Abstract
Auxin regulates many aspects of plant development, in part, through degradation of the Aux/IAA family of transcriptional repressors. Consequently, stabilizing mutations in several Aux/IAA proteins confer reduced auxin responsiveness. However, of the 29 apparent Aux/IAA proteins in Arabidopsis thaliana, fewer than half have roles established through mutant analysis. We identified iaa16-1, a dominant gain-of-function mutation in IAA16 (At3g04730), in a novel screen for reduced root responsiveness to abscisic acid. The iaa16-1 mutation also confers dramatically reduced auxin responses in a variety of assays, markedly restricts growth of adult plants, and abolishes fertility when homozygous. We compared iaa16-1 phenotypes with those of dominant mutants defective in the closely related IAA7/AXR2, IAA14/SLR, and IAA17/AXR3, along with the more distantly related IAA28, and found overlapping but distinct patterns of developmental defects. The identification and characterization of iaa16-1 provides a fuller understanding of the IAA7 / IAA14 / IAA16 / IAA17 clade of Aux/IAA proteins and the diverse roles of these repressors in hormone response and plant development.
Keywords: Auxin signaling, Abscisic Acid, Aux/IAA gain of function, IAA16
Introduction
The plant hormone auxin regulates both cell division and cell elongation and thus controls many aspects of plant growth and development (reviewed in Perrot-Rechenmann 2010). Auxin exerts its effects on these processes, at least in part, by regulating transcription through the activities of the AUXIN RESPONSE FACTOR (ARF) family of proteins (reviewed in Guilfoyle and Hagen 2007). Activity of these ARF transcription factors is impeded by dimerization with members of the Auxin/INDOLE-3-ACETIC ACID (Aux/IAA) repressor protein family (reviewed in Hagen and Guilfoyle 2002; Liscum and Reed 2002).
ARF proteins and Aux/IAA proteins share two motifs, domains III and IV, that mediate hetero- and homo-dimerization among members of these two protein families (reviewed in Guilfoyle and Hagen 2007). In addition to domains III and IV, most Aux/IAA proteins share two additional motifs, domains I and II. Domain I is a repression domain with a conserved LxLxL motif that allows for interaction with the TOPLESS corepressor (Szemenyei et al. 2008). Domain II contains a degron and confers instability to Aux/IAA proteins (Worley et al. 2000; Ramos et al. 2001; Dreher et al. 2006).
In the presence of auxin, the degron in domain II of the Aux/IAA protein interacts with the F-box E3 ubiquitin ligase TRANSPORT INHIBITOR RESPONSE1 (TIR1) and related F-box proteins (Kepinski and Leyser 2004; Dharmasiri et al. 2005b; Kepinski and Leyser 2005). This interaction, wherein auxin acts as a “molecular glue” (Tan et al. 2007), allows for the polyubiquitylation of the Aux/IAA repressor protein (dos Santos Maraschin et al. 2009). This polyubiquitylation targets Aux/IAA proteins to the 26S proteasome for degradation (Ramos et al. 2001; dos Santos Maraschin et al. 2009), thus relieving repression of ARF activity and allowing ARF-driven, auxin-responsive transcription (reviewed in Liscum and Reed 2002).
Because of the similarity exhibited by the 29 predicted Arabidopsis thaliana Aux/IAA proteins (Liscum and Reed 2002), some family members are expected to be at least partially redundant with one another. Indeed, plants lacking any one of 12 Aux/IAA genes examined do not suffer notable consequences (Overvoorde et al. 2005). In contrast, domain II mutations of various Aux/IAA proteins result in distinct and often dramatic gain-of-function phenotypes (reviewed in Mockaitis and Estelle 2008). To date, gain-of-function mutations predicted or demonstrated to increase protein stability have been reported for 10 of the 29 Arabidopsis Aux/IAA proteins: IAA1 (axr5/auxin-resistant 5; Yang et al. 2004), IAA3 (shy2/suppressor of hy2 2; Kim et al. 1996; Tian and Reed 1999), IAA6 (shy1/suppressor of hy2 1; Kim et al. 1996; Liscum and Reed 2002), IAA7 (axr2/auxin-resistant 2; Wilson et al. 1990; Nagpal et al. 2000), IAA12 (bdl/bodenlos; Hamann et al. 1999; Hamann et al. 2002), IAA14 (slr/solitary-root; Fukaki et al. 2002), IAA17 (axr3/auxin-resistant 3; Leyser et al. 1996; Rouse et al. 1998), IAA18 (iaa18; Uehara et al. 2008), IAA19 (msg2/massugu2; Tatematsu et al. 2004), and IAA28 (iaa28; Rogg et al. 2001). Remarkably, each of these mutants carries an amino acid substitution in the conserved GWPPV degron motif of domain II, which participates in binding auxin and TIR1 (Kepinski and Leyser 2004; Dharmasiri et al. 2005a; Kepinski and Leyser 2005). These single amino acid changes reduce TIR1 binding and thereby disrupt the auxin-responsive degradation of the Aux/IAA, resulting in increased protein accumulation and increased repression of the ARF transcription factors (reviewed in Calderon-Villalobos et al. 2010).
Several auxin response mutants, such as indole-3-butyric acid response5 (ibr5; Monroe-Augustus et al. 2003; Strader et al. 2008b), tir1 (Strader et al. 2008a), auxin-resistant1 (axr1; Monroe-Augustus et al. 2003), and auxin resistant1 (aux1; Strader et al. 2008a), display reduced responsiveness not only to auxin, but also to abscisic acid (ABA) in root elongation assays. ABA is a plant hormone that regulates many plant stress responses, including reducing growth in response to stress. The resistance of auxin response mutants to ABA-triggered root elongation inhibition suggests that ABA may slow root elongation via the auxin response pathway.
To identify additional components necessary for ABA-responsive inhibition of root elongation, we screened for mutants resistant to the inhibitory effects of ABA on root elongation. We found that one of these mutants was caused by a gain-of-function mutation in an Aux/IAA gene. iaa16-1 carries a mutation that alters the domain II degron motif of IAA16 and displays not only ABA resistance, but also dramatic auxin resistance and morphological phenotypes suggestive of reduced auxin signaling. Moreover, we found that additional Aux/IAA mutants, like iaa16-1, display resistance to ABA in root elongation and seedling establishment assays. Our identification and characterization of iaa16-1 provides the final gain-of-function mutation in the Aux/IAA clade that includes the previously identified gain-of-function mutations in IAA7/AXR2, IAA14/SLR, IAA17/AXR3, demonstrating that normal functioning of all of these proteins is needed for full auxin responsiveness.
Results
A gain-of-function mutation in IAA16 from an ABA resistance screen
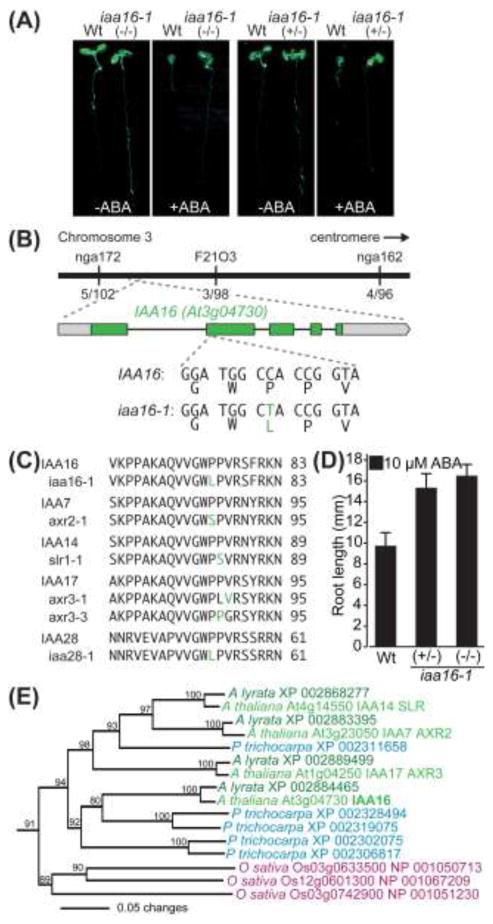
Exogenous abscisic acid inhibits seed germination, and many screens have used the inhibitory effects of ABA on germination to identify ABA signaling components (Koornneef et al. 1984; Finkelstein 1994; Cutler et al. 1996). ABA also inhibits root elongation, but perhaps because ABA inhibits germination at concentrations (1-3 μM) that do not dramatically impact root elongation, screens for mutants resistant to ABA in root elongation are rare. We used a transfer assay to uncover factors required for ABA response in root elongation. We grew EMS-mutagenized M2 seedlings on filter paper atop unsupplemented medium for 4 d to allow germination and seedling establishment prior to transfer to medium supplemented with 10 μM ABA for an additional 4 d of growth, after which we selected mutants with long roots. Isolate AR269 displayed strong resistance to the effects of ABA on root elongation (Fig. 1a). We used the ABA resistance in root elongation to map the causative mutation to the upper arm of chromosome 3 between molecular markers nga172 and F2103 (Fig. 1b). This region contains INDOLEACETIC ACID-INDUCED PROTEIN16 (IAA16 / At3g04730), a member of a family of genes in which gain-of-function mutations can result in dominant auxin resistance (reviewed in Mockaitis and Estelle 2008). Because AR269 displayed dominant resistance to ABA (Fig. 1a, d), and because several auxin-resistant mutants also display attenuated ABA responses (Wilson et al. 1990; Tian and Reed 1999; Beaudoin et al. 2000; Kwak et al. 2002; Monroe-Augustus et al. 2003; Strader et al. 2008a; Strader et al. 2008b; Belin et al. 2009), we PCR amplified and sequenced IAA16 from AR269 genomic DNA. We identified a C-to-T mutation at position 860 that results in a Pro65-to-Leu substitution that alters the first proline in the conserved GWPPV degron motif of IAA16 domain II (Fig. 1b), analogous to the gain-of-function mutations reported in other Aux/IAA proteins (Fig. 1c).
Fig. 1.
A gain-of-function mutation in IAA16 results in ABA resistance. a Photographs of Col-0 (Wt) and homozygous (−/−) or heterozygous (+/−) iaa16-1 seedlings 4 d after transfer of 4-d-old seedlings to media with or without 10 μM ABA. b Recombination mapping with the indicated markers localized the AR269 lesion to a region on Chromosome 3. Sequencing of the IAA16 (At3g04730) gene from AR269 DNA revealed a C-to-T mutation at position +860 from the IAA16 initiator codon that results in a Pro65-to-Leu substitution in domain II of IAA16. c The Pro65-to-Leu substitution in domain II of IAA16 is similar to gain-of-function mutations found in domain II of IAA7, IAA14, IAA17, and IAA28. Numbers on the right indicate the amino acid position of the last residue shown. d The iaa16-1 allele is dominant for ABA resistance. Mean root lengths (+SE; n ≥ 16) of Col-0 (Wt) and homozygous (−/−) or heterozygous (+/−) iaa16-1 seedlings 4 d after transfer of 4-d-old seedlings to medium supplemented with 10 μM ABA. e Phylogenetic tree, excised from the larger analysis of Aux/IAA proteins shown in Fig. 3, of Arabidopsis thaliana IAA16, IAA14/SLR, IAA7/AXR2, IAA17/AXR3, and relatives from Arabidopsis lyrata, Oryza sativa, and Populus trichocarpa.
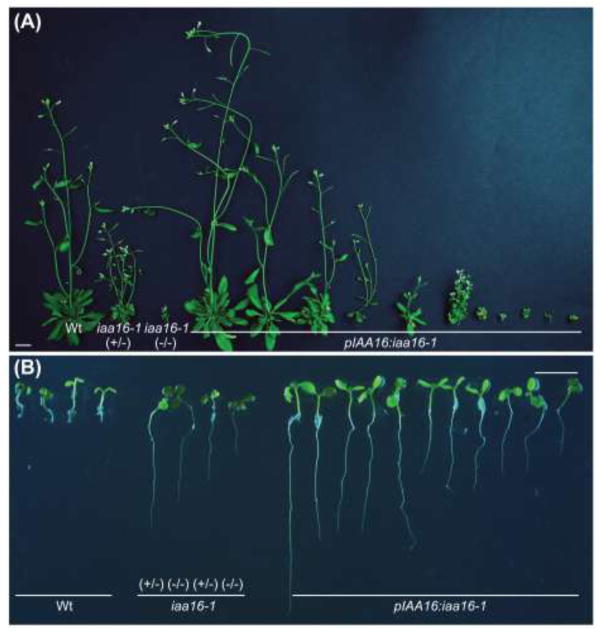
To confirm that the mutation we identified was responsible for the iaa16-1 mutant phenotypes, we tested whether we could recapitulate iaa16-1 phenotypes by introducing a genomic copy of iaa16-1 (pIAA16:iaa16-1) into a wild-type background. Out of 108 T1 seedlings selected for the Basta resistance conferred by the pIAA16:iaa16-1 transgene, only 23 survived after transfer to soil. These plants displayed a range of morphological defects, from a wild-type appearance to strikingly small plants resembling the iaa16-1 homozygote (Fig. 2a). The transformants that resembled the iaa16-1 mutant died without producing seeds, consistent with the possibility that, even when driven by the native IAA16 regulatory region, expression of iaa16-1 can be lethal. Because iaa16-1 also conferred dominant resistance to auxins, including the synthetic auxin 2,4-dichlorophenoxyacetic acid (2,4-D; see below), we also examined auxin response defects conferred by pIAA16:iaa16-1. We isolated pIAA16:iaa16-1 T1 individuals displaying long roots on an inhibitory concentration of 2,4-D (Fig. 2b) and determined that all of these individuals were Basta resistant, confirming that the iaa16-1 mutation confers dominant auxin resistance and that we had identified the causal mutation in AR269.
Fig. 2.
Expressing iaa16-1 using native flanking sequences in wild type recapitulates iaa16-1 phenotypes. a Photographs of 34-d-old Col-0 (Wt), heterozygous iaa16-1 (+/−), homozygous iaa16-1 (−/−) and pIAA16:iaa16-1 plants. Representatives of the 23 surviving transformants of the 108 selected for Basta resistance as seedlings are shown. Scale bar = 1 cm. b Photographs of 8-d-old Col-0 (Wt), heterozygous iaa16-1 (+/−), homozygous iaa16-1 (−/−) and pIAA16:iaa16-1-transformed Col-0 seedlings grown on 100 nM 2,4-D. Scale bar = 5 mm.
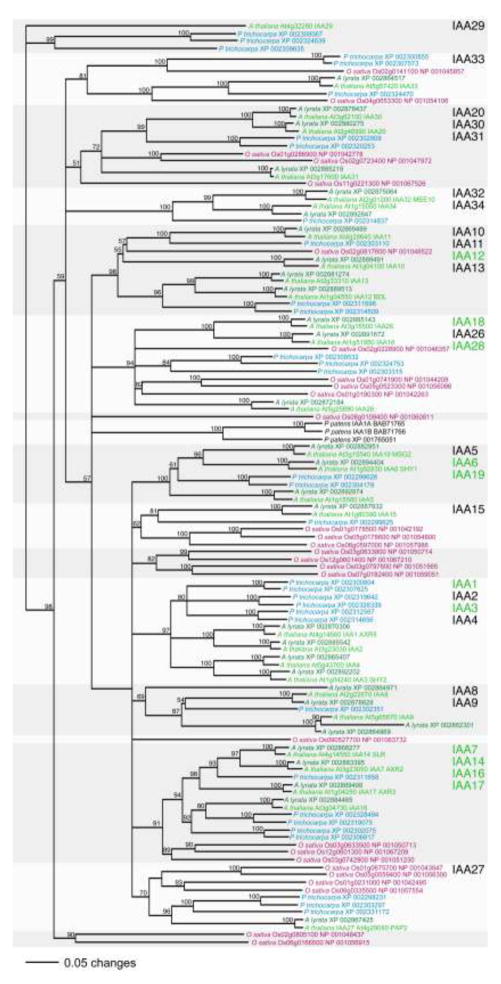
Arabidopsis IAA16 is most closely related (60 to 64% identical) to the Arabidopsis Aux/IAA proteins IAA7/AXR2, IAA14/SLR, and IAA17/AXR3 (Fig. 1e, 3; Remington et al. 2004). With the isolation of iaa16-1, gain-of-function mutations altering the GWPPV motif in each of the members of this clade have now been identified (Rouse et al. 1998; Nagpal et al. 2000; Fukaki et al. 2002). Using BLAST searches querying with the 29 predicted Arabidopsis thaliana Aux/IAA proteins (Liscum and Reed 2002), we identified 30 potential Aux/IAA proteins in Arabidopsis lyrata, 33 in Populus trichocarpa, and 28 in Oryza sativa (Fig. 3). In accordance with a previous report from Prigge et al. (2010), we found three predicted Aux/IAA proteins in Physcomitrella patens (Fig. 3). Phylogenetic analysis revealed that each of the four Aux/IAA proteins in the IAA16 clade correspond to an Aux/IAA protein in the close relative A. lyrata (Fig. 1e, 3). In addition, homologs of the IAA16 clade of Aux/IAA proteins are present in P. trichocarpa and O. sativa, suggesting that these proteins serve conserved roles in multiple plant species (Fig. 1e, 3).
Fig. 3.
Phylogenetic tree of Arabidopsis thaliana Aux/IAA proteins and relatives from Arabidopsis lyrata, Oryza sativa, Physcomitrella patens, and Populus trichocarpa. Proteins were aligned using ClustalW (alignment in Supplemental Dataset 1 online) and the unrooted phylogram was generated using PAUP 4.05b (Swofford 2001) using the bootstrap method with 1,000 replicates. Bootstrap values are displayed at the nodes. In the listing of Arabidopsis proteins at the right, green text indicates those for which dominant gain-of-function domain II mutations have been reported (including IAA16, reported here).
To compare the roles of the IAA16 clade of Aux/IAA proteins, we characterized phenotypes of iaa16-1 beside previously isolated gain-of-function mutations in the same clade, axr2-1 (IAA7; Wilson et al. 1990; Nagpal et al. 2000), axr3-1 (IAA17; Leyser et al. 1996; Rouse et al. 1998), slr-1 (IAA14; Fukaki et al. 2002) , and a gain-of-function mutation iaa28-1 (IAA28; Rogg et al. 2001) in a more distantly related Aux/IAA protein. Because the iaa16-1 mutation conferred sterility when homozygous (see below), we performed all assays on the progeny of an iaa16-1/IAA16 heterozygote and determined the genotypes of the plants following measurement or photography by using PCR. Thus we assessed the phenotype of not only the iaa16-1 homozygotes, but also individuals heterozygous for the iaa16-1 lesion. iaa16-1 is auxin resistant
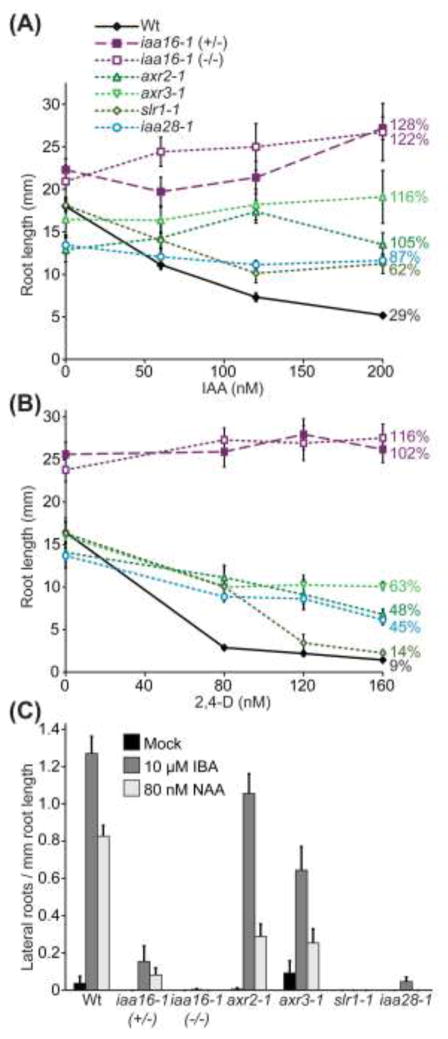
We expected iaa16-1 to display resistance to the phytohormone auxin, similar to the resistance conferred by other gain-of-function mutations in Aux/IAA genes. To assess auxin responsiveness in iaa16-1, we performed auxin-responsive root elongation assays. We found that, like the other Aux/IAA mutants from the IAA16 clade (Wilson et al. 1990; Leyser et al. 1996; Fukaki et al. 2002) and IAA28 (Rogg et al. 2001), iaa16-1 was resistant to the inhibitory effects of the naturally-occurring auxin indole-3-acetic acid (IAA; Fig. 4a) and the synthetic auxin 2,4-D (Fig. 4b) on primary root elongation. Indeed, iaa16-1 displayed greater resistance to 2,4-D than axr2-1, axr3-1, slr-1, or iaa28-1 (Fig. 4b). Even iaa16-1/IAA16 heterozygotes were uninhibited at 2,4-D concentrations that inhibited wild-type elongation by 90% (Fig. 4b). This auxin resistance supports a role for IAA16 in limiting auxin responses.
Fig. 4.
iaa16-1 displays auxin resistance in root elongation and lateral root initiation. a Mean root lengths (+/− SE; n 9) of 8-d-old Col-0 (Wt), heterozygous iaa16-1 (+/−), homozygous iaa16-1 (−/−), axr2-1, axr3-1, slr-1, and iaa28-1 seedlings grown on media supplemented with ethanol (0 nM IAA) or the IAA concentrations indicated. The ratio of the root length at the highest IAA concentration to the root length in the absence of IAA is expressed as a percentage next to each line. b Mean root lengths (+/− SE; n 13) of 8-d-old Col-0 (Wt), heterozygous iaa16-1 (+/−), homozygous iaa16-1 (−/−), axr2-1, axr3-1, slr-1, and iaa28-1 seedlings grown on medium supplemented with ethanol (0 nM 2,4-D) or the 2,4-D concentrations indicated. The ratio of the root length at the highest 2,4-D concentration to the root length in the absence of 2,4-D is expressed as a percentage next to each line. c iaa16-1 produces fewer lateral roots than wild type. Emerged lateral roots of Col-0 (Wt), heterozygous iaa16-1 (+/−), homozygous iaa16-1 (−/−), axr2-1, axr3-1, slr-1, and iaa28-1 were counted 4 d after transfer of 4-d-old seedlings to media supplemented with either ethanol (Mock), 10 μM IBA, or 80 nM NAA (mean +/− SE; n ≥ 12).
iaa16-1 is defective in lateral root production
In addition to inhibiting primary root elongation, auxin promotes lateral root production. We found that iaa16-1 had a dramatically reduced ability to make lateral roots. In the absence of exogenous hormone, 8-d-old iaa16-1, slr-1 (Fukaki et al. 2002), and iaa28-1 (Rogg et al. 2001) displayed no lateral roots and axr2-1 displayed fewer lateral roots than wild type (Fig. 4c). The naturally occurring auxin precursor indole-3-butyric acid (IBA) and the synthetic auxin 1-naphthaleneacetic acid (NAA) promoted lateral root production in wild type (Fig. 4c). iaa16-1, slr-1 (Fukaki et al. 2002), and iaa28-1 (Rogg et al. 2001) responded minimally to the effects of IBA and NAA on lateral root production, whereas axr3-1 and axr2-1 displayed intermediate responses (Fig. 4c). The decreased numbers of lateral roots in iaa16-1 is consistent with a role for IAA16 in inhibiting lateral root production.
iaa16-1 displays auxin-related developmental defects
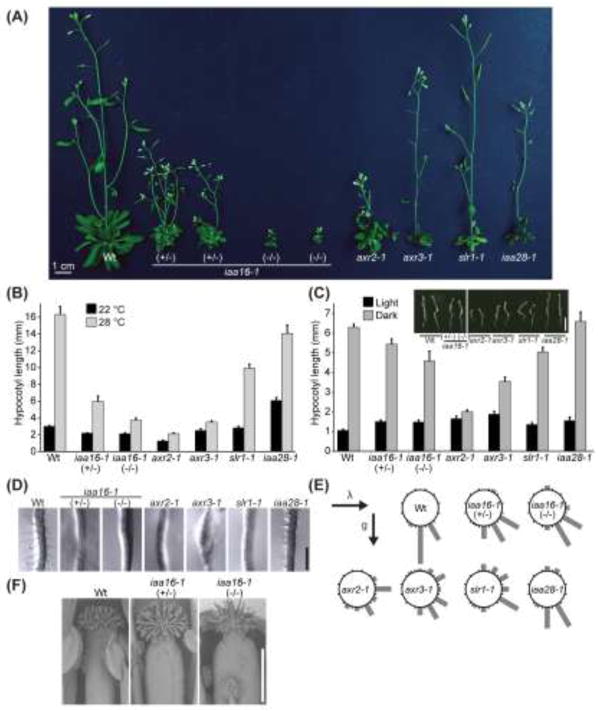
To reveal the respective roles of these Aux/IAA proteins in reducing auxin responses during normal growth and development, we compared the mutant phenotypes in the absence of exogenous hormone. Of the Aux/IAA mutants in the IAA16 clade, iaa16-1 displayed the most severe morphological and developmental defects as an adult, most notably the inability of homozygous iaa16-1 plants to produce seeds. Homozygous iaa16-1 plants were severely stunted, whereas iaa16-1/IAA16 heterozygotes, axr2-1, and iaa28-1 were shorter than wild type, and slr-1 height was similar to wild type (Fig. 5a). The severe defects of iaa16-1 adult plants are consistent with a role for IAA16 in inhibiting auxin responses in adult tissues.
Fig. 5.
iaa16-1 displays developmental defects. a Adult iaa16-1 plants are severely dwarfed and infertile. Photograph of 34-d-old Col-0 (Wt), heterozygous iaa16-1 (+/−), homozygous iaa16-1 (−/−), axr2-1, axr3-1, slr-1, and iaa28-1 plants. Scale bar = 1 cm. b iaa16-1 displays reduced high-temperature responsive hypocotyl elongation. Mean hypocotyl lengths (+SE, n 16) of Col-0 (Wt), heterozygous iaa16-1 (+/−), homozygous iaa16-1 (−/−), axr2-1, axr3-1, slr-1, and iaa28-1 seedlings grown for 8 d at 22 C or 28 C under yellow-filtered light. c iaa16-1 is partially de-etiolated in the dark. Mean hypocotyl lengths (+SE; n ≥ 14) of 4-d-old dark-grown Col-0 (Wt), heterozygous iaa16-1 (+/−), homozygous iaa16-1 (−/−), axr2-1, axr3-1, slr-1, and iaa28-1 seedlings. Inset: Photograph of 4-d-old dark-grown Col-0 (Wt), heterozygous iaa16-1 (+/−), homozygous iaa16-1 (−/−), axr2-1, axr3-1, slr-1, and iaa28-1 seedlings. Scale bar = 5 mm. d iaa16-1 displays decreased root hair lengths. Photographs of 5-d-old Col-0 (Wt), heterozygous iaa16-1 (+/−), homozygous iaa16-1 (−/−), axr2-1, axr3-1, slr-1, and iaa28-1 root hairs. Scale bar = 0.5 mm. e iaa16-1 displays reduced gravity-responsive root bending. Histogram showing distribution of root angles following 90-degree gravity reorientation (n ≥ 14). f iaa16-1 displays decreased anther filament elongation. Scanning electron micrographs of Col-0 (Wt), heterozygous iaa16-1 (+/−), and homozygous iaa16-1 (−/−) flowers. Scale bar = 0.5 mm.
Heterozygous iaa16-1/IAA16 individuals displayed decreased apical dominance, like axr2-1 (Wilson et al. 1990), whereas axr3-1 (Leyser et al. 1996), slr-1 (Fukaki et al. 2002), and iaa28-1 displayed increased apical dominance (Fig. 5a). In this respect, the phenotype of iaa28-1 introgressed into the Col-0 accession was different from that previously reported for the original iaa28-1 Ws allele, which displays reduced apical dominance (Rogg et al. 2001). This difference between iaa28-1 in the Col-0 and Ws-2 backgrounds suggests that certain phenotypes described for Aux/IAA mutants may be background-dependent, perhaps reflecting gene expression and/or interaction partner differences among various Arabidopsis accessions.
Growth at high temperatures increases free IAA levels and promotes hypocotyl elongation (Gray et al. 1998). We found that iaa16-1, like axr2-1, axr3-1, and slr-1, displayed shorter hypocotyls than wild type when grown in the light at elevated temperature (Fig. 5b), consistent with a role for IAA16 in decreasing auxin-responsive hypocotyl elongation.
Because some Aux/IAA mutants, such as axr2-1 and axr3-1 (Nagpal et al. 2000), are partially de-etiolated in the dark, we analyzed dark-grown iaa16-1 seedlings. Like wild type, dark-grown iaa16-1 seedlings had longer hypocotyls in the dark than in the light (Fig. 5c). However, iaa16-1 displayed slightly open apical hooks in darkness (Fig. 5c, inset), consistent with the possibility that IAA16 plays a role in inhibiting apical hook formation, an auxin-dependent process (Reed et al. 1998; Tian and Reed 1999; Nagpal et al. 2000; Friml et al. 2002; Vandenbussche et al. 2010; Žádníková et al. 2010).
Root hairs are outgrowths from certain root epidermal cell files that elongate in an auxin-dependent manner to increase water and nutrient uptake (reviewed in Grierson and Schiefelbein 2002). Like axr2-1 (Wilson et al. 1990), axr3-1 (Leyser et al. 1996), slr-1 (Fukaki et al. 2002), and iaa28-1 (Rogg et al. 2001), iaa16-1 displayed decreased root hair elongation (Fig. 5d). In addition, many iaa16-1 root epidermal cells appeared to be devoid of root hairs (Fig. 5d). The decreased root hair formation and elongation in iaa16-1 suggests a role for IAA16 in inhibiting this auxin-dependent cell elongation.
Root gravitropism is an auxin-dependent process (reviewed in Masson 1995). Because several Aux/IAA mutants display agravitropism in the hypocotyl and/or roots (Timpte et al. 1994; Leyser et al. 1996; Kim et al. 1998; Fukaki et al. 2002; Tatematsu et al. 2004; Yang et al. 2004), we assessed iaa16-1 root responsiveness to a change in the gravity vector. We grew seedlings vertically for 4 d, reoriented any seedlings whose roots were not growing downward, then turned the plates 90° for an additional 2 d of growth and measured the angle of growth. We found that iaa16-1 displayed moderately impaired graviresponsiveness (Fig. 5e), whereas axr2-1 (Timpte et al. 1994), axr3-1 (Leyser et al. 1996) and slr-1 (Fukaki et al. 2002) displayed strongly defective graviresponsiveness (Fig. 5e).
Because anther filament elongation is an auxin-regulated process (reviewed in Sundberg and Ostergaard 2009) and because the iaa16-1 homozygote was unable to produce seeds, we examined iaa16-1 flowers for filament defects. We found that heterozygous iaa16-1/IAA16 individuals, which produce seeds, had stamens that reach the stigma at anthesis, whereas homozygous iaa16-1, which produces no seeds, had stamens that fail to reach the stigma before dehiscence (Fig. 5f). Moreover, hand pollination of stigmas of homozygous iaa16-1 with pollen from homozygous iaa16-1 plants resulted in seed production (data not shown), suggesting that reduced filament elongation contributes to the infertility observed in homozygous iaa16-1 plants.
The breadth and severity of developmental defects in iaa16-1 are consistent with an important role for IAA16 in dampening auxin responses in a variety of tissues.
iaa16-1 displays resistance to ABA in root elongation and seed germination
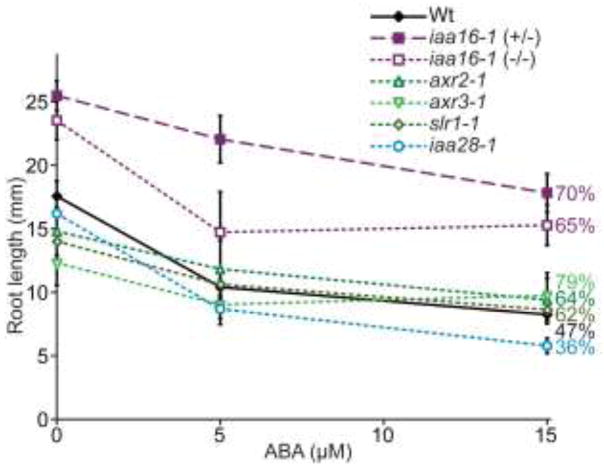
Because iaa16-1 was isolated as an ABA resistant mutant, we examined the responsiveness of axr2-1, axr3-1, slr-1, and iaa28-1 to the effects of ABA in both root elongation and seed germination. We found that, like iaa16-1, axr2-1 (Wilson et al. 1990), axr3-1, and slr-1 displayed some resistance to the effects of ABA on root elongation (Fig. 6). In contrast to mutants in the IAA16 clade, iaa28-1 responses to the inhibitory effects of ABA on root elongation were similar to those of wild type (Fig. 6). In addition to the iaa16-1 ABA resistance, the fact that iaa16-1 displayed a markedly longer root than wild type even in the absence of added hormone (Fig. 4a, 4b, 6) may have contributed to our initial isolation of this mutant.
Fig. 6.
ABA responsiveness conferred by gain-of-function Aux/IAA mutations. iaa16-1 is resistant to the inhibitory effects of ABA on root elongation. Mean root lengths (+SE; n ≥ 11) of Col-0 (Wt), heterozygous iaa16-1 (+/−), homozygous iaa16-1 (−/−), axr2-1, axr3-1, slr-1, and iaa28-1 seedlings 4 d after transfer of 4-d-old seedlings to media supplemented with ethanol (0 μM ABA) or the indicated ABA concentration. Numbers beside graph represent the percentage of root length on the highest level of ABA compared to the mock treatment for each genotype.
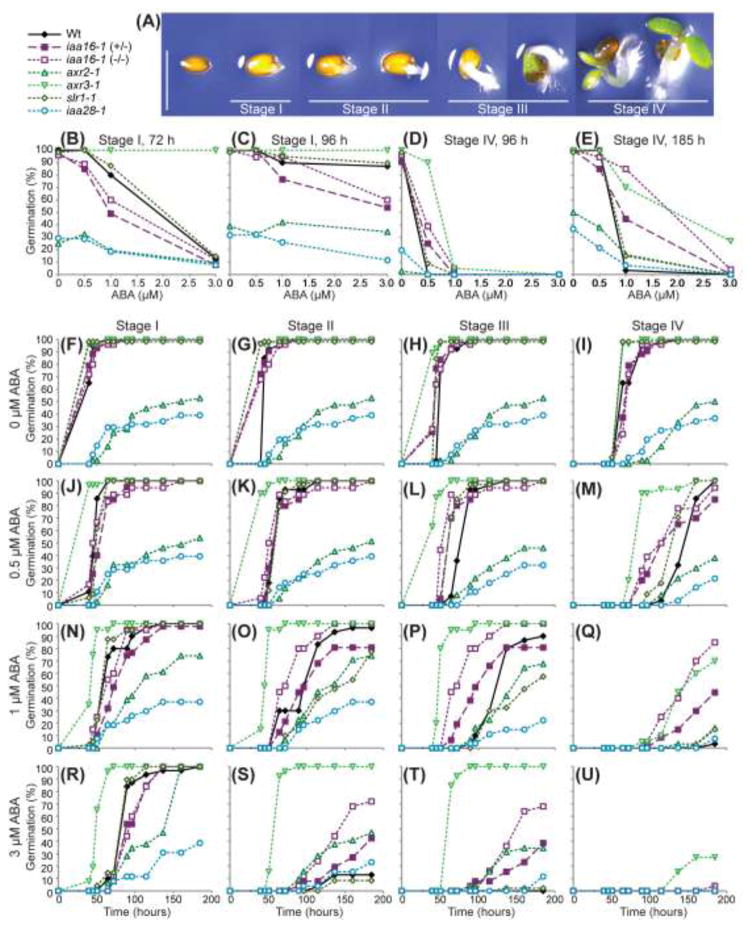
In addition to inhibiting root elongation, ABA inhibits seed germination and seedling establishment (reviewed in Himmelbach et al. 1998). We therefore examined these processes in iaa16-1, axr2-1, axr3-1, slr-1, and iaa28-1. Because seed germination and seedling establishment are a continuum, we scored the progress of individual seedlings over time: seeds with no emerged radicle were in Stage 0, seedlings with an emerged radicle without obvious elongation were in Stage I, seedlings with an elongated radicle that was less than the seed width were in Stage II, seedlings with a radicle longer than the seed width but with etiolated cotyledons were in Stage III, and seedlings with opened and greened cotyledons were in Stage IV (Fig. 7a). We observed various levels of ABA responsiveness in seed germination and seedling establishment in the examined Aux/IAA mutants. In the absence of ABA, axr2-1 and iaa28-1 displayed delayed and decreased germination levels, whereas axr3-1 and slr-1 progressed through the stages of seedling establishment more quickly than wild type (Fig. 7f-i). The axr3-1, axr2-1, and iaa28-1 mutants displayed the strongest resistance to the inhibitory effects of ABA on radicle emergence, whereas iaa16-1 and slr-1 responses to the effects of ABA on radicle emergence resembled those of wild type (Fig. 7b, c, f, j, n, r). Intriguingly, iaa16-1 resembled wild type in response to the effects of ABA on radicle emergence, yet was resistant to the effects of ABA on seedling establishment and displayed opened, greened cotyledons at inhibitory ABA concentrations (Fig. 7d, e, i, m, o, u). The ABA resistance displayed by the examined Aux/IAA mutants is consistent with a model in which auxin response acts downstream of ABA in regulating the processes of root elongation, radicle emergence, and seedling development.
Fig. 7.
iaa16-1 is resistant to ABA in germination. a Photographs of Col-0 seedlings in various stages of germination and seedling establishment. Stage I includes seeds with a protruding radicle that has not elongated. Stage II includes seedlings with a radicle that has elongated less than the width of the seed. Stage III denotes seedlings with a radicle longer than the width of the seed, but with cotyledons that remain etiolated. Stage IV includes seedlings with green cotyledons. Scale bar = 1 mm. b, c Percentage of Col-0 (Wt), heterozygous iaa16-1 (+/−), homozygous iaa16-1 (−/−), axr2-1, axr3-1, slr-1, and iaa28-1 seedlings at germination Stage I on the indicated ABA concentrations after 72 h (b) or 96 h (c) of incubation. d, e Percentage of Col-0 (Wt), heterozygous iaa16-1 (+/−), homozygous iaa16-1 (−/−), axr2-1, axr3-1, slr-1, and iaa28-1 seedlings at germination Stage IV (greened cotyledons) at the indicated ABA concentrations after 96 h (d) or 185 h (e) of incubation. f-u, Percentage of Col-0 (Wt), heterozygous iaa16-1 (+/−), homozygous iaa16-1 (−/−), axr2-1, axr3-1, slr-1, and iaa28-1 seedlings at germination Stage I (f, j, n, r), Stage II (g, k, o, s), Stage III (h, l, p, t) or Stage IV (i, m, q, u) on mock treatment (f, g, h, i), 0.5 μM ABA (j, k, l, m), 1 μM ABA (n, o, p, q), or 3 μM ABA (r, s, t, u) at the indicated time points (n ≥ 18).
Discussion
The 29-member Aux/IAA family of transcriptional repressors provides an intricate mechanism for attenuating auxin-responsive transcription by dimerizing with and inhibiting ARF family transcription factors (reviewed in Liscum and Reed 2002). Our identification of a gain-of-function mutation in IAA16 completes the collection of analogous mutants in the cluster of closely related Aux/IAA genes that includes IAA16, IAA7/AXR2, IAA14/SLR and IAA17/AXR3 and marks the eleventh Aux/IAA gene with an identified gain-of-function mutation in the highly conserved domain II degron.
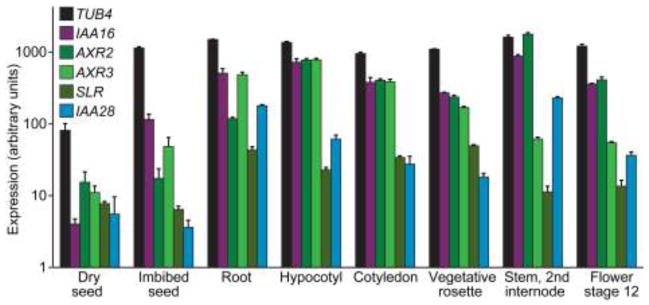
The distinct but overlapping phenotypes conferred by dominant mutations in various Aux/IAA genes may be explained by distinct but overlapping tissue and developmental expression patterns and protein binding partners. As expected based on the wide-ranging iaa16-1 developmental defects, analysis of compiled microarray data from the Arabidopsis eFP Browser (http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi) revealed robust IAA16 expression in all tissues where the iaa16-1 mutant displayed abnormalities, including cotyledons, roots, and hypocotyls of seedlings, and rosette leaves, stems, and flowers of adult plants (Fig. 8; Schmid et al. 2005). The Aux/IAA genes studied here display various expression patterns (Fig. 8; Che et al. 2002; Goda et al. 2004; Overvoorde et al. 2005; Vernoux et al. 2011) that likely contribute to phenotypic differences among the mutants. For example, IAA28 is most highly expressed in roots and stems (Fig. 8; Rogg et al. 2001), tissues in which iaa28-1 phenotypes are most apparent. Moreover, SLR and IAA28 display relatively little expression in hypocotyls (Fig. 8) and the slr-1 and iaa28-1 mutants display less abnormality in this organ than the other mutants (Fig. 5, b-d). IAA16 is expressed throughout seedlings and adult plants (Fig. 8), consistent with the severe phenotypes that we observed in all examined tissues.
Fig. 8.
IAA16 is expressed in many tissues and throughout development. Expression profiles were retrieved from microarray data provided at http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi for TUB4, IAA16, AXR2, AXR3, SLR and IAA28. Data are shown for dry seeds, seeds imbibed for 24 h, roots of 17-d-old seedlings, hypocotyls of 7-d-old seedlings, cotyledons of 7-d-old seedlings, vegetative rosettes from 14-d-old plants, second internodes of stems, and stage 12 flowers (Schmid et al. 2005).
In addition to differences in expression patterns (Fig. 8; Winter et al. 2007), distinct phenotypes in Aux/IAA mutants are likely affected by differences in interacting proteins. Aux/IAA proteins can homodimerize (Kim et al. 1997), heterodimerize with other Aux/IAA proteins (Kim et al. 1997), interact with members of the ARF family of auxin-responsive transcription factors that promote gene expression (Ouellet et al. 2001), interact with members of the ARF family that repress transcription (Ulmasov et al. 1997), and recruit co-repressors of the TOPLESS family (Szemenyei et al. 2008; Li et al. 2011). This complex network of interactions provides additional regulation of Aux/IAA activity and auxin-responsive transcription. The Aux/IAA proteins in this study have distinct and overlapping sets of interaction partners (Supplemental Table 1; Vernoux et al. 2011). These different interaction partners likely play a large role in the phenotypes conferred by the various gain-of-function mutations. Performing promoter-swapping experiments for members of the IAA16 clade of Aux/IAA proteins, as was done with MSG2, AXR2, and SLR (Muto et al. 2007), and in other studies (Knox et al. 2003; Weijers et al. 2005; Muto et al. 2007), would help determine the level of phenotypic input from expression versus protein-protein affinity differences.
iaa16-1 displays many phenotypes associated with reduced auxin response. Adult iaa16-1 plants are tiny and display decreased apical dominance (Fig. 5a) and reduced anther filament elongation (Fig. 5e), iaa16-1 seedlings display markedly increased primary root elongation (Fig. 4a, 4b, 6), iaa16-1 root hairs are much shorter than those of wild type (Fig. 5d), and iaa16-1 roots produce fewer lateral roots than wild type (Fig. 4c) and display mildly altered response to gravity-induced root reorientation (Fig. 5e). In addition, dark-grown iaa16-1 seedlings do not fully etiolate (Fig. 5c). The array and severity of iaa16-1 phenotypes suggest that the IAA16 protein plays important roles in inhibiting auxin responses in a variety of tissues and developmental programs.
Intriguingly, homozygous iaa16-1 mutants display stronger phenotypes than heterozygous iaa16-1/IAA16 mutants in only a subset of assays, suggesting that the level of iaa16-1 protein required to maximally inhibit auxin responses is context-dependent. For example, homozygous iaa16-1 mutants clearly have more severe defects than the heterozygotes in adult morphology, as demonstrated by the greatly reduced stature (Fig. 5a) and infertility. In addition, homozygous iaa16-1 is more ABA resistant than heterozygous iaa16-1/IAA16 in cotyledon greening assays (Fig. 7) and more auxin resistant in lateral root initiation assays (Fig. 4c). However, in ABA-responsive root elongation inhibition (Fig. 6) and auxin-responsive root elongation inhibition (Fig. 4a, b), heterozygous iaa16-1 defects are indistinguishable from those of homozygous iaa16-1. The semi-dominant nature of the iaa16-1 mutant may be partially masked in assays in which robust growth is a requirement.
We isolated iaa16-1, defective in a member of a family of proteins with known roles in auxin signaling, from a mutant screen for ABA resistance in root elongation, adding to a growing body of evidence that auxin responsiveness is required for a full ABA response. ABA regulates many plant responses to stress (reviewed in Wasilewska et al. 2008), often by reducing growth, including decreasing seed germination, post-germinative growth, and root elongation. Several auxin response mutants, including indole-3-butyric acid response5 (ibr5; Monroe-Augustus et al. 2003; Strader et al. 2008b), transport inhibitor response 1 (tir1; Strader et al. 2008a), auxin-resistant 1 (axr1; Monroe-Augustus et al. 2003), auxin resistant1 (aux1; Strader et al. 2008a), shy2 (Tian and Reed 1999), and axr2 (Wilson et al. 1990) display reduced responsiveness to ABA in root elongation assays, consistent with a role for auxin downstream of ABA in root elongation inhibition. Further, auxin treatment induces the ABA-responsive transcriptional reporter Dc3:GUS, even in the ABA insensitive abi1-1 and abi2-1 mutant backgrounds (Rock and Sun 2005), consistent with a role for auxin stimulating ABA responses in the root and potentially reinforcing the ABA response.
In addition to mutants displaying ABA resistant root elongation, many auxin response mutants, such as ibr5 (Monroe-Augustus et al. 2003), axr2 (Beaudoin et al. 2000; Belin et al. 2009), rcn1 (Kwak et al. 2002), aux1 (Belin et al. 2009), eir1/pin2 (Belin et al. 2009), and axr3 (Belin et al. 2009) display reduced responsiveness to ABA in seed germination and seedling establishment assays. Because both aux1 and eir1/pin2 mutants are resistant to the inhibitory effects of ABA on postembryonic growth, AUX1 and PIN2 auxin carrier activity, and thus auxin transport from the root apex to the elongation zone, is likely required for ABA-responsive inhibition of the embryonic axis during seedling establishment (Belin et al. 2009).
Conversely, some ABA signaling components are needed for full auxin responsiveness. The B3 transcription factor ABI3 plays a role in auxin-induced lateral root formation; the abi3 mutant displays reduced sensitivity to auxin in lateral root induction assays (Brady et al. 2003). Indeed, ABA potentiates auxin signaling during seedling establishment, possibly by downregulating AXR2 and AXR3 gene expression (Belin et al. 2009). These observations demonstrate the close connection between auxin and ABA response. However, the molecular nature of this interaction is not fully understood. Future studies may reveal whether modulation of auxin response by ABA is primarily dependent on altered Aux/IAA gene expression (Belin et al. 2009) or on altered gene expression in combination with additional factors.
Comparison of iaa16-1 phenotypes to those conferred by other gain-of-function mutations in closely related genes and in the more distantly related IAA28 gene reveals distinct and overlapping defects in development and in response to auxin and ABA. In many but not all assays, iaa16-1 displays more severe defects than axr2-1, slr-1, axr3-1, and iaa28-1. Most notably, homozygous iaa16-1 plants are extreme dwarfs that fail to produce progeny (Fig. 5a). This severe defect raises the possibility that similar mutations in additional Aux/IAA family members also may result in infertility that would hamper isolation. Given the fertility and extreme auxin resistance of the iaa16-1/IAA16 heterozygote, however, it is surprising that this mutant has not been isolated from previous screens for auxin-resistant root elongation, and suggests that screens for auxin-resistant Arabidopsis mutants are not yet saturated more than 30 years after the first report (Maher and Martindale 1980).
Materials and methods
Plant materials and genetic analysis
Arabidopsis thaliana lines were in the Columbia (Col-0) background, which was used as the wild type. iaa28-1, originally obtained in the Wassilewskija background (Rogg et al. 2001), was introgressed into the Col-0 background by crossing iaa28-1 to Col-0 three times and monitoring introgression using PCR-based markers to differentiate between the Col-0 and Wassilewskija backgrounds (Rogg 2001).
AR269 was isolated from the progeny of ethyl methanesulfonate (EMS) treated Col-0 seeds and was crossed to Landsberg erecta for recombination mapping. F2 plants were moved to soil and their progeny tested for ABA resistance in root elongation. DNA from both heterozygous ABA resistant and homozygous ABA sensitive progeny pools was scored using PCR-based polymorphic molecular markers (Konieczny and Ausubel 1993; Bell and Ecker 1994). The IAA16 gene (At3g04730) within the AR269 mapping interval was PCR amplified and sequenced from AR269 genomic DNA.
The iaa16-1 mutant was backcrossed to wild-type Col-0 two or three times before phenotypic analysis. All iaa16-1 measurements were made on the segregating progeny of an iaa16-1/IAA16 heterozygote. After measurement or photography, DNA was prepared from individual seedlings or leaves (Celenza et al. 1995), and PCR analysis was used to determine genotype of each individual. PCR amplification with 5 ′-AAATTTCAGGGCACAAGTTGTGGGATAGC-3′ and 5′-TGCACCGTCCATGCTAACCTTCACATAAGC-3′ yields a 190-bp product with one AluI restriction site in iaa16-1 and none in wild type.
Growth conditions and phenotypic analysis
Seeds were surface sterilized with 30% bleach and 0.1% Triton X-100 for 10 minutes, then rinsed with sterile water twice before suspension in 0.1% agar. To reduce Agrobacterium contamination, T1 seed sterilization included an additional 5-minute incubation with 70% ethanol prior to bleach treatment. Seeds were then stratified in 0.1% agar at 4 °C for 1 d. Surface-sterilized seeds were plated on Plant Nutrient (PN) medium (Haughn and Somerville 1986) supplemented with 0.5% sucrose (PNS) and solidified with 0.6% (w/v) agar. Plates were oriented horizontally in incubators at 22 °C under continuous illumination unless otherwise noted.
For auxin-responsive root elongation experiments, seeds were plated on PNS supplemented with the indicated auxin and grown for 8 d under yellow long-pass plexiglass filters to slow indolic compound breakdown (Stasinopoulos and Hangarter 1990).
For ABA-responsive root elongation experiments, seeds were plated on unsupplemented PNS and grown for 4 d under yellow long-pass plexiglass filters before transfer to PNS supplemented with ABA and grown for an additional 4 d under yellow filters.
For ABA-responsive germination and seedling development experiments, seeds were plated on PNS supplemented with the indicated concentrations of ABA and grown under white light. Seeds were examined at the indicated time points and the stage of germination and seedling development were noted. Stage I includes seeds with a protruding radicle that has not elongated. Stage II includes seedlings with a radicle that has elongated less than the width of the seed. Stage III denotes seedlings with a radicle longer than the width of the seed, but with cotyledons that remain etiolated. Stage IV includes all seedlings with green, opened cotyledons.
For dark-grown hypocotyl length experiments, seeds were plated on PNS and incubated for 1 d under white light to stimulate germination and then for an additional 4 d in the dark.
For temperature-dependent light-grown hypocotyl length experiments, seeds were plated on PNS and incubated under yellow light at either 22 °C or 28 °C.
For gravitropism experiments, seeds were plated on PNS and grown horizontally for 1 d followed by an additional 4 d of vertical growth. Seedlings were repositioned such that their roots were directed toward the gravity vector then the plates were turned 90° and grown for 2 d. Seedlings were then imaged with a Leica DFC295 camera attached to a Leica MZ10F dissecting microscope and NIH ImageJ was used to measure root angles.
For root hair elongation experiments, seeds were plated on PNS and grown horizontally for 1 d followed by additional vertical growth for 4 d. Root segments within 4 mm of the root-shoot junction were imaged with a Leica DFC295 camera attached to a Leica MZ10F dissecting microscope and representative images shown.
For lateral root experiments, seeds were plated on PNS and grown under yellow light for 4 d prior to transfer to media containing the indicated amounts of auxin and grown for an additional 4 d under yellow light. Emerged lateral roots were counted using a dissecting microscope.
For adult plant phenotype experiments, seeds were plated on PNS and grown under white light for 12 d before transfer to soil. Soil-grown plants were grown under continuous illumination at 22 °C for the indicated number of days prior to being photographed.
To examine flower morphology, flowers from adult plants were mounted on scanning electron microscopy stubs, frozen in liquid nitrogen and imaged in a Hitachi TM-1000 scanning electron microscope.
Vector construction and plant transformation
The pIAA16:iaa16-1 region was amplified from iaa16-1 genomic DNA using Pfx Platinum Taq (Invitrogen) using 5′-CACCGGTTGTGTTTAAGCCATGTTGTTCTC-3′ and 5′-AGATGCCTTACGGACACAGACC-3′, which included the region 1973 bp upstream of the iaa16-1 start codon to 316 bp downstream of the iaa16-1 stop codon. The resulting PCR product was cloned into the pENTR/D-TOPO vector (Invitrogen). The iaa16-1 genomic fragment was recombined into the promoterless destination vector pDMC123 (Curtis and Grossniklaus 2003) using LR Clonase (Invitrogen) to form pDMC123-iaa16-1. pDMC123-iaa16-1 was then transformed into Agrobacterium tumefaciens strain GV3101 (Koncz and Schell 1986), which was used to transform wild-type Arabidopsis using the floral dip method (Clough and Bent 1998). Transformants were selected on PNS supplemented with 7.5 g/mL Basta (phosphinothricin).
Phylogenetic analysis
Sequences from Arabidopsis thaliana Aux/IAA proteins were used to identify homologs in Arabidopisis lyrata, Oryza sativa, and Populus trichocarpa using NCBI BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi; Altschul et al. 1990). Identified protein sequences, along with previously identified Aux/IAA proteins from Physcomitrella patens (Prigge et al. 2010) were aligned using Lasergene MegAlign (DNASTAR) using the ClustalW default settings with the Gonnet series protein weight matrix. The alignment was manually adjusted for optimization. An unrooted phylogram was generated using PAUP 4.05b (Swofford 2001) by performing the bootstrap method with 1,000 replicates using distance as the optimality criterion and all characters equally weighted.
Expression data
Expression data was collected from the Arabidopsis eFP Browser at the University of Toronto, http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi (Winter et al. 2007).
Supplementary Material
Acknowledgments
We thank Luise Rogg for introgressing the iaa28-1 mutation into the Col-0 accession, Mark Estelle for axr2-1 and axr3-1, Hidehiro Fukaki for slr-1, and Lauren Gunther, David Korasick, and Julie Thole for critical comments on the manuscript. This research was supported by the National Institutes of Health (R00 GM089987-03 to L.C.S.), the National Science Foundation (MCB-0745122 to BB), and the Robert A. Welch Foundation (C-1309 to B.B.).
Contributor Information
Mauro A. Rinaldi, Department of Biochemistry and Cell Biology, Rice University, Houston, Texas 77005, USA
James Liu, Department of Biochemistry and Cell Biology, Rice University, Houston, Texas 77005, USA.
Tara A. Enders, Department of Biology, Washington University, St. Louis, Missouri 63130, USA
Bonnie Bartel, Department of Biochemistry and Cell Biology, Rice University, Houston, Texas 77005, USA.
Lucia C. Strader, Email: strader@wustl.edu, Department of Biology, Washington University, St. Louis, Missouri 63130, USA
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Beaudoin N, Serizet C, Gosti F, Giraudat J. Interactions between abscisic acid and ethylene signaling cascades. Plant Cell. 2000;12:1103–1115. doi: 10.1105/tpc.12.7.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin C, Megies C, Hauserova E, Lopez-Molina L. Abscisic acid represses growth of the Arabidopsis embryonic axis after germination by enhancing auxin signaling. Plant Cell. 2009;21:2253–2268. doi: 10.1105/tpc.109.067702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Brady SM, Sarkar SF, Bonetta D, McCourt P. The ABSCISIC ACID INSENSITIVE 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. Plant J. 2003;34:67–75. doi: 10.1046/j.1365-313x.2003.01707.x. [DOI] [PubMed] [Google Scholar]
- Calderon-Villalobos LI, Tan X, Zheng N, Estelle M. Auxin perception--structural insights. Cold Spring Harb Perspect Biol. 2010;2:a005546. doi: 10.1101/cshperspect.a005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza JL, Grisafi PL, Fink GR. A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev. 1995;9:2131–2142. doi: 10.1101/gad.9.17.2131. [DOI] [PubMed] [Google Scholar]
- Che P, Gingerich DJ, Lall S, Howell SH. Global and hormone-induced gene expression changes during shoot development in Arabidopsis. Plant Cell. 2002;14:2771–2785. doi: 10.1105/tpc.006668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S, Ghassemian M, Bonetta D, Cooney S, McCourt P. A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science. 1996;273:1239–1241. doi: 10.1126/science.273.5279.1239. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005a;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005b;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- dos Santos Maraschin F, Memelink J, Offringa R. Auxin-induced, SCF(TIR1)-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J. 2009;59:100–109. doi: 10.1111/j.1365-313X.2009.03854.x. [DOI] [PubMed] [Google Scholar]
- Dreher KA, Brown J, Saw RE, Callis J. The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell. 2006;18:699–714. doi: 10.1105/tpc.105.039172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR. Maternal effects govern variable dominance of two abscisic acid response mutations in Arabidopsis thaliana. Plant Physiol. 1994;105:1203–1208. doi: 10.1104/pp.105.4.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature. 2002;415:806–809. doi: 10.1038/415806a. [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 2002;29:153–168. doi: 10.1046/j.0960-7412.2001.01201.x. [DOI] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2004;134:1555–1573. doi: 10.1104/pp.103.034736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Östin A, Sandberg G, Romano CP, Estelle M. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:7197–7202. doi: 10.1073/pnas.95.12.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grierson C, Schiefelbein J. The Arabidopsis Book. American Society of Plant Biologists; Rockville, MD: 2002. Root Hairs: April 4, 2002; pp. 1–e0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G. Auxin response factors. Curr Opin Plant Biol. 2007;10:453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol. 2002;49:373–385. [PubMed] [Google Scholar]
- Hamann T, Benkova E, Bäurle I, Kientz M, Jürgens G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002;16:1610–1615. doi: 10.1101/gad.229402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T, Mayer U, Jürgens G. The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development. 1999;126:1387–1395. doi: 10.1242/dev.126.7.1387. [DOI] [PubMed] [Google Scholar]
- Haughn GW, Somerville C. Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet. 1986;204:430–434. [Google Scholar]
- Himmelbach A, Iten M, Grill E. Signalling of abscisic acid to regulate plant growth. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 1998;353:1439–1444. doi: 10.1098/rstb.1998.0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. Auxin-induced SCFTIR1-Aux/IAA interaction involves stable modification of the SCFTIR1 complex. Proc Natl Acad Sci USA. 2004;101:12381–12386. doi: 10.1073/pnas.0402868101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- Kim BC, Soh MS, Hong SH, Furuya M, Nam HG. Photomorphogenic development of the Arabidopsis shy2-1D mutation and its interaction with phytochromes in darkness. Plant J. 1998;15:61–68. doi: 10.1046/j.1365-313x.1998.00179.x. [DOI] [PubMed] [Google Scholar]
- Kim BC, Soh MS, Kang BJ, Furuya M, Nam HG. Two dominant photomorphogenic mutations of Arabidopsis thaliana identified as suppressor mutations of hy2. Plant J. 1996;9:441–456. doi: 10.1046/j.1365-313x.1996.09040441.x. [DOI] [PubMed] [Google Scholar]
- Kim J, Harter K, Theologis A. Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci USA. 1997;94:11786–11791. doi: 10.1073/pnas.94.22.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox K, Grierson CS, Leyser O. AXR3 and SHY2 interact to regulate root hair development. Development. 2003;130:5769–5777. doi: 10.1242/dev.00659. [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J. The promoter of the TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plant. 1984;61:377–383. [Google Scholar]
- Kwak JM, Moon JH, Murata Y, Kuchitsu K, Leonhardt N, DeLong A, Schroeder JI. Disruption of a guard cell-expressed protein phosphatase 2A regulatory subunit, RCN1, confers abscisic acid insensitivity in Arabidopsis. Plant Cell. 2002;14:2849–2861. doi: 10.1105/tpc.003335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser HMO, Pickett FB, Dharmasiri S, Estelle M. Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 1996;10:403–413. doi: 10.1046/j.1365-313x.1996.10030403.x. [DOI] [PubMed] [Google Scholar]
- Li H, Tiwari SB, Hagen G, Guilfoyle TJ. Identical amino acid substitutions in the repression domain of auxin/indole-3-acetic acid proteins have contrasting effects on auxin signaling. Plant Physiol. 2011;155:1252–1263. doi: 10.1104/pp.110.171322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Reed JW. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol. 2002;49:387–400. [PubMed] [Google Scholar]
- Maher EP, Martindale SJB. Mutants of Arabidopsis thaliana with altered responses to auxins and gravity. Biochem Genet. 1980;18:1041–1053. doi: 10.1007/BF00484337. [DOI] [PubMed] [Google Scholar]
- Masson PH. Root gravitropism. BioEssays : news and reviews in molecular, cellular and developmental biology. 1995;17:119–127. doi: 10.1002/bies.950170207. [DOI] [PubMed] [Google Scholar]
- Mockaitis K, Estelle M. Auxin receptors and plant development: a new signaling paradigm. Annual review of cell and developmental biology. 2008;24:55–80. doi: 10.1146/annurev.cellbio.23.090506.123214. [DOI] [PubMed] [Google Scholar]
- Monroe-Augustus M, Zolman BK, Bartel B. IBR5, a dual-specificity phosphatase-like protein modulating auxin and abscisic acid responsiveness in Arabidopsis. Plant Cell. 2003;15:2979–2991. doi: 10.1105/tpc.017046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto H, Watahiki MK, Nakamoto D, Kinjo M, Yamamoto KT. Specificity and similarity of functions of the Aux/IAA genes in auxin signaling of Arabidopsis revealed by promoter-exchange experiments among MSG2/IAA19, AXR2/IAA7, and SLR/IAA14. Plant Physiol. 2007;144:187–196. doi: 10.1104/pp.107.096628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW. AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 2000;123:563–573. doi: 10.1104/pp.123.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet F, Overvoorde PJ, Theologis A. IAA17/AXR3: biochemical insight into an auxin mutant phenotype. Plant Cell. 2001;13:829–841. doi: 10.1105/tpc.13.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde PJ, Okushima Y, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Liu A, Onodera C, Quach H, Smith A, Yu G, Theologis A. Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell. 2005;17:3282–3300. doi: 10.1105/tpc.105.036723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrot-Rechenmann C. Cellular responses to auxin: division versus expansion. Cold Spring Harb Perspect Biol. 2010;2:a001446. doi: 10.1101/cshperspect.a001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Lavy M, Ashton NW, Estelle M. Physcomitrella patens auxin-resistant mutants affect conserved elements of an auxin-signaling pathway. Curr Biol. 2010;20:1907–1912. doi: 10.1016/j.cub.2010.08.050. [DOI] [PubMed] [Google Scholar]
- Ramos JA, Zenser N, Leyser O, Callis J. Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell. 2001;13:2349–2360. doi: 10.1105/tpc.010244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Elumalai RP, Chory J. Suppressors of an Arabidopsis thaliana phyB mutation identify genes that control light signaling and hypocotyl elongation. Genetics. 1998;148:1295–1310. doi: 10.1093/genetics/148.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington DL, Vision TJ, Guilfoyle TJ, Reed JW. Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol. 2004;135:1738–1752. doi: 10.1104/pp.104.039669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock CD, Sun X. Crosstalk between ABA and auxin signaling pathways in roots of Arabidopsis thaliana (L.) Heynh. Planta. 2005;222:98–106. doi: 10.1007/s00425-005-1521-9. [DOI] [PubMed] [Google Scholar]
- Rogg LE. PhD. Rice University; Houston: 2001. Cloning and characterization of IAA28, a gene involved in suppressing lateral root development and mediating auxin responses in Arabidopsis thaliana. [Google Scholar]
- Rogg LE, Lasswell J, Bartel B. A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell. 2001;13:465–480. doi: 10.1105/tpc.13.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse D, Mackay P, Stirnberg P, Estelle M, Leyser O. Changes in auxin response from mutations in an AUX/IAA gene. Science. 1998;279:1371–1373. doi: 10.1126/science.279.5355.1371. [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nature genetics. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Stasinopoulos TC, Hangarter RP. Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol. 1990;93:1365–1369. doi: 10.1104/pp.93.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Monroe-Augustus M, Bartel B. The IBR5 phosphatase promotes Arabidopsis auxin responses through a novel mechanism distinct from TIR1-mediated repressor degradation. BMC Plant Biol. 2008a;8:41. doi: 10.1186/1471-2229-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Monroe-Augustus M, Rogers KC, Lin GL, Bartel B. Arabidopsis iba response5 (ibr5) suppressors separate responses to various hormones. Genetics. 2008b;180:2019–2031. doi: 10.1534/genetics.108.091512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg E, Ostergaard L. Distinct and dynamic auxin activities during reproductive development. Cold Spring Harb Perspect Biol. 2009;1:a001628. doi: 10.1101/cshperspect.a001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. Phylogenetic analysis using parsimony (and other methods) 4. Sinauer Associates; Sunderland, MA: 2001. PAUP*. [Google Scholar]
- Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science. 2008;319:1384–1386. doi: 10.1126/science.1151461. [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT. MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell. 2004;16:379–393. doi: 10.1105/tpc.018630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Reed JW. Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development. 1999;126:711–721. doi: 10.1242/dev.126.4.711. [DOI] [PubMed] [Google Scholar]
- Timpte C, Wilson AK, Estelle M. The axr2-1 mutation of Arabidopsis thaliana is a gain-of-function mutation that disrupts an early step in auxin response. Genetics. 1994;138:1239–1249. doi: 10.1093/genetics/138.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Okushima Y, Mimura T, Tasaka M, Fukaki H. Domain II mutations in CRANE/IAA18 suppress lateral root formation and affect shoot development in Arabidopsis thaliana. Plant Cell Physiol. 2008;49:1025–1038. doi: 10.1093/pcp/pcn079. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Petrášek J, Žádníková P, Hoyerová K, Pešek B, Raz V, Swarup R, Bennett M, Zažímalová E, Benková E, Van Der Straeten D. The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development. 2010;137:597–606. doi: 10.1242/dev.040790. [DOI] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, Legrand J, Oliva M, Das P, Larrieu A, Wells D, Guedon Y, Armitage L, Picard F, Guyomarc'h S, Cellier C, Parry G, Koumproglou R, Doonan JH, Estelle M, Godin C, Kepinski S, Bennett M, De Veylder L, Traas J. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Molecular systems biology. 2011;7:508. doi: 10.1038/msb.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasilewska A, Vlad F, Sirichandra C, Redko Y, Jammes F, Valon C, Frei dit Frey N, Leung J. An update on abscisic acid signaling in plants and more. Mol Plant. 2008;1:198–217. doi: 10.1093/mp/ssm022. [DOI] [PubMed] [Google Scholar]
- Weijers D, Benkova E, Jager KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jurgens G. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J. 2005;24:1874–1885. doi: 10.1038/sj.emboj.7600659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AK, Pickett FB, Turner JC, Estelle M. A dominant mutation in Arabidopsis confers resistance to auxin, ethylene, and abscisic acid. Mol Gen Genet. 1990;222:377–383. doi: 10.1007/BF00633843. [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An "Electronic Fluorescent Pictograph" browser for exploring and analyzing large-scale biological data sets. PloS one. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley CK, Zenser N, Ramos J, Rouse D, Leyser O, Theologis A, Callis J. Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J. 2000;21:553–562. doi: 10.1046/j.1365-313x.2000.00703.x. [DOI] [PubMed] [Google Scholar]
- Yang X, Lee S, So JH, Dharmasiri S, Dharmasiri N, Ge L, Jensen C, Hangarter R, Hobbie L, Estelle M. The IAA1 protein is encoded by AXR5 and is a substrate of SCFTIR1. Plant J. 2004;40:772–782. doi: 10.1111/j.1365-313X.2004.02254.x. [DOI] [PubMed] [Google Scholar]
- Žádniková P, Petraek J, Marhavý P, Raz V, Vandenbussche F, Ding Z, Schwarzerová K, Morita MT, Tasaka M, Hejátko J, Van Der Straeten D, Friml J, Benková E. Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development. 2010;137:607–617. doi: 10.1242/dev.041277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.