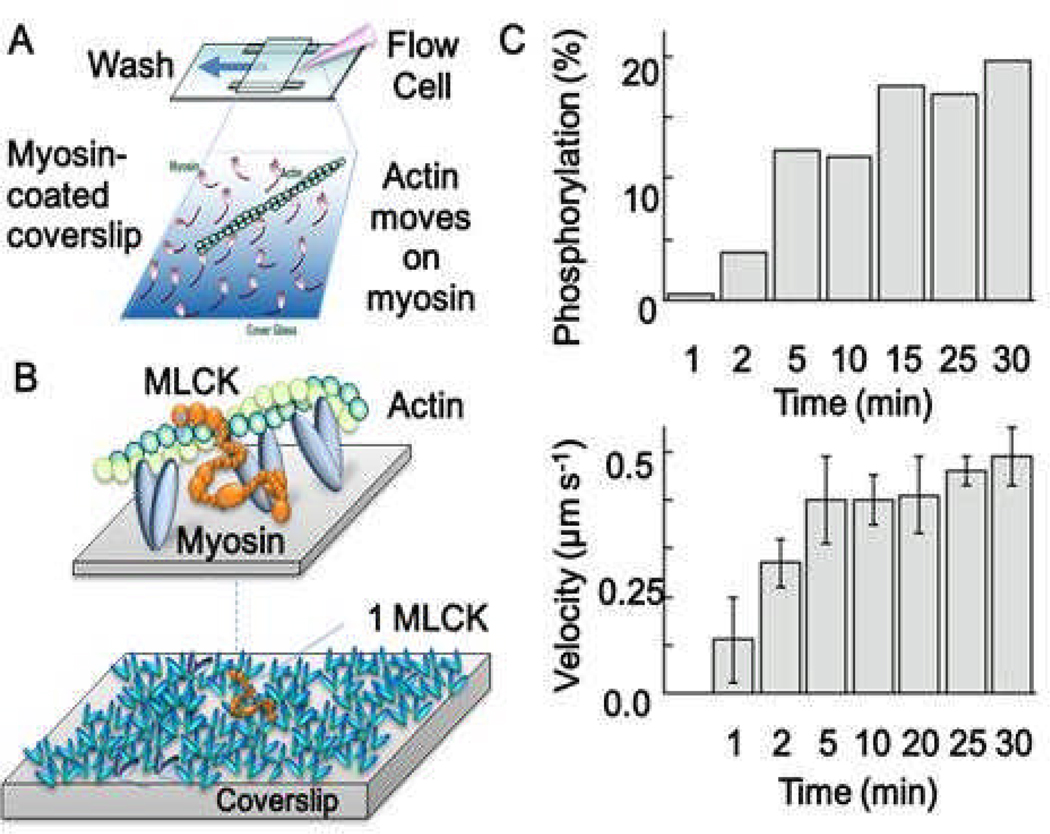
We have initiated a study of MLCK interactions with actin and myosin (21) using an in vitro motility assay (Figure 8A). Typically, this assay consists of a bed of phosphorylated myosin attached to a microscope coverslip over which fluorescently labeled actin filaments slide in the presence of ATP. We have modified this system by starting with unphosphorylated myosin containing co-purified MLCK-CaM. We have shown that phosphorylation of SMM by the interacting MLCK and the resulting activation of actin sliding exhibit a Ca+2-CaM dependence similar to that observed in smooth muscle cells (21). The advantage of this model system over studying regulation in smooth muscle cells is that the constituents and geometry can be controlled while measuring mechanics (using fluorescence imaging of actin sliding), SMM phosphorylation (using an on-coverslip ELISA assay), and single molecule dynamics (using total internal reflectance fluorescence, TIRF, microscopy). In experiments where Ca2+ was added (at time=0) to a system containing unphosphorylated SMM/MLCK/CaM (no actin) bound to the coverslip, the subsequent phosphorylation of SMM (Figure 7C, top) and activation of actin-SMM mechanics (Figure 8C, bottom) both occurred on a relatively slow (~120 s) time scale. Although the ratio of SMM heads to MLCK was roughly 200:1, nearly 20% of SMM were phosphorylated in this time (Figure 8B, bottom). To illustrate the approximate stoichiometry of the experiment, all of the myosin heads in Figure 8B (bottom) were eventually accessible to one MLCK. Actin was not required for this process. We do not know what limits the slow rate of activation in this assay, but preliminary work suggests that a free-diffusional process would be much faster than we observe here. This is consistent with the fact that the MLCK appears to be tightly associated with the myosin surface and cannot be washed away in these experiments. Studies are in progress to observe the behaviors of single MLCK molecules in this assay to see if MLCK-SMM interactions may be limiting the rate of myosin phosphorylation.
Fig. 8.
A. In vitro motility assay set-up. B (top), approximate dimensions of MLCK relative to actin and myosin. MLCK (orange) is shown simultaneously interacting with SMM heads (blue ovals) and actin (green). B (bottom), Illustration drawn approximately to scale showing one MLCK molecule surrounded by ~ 200 SMM molecules. Given the density of SMM and the stoichiometry of MLCK/SMM in the experiment in C, one stationary or attached MLCK must phosphorylate all heads in this image to account for phosphorylation of 20% of the total SMM on the coverslip. C. At time = 0, Ca2+-CaM was added to 100 µg/ml to unphosphorylated SMM containing co-purified MLCK-CaM on the coverslip surface. This initiated SMM phosphorylation and caused an increase actin sliding velocities. MLCK/SMM heads is ~1/200.
Can MLCK tether actin and myosin together in vitro?
Since MLCK has actin and myosin binding activity on opposite ends of the molecule, it is possible that conditions could be found where MLCK could serve as a “brake” on contraction. Kohama’s group has tested this using a standard motility assay in which the velocity of fluorescently-labeled F-actin filaments propelled by myosin can be measured. They found that less than 5 nM bovine MLCK or 20 nM of a peptide 1–114 from chicken MLCK almost completely inhibited the motion of actin filaments moving on a surface of pre-phosphorylated SMM monomers (14, 16, 121). Because the peptide 1–114 caused the inhibition, the mechanism cannot be a direct application of a drag force by simultaneous actin and myosin binding. Interestingly, for both proteins the addition of Ca2+-CaM to the assay relieved the inhibition of motion. The MLCK concentrations used for inhibiting motion are similar to those used in frictional loading experiments induced by alpha-actinin (122). Interestingly, the N-terminal region of twitchin kinase, which has some similarities to MLCK, binds thick and thin contractile filaments We have tried to reproduce some of the finding described above. We do not observe any drag force as measured by reduction in actin sliding velocity or by fragmentation of actin upon adding either tissue-purified chicken gizzard or recombinant rabbit MLCK, or peptide 1–75 of chicken MLCK to actin moving over pre-phosphorylated SMM in a motility assay. This was true even at concentrations far exceeding those used by Kohama’s group. In fact, we find that their conditions of loading MLCK/actin mixtures onto the coverslip in the presence of ATP leads to very poor actin binding to myosin. We do not know the reasons for these discrepancies. Our preparations bind normally to actin and myosin and have normal kinase activity.
Perspectives
In this review we have tried to point out aspects of MLCK biochemistry that need clarification and further study. Several of these stand out among the most important with potential for high impact on MLCK function in the muscle. The question of access of MLCK to myosin in the muscle is important because this could ultimately limit the rate of the interaction of MLCK with its substrate. Studies of disease states discussed above suggest that too much MLCK, perhaps if it mislocalized, may lead to dysfunction. Topics related to this are the functional significance of the flexibility of MLCK, the role of the potentially flexible proline-rich region, the importance of the IgG and Fn domains to actin or other protein binding, inter-domain interactions, the effect of contraction on MLCK interactions with the contractile apparatus, and the reasons for low fractional activation of MLCK in the muscle. Importantly, the kinetics of MLCK interactions need to be explored and focus should be placed on using more physiological ionic strengths. Finally, future studies could add to the field by focusing on the mammalian MLCKs versus the avian MLCK.
Abbreviations
- MLCK
smooth muscle myosin light chain kinase (other isoforms are specified)
- CaM
calmodulin
- RLC
regulatory light chain of smooth muscle myosin
- SMM
smooth muscle myosin
- NMM
nonmuscle myosin
- HMM
heavy meromyosin subfragment of SMM
- IgG
immunoglobulin G-like domain
- Fn
fibronectin-like domain
- IgT
C-terminal immunoglobulin domain of MLCK
- GST
glutathione S-transferase
References
- 1.Herring BP, El-Mounayri O, Gallagher PJ, Yin F, Zhou J. Regulation of myosin light chain kinase and telokin expression in smooth muscle tissues. Am J Physiol Cell Physiol. 2006;291:C817–C827. doi: 10.1152/ajpcell.00198.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan JY, Takeda M, Briggs LE, Graham ML, Lu JT, Horikoshi N, Weinberg EO, Aoki H, Sato N, Chien KR, Kasahara H. Identification of cardiac-specific myosin light chain kinase. Circ Res. 2008;102:571–580. doi: 10.1161/CIRCRESAHA.107.161687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lazar V, Garcia JG. A single human myosin light chain kinase gene (MLCK; MYLK) Genomics. 1999;57:256–267. doi: 10.1006/geno.1999.5774. [DOI] [PubMed] [Google Scholar]
- 4.Adelstein RS, Klee CB. Purification and characterization of smooth muscle myosin light chain kinase. J Biol Chem. 1981;256:7501–7509. [PubMed] [Google Scholar]
- 5.Adelstein RS, Klee CB. Purification of smooth muscle myosin light-chain kinase. Methods Enzymol. 1982;85(Pt B):298–308. doi: 10.1016/0076-6879(82)85029-5. [DOI] [PubMed] [Google Scholar]
- 6.Ngai PK, Carruthers CA, Walsh MP. Isolation of the native form of chicken gizzard myosin light-chain kinase. Biochem J. 1984;218:863–870. doi: 10.1042/bj2180863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conti MA, Adelstein RS. Purification and properties of myosin light chain kinases. Methods Enzymol. 1991;196:34–47. doi: 10.1016/0076-6879(91)96006-d. [DOI] [PubMed] [Google Scholar]
- 8.Pato MD, Kerc E. Purification of smooth muscle myosin phosphatase from turkey gizzard. Methods Enzymol. 1988;159:446–453. doi: 10.1016/0076-6879(88)59044-4. [DOI] [PubMed] [Google Scholar]
- 9.Walsh MP, Hinkins S, Flink IL, Hartshorne DJ. Bovine stomach myosin light chain kinase: purification, characterization, and comparison with the turkey gizzard enzyme. Biochemistry. 1982;21:6890–6896. doi: 10.1021/bi00269a041. [DOI] [PubMed] [Google Scholar]
- 10.Olson NJ, Pearson RB, Needleman DS, Hurwitz MY, Kemp BE, Means AR. Regulatory and structural motifs of chicken gizzard myosin light chain kinase. Proc Natl Acad Sci U S A. 1990;87:2284–2288. doi: 10.1073/pnas.87.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanoh S, Ito M, Niwa E, Kawano Y, Hartshorne DJ. Actin-binding peptide from smooth muscle myosin light chain kinase. Biochemistry. 1993;32:8902–8907. doi: 10.1021/bi00085a023. [DOI] [PubMed] [Google Scholar]
- 12.Smith L, Su X, Lin P, Zhi G, Stull JT. Identification of a novel actin binding motif in smooth muscle myosin light chain kinase. J Biol Chem. 1999;274:29433–29438. doi: 10.1074/jbc.274.41.29433. [DOI] [PubMed] [Google Scholar]
- 13.Gallagher PJ, Stull JT. Localization of an actin binding domain in smooth muscle myosin light chain kinase. Mol Cell Biochem. 1997;173:51–57. doi: 10.1023/a:1006876318155. [DOI] [PubMed] [Google Scholar]
- 14.Ye LH, Hayakawa K, Kishi H, Imamura M, Nakamura A, Okagaki T, Takagi T, Iwata A, Tanaka T, Kohama K. The structure and function of the actin-binding domain of myosin light chain kinase of smooth muscle. J Biol Chem. 1997;272:32182–32189. doi: 10.1074/jbc.272.51.32182. [DOI] [PubMed] [Google Scholar]
- 15.Sellers JR, Pato MD. The binding of smooth muscle myosin light chain kinase and phosphatases to actin and myosin. J Biol Chem. 1984;259:7740–7746. [PubMed] [Google Scholar]
- 16.Xie C, Zhang Y, Wang HH, Matsumoto A, Nakamura A, Ishikawa R, Yoshiyama S, Hayakawa K, Kohama K, Gao Y. Calcium regulation of non-kinase and kinase activities of recombinant myosin light-chain kinase and its mutants. IUBMB Life. 2009;61:1092–1098. doi: 10.1002/iub.266. [DOI] [PubMed] [Google Scholar]
- 17.Silver DL, Vorotnikov AV, Watterson DM, Shirinsky VP, Sellers JR. Sites of interaction between kinase-related protein and smooth muscle myosin. J Biol Chem. 1997;272:25353–25359. doi: 10.1074/jbc.272.40.25353. [DOI] [PubMed] [Google Scholar]
- 18.Shirinsky VP, Vorotnikov AV, Birukov KG, Nanaev AK, Collinge M, Lukas TJ, Sellers JR, Watterson DM. A kinase-related protein stabilizes unphosphorylated smooth muscle myosin minifilaments in the presence of ATP. J Biol Chem. 1993;268:16578–16583. [PubMed] [Google Scholar]
- 19.Numata T, Katoh T, Yazawa M. Functional role of the C-terminal domain of smooth muscle myosin light chain kinase on the phosphorylation of smooth muscle myosin. J Biochem (Tokyo) 2001;129:437–444. doi: 10.1093/oxfordjournals.jbchem.a002875. [DOI] [PubMed] [Google Scholar]
- 20.Sobieszek A. Phosphorylation reaction of vertebrate smooth muscle myosin: an enzyme kinetic analysis. Biochemistry. 1985;24:1266–1274. doi: 10.1021/bi00326a032. [DOI] [PubMed] [Google Scholar]
- 21.Hong F, Haldeman BD, John OA, Brewer PD, Wu YY, Ni S, Wilson DP, Walsh MP, Baker JE, Cremo CR. Characterization of tightly associated smooth muscle myosin-myosin light-chain kinase-calmodulin complexes. J Mol Biol. 2009;390:879–892. doi: 10.1016/j.jmb.2009.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cross RA, Sobieszek A. Influence of smooth muscle myosin conformation on myosin light chain kinase binding and on phosphorylation. FEBS Lett. 1985;188:367–374. doi: 10.1016/0014-5793(85)80404-x. [DOI] [PubMed] [Google Scholar]
- 23.Kobe B, Heierhorst J, Feil SC, Parker MW, Benian GM, Weiss KR, Kemp BE. Giant protein kinases: domain interactions and structural basis of autoregulation. Embo J. 1996;15:6810–6821. [PMC free article] [PubMed] [Google Scholar]
- 24.Krueger JK, Olah GA, Rokop SE, Zhi G, Stull JT, Trewhella J. Structures of calmodulin and a functional myosin light chain kinase in the activated complex: a neutron scattering study. Biochemistry. 1997;36:6017–6023. doi: 10.1021/bi9702703. [DOI] [PubMed] [Google Scholar]
- 25.Mabuchi Y, Mabuchi K, Stafford WF, Grabarek Z. Modular structure of smooth muscle Myosin light chain kinase: hydrodynamic modeling and functional implications. Biochemistry. 2010;49:2903–2917. doi: 10.1021/bi901963e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bagshaw CR. Muscle Contraction. London: Chapman and Hall; 1993. [Google Scholar]
- 27.Gallagher PJ, Herring BP, Stull JT. Myosin light chain kinases. J Muscle Res Cell Motil. 1997;18:1–16. doi: 10.1023/a:1018616814417. [DOI] [PubMed] [Google Scholar]
- 28.Yuen SL, Ogut O, Brozovich FV. Nonmuscle myosin is regulated during smooth muscle contraction. Am J Physiol Heart Circ Physiol. 2009;297:H191–H199. doi: 10.1152/ajpheart.00132.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang WC, Peng YJ, Zhang GS, He WQ, Qiao YN, Dong YY, Gao YQ, Chen C, Zhang CH, Li W, Shen HH, Ning W, Kamm KE, Stull JT, Gao X, Zhu MS. Myosin light chain kinase is necessary for tonic airway smooth muscle contraction. J Biol Chem. 2010;285:5522–5531. doi: 10.1074/jbc.M109.062836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He WQ, Peng YJ, Zhang WC, Lv N, Tang J, Chen C, Zhang CH, Gao S, Chen HQ, Zhi G, Feil R, Kamm KE, Stull JT, Gao X, Zhu MS. Myosin light chain kinase is central to smooth muscle contraction and required for gastrointestinal motility in mice. Gastroenterology. 2008;135:610–620. doi: 10.1053/j.gastro.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kovacs M, Thirumurugan K, Knight PJ, Sellers JR. Load-dependent mechanism of nonmuscle myosin 2. Proc Natl Acad Sci U S A. 2007;104:9994–9999. doi: 10.1073/pnas.0701181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang F, Kovacs M, Hu A, Limouze J, Harvey EV, Sellers JR. Kinetic mechanism of non-muscle myosin IIB: functional adaptations for tension generation and maintenance. J Biol Chem. 2003;278:27439–27448. doi: 10.1074/jbc.M302510200. [DOI] [PubMed] [Google Scholar]
- 33.Lofgren M, Ekblad E, Morano I, Arner A. Nonmuscle Myosin motor of smooth muscle. J Gen Physiol. 2003;121:301–310. doi: 10.1085/jgp.200208720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morano I, Chai GX, Baltas LG, Lamounier-Zepter V, Lutsch G, Kott M, Haase H, Bader M. Nat Cell Biol. 2000;Vol. 2:371–375. doi: 10.1038/35014065. [DOI] [PubMed] [Google Scholar]
- 35.Rhee AY, Ogut O, Brozovich FV. Nonmuscle myosin, force maintenance, and the tonic contractile phenotype in smooth muscle. Pflugers Arch. 2006;452:766–774. doi: 10.1007/s00424-006-0091-4. [DOI] [PubMed] [Google Scholar]
- 36.Sherry JMF, Gorecka AAMO, Dabrowska R, Hartshorne DJ. Roles of Calcium and Phosphorylation in the Regulation of the Activity of Gizzard Myosin. Biochemistry. 1978;17:4411–4418. doi: 10.1021/bi00614a009. [DOI] [PubMed] [Google Scholar]
- 37.Pearson RB, Ito M, Morrice NA, Smith AJ, Condron R, Wettenhall RE, Kemp BE, Hartshorne DJ. Proteolytic cleavage sites in smooth muscle myosin-light-chain kinase and their relation to structural and regulatory domains. Eur J Biochem. 1991;200:723–730. doi: 10.1111/j.1432-1033.1991.tb16237.x. [DOI] [PubMed] [Google Scholar]
- 38.Lin P, Luby-Phelps K, Stull JT. Properties of filament-bound myosin light chain kinase. J Biol Chem. 1999;274:5987–5994. doi: 10.1074/jbc.274.9.5987. [DOI] [PubMed] [Google Scholar]
- 39.Lin P-j, Luby-Phelps K, Stull JT. Binding fo Myosin Light Chain Kinase to Cellular Actin-Myosin Filaments. J Biol Chem. 1997;272:7412–7420. doi: 10.1074/jbc.272.11.7412. [DOI] [PubMed] [Google Scholar]
- 40.Foyt HL, Guerriero V, Jr, Means AR. Functional domains of chicken gizzard myosin light chain kinase. J Biol Chem. 1985;260:7765–7774. [PubMed] [Google Scholar]
- 41.Walsh MP, Dabrowska R, Hinkins S, Hartshorne DJ. Calcium-independent myosin light chain kinase of smooth muscle. Preparation by limited chymotryptic digestion of the calcium ion dependent enzyme, purification, and characterization. Biochemistry. 1982;21:1919–1925. doi: 10.1021/bi00537a034. [DOI] [PubMed] [Google Scholar]
- 42.Ikebe M, Stepinska M, Kemp BE, Means AR, Hartshorne DJ. Proteolysis of smooth muscle myosin light chain kinase. Formation of inactive and calmodulin-independent fragments. J Biol Chem. 1987;262:13828–13834. [PubMed] [Google Scholar]
- 43.Deng JT, Van Lierop JE, Sutherland C, Walsh MP. Ca2+-independent smooth muscle contraction. a novel function for integrin-linked kinase. J Biol Chem. 2001;276:16365–16373. doi: 10.1074/jbc.M011634200. [DOI] [PubMed] [Google Scholar]
- 44.Niiro N, Ikebe M. Zipper-interacting protein kinase induces Ca(2+)-free smooth muscle contraction via myosin light chain phosphorylation. J Biol Chem. 2001;276:29567–29574. doi: 10.1074/jbc.M102753200. [DOI] [PubMed] [Google Scholar]
- 45.Ikebe M, Inagaki M, Naka M, Hidaka H. Correlation of conformation and phosphorylation and dephosphorylation of smooth muscle myosin. J Biol Chem. 1988;263:10698–10704. [PubMed] [Google Scholar]
- 46.Kamm KE, Hsu LC, Kubota Y, Stull JT. Phosphorylation of smooth muscle myosin heavy and light chains. Effects of phorbol dibutyrate and agonists. J Biol Chem. 1989;264:21223–21229. [PubMed] [Google Scholar]
- 47.Geuss U, Mayr GW, Heilmeyer LM., Jr Steady-state kinetics of skeletal muscle myosin light chain kinase indicate a strong down regulation by products. Eur J Biochem. 1985;153:327–334. doi: 10.1111/j.1432-1033.1985.tb09305.x. [DOI] [PubMed] [Google Scholar]
- 48.Krisanda JM, Paul RJ. Phosphagen and metabolite content during contraction in porcine carotid artery. Am J Physiol. 1983;244:C385–C390. doi: 10.1152/ajpcell.1983.244.5.C385. [DOI] [PubMed] [Google Scholar]
- 49.Wirth A, Schroeter M, Kock-Hauser C, Manser E, Chalovich JM, de Lanerolle P, Pfitzer G. Inhibition of contraction and myosin light chain phosphorylation in guinea-pig smooth muscle by p21-activated kinase 1. J Physiol (Paris) 2003;549:489–500. doi: 10.1113/jphysiol.2002.033167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gallagher PJ, Herring BP, Trafny A, Sowadski J, Stull JT. A molecular mechanism for autoinhibition of myosin light chain kinases. J Biol Chem. 1993;268:26578–26582. [PMC free article] [PubMed] [Google Scholar]
- 51.Kamm KE, Stull JT. Dedicated myosin light chain kinases with diverse cellular functions. J Biol Chem. 2001;276:4527–4530. doi: 10.1074/jbc.R000028200. [DOI] [PubMed] [Google Scholar]
- 52.Stull JT, Tansey MG, Tang DC, Word RA, Kamm KE. Phosphorylation of myosin light chain kinase: a cellular mechanism for Ca2+ desensitization. Mol Cell Biochem. 1993;127–128:229–237. doi: 10.1007/BF01076774. [DOI] [PubMed] [Google Scholar]
- 53.Tansey MG, Luby-Phelps K, Kamm KE, Stull JT. Ca(2+)-dependent phosphorylation of myosin light chain kinase decreases the Ca2+ sensitivity of light chain phosphorylation within smooth muscle cells. J Biol Chem. 1994;269:9912–9920. [PubMed] [Google Scholar]
- 54.Isotani E, Zhi G, Lau KS, Huang J, Mizuno Y, Persechini A, Geguchadze R, Kamm KE, Stull JT. Real-time evaluation of myosin light chain kinase activation in smooth muscle tissues from a transgenic calmodulin-biosensor mouse. Proc Natl Acad Sci U S A. 2004;101:6279–6284. doi: 10.1073/pnas.0308742101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Geguchadze R, Zhi G, Lau KS, Isotani E, Persechini A, Kamm KE, Stull JT. Quantitative measurements of Ca(2+)/calmodulin binding and activation of myosin light chain kinase in cells. FEBS Lett. 2004;557:121–124. doi: 10.1016/s0014-5793(03)01456-x. [DOI] [PubMed] [Google Scholar]
- 56.Fajmut A, Jagodic M, Brumen M. Mathematical modeling of the myosin light chain kinase activation. J Chem Inf Model. 2005;45:1605–1609. doi: 10.1021/ci050177i. [DOI] [PubMed] [Google Scholar]
- 57.Injeti ER, Sandoval RJ, Williams JM, Smolensky AV, Ford LE, Pearce WJ. Maximal stimulation-induced in situ myosin light chain kinase activity is upregulated in fetal compared with adult ovine carotid arteries. Am J Physiol Heart Circ Physiol. 2008;295:H2289–H2298. doi: 10.1152/ajpheart.00606.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walsh MP, Hinkins S, Dabrowska R, Hartshorne DJ. Smooth muscle myosin light chain kinase. Methods Enzymol. 1983;99:279–288. doi: 10.1016/0076-6879(83)99063-8. [DOI] [PubMed] [Google Scholar]
- 59.Dabrowska R, Hinkins S, Walsh MP, Hartshorne DJ. The binding of smooth muscle myosin light chain kinase to actin. Biochem Biophys Res Commun. 1982;107:1524–1531. doi: 10.1016/s0006-291x(82)80172-1. [DOI] [PubMed] [Google Scholar]
- 60.Hartshorn DJ. Physiology of the Gastrointestinal Tract. New York: Raven Press; 1987. [Google Scholar]
- 61.Murphy RA, Driska SP, Cohen DM. Excitation-Contraction Coupling in Smooth Muscles. Amsterdam: Elsevier-North Holland Biomedical Press; 1977. [Google Scholar]
- 62.Murphy RA, Herlihy JT, Megerman J. Force-generating capacity and contractile protein content of arterial smooth muscle. J Gen Physiol. 1974;64:691–705. doi: 10.1085/jgp.64.6.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walcott S, Fagnant PM, Trybus KM, Warshaw DM. Smooth muscle heavy meromyosin phosphorylated on one of its two heads supports force and motion. J Biol Chem. 2009;284:18244–18251. doi: 10.1074/jbc.M109.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cremo CR, Hartshorne DJ. In: Myosins: A Superfamily of Molecular Motors. Coluccio LM, editor. Vol. 7. The Netherlands: Springer, Dordrecht; 2008. pp. 171–222. [Google Scholar]
- 65.Kenney RE, Hoar PE, Kerrick GL. The Relationship between ATPase Activity, Isometric Force, and Myosin Light-chain Phosphorylation and Thiophosphorylation in Skinned Smooth Muscle Fiber Bundles from Chicken Gizzard. J. Biol. Chem. 1990;265:8642–8649. [PubMed] [Google Scholar]
- 66.Bagshaw CR, Eccleston JF, Trentham DR, Yates DW. Transient kinetic studies of the Mg++-dependent ATPase of myosin and its proteolytic subfragments. Cold Spring Harbor Symp. Quant. Biol. 1972;37:127–135. [Google Scholar]
- 67.Barrington-Leigh J, Holmes KC, Mannherz HG, Rosenbaum G, Eckstein F, Goody RS. Effects of ATP analogs on the lowangle x-ray diffraction pattern of insect flight muscle. Cold SpringHarbor Symp. Quant. Biol. 1972;37:443–448. [Google Scholar]
- 68.Kossmann T, Furst D, Small JV. Structural and biochemical analysis of skinned smooth muscle preparations. J Muscle Res Cell Motil. 1987;8:135–144. doi: 10.1007/BF01753989. [DOI] [PubMed] [Google Scholar]
- 69.Sobieszek A. Vectorial activation of smooth muscle myosin filaments and its modulation by telokin Can. J Physiol Pharmacol. 2005;83:899–912. doi: 10.1139/y05-053. [DOI] [PubMed] [Google Scholar]
- 70.Sobieszek A, Andruchov OY, Grabarek Z, Kulikova N, Liebetrau C, Matusovsky OS. Modulation of myosin filament activation by telokin in smooth muscle liberation of myosin kinase and phosphatase from supramolecular complexes. Biophys Chem. 2005;113:25–40. doi: 10.1016/j.bpc.2004.07.038. [DOI] [PubMed] [Google Scholar]
- 71.Leguillette R, Laviolette M, Bergeron C, Zitouni N, Kogut P, Solway J, Kachmar L, Hamid Q, Lauzon A-M. Myosin, Transgelin, and Myosin Light Chain Kinase: Expression and Function in Asthma. Am. J. Respir. Crit. Care Med. 2009;179:194–204. doi: 10.1164/rccm.200609-1367OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang H, Rao K, Halayko AJ, Liu X, Stephens NL. Ragweed sensitization-induced increase of myosin light chain kinase content in canine airway smooth muscle. Am J Respir Cell Mol Biol. 1992;7:567–573. doi: 10.1165/ajrcmb/7.6.567. [DOI] [PubMed] [Google Scholar]
- 73.Flores C, Ma SF, Maresso K, Ober C, Garcia JG. A variant of the myosin light chain kinase gene is associated with severe asthma in African Americans. Genet Epidemiol. 2007;31:296–305. doi: 10.1002/gepi.20210. [DOI] [PubMed] [Google Scholar]
- 74.Gao L, Grant AV, Rafaels N, Stockton-Porter M, Watkins T, Gao P, Chi P, Munoz M, Watson H, Dunston G, Togias A, Hansel N, Sevransky J, Maloney JP, Moss M, Shanholtz C, Brower R, Garcia JG, Grigoryev DN, Cheadle C, Beaty TH, Mathias RA, Barnes KC. Polymorphisms in the myosin light chain kinase gene that confer risk of severe sepsis are associated with a lower risk of asthma. J Allergy Clin Immunol. 2007;119:1111–1118. doi: 10.1016/j.jaci.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 75.Moitra J, Evenoski C, Sammani S, Wadgaonkar R, Turner JR, Ma SF, Garcia JG. A transgenic mouse with vascular endothelial over-expression of the non-muscle myosin light chain kinase-2 isoform is susceptible to inflammatory lung injury: role of sexual dimorphism and age. Transl Res. 2008;151:141–153. doi: 10.1016/j.trsl.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu G, Liu X, Rao K, Jiang H, Stephens NL. Increased myosin light chain kinase content in sensitized canine saphenous vein. J Appl Physiol. 1996;80:665–669. doi: 10.1152/jappl.1996.80.2.665. [DOI] [PubMed] [Google Scholar]
- 77.Turner JR. Molecular basis of epithelial barrier regulation: from basic mechanisms to clinical application. Am J Pathol. 2006;169:1901–1909. doi: 10.2353/ajpath.2006.060681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shen L, Black ED, Witkowski ED, Lencer WI, Guerriero V, Schneeberger EE, Turner JR. Myosin light chain phosphorylation regulates barrier function by remodeling tight junction structure. J Cell Sci. 2006;119:2095–2106. doi: 10.1242/jcs.02915. [DOI] [PubMed] [Google Scholar]
- 79.Shen L, Su L, Turner JR. Mechanisms and functional implications of intestinal barrier defects. Dig Dis. 2009;27:443–449. doi: 10.1159/000233282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Garcia JG, Verin AD, Schaphorst K, Siddiqui R, Patterson CE, Csortos C, Natarajan V. Regulation of endothelial cell myosin light chain kinase by Rho, cortactin, and p60(src) Am J Physiol. 1999;276:L989–L998. doi: 10.1152/ajplung.1999.276.6.L989. [DOI] [PubMed] [Google Scholar]
- 81.Su L, Shen L, Clayburgh DR, Nalle SC, Sullivan EA, Meddings JB, Abraham C, Turner JR. Targeted epithelial tight junction dysfunction causes immune activation and contributes to development of experimental colitis. Gastroenterology. 2009;136:551–563. doi: 10.1053/j.gastro.2008.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weber CR, Turner JR. Inflammatory bowel disease: is it really just another break in the wall? Gut. 2007;56:6–8. doi: 10.1136/gut.2006.104182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Blair SA, Kane SV, Clayburgh DR, Turner JR. Epithelial myosin light chain kinase expression and activity are upregulated in inflammatory bowel disease. Lab Invest. 2006;86:191–201. doi: 10.1038/labinvest.3700373. [DOI] [PubMed] [Google Scholar]
- 84.Clayburgh DR, Barrett TA, Tang Y, Meddings JB, Van Eldik LJ, Watterson DM, Clarke LL, Mrsny RJ, Turner JR. Epithelial myosin light chain kinase-dependent barrier dysfunction mediates T cell activation-induced diarrhea in vivo. J Clin Invest. 2005;115:2702–2715. doi: 10.1172/JCI24970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reynoso R, Perrin RM, Breslin JW, Daines DA, Watson KD, Watterson DM, Wu MH, Yuan S. A role for long chain myosin light chain kinase (MLCK-210) in microvascular hyperpermeability during severe burns. Shock. 2007;28:589–595. doi: 10.1097/SHK.0b013e31804d415f. [DOI] [PubMed] [Google Scholar]
- 86.Zolotarevsky Y, Hecht G, Koutsouris A, Gonzalez DE, Quan C, Tom J, Mrsny RJ, Turner JR. A membrane-permeant peptide that inhibits MLC kinase restores barrier function in in vitro models of intestinal disease. Gastroenterology. 2002;123:163–172. doi: 10.1053/gast.2002.34235. [DOI] [PubMed] [Google Scholar]
- 87.Ito M, Dabrowska R, Guerriero V, Hartshorne DJ. Identification in Turkey Gizzard of an Acidic Protein Related to the C-terminal Portion of Smooth Muscle Myosin Light Chain Kinase. J. Biol. Chem. 1989;264:13971–13974. [PubMed] [Google Scholar]
- 88.Shcherbakova OV, Serebryanaya DV, Postnikov AB, Schroeter MM, Zittrich S, Noegel AA, Shirinsky VP, Vorotnikov AV, Pfitzer G. Kinase-related protein/telokin inhibits Ca2+-independent contraction in Triton-skinned guinea pig taenia coli. Biochem J. 2010;429:291–302. doi: 10.1042/BJ20090819. [DOI] [PubMed] [Google Scholar]
- 89.Houmeida A, Holt J, Tskhovrebova L, Trinick J. Studies of the interaction between titin and myosin. J Cell Biol. 1995;131:1471–1481. doi: 10.1083/jcb.131.6.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Levine RJ, Chantler PD, Kensler RW, Woodhead JL. Effects of phosphorylation by myosin light chain kinase on the structure of Limulus thick filaments. J Cell Biol. 1991;113:563–572. doi: 10.1083/jcb.113.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Levine RJ, Kensler RW, Yang Z, Stull JT, Sweeney HL. Myosin light chain phosphorylation affects the structure of rabbit skeletal muscle thick filaments. Biophys J. 1996;71:898–907. doi: 10.1016/S0006-3495(96)79293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Levine RJ, Yang Z, Epstein ND, Fananapazir L, Stull JT, Sweeney HL. Structural and functional responses of mammalian thick filaments to alterations in myosin regulatory light chains. J Struct Biol. 1998;122:149–161. doi: 10.1006/jsbi.1998.3980. [DOI] [PubMed] [Google Scholar]
- 93.Masato T, Numata T, Katoh T, Morita F, Yazawa M. Crosslinking of telokin to chicken gizzard smooth muscle myosin. J Biochem (Tokyo) 1997;121:225–230. [PubMed] [Google Scholar]
- 94.Holden HM, Ito M, Hartshorne DJ, Rayment I. X-ray Structure Determination of Telokin, the C-terminal Domain of Myosin Light Chain Kinase at 2.8 A Resolution. J. Mol. Biol. 1992;227:840–851. doi: 10.1016/0022-2836(92)90226-a. [DOI] [PubMed] [Google Scholar]
- 95.Rogowski K, van Dijk J, Magiera MM, Bosc C, Deloulme JC, Bosson A, Peris L, Gold ND, Lacroix B, Grau MB, Bec N, Larroque C, Desagher S, Holzer M, Andrieux A, Moutin MJ, Janke C. A family of protein-deglutamylating enzymes associated with neurodegeneration. Cell. 2010;143:564–578. doi: 10.1016/j.cell.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 96.Rusconi F, Potier MC, Le Caer JP, Schmitter JM, Rossier J. Characterization of the chicken telokin heterogeneity by time-of-flight mass spectrometry. Biochemistry. 1997;36:11021–11026. doi: 10.1021/bi970752e. [DOI] [PubMed] [Google Scholar]
- 97.Milton DL, Schneck AN, Ziech DA, Ba M, Facemyer KC, Halayko AJ, Baker JE, Gerthoffer WT, Cremo CR. Direct evidence for functional smooth muscle myosin II in the 10S self-inhibited monomeric conformation in airway smooth muscle cells. Proc Natl Acad Sci U S A. 2011;108:1421–1426. doi: 10.1073/pnas.1011784108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ngai PK, Walsh MP. Purification of smooth-muscle myosin free of calmodulin and myosin light-chain kinase. Susceptibility to oxidation. Biochem J. 1987;246:205–211. doi: 10.1042/bj2460205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hatch V, Zhi G, Smith L, Stull JT, Craig R, Lehman W. Myosin light chain kinase binding to a unique site on F-actin revealed by three-dimensional image reconstruction. J Cell Biol. 2001;154:611–617. doi: 10.1083/jcb.200105079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McGough A. F-actin-binding proteins. Curr Opin Struct Biol. 1998;8:166–176. doi: 10.1016/s0959-440x(98)80034-1. [DOI] [PubMed] [Google Scholar]
- 101.Dominguez R. Actin-binding proteins--a unifying hypothesis. Trends Biochem Sci. 2004;29:572–578. doi: 10.1016/j.tibs.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 102.Smith L, Stull JT. Myosin light chain kinase binding to actin filaments. FEBS Lett. 2000;480:298–300. doi: 10.1016/s0014-5793(00)01931-1. [DOI] [PubMed] [Google Scholar]
- 103.Otey CA, Dixon R, Stack C, Goicoechea SM. Cytoplasmic Ig-domain proteins: cytoskeletal regulators with a role in human disease. Cell Motil Cytoskeleton. 2009;66:618–634. doi: 10.1002/cm.20385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Galler S. Molecular basis of the catch state in molluscan smooth muscles: a catchy challenge. J Muscle Res Cell Motil. 2008;29:73–99. doi: 10.1007/s10974-008-9149-6. [DOI] [PubMed] [Google Scholar]
- 105.Marston SB, Lehman W. Caldesmon is a Ca2+-regulatory component of native smooth-muscle thin filaments. Biochem J. 1985;231:517–522. doi: 10.1042/bj2310517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Marston SB, Smith CW. Purification and properties of Ca2+-regulated thin filaments and F-actin from sheep aorta smooth muscle. J Muscle Res Cell Motil. 1984;5:559–575. doi: 10.1007/BF00713261. [DOI] [PubMed] [Google Scholar]
- 107.Driska S, Hartshorne DJ. The contractile proteins of smooth muscle. Properties and components of a Ca2+-sensitive actomyosin from chicken gizzard. Arch Biochem Biophys. 1975;167:203–212. doi: 10.1016/0003-9861(75)90457-9. [DOI] [PubMed] [Google Scholar]
- 108.Walters M, Marston SB. Phosphorylation of the calcium ion-regulated thin filaments from vascular smooth muscle. A new regulatory mechanism? Biochem J. 1981;197:127–139. doi: 10.1042/bj1970127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Marston SB, Smith CW. The thin filaments of smooth muscles. J Muscle Res Cell Motil. 1985;6:669–708. doi: 10.1007/BF00712237. [DOI] [PubMed] [Google Scholar]
- 110.Hodgkinson JL, el-Mezgueldi M, Craig R, Vibert P, Marston SB, Lehman W. 3-D image reconstruction of reconstituted smooth muscle thin filaments containing calponin: visualization of interactions between F-actin and calponin. J Mol Biol. 1997;273:150–159. doi: 10.1006/jmbi.1997.1307. [DOI] [PubMed] [Google Scholar]
- 111.Babiychuk EB, Babiychuk VS, Sobieszek A. Modulation of smooth muscle myosin light chain kinase activity by Ca2+/calmodulin-dependent, oligomeric-type modifications. Biochemistry. 1995;34:6366–6372. doi: 10.1021/bi00019a015. [DOI] [PubMed] [Google Scholar]
- 112.Nieznanski K, Sobieszek A. Telokin (kinase-related protein) modulates the oligomeric state of smooth-muscle myosin light-chain kinase and its interaction with myosin filaments. Biochem J. 1997;322(Pt 1):65–71. doi: 10.1042/bj3220065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Okagaki T, Hayakawa K, Samizo K, Kohama K. Inhibition of the ATP-dependent interaction of actin and myosin by the catalytic domain of the myosin light chain kinase of smooth muscle: possible involvement in smooth muscle relaxation. J Biochem (Tokyo) 1999;125:619–626. doi: 10.1093/oxfordjournals.jbchem.a022328. [DOI] [PubMed] [Google Scholar]
- 114.Ye LH, Kishi H, Nakamura A, Okagaki T, Tanaka T, Oiwa K, Kohama K. Myosin light-chain kinase of smooth muscle stimulates myosin ATPase activity without phosphorylating myosin light chain. Proc Natl Acad Sci U S A. 1999;96:6666–6671. doi: 10.1073/pnas.96.12.6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nakamura A, Xie C, Zhang Y, Gao Y, Wang HH, Ye LH, Kishi H, Okagaki T, Yoshiyama S, Hayakawa K, Ishikawa R, Kohama K. Role of non-kinase activity of myosin light-chain kinase in regulating smooth muscle contraction, a review dedicated to Dr. Setsuro Ebashi. Biochem Biophys Res Commun. 2008;369:135–143. doi: 10.1016/j.bbrc.2007.11.096. [DOI] [PubMed] [Google Scholar]
- 116.Gao Y, Kawano K, Yoshiyama S, Kawamichi H, Wang X, Nakamura A, Kohama K. Myosin light chain kinase stimulates smooth muscle myosin ATPase activity by binding to the myosin heads without phosphorylating the myosin light chain. Biochem Biophys Res Commun. 2003;305:16–21. doi: 10.1016/s0006-291x(03)00690-9. [DOI] [PubMed] [Google Scholar]
- 117.Wang HH, Nakamura A, Matsumoto A, Yoshiyama S, Qin X, Ye LH, Xie C, Zhang Y, Gao Y, Ishikawa R, Kohama K. Nonkinase activity of MLCK in elongated filopodia formation and chemotaxis of vascular smooth muscle cells toward sphingosylphosphorylcholine. Am J Physiol Heart Circ Physiol. 2009;296:H1683–H1693. doi: 10.1152/ajpheart.00965.2008. [DOI] [PubMed] [Google Scholar]
- 118.Shimizu H, Ito M, Miyahara M, Ichikawa K, Okubo S, Konishi T, Naka M, Tanaka T, Hirano K, Hartshorne DJ, et al. Characterization of the myosin-binding subunit of smooth muscle myosin phosphatase. J Biol Chem. 1994;269:30407–30411. [PubMed] [Google Scholar]
- 119.Kolega J. Fluorescent analogues of myosin II for tracking the behavior of different myosin isoforms in living cells. J Cell Biochem. 1998;68:389–401. doi: 10.1002/(sici)1097-4644(19980301)68:3<389::aid-jcb9>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 120.Blue EK, Goeckeler ZM, Jin Y, Hou L, Dixon SA, Herring BP, Wysolmerski RB, Gallagher PJ. 220- and 130-kDa MLCKs have distinct tissue distributions and intracellular localization patterns. Am J Physiol Cell Physiol. 2002;282:C451–C460. doi: 10.1152/ajpcell.00333.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kohama K, Okagaki T, Hayakawa K, Lin Y, Ishikawa R, Shimmen T, Inoue A. A novel regulatory effect of myosin light chain kinase from smooth muscle on the ATP -dependent interaction between actin and myosin. Biochem Biophys Res Commun. 1992;184:1204–1211. doi: 10.1016/s0006-291x(05)80010-5. [DOI] [PubMed] [Google Scholar]
- 122.Greenberg MJ, Moore JR. The molecular basis of frictional loads in the in vitro motility assay with applications to the study of the loaded mechanochemistry of molecular motors. Cytoskeleton (Hoboken) 67:273–285. doi: 10.1002/cm.20441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Olney JJ, Sellers JR, Cremo CR. Structure and Function of the 10 S Conformation of Smooth Muscle Myosin. J Biol Chem. 1996;271:20375–20384. doi: 10.1074/jbc.271.34.20375. [DOI] [PubMed] [Google Scholar]
- 124.Burgess SA, Yu S, Walker ML, Hawkins RJ, Chalovich JM, Knight PJ. Structures of smooth muscle myosin and heavy meromyosin in the folded, shutdown state. J Mol Biol. 2007;372:1165–1178. doi: 10.1016/j.jmb.2007.07.014. [DOI] [PubMed] [Google Scholar]