Abstract
Celiac sprue is an inflammatory disease of the small intestine caused by dietary gluten and treated by adherence to a lifelong gluten-free diet. The recent identification of immunodominant gluten peptides, the discovery of their cogent properties, and the elucidation of the mechanisms by which they engender immunopathology in genetically-susceptible individuals have advanced our understanding of the molecular pathogenesis of this complex disease, enabling the rational design of new therapeutic strategies. The most clinically advanced of these is oral enzyme therapy, in which enzymes capable of proteolyzing gluten (i.e. glutenases) are delivered to the alimentary tract of a celiac sprue patient to detoxify ingested gluten in situ. In this chapter, we discuss the key challenges for discovery and preclinical development of oral enzyme therapies for celiac sprue. Methods for lead identification, assay development, gram-scale production and formulation, and lead optimization for next-generation proteases are described and critically assessed.
1. Introduction
Celiac sprue is a complex inflammatory disease of the small intestine involving both genetic and environmental factors. It is unique among chronic bowel disorders in that a specific dietary component – gluten from wheat and related protein content from rye and barley – has been identified as the proximate environmental cause of inflammation (Dicke et al., 1953). Gluten is a heterogeneous mixture of water insoluble storage proteins comprising monomeric gliadins and polymeric glutenin aggregates up to 10 MDa in size (Wieser, 2007). Both gliadins and glutenins contain repetitive sequences that are rich in proline (Pro; 15%) and glutamine (Gln; 35%), residues that are not preferred substrates for any human digestive enzyme. Consequently, gluten is relatively resistant to gastrointestinal proteolysis, releasing metastable Pro/Gln-rich peptides up to 30-40 amino acids in length into the gut lumen following digestion (Figure 1) (Shan et al., 2002; Shan et al., 2005b). Intestinal transglutaminase 2 (TG2) deamidates some of these peptides at specific Gln residues (Molberg et al., 1998; Shan et al., 2002), thereby increasing their affinity for human leukocyte antigen (HLA) DQ2 (Quarsten et al., 1999), a major histocompatibility (MHC) class II molecule associated with over 90% of celiac patients (Sollid et al., 1989). Deamidated gluten peptide-DQ2 complexes on the surface of antigen-presenting cells (APCs) elicit a potent TH1 inflammatory response from gluten-specific intestinal T cells (Nilsen et al., 1998; Nilsen et al., 1995). Ultimately, this inflammatory response causes destruction of the intestinal architecture, malabsorption of nutrients, and, in many patients, numerous secondary symptoms including diarrhea, wasting, and anemia (Alaedini and Green, 2005). Complete and life-long adherence to a gluten-free diet reverses the signs and symptoms of celiac sprue in most patients. However, the ubiquity of gluten in human diets renders this an extraordinarily difficult prescription to follow, leading to frequent lapses and chronic morbidity in many patients (Ciacci et al., 2002; Cornell et al., 2005; Mayer et al., 1991; Pietzak, 2005). Non-dietary therapies that detoxify ingested gluten would therefore improve quality of life considerably for the 1-2% of people afflicted with this life-long disease.
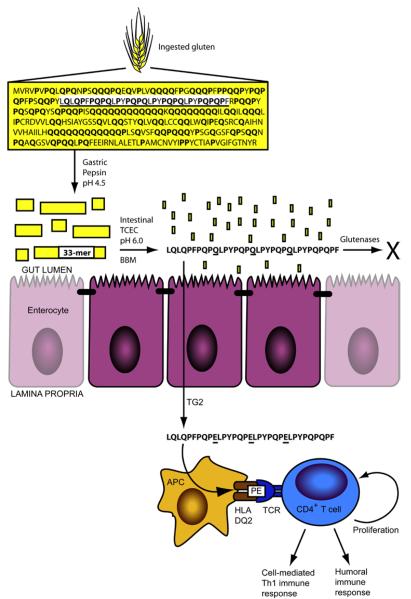
Figure 1.
Gastrointestinal digestion of gluten releases metastable immunogenic peptides. The sequence for α2-gliadin, a representative gluten protein, is shown with proline (17%) and glutamine (36%) content in bold. The 33-mer sequence is underlined. Pepsin cleaves α2-gliadin in the stomach into large peptides, which empty into the upper small intestine. In the intestinal lumen, pancreatic proteases (trypsin, chymotrypsin, elastase, and carboxypeptidase A; TCEC) and intestinal brush border membrane (BBM) peptidases digest most peptides to single amino acids, di-, and tri-peptides for absorption. The 33-mer, however, persists through digestion to traverse the epithelial barrier, becoming deamidated by transglutaminase 2 (TG2) at select glutamine residues (underlined). In the underlying lamina propria, epitopes derived from the deamidated 33-mer (schematized as “PE” to emphasize their proline and deamidated glutamine content) are presented by HLA-DQ2+ antigen-presenting cells (APC), eliciting proliferation and deleterious immune activation from gluten-specific, DQ2-restricted CD4+ T cells. Gastrointestinal glutenases administered during oral enzyme therapy are proposed to digest gluten in the stomach or gut, thereby preventing immune activation.
A number of therapeutic alternatives to the gluten-free diet are under development, including enzymatic detoxification of gluten, inhibition of TG2 to prevent peptide deamidation, antagonism of deamidated peptide binding to HLA-DQ2, or suppression of deleterious immune responses (reviewed in (Schuppan et al., 2009)). In this chapter we focus on enzymatic detoxification of gluten by oral administration of enzymes capable of digesting gluten (i.e. glutenases).
Oral enzyme therapy is attractive for several reasons:
First, it follows the lead of the established gluten-free diet by targeting the exogenous pathogen, gluten, rather than targeting endogenous effectors. During clinical development, gluten intake and glutenase dose can be titrated in a complementary, dynamic, and individualized manner, thereby enabling systematic analysis of drug pharmacodynamics.
Second, by targeting ingested gluten, oral enzyme therapy avoids the shortcomings of strategies that target gluten at the level of exposure, including the gluten-free diet, selection or genetic engineering of less toxic grains (Molberg et al., 2005; Spaenij-Dekking et al., 2005), or pretreatment of gluten with glutenases, analogous to pretreatment of dairy products with lactase (De Angelis et al., 2006). Because the same proteins that cause toxicity in patients impart the desirable viscoelastic properties of dough, genetic or enzymatic removal of toxic epitopes from gluten will likely reduce baking quality. More importantly, such strategies do not address inadvertent exposure to ubiquitous gluten.
Third, the absence of a bona fide animal model for gluten sensitivity precludes preclinical safety and efficacy testing of drugs targeting endogenous effectors (Bethune and Khosla, 2008). By contrast, oral enzyme therapy can be evaluated in vivo using healthy or existing gluten-sensitive animal models (Bethune et al., 2008; Gass et al., 2006).
Fourth, glutenases that incompletely detoxify gluten as monotherapies can be co-administered with enzymes of complementary specificity to synergistic benefit (Gass et al., 2007a; Siegel et al., 2006).
This chapter describes methods for preclinical evaluation of oral enzyme therapy for celiac sprue. In Section 2, we provide strategies for identifying novel glutenase candidates. In Section 3, we provide protocols for characterizing the enzymatic properties of these candidates using gluten-derived and surrogate substrates. In Section 4, we describe methods for gram-scale production and formulation of promising candidates for delivery to alimentary sites of action. Finally, in Section 5, we discuss prospects for evolving next-generation glutenases and delivering them to the gut by various mechanisms.
2. Lead identification
Oral enzyme therapy requires proteases that can digest the uniquely Pro- and Gln-rich sequences constituting immunogenic gluten epitopes. Moreover, these enzymes must be stable and active in the harsh environments of the stomach and/or upper small intestine. In this section, we describe strategies for identifying novel glutenases for oral enzyme therapy. These strategies conform to two general approaches. The first approach is to rationally select glutenase candidates based on therapeutically relevant properties reported in the literature. Such properties include sub-site and chain length specificity, pH profile, stability in the presence of gastrointestinal proteases and bile acids, and evolved ability to proteolyze gluten. The second approach is to screen for glutenase candidates in vitro, based on activity in complex biological mixtures, or in silico, based on sequence homology with known glutenases.
2.1. Rational identification of glutenases
In this section we discuss two strategies for rational selection of glutenase candidates: property-directed selection of biochemically characterized proteases reported in the literature or substrate-directed selection of naturally-evolved glutenases.
2.1.1. Biochemically characterized proteases
Due to the high proline content of gluten peptides, our early investigations into oral protease therapy focused on microbial prolyl endopeptidases (PEP) (Hausch et al., 2002; Marti et al., 2005; Piper et al., 2004; Pyle et al., 2005; Shan et al., 2002). These studies demonstrated that recombinant PEP derived from Flavobacterium meningosepticum (FM) catalyzes post-proline endoproteolytic cleavage of metastable gluten peptides in vitro, mitigating their immunogenicity toward cultured celiac sprue patient-derived T lymphocytes. Subsequent work with this and related enzymes from Myxococcus xanthus (MX) and Sphingomonas capsulata (SC) highlighted the diversity of PEP family enzymes in terms of their sequence and chain length specificity and their stability in the presence of gastrointestinal proteases (Gass et al., 2007b; Shan et al., 2004). Together, these studies underscored the importance of selecting candidate glutenases that: i) cleave proline-containing sequences similar to those present in gluten; ii) accept a broad range of substrate chain lengths; and iii) remain stable and active in and en route to their targeted physiological site of action.
Because they exhibit optimal activity at pH 7-8, the PEPs used in the above studies were investigated initially for application as oral proteases targeted to the gut. However, FM PEP and MX PEP are rapidly proteolyzed by gastric pepsin (Shan et al., 2004; Stepniak et al., 2006), complicating oral delivery, and they are inactivated by trypsin and chymotrypsin in the presence of intestinal bile salts (Gass et al., 2007b). Targeting acid-active proteases to the stomach obviates these issues and, importantly, enables detoxification of gluten upstream of its intestinal site of toxicity. Consequently, recent efforts focus on acid-active PEPs that digest gluten alone (Stepniak et al., 2006) or in combination with a naturally-evolved glutenase of complementary specificity (Gass et al., 2007a).
2.1.2. Naturally-evolved glutenases
In the preceding section, we described a property-directed strategy for selecting lead proteases, whereby a candidate’s activity, specificity, stability, and other properties relevant to its therapeutic application form the criteria for selection. An alternative is a substrate-directed strategy, whereby we look to Nature for enzymes that have evolved to digest gluten as their natural substrate and select candidates from these that meet the criteria discussed above. This strategy led us to investigate the barley (Hordeum vulgare) endoprotease, EP-B2, as a naturally-evolved glutenase candidate. Its physiological substrates are hordeins, the similarly immunogenic and Pro/Gln-rich barley orthologs of wheat gliadins (Jacobsen and Varner, 1967; Mikkonen et al., 1996). Accordingly, EP-B2 cleaves preferentially after Gln residues, often with Pro at the P2 and P2′ positions (Bethune et al., 2006; Davy et al., 1998). This sub-site specificity overlaps with that of TG2, the enzyme that increases the immunogenicity of gluten by deamidating it at QXP motifs (Figure 2) (Fleckenstein et al., 2002; Vader et al., 2002). Thus, EP-B2-mediated cleavage of gluten prevents TG2-mediated deamidation at these sites. Additionally, EP-B2 is auto-activated to its catalytic form following its secretion into the acidified endosperm of a germinating barley seed (Koehler and Ho, 1990). Thus, EP-B2 has evolved to digest gluten proteins at a pH similar to that in the human stomach. Characterization of recombinant EP-B2 demonstrated it is stable in the presence of pepsin and it detoxifies gluten in vitro and in vivo (Bethune et al., 2008; Bethune et al., 2006; Gass et al., 2006; Siegel et al., 2006). Due to their complementary specificity, a combination of EP-B2 and a suitable PEP constitutes a particularly effective glutenase (Gass et al., 2007a; Siegel et al., 2006). Plants encode hundreds of proteases, many of which play a role in digesting storage proteins such as gluten during seed germination (Simpson, 2001; van der Hoorn, 2008). This strategy is therefore likely to yield additional cereal-derived enzymes with utility for oral enzyme therapy (Gessendorfer et al., 2011; Hartmann et al., 2006; Kiyosaki et al., 2007; Stenman et al., 2009). A related strategy - purifying glutenases from yeast and Lactobacillus spp. that have been propagated at length with dough - holds similar promise (De Angelis et al., 2010; Di Cagno et al., 2008).
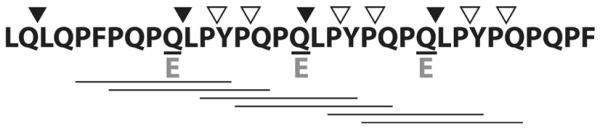
Figure 2.
Features of the immunogenic 33-mer gluten peptide. Prolines (P; 39%) and glutamines (Q; 30%) are over-represented, imparting resistance to gastrointestinal digestion. Transglutaminase 2-mediated deamidation of QXP motifs at underlined glutamines introduces negatively-charged glutamates (E), thereby unmasking six overlapping HLA-DQ2-restricted T cell epitopes (underlined) present in the multivalent 33-mer. Cleavage sites for glutenase candidates EP-B2 (filled arrowheads) and SC PEP (open arrowheads) are indicated.
2.2. Screening for glutenases
In section 2.1 we outlined strategies for selecting enzymes from the published literature as lead candidates for enzyme therapy. In this section we outline strategies for identifying novel candidates by screening complex biological samples for glutenase activity or by searching protein databases for sequences that are similar to known glutenases.
2.2.1. Activity-based purification of glutenases from complex biological samples
A number of microbes and crude enzyme extracts are reported to exhibit glutenase activity. Probiotic preparation VSL#3 hydrolyzes recombinant gliadin proteins, reducing gliadin-induced infiltration of CD3+ intraepithelial lymphocytes in celiac patient jejunal biopsies (De Angelis et al., 2006). Another probiotic, Bifidobacterium lactis, appears to counteract gliadin-induced cytoskeletal rearrangements in the cultured human colon cell line Caco-2 (Lindfors et al., 2008). Microbial enzymes derived from dental plaque in the human oral cavity hydrolyze proline-containing chromogenic substrates, especially Cbz-YPQ-pNA, as well as the gluten-derived 33-mer and 26-mer peptides (Helmerhorst et al., 2010). Porcine intestinal extracts have been tested clinically, to modest effect, in celiac sprue patients (Cornell et al., 2005). Finally, as mentioned in Section 2.1.2, extracts from germinating cereals and sourdough lactobacilli are likely to be a rich source for as-yet uncharacterized glutenases.
To be candidates for oral enzyme therapy, glutenases from these heterogeneous sources must be purified and well-characterized. In Section 3, we will describe methods that can be used toward this end. Several of these, including chromogenic assays for prolyl endopeptidase activity (Edens et al., 2005), competitive ELISA (De Angelis et al., 2010), and HPLC-based gluten peptide digestion assays (Gessendorfer et al., 2011) have demonstrated success in isolating novel enzymes of potential utility. In addition to these screening techniques, glutenase activity can be used directly as a handle for purification. A biotinylated derivative of the cysteine endoprotease inhibitor, E-64, enables activity-based purification of plant cysteine endoproteases (van der Hoorn et al., 2004). Biotinylated gluten peptide-based derivatives of irreversible inhibitors like E-64 could select from a diverse collection of enzymes those most relevant for oral enzyme therapy.
2.2.2. In silico screening
Prolyl endopeptidases in the S9 family of enzyme clan SC possess a unique β-propeller domain, which imposes the chain length specificity on enzymes of this family, and a catalytic domain containing the Ser-Asp-His catalytic triad. Within the catalytic domain, 18% of residues, including the catalytic triad and the S1 substrate binding pocket, are invariant across 28 aligned PEPs. Ten of these residues are invariant across 72 enzymes representing PEPs and 3 related SC clan proline-specific enzyme groups: dipeptidyl peptidase IV, oligopeptidase B, and acylaminoacyl peptidase (Venalainen et al., 2004). More distantly related prolyl endoproteases, such as AN-PEP from Aspergillus niger, feature lower sequence identity but share the Ser-Asp-His topology.
Cereal cysteine endoproteases are even better conserved, with identitites of 70-90% common in pairwise protein sequence alignments. These papain-like proteases share a signal peptide, an auto-inhibitory pro-peptide domain that is removed upon activation, and the catalytic domain containing three conserved disulfides and a Cys-His-Asn catalytic triad. The high degree of conservation in this family is underscored by the successful use of oligonucleotide probes based on known rice glutenases (oryzains) to clone novel glutenases from a germinating wheat cDNA library (Kiyosaki et al., 2009; Kiyosaki et al., 2007).
The high degree of conservation within these enzyme families suggests that current lead candidates can inform in silico approaches for identifying additional enzymes of therapeutic interest. Such approaches will rely on databases for detecting homology in protein sequence, structure (e.g. 3D-BLAST), or proteolytic specificity (e.g. MEROPS, CutDB). (Igarashi et al., 2007; Rawlings et al., 2010; Yang and Tung, 2006)
3. Assay development
A bona fide animal model for celiac sprue has not been discovered or engineered. Assays for glutenase efficacy are therefore conducted in vitro, ex vivo, and in surrogate animal models (e.g. in gluten-tolerant animals or in incomplete animal models of gluten sensitivity). Primary assays utilize biochemical tools such as spectrophotometry, reversed-phase high performance liquid chromatography (HPLC), and liquid chromatography-assisted mass spectrometry (LC-MS) to identify disease-relevant gluten peptides and to preliminarily assess glutenase activity against these and surrogate peptide substrates. Secondary assays require more advanced mass spectrometric and immunological techniques to quantify the ability of glutenases to detoxify the more complex substrate of whole gluten. Tertiary assays use these same tools to measure glutenase-mediated digestion of gluten in vivo, in gluten-tolerant (Gass et al., 2007a; Gass et al., 2006) or gluten-sensitive (Bethune et al., 2008) animal models of digestion. Extensive discussion of in vivo assays is beyond the scope of this review, but at relevant points we will note how those techniques discussed can be modified to accommodate complex biological samples derived from such assays.
3.1. Primary assays: peptide substrates
Synthetic dipeptides with chromogenic or fluorogenic labels are used as a convenient substrate for rapid biochemical characterization of candidate glutenases. Recombinant gluten proteins and synthetic peptides based on immunodominant gluten epitopes are used to assess enzyme specificity with more therapeutically relevant substrates.
3.1.1. Chromogenic or fluorogenic substrate assays
Peptide-paranitroanilide (pNA) conjugates have enabled chromogenic measurement of protease activity and selectivity since the early Sixties (Erlanger et al., 1961). Typically, current research employs commercially-available di-, tri-, and tetra-peptide substrates conjugated to an N-terminal benzyloxycarbonyl (Z) or succinyl (Suc) protecting group and C-terminal pNA (Figure 3). Protease-catalyzed cleavage of the C-terminal amide bond releases paranitroaniline, which can be monitored spectrophotometrically at 410 nm (ε410 = 8800 M−1cm−1). These substrates thus allow for high-throughput, quantitative measurement of enzyme activity. The identities of the substrate residues can be selected to purify enzymes of a desired specificity from complex mixtures or they can be varied systematically to assess the subsite specificity of a protease under study. By varying other components of the reaction, peptide-pNA conjugates can be used to characterize an enzyme’s catalytic mechanism, pH profile over a physiologically relevant range (pKa of free paranitroaniline is 1.0), stability in the presence of bile or other proteases, or activation from an inactive zymogen.
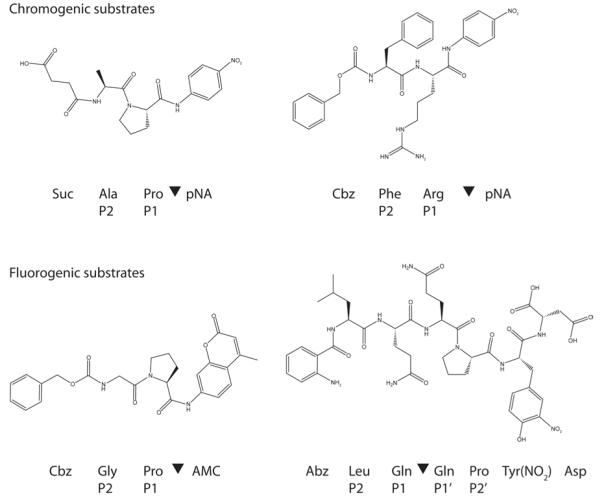
Figure 3.
Chromogenic and fluorogenic substrates. Substrate positions flanking the cleavage site (▼) are indicated as …P2-P1-▼-P1′-P2′… according to standard protease nomenclature (Berger and Schechte.I, 1970).
Fluorogenic assays are similar but employ peptides that are conjugated to a fluorophore, typically C-terminal 7-amino-4-methylcoumarin (AMC; Figure 3). The excitation and emission maxima of proteolytically-released AMC are higher than those of the peptide-conjugated fluorophore, allowing protease activity to be measured. However, excitation and emission wavelengths must be optimized and a cutoff filter employed to minimize background from the substrate. Typical values for excitation and emission are 360-380 nm and 440-460 nm, respectively. Because the pKa of AMC is 7.8, this fluorophore cannot be used below pH 5.0.
In an alternative fluorogenic assay, peptides are conjugated to an internally-quenched donor/acceptor pair, such as N-terminal 2-aminobenzoyl (Abz) and C-terminal 3-nitrotyrosine (Tyr(NO2)) (Figure 3). Internal cleavage relieves the quench, resulting in increased fluorescence. Notably, this assay allows residues on either side of the cleavage site to be varied, providing a more detailed description of subsite specificity (Davy et al., 1998).
Chromogenic and fluorogenic substrates can be purchased from Bachem (Torrance, CA). We use Cbz-Phe-Arg-pNA for assaying EP-B2 activity and both Cbz-Gly-Pro-pNA and Suc-Ala-Pro-pNA for assaying activity of various PEPs. Whereas Suc-Ala-Pro-pNA is soluble in water, Cbz-Phe-Arg-pNA requires dissolution in 5% dimethylsulfoxide and Cbz-Gly-Pro-pNA requires 8% dioxane. Substrate stocks are stored at −20 °C. All activity assays utilizing Cbz-Phe-Arg-pNA contain 5% (v/v) DMSO in the final reaction mixture to maintain the solubility of the substrate. DMSO at this concentration does not impair the enzymatic activity of EP-B2. Reactions typically contain 50 μM peptide-pNA substrate and 0.1 to 2 μM enzyme. Optimal pH can be determined in a citrate-phosphate buffer between pH 2.2 and 8.0. Reaction progress in the pNA chromogenic assay is monitored by measuring A410 with a UV/Vis spectrometer (410 nm, 4-nm slit width; model Lambda 35, Perkin-Elmer). The reaction rate is calculated from the initial slope of A410 versus time.
3.1.2. LC-MS detection of degraded peptide fragments
A critical feature of those peptides that are immunogenic in celiac sprue is that they persist through gastrointestinal digestion intact. To identify such peptides, recombinant gliadin proteins are treated with gastric, pancreatic, and intestinal proteases and remaining metastable peptides are identified by LC-MS.
Gastric and pancreatic proteases are purchased from Sigma-Aldrich (St. Louis, MO). Brush border membranes are purified from rat intestine as previously described (Kessler et al., 1978) and stored at −80 °C until use. Recombinant α2-gliadin (GenBank accession number AJ133612), a representative gluten protein, is expressed in E. coli, purified, lyophilized, and stored at −20°C as described (Shan et al., 2002). Prior to use, α2-gliadin is dissolved in 0.1 M acetic acid and stored on ice.
To simulate gastric digestion, 1.2 mg/mL (~35 μM) α2-gliadin is incubated at 37 °C in a 10 mM sodium acetate buffer, pH 4.5 with 1:10 (w/w) pepsin (120 μg/ml). After variable durations up to 60 min, digests are adjusted to pH 6.0 with sodium phosphate buffer (50 mM, final concentration) and commercially-available pancreatic proteases trypsin (30 μg/ml), chymotrypsin (30 μg/ml), elastase (6 μg/ml), and carboxypeptidase A (6 μg/ml), as well as 27 μg/ml rat intestinal brush border membrane, are added to simulate digestion in the proximal small intestine. The brush border is added because it contains membrane exopeptidases that perform the final stages of digestion. Simulated intestinal digests are performed at 37 °C for 60 min after which enzymes are denatured at 95 °C for 5 min. Samples are filtered (0.2 μm), diluted 1:5 in HPLC Solvent A (95% H2O, 5% acetonitrile, 0.1% trifluoroacetic acid), and analyzed by LC-MS. Samples (50 μl) processed for LC-MS are injected on a reversed-phase C18 HPLC system (Waters Corporation, Milford, MA) coupled to a UV/Vis detector and a ZQ single quadrupole mass spectrometer with an electrospray ionization source. Samples are eluted with a water-acetonitrile gradient in the presence of 0.1% formic acid. Absorbance at 214/254 nm and total ion current are monitored, and spectra corresponding to major chromatographic peaks are examined.
This method revealed the presence of a 33-mer peptide that is highly resistant to gastrointestinal digestion, deamidated at multiple Gln residues by TG2, and immunogenic toward most celiac patient biopsy-derived intestinal T cell lines when presented on DQ2+ antigen-presenting cells (Figure 2) (Shan et al., 2002). Similar mock digestion of another representative gluten protein, γ5-gliadin, revealed a 26-mer peptide with these same properties (Shan et al., 2005b). As such, simulated gastrointestinal digestion of gluten is an effective method for identifying disease-relevant peptide substrates for preclinical testing of glutenases. Additionally, varied concentrations of glutenase candidates can be added to the gastric and/or intestinal phase of this protocol to evaluate their ability to digest select gluten proteins in vitro.
3.1.3. Gluten-derived peptide substrates
Peptides identified as proteolytically-resistant through gluten digestion assays are convenient and therapeutically-relevant substrates for evaluating glutenase candidates. Peptides are synthesized using Boc/HBTU chemistry on solid-phase as previously described (Xia et al., 2005), purified by reversed-phase HPLC, and stored at −20°C following lyophilization. The expected mass and retention time of peptides is confirmed by LC-MS. Peptides are resuspended in 50 mM sodium phosphate, pH 7.0 supplemented with 0.02% NaN3. Some peptides may require acidic or basic pH for resuspension. The concentration of resuspended peptide is determined by spectrophotometric measurement of A280. Working peptide stocks are stored at 4 °C and their integrity confirmed periodically by reversed-phase HPLC.
Digestion of individual gluten peptides is performed as described for proteins in Section 3.1.2, with peptides typically used at 1.2 mg/mL (300 μM for the 33-mer), gastrointestinal enzyme concentrations as described, and candidate glutenases added to the gastric and/or intestinal phase at varied concentrations. Prominent products of digestion are identified by LCMS as described. Digestion progress can be monitored quantitatively by HPLC after diluting samples taken from digests at varied time points with HPLC Solvent A supplemented with an internal standard (Nα-p-tosyl-L-arginine methyl ester hydrochloride (TAME)). The area-under-the-curve for the intact peptide peak as well as prominent product peaks are calculated and normalized to the area-under-the-curve for TAME.
3.2. Secondary assays: whole gluten
As the ultimate goal of oral enzyme therapy is to enable digestion of dietary gluten, candidate glutenases that exhibit promising activity against peptide substrates in primary assays must be validated against this more complex substrate. In a typical experiment, whole gluten (15 mg/mL) is digested under simulated gastric conditions (50 mM sodium acetate buffer, pH 4.5 at 37°C) for 60 minutes by 0.6 mg/mL pepsin and varied concentrations of gastric glutenase(s). The tube is inverted every 10 min to agitate insoluble gluten. Each simulated gastric reaction is then adjusted to pH 6.0 with an equal volume of 500 mM sodium phosphate buffer containing intestinal proteases at final concentrations of 0.375 mg/mL trypsin, 0.375 mg/mL chymotrypsin, 0.075 mg/mL elastase, and 0.075 mg/mL carboxypeptidase A. If applicable, varied concentrations of intestinal glutenase are added as well. Digests are further incubated under mock intestinal conditions (pH 6.0 at 37 °C). At desired time-points, reactions are quenched by boiling for 10 min. Some insoluble material will remain, the amount of which will be visibly reduced in the presence of glutenase. After clarifying digests by centrifugation for 10 min at 9,300 x g, the supernatant is collected, filtered (0.45 μm), and stored at −20 °C until use. In some experiments, we use whole-wheat bread (Alvarado St Bakery, Rohnert Park, CA) in lieu of whole gluten powder (Bob’s Red Mill, Milwaukie, OR) (Ehren et al., 2009; Gass et al., 2007a).
In this section, we describe two methods - competitive ELISA and triple quadrupole mass spectrometry - for quantifying specific immunogenic peptides present in complex gluten digests. Additionally, we describe the T cell proliferation assay, a standard assay for measuring immunogenicity remaining after gluten digestion.
3.2.1. Competitive ELISA
Filtered supernatants from gluten or whole-wheat bread digests prepared as in Section 3.2 are tested for remaining 33-mer (and similar epitopes) using a competitive enzyme-linked immunosorbent assay (ELISA). In our experiments, we use either A1 or G12, monoclonal antibodies which bind epitopes QLPYPQP and QPQLPY, respectively (Moron et al., 2008a; Moron et al., 2008b). In a typical experiment, gliadin (Sigma) or synthetic 33-mer peptide is diluted to 5 μg/ml in 100 μL coating solution (50 mM sodium carbonate/bicarbonate buffer, pH 9.6, 0.02% NaN3) and 100 μL/well is incubated 1 h at 37 °C and overnight at 4 °C on Maxisorp plates (Nunc). The next day, gliadin-coated plates are washed twice with washing buffer (PBS, pH 7.2 containing 0.05% Tween-20) then blocked with blocking buffer (5% (w/v) nonfat milk in PBS, pH 7.2) for 2 h at room temperature. Synthetic 33-mer standard (10−5 – 10 μg/ml) or filtered gluten digest supernatants are serially diluted in assay buffer (3% (w/v) bovine serum albumin in PBS, pH 7.2). An equal volume of G12-HRP mAb-horseradish peroxidase conjugate (Biomedal, Seville, Spain) diluted 1:10,000 in assay buffer is added to each standard or sample dilution. Mixtures are gently agitated for 2 h at room temperature and then added (100 μL/well) to plates in triplicate. After 30 min incubation at room temperature, wells are washed 5 times with washing buffer, and TMB liquid substrate solution (Sigma) is added to wells. The reaction is incubated at room temperature until wells with no competing digest or standard are nearly saturated (usually 30 min). Reactions are quenched by addition of 100 μL/well 2 N sulfuric acid and the absorbance at 450 nm is measured. The 33-mer standard curve is fit to the sigmoidal model:
in which x is the peptide concentration, IC50 is the 33-mer concentration at which competition is half-maximal, and n is the Hill slope. We use Origin 6.0 (OriginLab, Northampton, MA) for non-linear regression. The concentration of peptides containing the sequence QPQLPY in gluten or whole-wheat bread digests is determined by comparison to the linear portion of the 33-mer standard curve.
This protocol can be adapted to testing gastric contents from in vivo (rat or macaque) gluten digestion experiments. Gastric samples (100 mg) are solubilized in 1 mL 70% (v/v) ethanol supplemented with appropriate protease inhibitors to prevent ex vivo digestion. For example, 500 μM leupeptin is included when cysteine protease EP-B2 is dosed and pepstatin A can be included to inhibit endogenous pepsin. After 10 min incubation at 37 °C, samples are centrifuged for 10 min at 16,100 x g and the supernatant is filtered (0.45 μm). Various dilutions of the supernatant are tested for 33-mer by competitive ELISA as above.
3.2.2. Triple quadrupole (3Q) LC-MS quantification of specific peptides
The concentrations of specific immunogenic gluten peptides present in gluten or whole-wheat bread digests are quantified using a Micromass Quattro Premier (Waters) triple quadrupole (3Q) LC-MS system. Biological samples, such as serum and gastrointestinal contents from in vivo digestion experiments, can be similarly assayed (Bethune et al., 2009; Bethune et al., 2008; Gass et al., 2006). Syringe-filtered (0.45 μm) biological samples or gluten digests prepared as in Section 3.2 are depleted of larger proteins by addition of acetonitrile. Samples are mixed with an equal volume of cold acetonitrile containing 0.1% formic acid and an internal standard (typically an isotope-labeled version of the peptide being measured). Samples are vortexed, incubated for 2 h at 4 °C, and then centrifuged for 10 min at 4 °C, 16,100 x g. Supernatants are mixed with an equal volume of 0.1% formic acid in water to dilute the acetonitrile to a final concentration of 25% and then injected in triplicate (80 μl each) on 3Q LCMS. Injected samples are eluted with a water-acetonitrile gradient in the presence of 0.1% formic acid.
The 3Q LC-MS is configured to run in selected (or multiple) reaction monitoring (SRM) mode (Figure 4). In SRM mode, the first quadrupole is configured to select precursor ions of a known mass (i.e. that of the intact peptide). Selected ions are fragmented by collision with an inert gas in the second quadrupole. The third quadrupole is configured to select prominent fragment ions produced by this collision. Thus, to constitute a signal, a transition from a selected precursor ion to a selected fragment ion must be detected at a specified retention time. When multiple transitions are monitored, the SRM configuration is called multiple reaction monitoring (MRM). Monitoring multiple transitions improves sensitivity and specificity and enables quantification. For 33-mer, typical transitions monitored are m/z 978.84+→263.1+ (30V cone voltage, 27eV collision energy) for the quantitation assay and m/z 1304.73+→263.1+ (40V cone voltage, 50eV collision energy) as a confirmatory transition. For an internal standard of 33-mer with nine deuterium atoms incorporated (D933-mer), the transitions monitored are m/z 981.04+→263.1+ (30V cone voltage, 27eV collision energy.) for the quantitation assay and m/z 1307.73+→263.1+ (40V cone voltage, 50eV collision energy) as a confirmatory transition. Levels of 33-mer in each sample are determined by comparison of the area under its transition peak to the area under the D933-mer internal standard transition peak and to a calibration curve corresponding to each peptide.
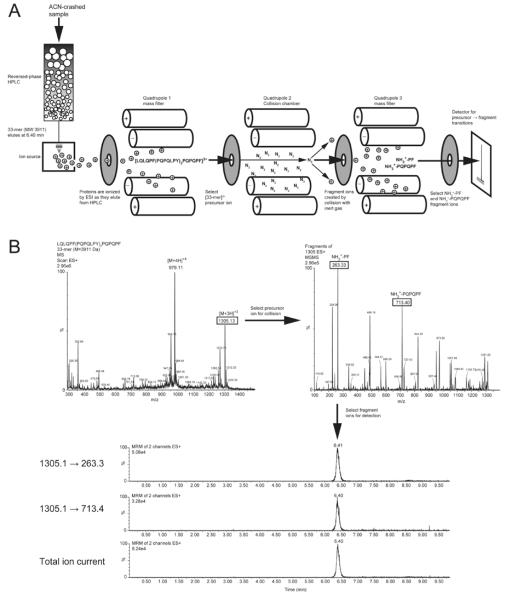
Figure 4.
Detection of 33-mer by liquid chromatography-assisted triple quadrupole mass spectrometry (3Q LC-MS). A) Schematic of the 3Q LC-MS instrument, configured to run in multiple reaction monitoring (MRM) mode. Gluten digests, serum samples, or other complex biological samples are depleted of proteins (>10 kDa) by precipitation with acetonitrile (ACN). Peptides remaining in the supernatant are injected on reversed-phase HPLC and separated according to hydrophobicity. The retention time for 33-mer is 6.4 minutes in the example shown. Peptides eluted from the column are ionized by electrospray ionization and precursor ions enter the first quadrupole. Undesired ion species are filtered out, whereas [33-mer+3H]3+ is selected for fragmentation by collision with an inert gas in the collision chamber. Fragment ions then pass through the third quadrupole, which again filters out undesired species and transmits only those selected ions to the detector. B) Spectra showing the precursor ion selected in the first quadrupole (left) and the fragment ions selected in the third quadrupole (right). Note that another precursor ion, [33-mer+4H]4+ is prominent in the first quadrupole. This precursor ion can also be selected for fragmentation in MRM mode, providing additional precursor → fragment transitions. Chromatograms (bottom) show peaks at the expected retention time for the two transitions monitored, as well as for the total ion current. The analyte is quantified by integrating an individual transition or the TIC chromatogram peak, normalizing to an internal control (e.g. isotope-labeled 33-mer), and comparing normalized area-under-the-curve to a standard curve.
3.2.3. T cell proliferation assay
The immunogenicity remaining in gluten or whole-wheat bread following digestion is quantified using celiac patient intestinal biopsy-derived T cells in a 3H-thymidine T Cell proliferation assay. To unmask this immunogenicity, gluten peptides remaining in digest supernatants from Section 3.2 are deamidated by treatment with 100 μg/mL recombinant TG2 for 2 hours at 37 °C in the presence of 2 mM CaCl2. Deamidation is quenched by heating reactions to 95 °C for 5 min. Deamidated digests are centrifuged for 2 minutes at 16,100 x g to pellet precipitated calcium and stored at −20 °C until use in T cell proliferation assays.
For the T cell proliferation assay, DQ2+ homozygous APCs (e.g. Epstein-Barr virus-transformed B lymphoblastoid cell lines CD114 or VAVY) are resuspended in T cell media, γ-irradiated with 12,000 rads, and 60,000 cells/well are added to U-bottom, 96-well plates. Varied dilutions of deamidated gluten digests are prepared in T cell media and incubated with APCs overnight. Gluten digested without glutenases is added to APCs as a positive control and only T cell media is added to APCs as a negative control. The next day, T cell lines and/or clones isolated from intestinal biopsies of HLA-DQ2+ celiac patients are thawed and added (40,000 cells/well) to triplicate wells containing peptide-loaded APCs. Cells are co-incubated for 48 hours to allow T cells to proliferate in response to DQ2-peptide complex stimulation. After 48 hours, 1 μCi [3H]-thymidine is added to each well and cells were incubated for an additional 12-16 hours. Finally, DNA is collected on a filter mat using a cell harvester and radioactive counts-per-minute (cpm) resulting from incorporated [3H]-thymidine is measured with a liquid scintillation counter. The stimulation index for each sample can be calculated by dividing its cpm by that of the negative control.
Recombinant TG2 is expressed in E. coli and purified as previously described (Piper et al., 2002). Antigen-presenting cells are grown in RPMI media supplemented with antibiotics and 5% (v/v) fetal bovine serum. Cells are split to 0.4 × 106 cells/ml every other day. Gluten-specific, DQ2-restricted T cell lines and clones are isolated from celiac patient intestinal biopsies, expanded, and frozen as previously described (Molberg et al., 2000; Siegel et al., 2006). The T cell proliferation assay is performed in T cell media (RPMI 1640 supplemented with antibiotics and 10% (v/v) human serum). All cells are grown and assayed at 37 °C with 5% atmospheric CO2.
3.3. In vitro systems that mimic human digestion
The absence of an animal model for celiac sprue necessitates surrogate systems for preclinical evaluation of glutenase therapies. We have employed several such systems, including live rat intestinal perfusion (Piper et al., 2004; Shan et al., 2002), oral administration to both gluten-tolerant rats and gluten-sensitive macaques (Bethune et al., 2009; Bethune et al., 2008; Gass et al., 2007a; Gass et al., 2006), and in vitro models of varying sophistication. In vivo models are described in detail in the cited publications. In this review, we will focus on two in vitro systems for modeling digestion: minimal reconstitution in a beaker and the dynamic gastrointestinal model.
3.3.1. Reconstitution of gastrointestinal digestion in a beaker
We perform most of our gluten digestion experiments in beakers or polypropylene tubes. The only parameters of in vivo digestion that are modeled in this minimal reconstitution are physiological temperature, pH, enzyme (and bile) levels, and duration of residence in the gastrointestinal tract. Thus, all experiments are performed in a 37 °C water bath; simulated gastric digestion includes pepsin and is performed for 30-60 min in hydrochloric acid or acetate buffer at pH 4.5; and simulated intestinal digestion includes pancreatic enzymes, supplemented in some experiments with intestinal brush border membrane and bile acids, and is performed for up to 120 min in bicarbonate or phosphate buffer at pH 6.0. These parameters approximate the measured postprandial gastric pH range of 3-5 and mean intestinal pH of 6.0 in humans (Dressman, 1986; Lui et al., 1986), as well as representative gastric emptying and intestinal transit times for a solid meal (Malagelada and Azpiroz, 2010). The advantages of minimal in vitro reconstitution include high reproducibility, absolute control over defined conditions, small volume and sample requirements (milligram-scale quantities of glutenase are sufficient), and ease of implementation due to low cost of universally obtainable materials. Notably, this simple in vitro system accurately identified the 33-mer as a stable product of gluten digestion (Shan et al., 2002), a finding subsequently validated in both rats and cynomolgus monkeys fed gluten (Gass et al., 2006)(Bethune and Khosla, unpublished results). However, as reductionist models must, simple reconstitution in vitro neglects many factors impacting gastrointestinal digestion in vivo (e.g. salivary enzymes, gut microbiota, other dietary components). A potentially important omission is that, although reactions are periodically mixed by inversion, the motor activity of the digestive system is not modeled in this system. In the next section, we discuss a more sophisticated in vitro system that incorporates this aspect of digestion.
3.3.2. Dynamic gastrointestinal model
Regular peristaltic contractions of the stomach and small intestine enhance digestion by triturating, mixing, and moving dietary nutrients along the alimentary tract, thereby maximizing their absorption (Malagelada and Azpiroz, 2010). The predictive power of in vitro gluten digestion models may be improved by incorporating this extensively-characterized mechanical action. A dynamic gastrointestinal model, TIM-1 (TNO intestinal model-1), has been developed for this purpose (Minekus et al., 1995). The TIM-1 system comprises four compartments modeling the stomach and each segment of the small intestine: the duodenum, jejunum, and ileum. Each compartment has flexible walls and is contained in a temperature-controlled water jacket. By manipulating the jacket water pressure, post-prandial gastric contractions and peristaltic intestinal mixing are simulated according to pre-set curves based on physiological data. Enzyme composition, pH, gastric emptying, and transit times are also under computerized control. Several studies suggest this model is predictive of in vivo data (Blanquet et al., 2004; Souliman et al., 2006).
The TIM-1 model has been used to test the activity of a candidate glutenase, AN-PEP, against both bread and a more complex (fast food) meal (Mitea et al., 2007). The verisimilitude of this dynamic in vitro model positions it to be of great use in future testing of glutenases, particularly against actual food products. There are three prominent disadvantages of this model relative to simple reconstitution in a beaker. First, the specialized equipment required for the dynamic gastric model hinders broad implementation. Second, the method is low-throughput, limiting its utility to late-stage preclinical testing. Third, the larger reaction volumes employed (hundreds of milliliters) necessitate gram-scale quantities of glutenase (Mitea et al., 2007), similar to in vivo assays.
4. Gram-scale production and formulation
Lead glutenases that show promise in early (i.e. in vitro) experiments must be evaluated in more advanced gastrointestinal models and/or animal surrogates prior to human clinical trials. Such studies require gram-scale quantities of glutenase, necessitating high-yield production methods. In this section, we describe the heterologous expression, purification, formulation, and storage of gram-scale quantities of recombinant glutenases.
4.1. Choice of heterologous host and expression system
We use Escherichia coli for heterologous expression of glutenases because this host grows rapidly on inexpensive media and can be induced with minimal optimization to express recombinant protein at high levels (several hundred milligrams to grams). Additionally, the well-developed molecular biology of E. coli make generating, screening, and isolating mutants straightforward, facilitating engineering of next-generation proteases.
Candidate glutenases are expressed initially in shake-flask cultures. Competent E. coli BL21(DE3) cells are transformed with a plasmid encoding the an antibiotic resistance marker and the glutenase gene under the inducible control of a T7 promoter. Transformed cells are plated overnight at 37 °C on Luria-Bertani (LB)/agar plates containing appropriate antibiotics. A 5 mL overnight starter culture is inoculated with a single colony and is grown for 12-16 hours by shaking at 37 °C. One liter of LB broth containing antibiotics is inoculated with the starter culture and grown at 37 °C. When the OD600 is 0.6, expression of recombinant glutenase is induced by addition of 0.2-1mM isopropyl β-D-thiogalactoside (IPTG). The induced culture is incubated for 5-20 hours at optimal temperature and then cells are harvested by centrifugation at 5,000 x g for 20 minutes. Concentration of IPTG, induction time, duration, and temperature are all optimized for maximum (soluble) protein expression. Proenzymes often express in inclusion bodies, in which case total expression is optimized and soluble protein is purified and refolded from inclusion bodies as previously described (Bethune et al., 2006).
The expression of promising lead candidates is scaled-up in fed-batch fermentations. Fermentation conditions are optimized for each glutenase in a B. Braun Biostat 10 L fermenter, as described in detail for MX PEP and EP-B2 (Gass et al., 2005; Vora et al., 2007). Generally, a 100-250 mL overnight shake-flask starter is used to inoculate 8.5 L fermentation medium (Gass et al., 2005). Typical fermentation parameters are 30 °C, 400 RPM agitation, pH 7.0, and 1 VVM sterile-filtered air. When the OD600 is 6-7, a sterile glucose feed solution is added at 0.3 mL/min. When the OD600 is 12-15, 0.8 mL of IPTG is added to induce protein expression. Induction duration is a critical parameter for maximizing yield while minimizing degradation; expression of MX PEP is induced over 10-12 h whereas optimal production of the EP-B2 proenzyme is achieved at 2-4 h (Vora et al., 2007). Following induction, cells are harvested by centrifugation at 5,000 x g for 20 minutes in a J2-21M Beckman centrifuge. If cytoplasmic proteins are to be purified, the pellet is rinsed once with buffered saline prior to freezing or processing.
Once optimal expression conditions are determined, fermentations can be scaled-up further if needed. For example, expression of EP-B2 can be produced in a 100 L fed-batch fermentation format with minimal modification of the optimized conditions for 10 L scale fermentation (Vora et al., 2007).
4.2. Purification strategies
Prolyl endopeptidases are purified as soluble proteins from the cytoplasm of E. coli whereas the proenzyme form of EP-B2 is purified from inclusion bodies and refolded. Chromogenic substrates Cbz-Gly-Pro-pNA and Cbz-Phe-Arg-pNA are used respectively to monitor the activity of PEPs throughout purification and of EP-B2 following refolding. We generally employ a two-step purification strategy, comprising an initial affinity-based step followed by a second chromatographic separation to ensure proper folding. Glutenases are genetically fused to a C-terminal His6 tag to facilitate affinity purification with Ni-NTA resin (Qiagen). Nickel-NTA purification is a relatively inexpensive means to achieve >90% purity in the first purification step using a generally unobtrusive tag. Additionally, Ni-NTA purification can be performed under both native and denaturing conditions, enabling partial purification of denatured EP-B2 prior to refolding. Typically, anion exchange is favored over gel filtration for the second chromatographic separation due to its lower expense and higher capacity. Purification of soluble PEPs (Gass et al., 2005; Shan et al., 2004) and purification and refolding of pro-EP-B2 from inclusion bodies have been described in detail (Bethune et al., 2006; Vora et al., 2007). Typical purified yields are 300-500 mg MX PEP and 250-300 mg pro-EP-B2 per liter of fermentation.
4.3. Formulation and storage considerations
Purified glutenases intended for gastric delivery (e.g. pro-EP-B2) are buffer-exchanged into lyophilization buffer (for pro-EP-B2: 100 mM Tris-Cl, pH 8.5, 5 mM EDTA, 2 mM 1-thioglycerol, 4% (w/v) mannitol, 1% (w/v) sucrose) and concentrated with a 10 kDa MWCO membrane. For small-scale production, buffer exchange and concentration are performed with a centrifugal filtration device; for gram-scale production, higher throughput tangential flow filtration (Pellicon XL, Millipore) is required. The final enzyme concentration is determined by the Bradford or bicinchoninic acid (BCA) assay. Concentrated enzyme aliquots are stored at −20 °C or flash-frozen in liquid nitrogen and lyophilized.
Purified glutenases intended for intestinal delivery (e.g. MX PEP) are buffer-exchanged into lyophilization buffer (for MX PEP: 50 mM Na2HPO4, pH 7.5; 3% (w/v) sucrose), concentrated and quantified as described above, and lyopilized. Intestinally-targeted proteases are delivered in enteric coated capsules, which remain intact through the acidic stomach and dissolve in the more neutral pH of the proximal bowel, releasing their payload therein. Detailed methods for blending lyophilized PEP with excipients, as well as for filling and coating capsules, have been described (Gass et al., 2005). Enteric coated capsules containing lyophilized PEP are stored in capped bottles at 4 °C.
5. Lead optimization for next-generation proteases
In previous sections, we described strategies for identifying and characterizing existing proteases for use in oral enzyme therapy. In this section, we discuss strategies for improving on these enzymes, either by simply co-administering them as a combination glutenase or by augmenting their therapeutically-desirable properties through protein engineering. Additionally, we discuss potential modes of delivery for enzyme therapy, oral and otherwise.
5.1. Improved combination therapies
Two glutenases - EP-B2 and AN-PEP - have been identified that digest gluten effectively as single enzyme treatments in preliminary assays (Bethune et al., 2006; Gass et al., 2006; Mitea et al., 2007; Stepniak et al., 2006). However, gluten is a complex substrate, containing scores of potential epitopes (Shan et al., 2005b), and these epitopes must be digested in vivo with sufficient rapidity to preempt immune responses in the gut. Rapid detoxification of the diverse forms of gluten present in human diets may therefore require a combination of glutenases with complementary specificities.
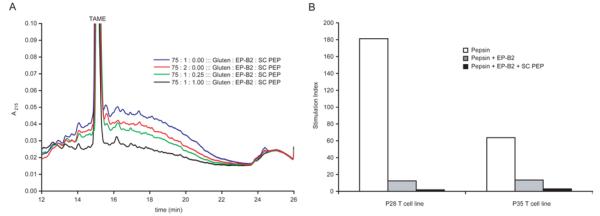
We investigated the potential for two PEPs to potentiate EP-B2-mediated gluten detoxification. The first, FM PEP, enhances gluten digestion in vitro (Siegel et al., 2006), but its pH profile would require targeting it to the duodenum in vivo, a significant obstacle in terms of its stability and the kinetics of its effect on gluten (Gass et al., 2007b). The second, SC PEP, is active down to pH 4, suggesting it can act alongside EP-B2 in the stomach. Indeed, the combination of EP-B2 and SC PEP completely detoxifies whole-wheat bread under acidic conditions in vitro, as assessed by HPLC and T cell proliferation assays (Figure 5) (Gass et al., 2007a). This combination therapy, branded ALV003, is in Phase II trials to evaluate its clinical benefit. Similarly, a combination of commercially-available food-grade enzymes, STAN1, also exhibits modest glutenase activity and is now in clinical trials (Ehren et al., 2009).
Figure 5.
Detoxification of gluten from whole-wheat bread using an acid-active two-enzyme glutenase. Whole-wheat bread was digested with pepsin supplemented with varied concentrations of EP-B2 ± SC PEP for 60 min at pH 4.5 at 37 °C. A. Reversed-phase HPLC analysis of effect of EP-B2 ± SC PEP on gluten digestion. The broad peak between 12.5-22 minutes reflects immunogenic gluten peptides 9-22 residues in length. Note that ordinate axis is scaled to highlight the incremental effect of increasing SC PEP; this view truncates the TAME internal standard peak. B. T cell proliferation assays measuring antigen content of whole-wheat bread digests. Pepsin (0.6 mg/mL) was supplemented with EP-B2 (200 U) ± SC PEP (0.5 U). Gastric digests were treated for an additional 10 min under duodenal conditions (0.375 mg/mL trypsin and chymotrypsin and 0.075 mg/mL elastase and carboxypeptidase A at pH 6.0 at 37 °C) to solubilize all remaining T cell epitopes. P28 and P35 are T cell lines expanded from celiac patient intestinal biopsies. A stimulation index of 1 indicates background levels of proliferation.
Given the relatively limited number of plant and microbial proteases that have been tested for glutenase activity, it is likely more effective combinations can be identified.
5.2. Protein engineering
In addition to identifying novel glutenase candidates from Nature, the desirable properties of lead glutenases can be enhanced through protein engineering. Two general approaches, directed evolution and protein design, are employed. In directed evolution, a diverse library of protein variants is generated by random mutagenesis techniques such as error-prone PCR and DNA shuffling. An appropriate functional assay is then used to select or screen for variants with improved function. In protein design, variants are constructed rationally, using prior structural or functional knowledge of the protein. When a library of rationally-constructed variants is selected or screened for through functional assays, this hybrid approach is called semi-rational evolution.
To facilitate semi-rational evolution of glutenases, we solved the structures of several current lead enzymes, including MX PEP, SC PEP, and EP-B2 (Bethune et al., 2006; Shan et al., 2005a). Using a chromogenic assay to screen variants, we have since evolved one of these enzymes, SC PEP, to have 200-fold higher resistance to pepsin and 20% higher turnover at acidic pH (4.5) (Ehren et al., 2008). This study highlighted the potential for protein design to improve existing and future glutenase candidates. However, it also underscored the difficulty of rationalizing certain beneficial mutations, even when structural data is available.
5.3. Drug delivery
As gluten is a dietary antigen that elicits immunotoxicity in the gut, the obvious delivery route for glutenase therapy is oral administration. As discussed in Section 4.3, this is straightforward for glutenases targeted to the acidic environment of the stomach. For glutenases targeted to the duodenum, enteric coated capsules will be necessary to protect the drug during gastric transit. Either way, oral delivery is likely to require administration of glutenase with every meal in which dietary gluten is intentionally or inadvertently ingested.
More sophisticated delivery routes that may reduce the requisite frequency of glutenase administration include sustained-release capsules, engineered probiotics, and gene therapy.
Sustained-release capsules would involve encapsulating glutenases in a porous polymer capsule or embedding glutenases in an insoluble matrix such that release of glutenase is gradual and constant over time. A practical goal for such an approach is to reduce glutenase administration to a once-daily dose.
The strategy of engineering gut-colonizing probiotics to produce protein pharmaceuticals is under investigation for other inflammatory autoimmune diseases including diabetes (Duan et al., 2008), as well as for prevention of infection by rotavirus (Pant et al., 2006), HIV (Rao et al., 2005), and Vibrio cholerae (Duan and March, 2008). For celiac sprue, this approach would involve engineering an acid-tolerant probiotic that colonizes the stomach or upper small bowel to secrete glutenase continuously or upon introduction of dietary protein. Commercially-available probiotics such as E. coli Nissle 1917 (Mutaflor) and Bacillus coagulans (GanedenBC30), which are currently used as oral supplements for irritable bowel syndrome and lactose intolerance, are potential candidates for such an approach. Alternatively, prominent gut commensals such as Enterococcus faecalis, Bacteroides thetaiotaomicron, or Lactobacillus spp. can be engineered for acid-resistance via gene shuffling (Patnaik et al., 2002).
Finally, gene therapy has been proposed as an intervention in celiac sprue (Londei et al., 2003). Glutenase gene therapy would involve engineering gastrointestinal epithelial progenitors or functionally-specialized cells involved in zymogen secretion (e.g. gastric chief cells or pancreatic acinar cells) to express glutenase in a constitutive or hormonally-regulated manner. Historically, the inherent risks of gene therapy have restricted its use to diseases with high mortality and few effective treatment options. These include monogenic hereditary diseases, such as cystic fibrosis and severe combined immunodeficiencies, as well as cancer. Celiac sprue is not generally life-threatening and is effectively treated with a gluten-free diet in most compliant patients. Nonetheless, the burden of this life-long treatment, as well as the morbidity associated with frequent inadvertent exposure, may make gene therapy an attractive option, particularly as gene transfer techniques become safer.
6. Summary and outlook
Since the identification in 1953 of gluten as the primary environmental trigger of celiac sprue (Dicke et al., 1953), we have gained an expansive understanding of the genetic, immunological, and biochemical basis for the pathogenesis of this complex disease. Nonetheless, over this same timespan the prevalence of celiac sprue has increased 4-fold to 1-2% of humans (Lohi et al., 2007; Rubio-Tapia et al., 2009) and the only treatment available to this growing population remains a life-long gluten-free diet. Compliance with this treatment is poor due to inadvertent or intentional gluten ingestion, resulting in widespread chronic inflammation and increased morbidity and mortality (Catassi et al., 2002; Corrao et al., 2001). There is thus a compelling need to innovate better treatments for celiac sprue.
In this chapter, we described the development of oral protease therapy as an alternative (or adjunct) to the gluten-free diet. The therapeutic use of glutenases is under investigation by several research groups (Cerf-Bensussan et al., 2007; Stepniak and Koning, 2006) and three glutenase candidates have entered clinical trials (Sollid and Khosla, 2011; Tye-Din et al., 2010). Critical toward these efforts have been the design of in vitro activity assays that predict in vivo efficacy and the use of these to identify effective glutenases.
We intuited that the most predictive assays for glutenase efficacy would use proteolytically-resistant, immunogenic gluten peptides as substrates. We therefore conducted the experiment described in Section 3.1.2 to identify gluten peptides that remain intact after treatment with the normal complement of proteases present in the human gastrointestinal tract (Shan et al., 2002; Shan et al., 2005b). In identifying such peptides, this approach also elucidated the structural features underlying their immunogenicity and thereby informed subsequent selection of glutenase candidates. Alternative strategies to identify disease-relevant gluten peptides include mAb-based screening (Mitea et al., 2008), in silico proteome analysis (Shan et al., 2005b), and screening for gluten-peptides that induce immune responses from celiac patient-derived intestinal biopsies (Sturgess et al., 1994). Peptides discovered through these screens are invaluable tools for evaluating glutenase candidates.
The most fertile sources of glutenases to date are natural glutenases from germinating grains and microbial prolyl endopeptidases. We predicted that a combination glutenase comprising representatives of each of these complementary enzyme families would detoxify gluten more efficiently than either enzyme alone. Indeed, from the extensive toolbox of proteases available from plant and microbial sources, it has proven relatively straightforward to identify enzyme combinations that detoxify gluten in vitro and in vivo (Gass et al., 2007a; Siegel et al., 2006). In silico and activity-based screening are likely to identify more effective combinations and protein engineering can expand the utility of these enzymes further still.
Several points must be addressed as glutenases validated in preclinical studies progress through clinical trials. The first regards the amount of gluten that can be detoxified in vivo by a reasonable glutenase dose. This will determine whether oral enzyme therapy will enable celiac sprue patients to consume gluten ad libitum, to ingest sparing quantities only, or simply to avert inflammation when trace gluten is encountered inadvertently. Here again, the combined use of complementary glutenases is likely to reduce the dose of each enzyme needed to detoxify a given amount of gluten (Figure 5). A second point for clinical studies to address is the impact of other dietary proteins on the efficiency of glutenases. A limited number of experiments suggest that current lead candidates preferentially digest gluten (Bethune et al., 2006; Mitea et al., 2007), but further studies to assess the full implications of the myriad potentially competing proteins in the human diet are warranted. Finally, in initial studies, glutenases will likely be formulated for once-per-meal delivery via oral capsule; this dosage schedule could be improved upon. Future studies should investigate alternative routes for enzyme delivery that provide sustained protection for patients with celiac sprue.
7. References
- Alaedini A, Green PH. Narrative review: celiac disease: understanding a complex autoimmune disorder. Ann. Intern. Med. 2005;142:289–98. doi: 10.7326/0003-4819-142-4-200502150-00011. [DOI] [PubMed] [Google Scholar]
- Bethune MT, Crespo-Bosque M, Bergseng E, Mazumdar K, Doyle L, Sestak K, Sollid LM, Khosla C. Noninflammatory gluten peptide analogs as biomarkers for celiac sprue. Chem Biol. 2009;16:868–81. doi: 10.1016/j.chembiol.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethune MT, Khosla C. Parallels between pathogens and gluten peptides in celiac sprue. PLoS Pathog. 2008;4:e34. doi: 10.1371/journal.ppat.0040034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethune MT, Ribka E, Khosla C, Sestak K. Transepithelial transport and enzymatic detoxification of gluten in gluten-sensitive rhesus macaques. PLoS ONE. 2008;3:e1857. doi: 10.1371/journal.pone.0001857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethune MT, Strop P, Tang Y, Sollid LM, Khosla C. Heterologous expression, purification, refolding, and structural-functional characterization of EP-B2, a self-activating barley cysteine endoprotease. Chem. Biol. 2006;13:637–47. doi: 10.1016/j.chembiol.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Blanquet S, Zeijdner E, Beyssac E, Meunier JP, Denis S, Havenaar R, Alric M. A dynamic artificial gastrointestinal system for studying the behavior of orally administered drug dosage forms under various physiological conditions. Pharm Res. 2004;21:585–91. doi: 10.1023/b:pham.0000022404.70478.4b. [DOI] [PubMed] [Google Scholar]
- Catassi C, Fabiani E, Corrao G, Barbato M, De Renzo A, Carella AM, Gabrielli A, Leoni P, Carroccio A, Baldassarre M, Bertolani P, Caramaschi P, Sozzi M, Guariso G, Volta U, Corazza GR. Risk of non-Hodgkin lymphoma in celiac disease. JAMA. 2002;287:1413–9. doi: 10.1001/jama.287.11.1413. [DOI] [PubMed] [Google Scholar]
- Cerf-Bensussan N, Matysiak-Budnik T, Cellier C, Heyman M. Oral proteases: a new approach to managing coeliac disease. Gut. 2007;56:157–60. doi: 10.1136/gut.2005.090498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciacci C, Cirillo M, Cavallaro R, Mazzacca G. Long-term follow-up of celiac adults on gluten-free diet: prevalence and correlates of intestinal damage. Digestion. 2002;66:178–85. doi: 10.1159/000066757. [DOI] [PubMed] [Google Scholar]
- Cornell HJ, Macrae FA, Melny J, Pizzey CJ, Cook F, Mason S, Bhathal PS, Stelmasiak T. Enzyme therapy for management of coeliac disease. Scand. J. Gastroenterol. 2005;40:1304–12. doi: 10.1080/00365520510023855. [DOI] [PubMed] [Google Scholar]
- Corrao G, Corazza GR, Bagnardi V, Brusco G, Ciacci C, Cottone M, Sategna Guidetti C, Usai P, Cesari P, Pelli MA, Loperfido S, Volta U, Calabro A, Certo M. Mortality in patients with coeliac disease and their relatives: a cohort study. Lancet. 2001;358:356–61. doi: 10.1016/s0140-6736(01)05554-4. [DOI] [PubMed] [Google Scholar]
- Davy A, Svendsen I, Sorensen SO, Blom Sorensen M, Rouster J, Meldal M, Simpson DJ, Cameron-Mills V. Substrate specificity of barley cysteine endoproteases EP-A and EP-B. Plant Physiol. 1998;117:255–61. doi: 10.1104/pp.117.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angelis M, Cassone A, Rizzello CG, Gagliardi F, Minervini F, Calasso M, Di Cagno R, Francavilla R, Gobbetti M. Mechanism of degradation of immunogenic gluten epitopes from Triticum turgidum L. var. durum by sourdough lactobacilli and fungal proteases. Appl Environ Microbiol. 2010;76:508–18. doi: 10.1128/AEM.01630-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angelis M, Rizzello CG, Fasano A, Clemente MG, De Simone C, Silano M, De Vincenzi M, Losito I, Gobbetti M. VSL#3 probiotic preparation has the capacity to hydrolyze gliadin polypeptides responsible for Celiac Sprue. Biochim Biophys Acta. 2006;1762:80–93. doi: 10.1016/j.bbadis.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Di Cagno R, Rizzello CG, De Angelis M, Cassone A, Giuliani G, Benedusi A, Limitone A, Surico RF, Gobbetti M. Use of selected sourdough strains of Lactobacillus for removing gluten and enhancing the nutritional properties of gluten-free bread. J Food Prot. 2008;71:1491–5. doi: 10.4315/0362-028x-71.7.1491. [DOI] [PubMed] [Google Scholar]
- Dicke WK, Weijers HA, Van De Kamer JH. Coeliac disease. II. The presence in wheat of a factor having a deleterious effect in cases of coeliac disease. Acta. Paediatr. 1953;42:34–42. doi: 10.1111/j.1651-2227.1953.tb05563.x. [DOI] [PubMed] [Google Scholar]
- Dressman JB. Comparison of Canine and Human Gastrointestinal Physiology. Pharmaceutical Research. 1986;3:123–131. doi: 10.1023/A:1016353705970. [DOI] [PubMed] [Google Scholar]
- Duan F, Curtis KL, March JC. Secretion of insulinotropic proteins by commensal bacteria: rewiring the gut to treat diabetes. Appl Environ Microbiol. 2008;74:7437–8. doi: 10.1128/AEM.01019-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan FP, March JC. Interrupting Vibrio cholerae infection of human epithelial cells with engineered commensal bacterial signaling. Biotechnology and Bioengineering. 2008;101:128–134. doi: 10.1002/bit.21897. [DOI] [PubMed] [Google Scholar]
- Edens L, Dekker P, van der Hoeven R, Deen F, de Roos A, Floris R. Extracellular prolyl endoprotease from Aspergillus niger and its use in the debittering of protein hydrolysates. J Agric Food Chem. 2005;53:7950–7. doi: 10.1021/jf050652c. [DOI] [PubMed] [Google Scholar]
- Ehren J, Govindarajan S, Moron B, Minshull J, Khosla C. Protein engineering of improved prolyl endopeptidases for celiac sprue therapy. Protein Eng Des Sel. 2008;21:699–707. doi: 10.1093/protein/gzn050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehren J, Moron B, Martin E, Bethune MT, Gray GM, Khosla C. A food-grade enzyme preparation with modest gluten detoxification properties. PLoS One. 2009;4:e6313. doi: 10.1371/journal.pone.0006313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlanger BF, Kokowsky N, Cohen W. The preparation and properties of two new chromogenic substrates of trypsin. Arch Biochem Biophys. 1961;95:271–8. doi: 10.1016/0003-9861(61)90145-x. [DOI] [PubMed] [Google Scholar]
- Fleckenstein B, Molberg O, Qiao SW, Schmid DG, von der Mulbe F, Elgstoen K, Jung G, Sollid LM. Gliadin T cell epitope selection by tissue transglutaminase in celiac disease. Role of enzyme specificity and pH influence on the transamidation versus deamidation process. J. Biol. Chem. 2002;277:34109–16. doi: 10.1074/jbc.M204521200. [DOI] [PubMed] [Google Scholar]
- Gass J, Bethune MT, Siegel M, Spencer A, Khosla C. Combination enzyme therapy for gastric digestion of dietary gluten in patients with celiac sprue. Gastroenterology. 2007a;133:472–80. doi: 10.1053/j.gastro.2007.05.028. [DOI] [PubMed] [Google Scholar]
- Gass J, Ehren J, Strohmeier G, Isaacs I, Khosla C. Fermentation, purification, formulation, and pharmacological evaluation of a prolyl endopeptidase from Myxococcus xanthus: implications for Celiac Sprue therapy. Biotechnol Bioeng. 2005;92:674–84. doi: 10.1002/bit.20643. [DOI] [PubMed] [Google Scholar]
- Gass J, Vora H, Bethune MT, Gray GM, Khosla C. Effect of barley endoprotease EP-B2 on gluten digestion in the intact rat. J. Pharmacol. Exp. Ther. 2006;318:1178–86. doi: 10.1124/jpet.106.104315. [DOI] [PubMed] [Google Scholar]
- Gass J, Vora H, Hofmann AF, Gray GM, Khosla C. Enhancement of dietary protein digestion by conjugated bile acids. Gastroenterology. 2007b;133:16–23. doi: 10.1053/j.gastro.2007.04.008. [DOI] [PubMed] [Google Scholar]
- Gessendorfer B, Hartmann G, Wieser H, Koehler P. Determination of celiac disease-specific peptidase activity of germinated cereals. European Food Research and Technology. 2011;232:205–209. [Google Scholar]
- Hartmann G, Koehler P, Wieser H. Rapid degradation of gliadin peptides toxic for coeliac disease patients by proteases from germinating cereals. Journal of Cereal Science. 2006;44:368–371. [Google Scholar]
- Hausch F, Shan L, Santiago NA, Gray GM, Khosla C. Intestinal digestive resistance of immunodominant gliadin peptides. Am. J. Physiol. Gastrointest. Liver Physiol. 2002;283:G996–G1003. doi: 10.1152/ajpgi.00136.2002. [DOI] [PubMed] [Google Scholar]
- Helmerhorst EJ, Zamakhchari M, Schuppan D, Oppenheim FG. Discovery of a novel and rich source of gluten-degrading microbial enzymes in the oral cavity. PLoS One. 2010;5:e13264. doi: 10.1371/journal.pone.0013264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi Y, Eroshkin A, Gramatikova S, Gramatikoff K, Zhang Y, Smith JW, Osterman AL, Godzik A. CutDB: a proteolytic event database. Nucleic Acids Res. 2007;35:D546–9. doi: 10.1093/nar/gkl813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen JV, Varner JE. Gibberellic Acid-Induced Synthesis of Protease by Isolated Aleurone Layers of Barley. Plant Physiol. 1967;42:1596–1600. doi: 10.1104/pp.42.11.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler M, Acuto O, Storelli C, Murer H, Muller M, Semenza G. A modified procedure for the rapid preparation of efficiently transporting vesicles from small intestinal brush border membranes. Their use in investigating some properties of D-glucose and choline transport systems. Biochim Biophys Acta. 1978;506:136–54. doi: 10.1016/0005-2736(78)90440-6. [DOI] [PubMed] [Google Scholar]
- Kiyosaki T, Asakura T, Matsumoto I, Tamura T, Terauchi K, Funaki J, Kuroda M, Misaka T, Abe K. Wheat cysteine proteases triticain alpha, beta and gamma exhibit mutually distinct responses to gibberellin in germinating seeds. J Plant Physiol. 2009;166:101–6. doi: 10.1016/j.jplph.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Kiyosaki T, Matsumoto I, Asakura T, Funaki J, Kuroda M, Misaka T, Arai S, Abe K. Gliadain, a gibberellin-inducible cysteine proteinase occurring in germinating seeds of wheat, Triticum aestivum L., specifically digests gliadin and is regulated by intrinsic cystatins. FEBS J. 2007;274:1908–17. doi: 10.1111/j.1742-4658.2007.05749.x. [DOI] [PubMed] [Google Scholar]
- Koehler SM, Ho TH. Hormonal regulation, processing, and secretion of cysteine proteinases in barley aleurone layers. Plant Cell. 1990;2:769–83. doi: 10.1105/tpc.2.8.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindfors K, Blomqvist T, Juuti-Uusitalo K, Stenman S, Venalainen J, Maki M, Kaukinen K. Live probiotic Bifidobacterium lactis bacteria inhibit the toxic effects induced by wheat gliadin in epithelial cell culture. Clin Exp Immunol. 2008;152:552–8. doi: 10.1111/j.1365-2249.2008.03635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohi S, Mustalahti K, Kaukinen K, Laurila K, Collin P, Rissanen H, Lohi O, Bravi E, Gasparin M, Reunanen A, Maki M. Increasing prevalence of coeliac disease over time. Aliment Pharmacol Ther. 2007;26:1217–25. doi: 10.1111/j.1365-2036.2007.03502.x. [DOI] [PubMed] [Google Scholar]
- Londei M, Quaratino S, Maiuri L. Celiac disease: a model autoimmune disease with gene therapy applications. Gene Ther. 2003;10:835–43. doi: 10.1038/sj.gt.3302041. [DOI] [PubMed] [Google Scholar]
- Lui CY, Amidon GL, Berardi RR, Fleisher D, Youngberg C, Dressman JB. Comparison of gastrointestinal pH in dogs and humans: implications on the use of the beagle dog as a model for oral absorption in humans. J Pharm Sci. 1986;75:271–4. doi: 10.1002/jps.2600750313. [DOI] [PubMed] [Google Scholar]
- Malagelada J-R, Azpiroz F. Determinants of gastric emptying and transit in the small intestine. John Wiley & Sons, Inc.; 2010. [Google Scholar]
- Marti T, Molberg O, Li Q, Gray GM, Khosla C, Sollid LM. Prolyl endopeptidase-mediated destruction of T cell epitopes in whole gluten: chemical and immunological characterization. J Pharmacol Exp Ther. 2005;312:19–26. doi: 10.1124/jpet.104.073312. [DOI] [PubMed] [Google Scholar]
- Mayer M, Greco L, Troncone R, Auricchio S, Marsh MN. Compliance of adolescents with coeliac disease with a gluten free diet. Gut. 1991;32:881–5. doi: 10.1136/gut.32.8.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkonen A, Porali I, Cercos M, Ho TH. A major cysteine proteinase, EPB, in germinating barley seeds: structure of two intronless genes and regulation of expression. Plant Mol Biol. 1996;31:239–54. doi: 10.1007/BF00021787. [DOI] [PubMed] [Google Scholar]
- Minekus M, Marteau P, Havenaar R, Huis in ’T Veld JHJ. Fund for the Replacement of Animals in Medical Experiments. ROYAUME-UNI; Nottingham: 1995. A multicompartmental dynamic computer-controlled model simulating the stomach and small intestine. [Google Scholar]
- Mitea C, Havenaar R, Drijfhout JW, Edens L, Dekking L, Koning F. Efficient degradation of gluten by a prolyl endoprotease in a gastrointestinal model: implications for celiac disease. Gut. 2007 doi: 10.1136/gut.2006.111609. [DOI] [PubMed] [Google Scholar]
- Mitea C, Kooy-Winkelaar Y, van Veelen P, de Ru A, Drijfhout JW, Koning F, Dekking L. Fine specificity of monoclonal antibodies against celiac disease-inducing peptides in the gluteome. Am J Clin Nutr. 2008;88:1057–66. doi: 10.1093/ajcn/88.4.1057. [DOI] [PubMed] [Google Scholar]
- Molberg Ø, McAdam S, Lundin K, Sollid L. Studies of gliadin-specific T cells in celiac disease. In: Marsh MN, editor. Celiac disease. Methods and protocols. Humana; Totowa, New Jersey, USA: 2000. pp. 105–124. [DOI] [PubMed] [Google Scholar]
- Molberg O, McAdam SN, Korner R, Quarsten H, Kristiansen C, Madsen L, Fugger L, Scott H, Noren O, Roepstorff P, Lundin KE, Sjostrom H, Sollid LM. Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nat Med. 1998;4:713–7. doi: 10.1038/nm0698-713. [DOI] [PubMed] [Google Scholar]
- Molberg O, Uhlen AK, Jensen T, Flaete NS, Fleckenstein B, Arentz-Hansen H, Raki M, Lundin KE, Sollid LM. Mapping of gluten T-cell epitopes in the bread wheat ancestors: implications for celiac disease. Gastroenterology. 2005;128:393–401. doi: 10.1053/j.gastro.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Moron B, Bethune MT, Comino I, Manyani H, Ferragud M, Lopez MC, Cebolla A, Khosla C, Sousa C. Toward the assessment of food toxicity for celiac patients: characterization of monoclonal antibodies to a main immunogenic gluten peptide. PLoS ONE. 2008a;3:e2294. doi: 10.1371/journal.pone.0002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moron B, Cebolla A, Manyani H, Alvarez-Maqueda M, Megias M, Thomas Mdel C, Lopez MC, Sousa C. Sensitive detection of cereal fractions that are toxic to celiac disease patients by using monoclonal antibodies to a main immunogenic wheat peptide. Am J Clin Nutr. 2008b;87:405–14. doi: 10.1093/ajcn/87.2.405. [DOI] [PubMed] [Google Scholar]
- Nilsen EM, Jahnsen FL, Lundin KE, Johansen FE, Fausa O, Sollid LM, Jahnsen J, Scott H, Brandtzaeg P. Gluten induces an intestinal cytokine response strongly dominated by interferon gamma in patients with celiac disease. Gastroenterology. 1998;115:551–63. doi: 10.1016/s0016-5085(98)70134-9. [DOI] [PubMed] [Google Scholar]
- Nilsen EM, Lundin KE, Krajci P, Scott H, Sollid LM, Brandtzaeg P. Gluten specific, HLA-DQ restricted T cells from coeliac mucosa produce cytokines with Th1 or Th0 profile dominated by interferon gamma. Gut. 1995;37:766–76. doi: 10.1136/gut.37.6.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant N, Hultberg A, Zhao YF, Svensson L, Pan-Hammarstrom Q, Johansen K, Pouwels PH, Ruggeri FM, Hermans P, Frenken L, Boren T, Marcotte H, Hammarstrom L. Lactobacilli expressing variable domain of llama heavy-chain antibody fragments (lactobodies) confer protection against rotavirus-induced diarrhea. Journal of Infectious Diseases. 2006;194:1580–1588. doi: 10.1086/508747. [DOI] [PubMed] [Google Scholar]
- Patnaik R, Louie S, Gavrilovic V, Perry K, Stemmer WP, Ryan CM, del Cardayre S. Genome shuffling of Lactobacillus for improved acid tolerance. Nat Biotechnol. 2002;20:707–12. doi: 10.1038/nbt0702-707. [DOI] [PubMed] [Google Scholar]
- Pietzak MM. Follow-up of patients with celiac disease: achieving compliance with treatment. Gastroenterology. 2005;128:S135–41. doi: 10.1053/j.gastro.2005.02.025. [DOI] [PubMed] [Google Scholar]
- Piper JL, Gray GM, Khosla C. High selectivity of human tissue transglutaminase for immunoactive gliadin peptides: implications for celiac sprue. Biochemistry. 2002;41:386–93. doi: 10.1021/bi011715x. [DOI] [PubMed] [Google Scholar]
- Piper JL, Gray GM, Khosla C. Effect of prolyl endopeptidase on digestive-resistant gliadin peptides in vivo. J Pharmacol Exp Ther. 2004;311:213–9. doi: 10.1124/jpet.104.068429. [DOI] [PubMed] [Google Scholar]
- Pyle GG, Paaso B, Anderson BE, Allen DD, Marti T, Li Q, Siegel M, Khosla C, Gray GM. Effect of pretreatment of food gluten with prolyl endopeptidase on gluten-induced malabsorption in celiac sprue. Clin Gastroenterol Hepatol. 2005;3:687–94. doi: 10.1016/s1542-3565(05)00366-6. [DOI] [PubMed] [Google Scholar]
- Quarsten H, Molberg O, Fugger L, McAdam SN, Sollid LM. HLA binding and T cell recognition of a tissue transglutaminase-modified gliadin epitope. Eur. J. Immunol. 1999;29:2506–14. doi: 10.1002/(SICI)1521-4141(199908)29:08<2506::AID-IMMU2506>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Rao S, Hu S, McHugh L, Lueders K, Henry K, Zhao Q, Fekete RA, Kar S, Adhya S, Hamer DH. Toward a live microbial microbicide for HIV: Commensal bacteria secreting an HIV fusion inhibitor peptide. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11993–11998. doi: 10.1073/pnas.0504881102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38:D227–33. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Tapia A, Kyle RA, Kaplan EL, Johnson DR, Page W, Erdtmann F, Brantner TL, Kim WR, Phelps TK, Lahr BD, Zinsmeister AR, Melton LJ, 3rd, Murray JA. Increased prevalence and mortality in undiagnosed celiac disease. Gastroenterology. 2009;137:88–93. doi: 10.1053/j.gastro.2009.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuppan D, Junker Y, Barisani D. Celiac disease: from pathogenesis to novel therapies. Gastroenterology. 2009;137:1912–33. doi: 10.1053/j.gastro.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Shan L, Marti T, Sollid LM, Gray GM, Khosla C. Comparative biochemical analysis of three bacterial prolyl endopeptidases: implications for coeliac sprue. Biochem J. 2004;383:311–8. doi: 10.1042/BJ20040907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, Mathews II, Khosla C. Structural and mechanistic analysis of two prolyl endopeptidases: role of interdomain dynamics in catalysis and specificity. Proc Natl Acad Sci U S A. 2005a;102:3599–604. doi: 10.1073/pnas.0408286102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, Molberg O, Parrot I, Hausch F, Filiz F, Gray GM, Sollid LM, Khosla C. Structural basis for gluten intolerance in celiac sprue. Science. 2002;297:2275–9. doi: 10.1126/science.1074129. [DOI] [PubMed] [Google Scholar]
- Shan L, Qiao SW, Arentz-Hansen H, Molberg O, Gray GM, Sollid LM, Khosla C. Identification and analysis of multivalent proteolytically resistant peptides from gluten: implications for celiac sprue. J. Proteome Res. 2005b;4:1732–41. doi: 10.1021/pr050173t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel M, Bethune MT, Gass J, Ehren J, Xia J, Johannsen A, Stuge TB, Gray GM, Lee PP, Khosla C. Rational design of combination enzyme therapy for celiac sprue. Chem. Biol. 2006;13:649–58. doi: 10.1016/j.chembiol.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Simpson DJ. Proteolytic degradation of cereal prolamins - the problem with proline. Plant Science. 2001;161:825–838. [Google Scholar]
- Sollid LM, Khosla C. Novel Therapies for Coeliac Disease. J Intern Med. 2011 doi: 10.1111/j.1365-2796.2011.02376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollid LM, Markussen G, Ek J, Gjerde H, Vartdal F, Thorsby E. Evidence for a primary association of celiac disease to a particular HLA-DQ alpha/beta heterodimer. J. Exp. Med. 1989;169:345–50. doi: 10.1084/jem.169.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souliman S, Blanquet S, Beyssac E, Cardot JM. A level A in vitro/in vivo correlation in fasted and fed states using different methods: applied to solid immediate release oral dosage form. Eur J Pharm Sci. 2006;27:72–9. doi: 10.1016/j.ejps.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Spaenij-Dekking L, Kooy-Winkelaar Y, van Veelen P, Drijfhout JW, Jonker H, van Soest L, Smulders MJ, Bosch D, Gilissen LJ, Koning F. Natural variation in toxicity of wheat: potential for selection of nontoxic varieties for celiac disease patients. Gastroenterology. 2005;129:797–806. doi: 10.1053/j.gastro.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Stenman SM, Venalainen JI, Lindfors K, Auriola S, Mauriala T, Kaukovirta-Norja A, Jantunen A, Laurila K, Qiao SW, Sollid LM, Mannisto PT, Kaukinen K, Maki M. Enzymatic detoxification of gluten by germinating wheat proteases: implications for new treatment of celiac disease. Ann Med. 2009;41:390–400. doi: 10.1080/07853890902878138. [DOI] [PubMed] [Google Scholar]
- Stepniak D, Koning F. Enzymatic gluten detoxification: the proof of the pudding is in the eating. Trends Biotechnol. 2006;24:433–4. doi: 10.1016/j.tibtech.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Stepniak D, Spaenij-Dekking L, Mitea C, Moester M, de Ru A, Baak-Pablo R, van Veelen P, Edens L, Koning F. Highly efficient gluten degradation with a newly identified prolyl endoprotease: implications for celiac disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2006;291:G621–9. doi: 10.1152/ajpgi.00034.2006. [DOI] [PubMed] [Google Scholar]
- Sturgess R, Day P, Ellis HJ, Lundin KE, Gjertsen HA, Kontakou M, Ciclitira PJ. Wheat peptide challenge in coeliac disease. Lancet. 1994;343:758–61. doi: 10.1016/s0140-6736(94)91837-6. [DOI] [PubMed] [Google Scholar]
- Tye-Din JA, Anderson RP, Ffrench RA, Brown GJ, Hodsman P, Siegel M, Botwick W, Shreeniwas R. The effects of ALV003 pre-digestion of gluten on immune response and symptoms in celiac disease in vivo. Clin Immunol. 2010;134:289–95. doi: 10.1016/j.clim.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Vader LW, de Ru A, van der Wal Y, Kooy YM, Benckhuijsen W, Mearin ML, Drijfhout JW, van Veelen P, Koning F. Specificity of tissue transglutaminase explains cereal toxicity in celiac disease. J. Exp. Med. 2002;195:643–9. doi: 10.1084/jem.20012028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn RA. Plant proteases: from phenotypes to molecular mechanisms. Annu Rev Plant Biol. 2008;59:191–223. doi: 10.1146/annurev.arplant.59.032607.092835. [DOI] [PubMed] [Google Scholar]
- van der Hoorn RA, Leeuwenburgh MA, Bogyo M, Joosten MH, Peck SC. Activity profiling of papain-like cysteine proteases in plants. Plant Physiol. 2004;135:1170–8. doi: 10.1104/pp.104.041467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venalainen JI, Juvonen RO, Mannisto PT. Evolutionary relationships of the prolyl oligopeptidase family enzymes. Eur J Biochem. 2004;271:2705–15. doi: 10.1111/j.1432-1033.2004.04199.x. [DOI] [PubMed] [Google Scholar]
- Vora H, McIntire J, Kumar P, Deshpande M, Khosla C. A scaleable manufacturing process for pro-EP-B2, a cysteine protease from barley indicated for celiac sprue. Biotechnol Bioeng. 2007;98:177–85. doi: 10.1002/bit.21423. [DOI] [PubMed] [Google Scholar]
- Wieser H. Chemistry of gluten proteins. Food Microbiol. 2007;24:115–9. doi: 10.1016/j.fm.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Xia J, Sollid LM, Khosla C. Equilibrium and kinetic analysis of the unusual binding behavior of a highly immunogenic gluten peptide to HLA-DQ2. Biochemistry. 2005;44:4442–9. doi: 10.1021/bi047747c. [DOI] [PubMed] [Google Scholar]
- Yang JM, Tung CH. Protein structure database search and evolutionary classification. Nucleic Acids Res. 2006;34:3646–59. doi: 10.1093/nar/gkl395. [DOI] [PMC free article] [PubMed] [Google Scholar]