Abstract
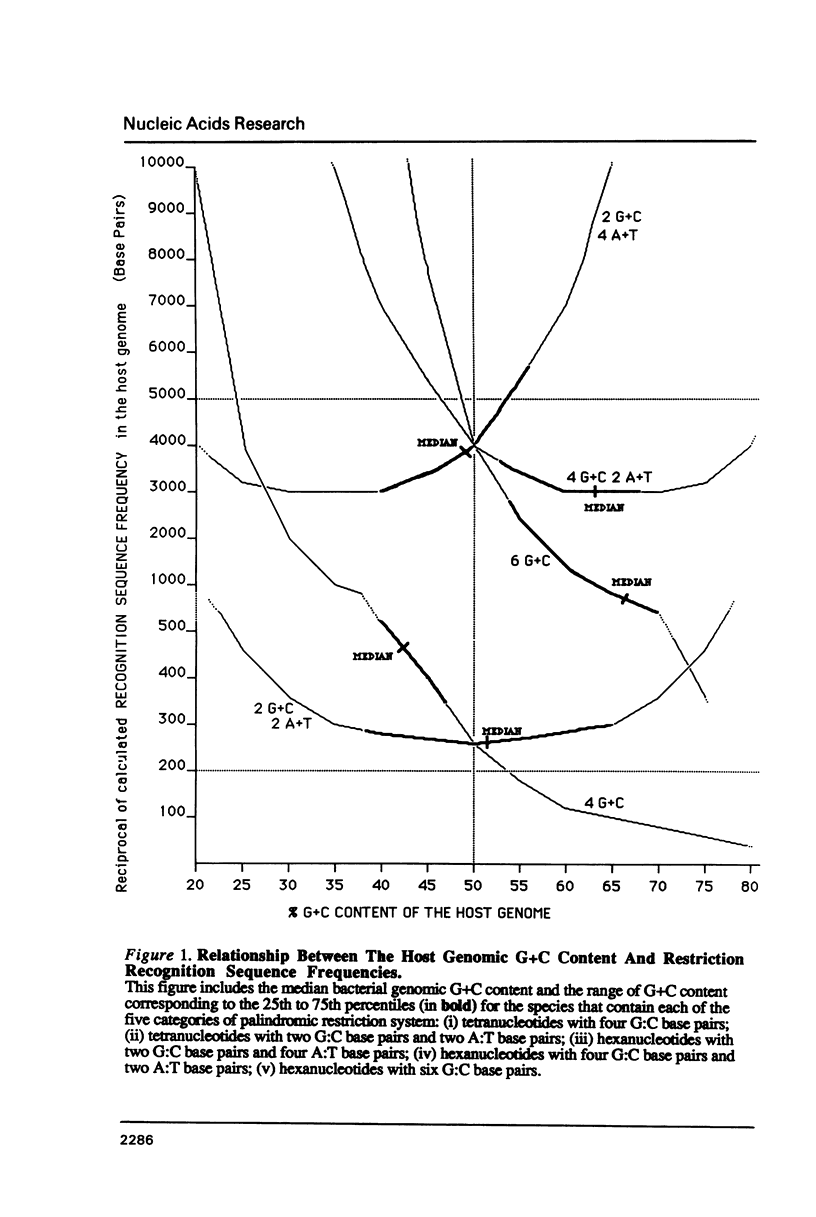
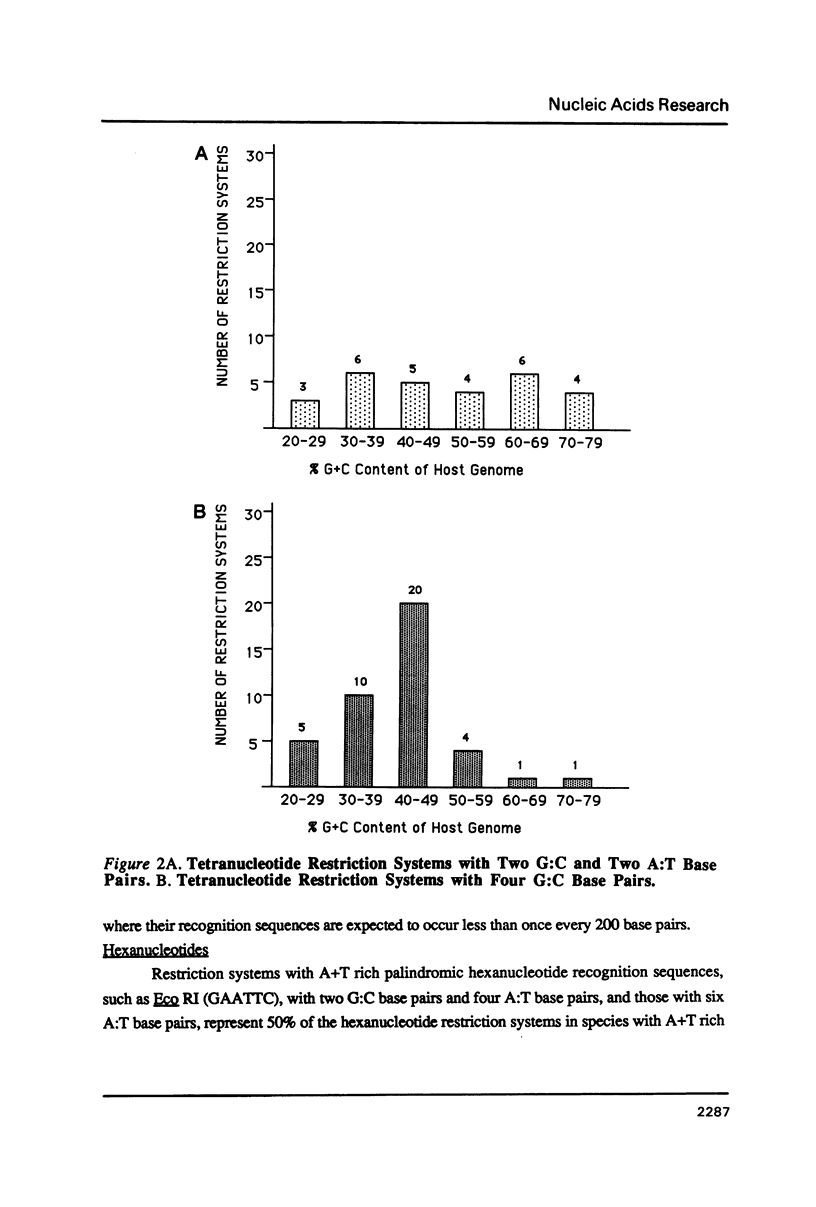
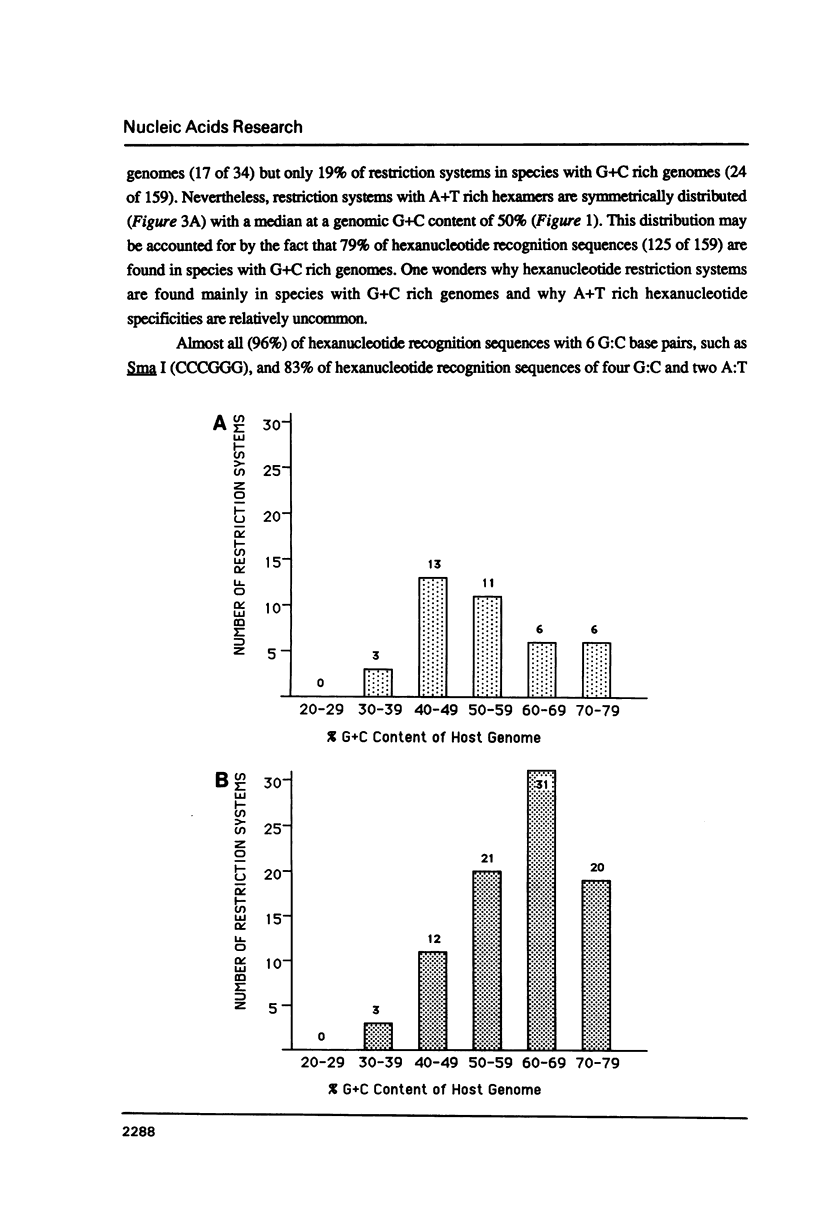
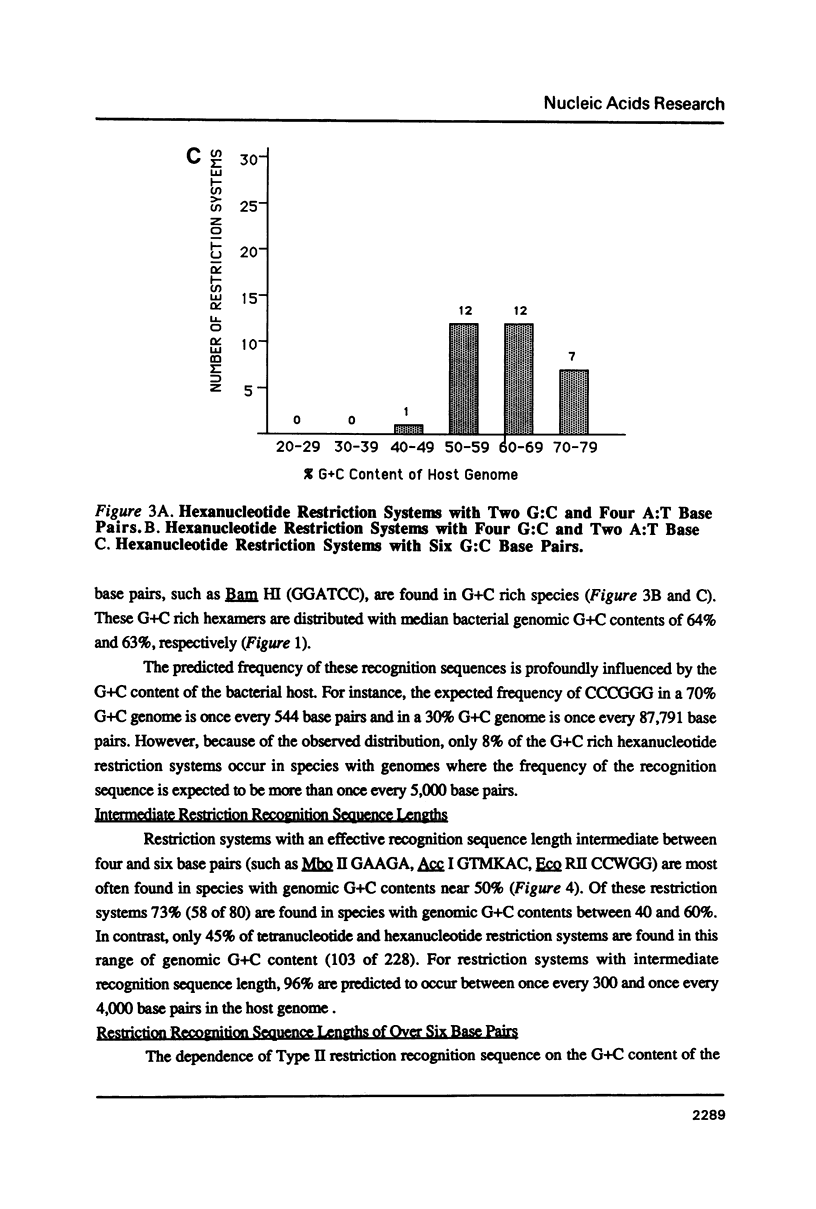
I show that the recognition sequences of Type II restriction systems are correlated with the G + C content of the host bacterial DNA. Almost all restriction systems with G + C rich tetranucleotide recognition sequences are found in species with A + T rich genomes, whereas G + C rich hexanucleotide and octanucleotide recognition sequences are found almost exclusively in species with G + C rich genomes. Most hexanucleotide recognition sequences found in species with A + T rich genomes are A + T rich. This distribution eliminates a substantial proportion of the potential variance in the frequency of restriction recognition sequences in the host genomes. As a consequence, almost all restriction recognition sequences, including those eight base pairs in length (Not I and Sfi I), are predicted to occur with a frequency ranging from once every 300 to once every 5,000 base pairs in the host genome. Since the G + C content of bacteriophage DNA and of the host genome are also correlated, the data presented is evidence that most Type II "restriction systems" are indeed involved in phage restriction.
Full text
PDF


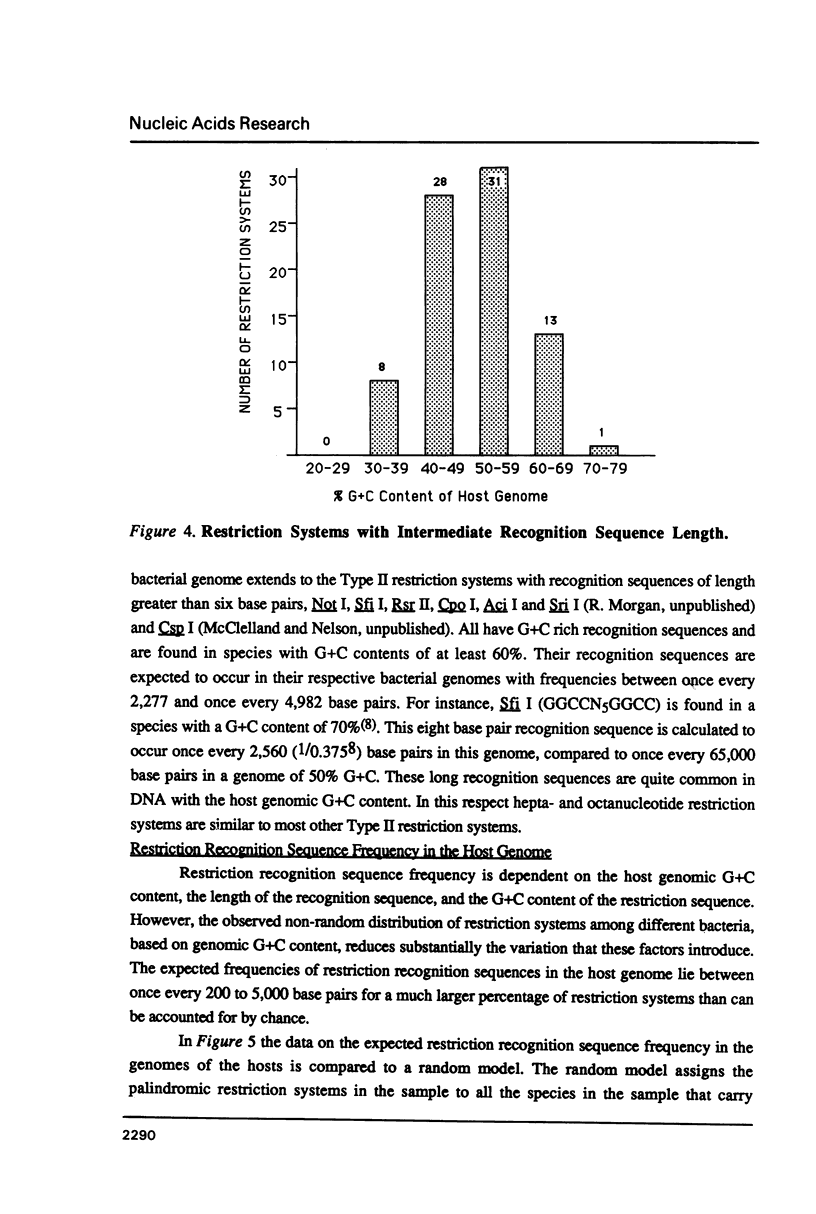
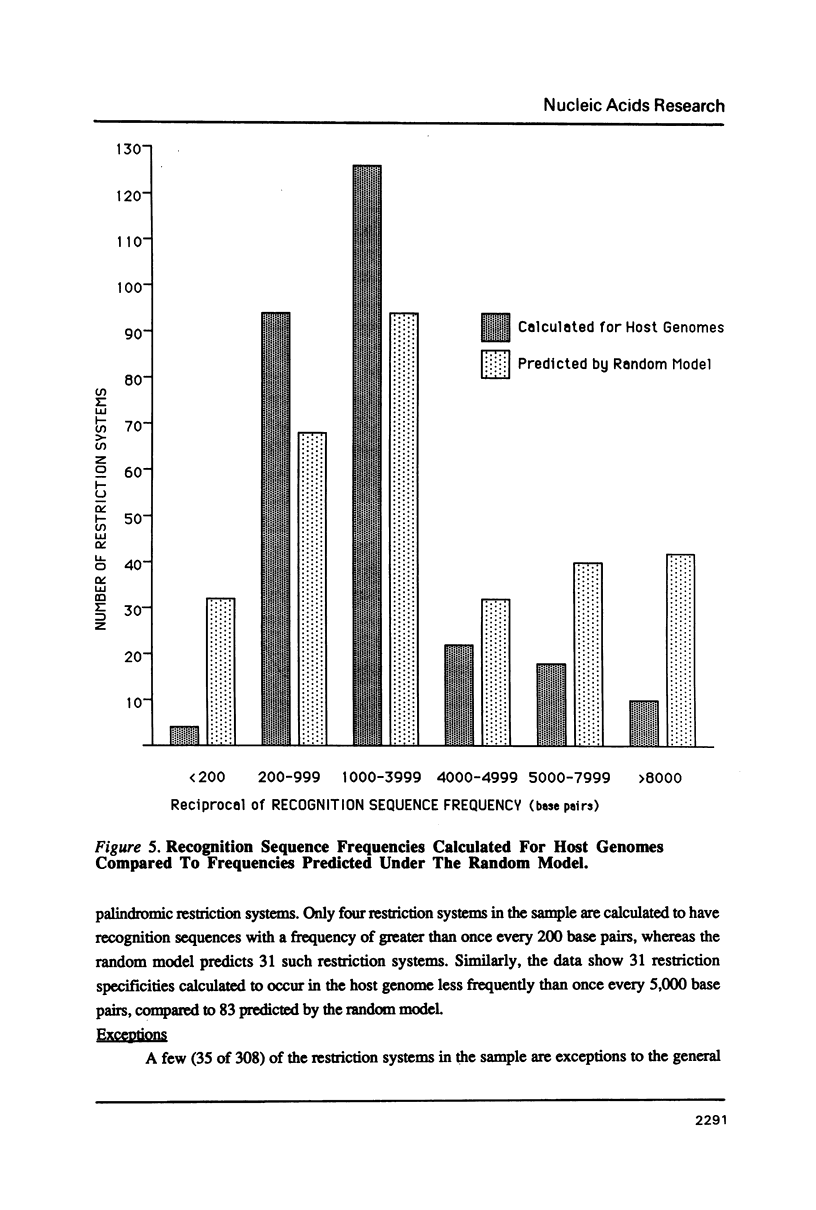



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Coulondre C., Miller J. H., Farabaugh P. J., Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978 Aug 24;274(5673):775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Gama-Sosa M. A., Carreira L. H., Ljungdahl L. G., Kuo K. C., Gehrke C. W. DNA methylation in thermophilic bacteria: N4-methylcytosine, 5-methylcytosine, and N6-methyladenine. Nucleic Acids Res. 1985 Feb 25;13(4):1399–1412. doi: 10.1093/nar/13.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. H., Farnet C. M., Ehrlich K. C., Ehrlich M. Digestion of highly modified bacteriophage DNA by restriction endonucleases. Nucleic Acids Res. 1982 Mar 11;10(5):1579–1591. doi: 10.1093/nar/10.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Johnson A., Stokes J. L. Deoxyribonucleic acid base composition of Sphaerotilus natans and Sphaerotilus discophorus. J Bacteriol. 1966 Apr;91(4):1657–1658. doi: 10.1128/jb.91.4.1657-1658.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Jones R., Patel Y., Nelson M. Restriction endonucleases for pulsed field mapping of bacterial genomes. Nucleic Acids Res. 1987 Aug 11;15(15):5985–6005. doi: 10.1093/nar/15.15.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang B. Q., Schildkraut I. A type II restriction endonuclease with an eight nucleotide specificity from Streptomyces fimbriatus. Nucleic Acids Res. 1984 Jun 11;12(11):4507–4516. doi: 10.1093/nar/12.11.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J. Restriction and modification enzymes and their recognition sequences. Nucleic Acids Res. 1985;13 (Suppl):r165–r200. doi: 10.1093/nar/13.suppl.r165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Sharp P. M. Molecular evolution of bacteriophages: evidence of selection against the recognition sites of host restriction enzymes. Mol Biol Evol. 1986 Jan;3(1):75–83. doi: 10.1093/oxfordjournals.molbev.a040377. [DOI] [PubMed] [Google Scholar]


