Abstract
Co-ordination between plastids and mitochondria is so essential that there should be extensive inter-organellar crosstalk during development of plant cells. Indeed, chloroplast dysfunction in white leaves of plastid ribosome-deficient mutant barley, albostrians, is reportedly accompanied by increases in the levels of mitochondrial DNA and mitochondrial transcripts, suggesting that (i) developmental/physiological status of plastids (or heterotrophic growth condition of albino leaves) can affect the status of mitochondrial genome, and (ii) the function of the affected mitochondria may also be up-regulated accordingly. However, functional aspects of the mitochondria affected by chloroplast dysfunction have not yet been examined in detail. Here, we examined the effects of chloroplast dysfunction on mitochondrial DNA level and dark respiration rate, by comparing white and green sectors within individual variegated leaves, using 12 ornamental plants as experimental materials. The pattern of leaf variegation differed from species to species, suggesting that different mechanisms were involved in the formation of white sectors in different species. Quantitative hybridization analysis revealed that mitochondrial DNA levels were generally higher in white sectors than in green sectors. In spite of the elevated mitochondrial DNA levels, however, dark respiration rates in white sectors were generally lower than those in green sectors. Several possible mechanisms for elevation of mitochondrial DNA level and suppression of dark respiration rates in white sectors are discussed.
Electronic supplementary material
The online version of this article (doi:10.1007/s00709-011-0325-y) contains supplementary material, which is available to authorized users.
Keywords: Chloroplast dysfunction, Inter-organellar crosstalk, Leaf variegation, Mitochondrial DNA, Ornamental plant, Respiration
Introduction
Plant cells possess two types of DNA-containing, energy-converting organelles: plastids and mitochondria. These two organelle types are tightly connected to each other in the metabolic network within plant cells. For example, plastids (chloroplasts) supply plant mitochondria with oxygen and carbohydrates necessary for energy conversion and metabolism (Noctor et al. 2007). Conversely, plant mitochondria play specific roles that support chloroplast functions, such as dissipation of excess energy through a cyanide-resistant respiratory pathway (Yoshida et al. 2007) and detoxification of glycolate through photorespiration pathway (Ogren 1984).
Because the metabolic associations between the two organelles are so tight, existence of mechanism(s) to co-ordinate development and function of the two organelles is(are) expected. For example, the components and function of mitochondria in leaf cells are modulated according to the status of plastids within the same cell. Alternative oxidase (AOX), which is a nuclear-encoded mitochondrial protein that plays an important role in the cyanide-resistant respiratory pathway, is up-regulated under high light conditions (Finnegan et al. 1997; Yoshida et al. 2007), and the extent of up-regulation is affected by developmental status of plastids (Yoshida et al. 2008). This situation, where plastids regulate expression of nuclear genes for mitochondrial proteins, is similar to retrograde regulation via “plastid signal”, in which plastids regulate nuclear genes for photosynthesis-related, chloroplast proteins (Jarvis 2003).
Another report by Hedtke et al. (1999) suggests that developmental/physiological status of plastids can also affect the mitochondrial genome. They reported that the levels of mitochondrial DNA and mitochondrial transcripts were elevated in white leaves of albostrians, a barley mutant that has deficient plastid ribosomes. They also reported that mitochondrial transcript levels were elevated similarly when wild-type barley plants were photo-bleached by treatment with a carotenoid biosynthesis inhibitor, norflurazon. These results suggest that chloroplast dysfunction causes elevation in levels of mitochondrial DNA and mitochondrial transcript. Their results further imply that, in the plastid-deficient leaf cells, mitochondrial function is enhanced accordingly. However, the physiological consequences, as well as the underlying mechanisms, of the elevation of mitochondrial DNA and transcript levels remain unclear.
Here, we examined the effects of chloroplast dysfunction on mitochondria, by comparing white and green sectors within individual variegated leaves from 12 ornamental plants. Leaf variegation is defined as patches of different colors in the same leaf, and is seen in a variety of ornamental plants (Sakamoto et al. 2009). There are various colors and patterns of leaf variegation, suggesting that various causes and mechanisms are responsible in the respective cases (Kirk and Tilney-Bassett 1978). In many variegated plants with green/white sectors, white sectors are composed of living cells that contain undifferentiated plastids (Kato et al. 2007; Sakamoto et al. 2009). Thus, we may be able to examine the effects of chloroplast dysfunction on mitochondria by comparing green and white sectors within individual leaves. Moreover, using various kinds of variegated plants with various causes for leaf variegation, we may be able to distinguish the effects of “chloroplast dysfunction”, which are common to all cases, from the effects that are specific to respective cases. Thus, variegated plants may be useful resources for the study of the effects of developmental/physiological status of plastids on mitochondria.
We examined morphological characters of leaves with green/white variegation in 12 ornamental plants, and compared mitochondrial DNA level and dark respiration rates between white and green sectors in each; plastid DNA levels and photosynthesis rates were also examined. Our results indicated that chloroplast dysfunction can cause higher mitochondrial DNA level and lower dark respiration rates. Several possible mechanisms for these changes are discussed.
Materials and methods
Plant materials
Plants used in this study were dwarf umbrella tree (Schefflera arboricola, Araliaceae), coleus (Solenostemon scutellarioides “Peter Green”, Lamiaceae), sunpatiens (Impatiens hybrids “Fuiri–Salmon”, Balsaminaceae), plantain lily (Hosta sp. “Reversed”, Agavaceae), Japanese morning glory (Ipomea nil “Hamanokagayaki”, Convolvulaceae), blue daisy bush (Felicia amelloides “Cassiopeia”, Asteraceae), canary ivy (Hedera canariensis “Shiro-okame”, Araliaceae), pothos (Epipremnum aureum “N’ JOY”, Araceae), abutilon (Abutilon variegatum, Malvaceae), strawberry (Fragaria × ananassa, “marble strawberry”, Rosaceae), geranium (Pelargonium zonale, Geraniaceae), and dracaena (Dracaena sanderiana, Agavaceae). Morning glory was grown from seeds in soil-containing pots for approximately 2 months under natural conditions. Other plants were purchased as pot cultures from several local markets, and maintained briefly under natural conditions. Matured, but not senescent, leaves were collected and used for the experiments.
Microscopic observations
For microscopic observations, leaf pieces (approximately 3 × 3 mm) were carefully dissected from green and white sectors, fixed in 1% (w/v) glutaraldehyde and 2% (w/v) formaldehyde dissolved in 100 mM sodium phosphate buffer (pH 7.0), embedded in 5% (w/v) agar, sectioned to 40- to 120-μm thickness with a micro-slicer (DTK-1500; DSK, Kyoto, Japan), and observed with a microscope (BX-60, Olympus, Tokyo, Japan).
Measurement of chlorophyll content
For extraction of chlorophyll, leaf discs (38.6 mm2) were carefully punched out from green and white sectors with a cork borer, ground to a powder in liquid nitrogen with a sample crusher (Multi-beads shocker; Yasui-Kikai Co. Ltd, Osaka, Japan), suspended in 1 ml of 80% (v/v) acetone, homogenized again using the sample crusher, and then these suspensions were centrifuged. The supernatant was used for spectrophotometric quantification of chlorophyll, according to Porra et al. (1989).
Determination of organelle DNA level
Organelle DNA level was examined by quantitative dot-bot hybridization. Leaf discs (approximately 60 mm2) were carefully punched out from green and white sectors with a cork borer and ground to a powder in liquid nitrogen with a sample crusher (Multi-beads shocker; Yasui-Kikai Co.). Total cellular DNA was purified with DNeasy Plant Mini Kit (Qiagen, Maryland, USA). The concentrations of extracted DNA were determined with DNA fluorometer DyNA Quant (GE Healthcare, Backinghamshir, England). The DNA samples were heat-denatured, dot-blotted onto nylon membrane sheets (Biodyne B, Pall, NY, USA) as a dilution series (300, 150, and 75 ng/dot or 200, 100, and 50 ng/dot) using a dot-blot apparatus (Bio-Dot; Bio-Rad, Hercules, CA, USA), and fixed by successive treatments with denaturing solution (1.5 M NaCl, 0.5 M NaOH) for 5 min, neutralizing solution (1.5 M NaCl, 0.5 M Tris–HCl pH 7.2, 1 mM EDTA) for 1 min, and alkali-fixation solution (0.4 M NaOH) for 20 min. The membrane sheets were rinsed with 2× SSC solution, blot-dried, wrapped, and stored at 4°C until use. For detection of nuclear, mitochondrial, and plastid DNAs by hybridization, cloned DNA fragments containing sequences for the cytoplasmic 26S/18S rRNA genes (rDNA) from rice, cytochrome oxidase gene (coxI) from rice, and the ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit gene (rbcL) from tobacco, respectively, were used as probes (Suzuki et al. 1992; Sakai et al. 1998). For detection of P. zonale mitochondrial DNA, total mitochondrial DNA from tobacco was used as a probe instead of rice coxI. Probes were labeled with [α-32P] dCTP using DNA labeling kit Megaprime (GE healthcare) and hybridized to the dot blots according to the standard procedures. Intensities of hybridization signals were quantified by densitometric scanning of the X-ray films using Densitograph and Lane Analyzer systems (ATTO Co. Ltd., Tokyo, Japan).
Measurement of gross photosynthesis and dark respiration rates
Photosynthesis and dark respiration rates were measured with a liquid-phase O2 electrode system (DW1 + CB1D; WALZ, Effeltrich, Germany). Small leaf pieces (5 × 5 mm square) were carefully cut out from green and white sectors and further cut into smaller sizes (1–1.25 mm squares) with a sharp razor. They were suspended in 0.9 ml bathing solution (50 mM Hepes–KOH pH 7.0, 0.5 mM CaSO4) within a closed DW1 O2 electrode cuvette maintained at 25°C, stirred, and pre-illuminated for 5 min at 1,600 μmol m−2 s−1 with a halogen lamp. Then, 0.1 ml of 0.625 M NaHCO3 was injected into the cuvette to stimulate photosynthesis, and the changes in O2 concentration in the bathing solution due to photosynthetic O2 evolution were monitored. When steady changes in O2 concentration were recorded (within 15–20 min measurement), the cuvette was covered with a blackout curtain, and the changes in O2 concentration due to respiratory O2 consumption was monitored until steady rates of the change were obtained.
Statistical analysis
Statistical analyses were conducted for each of the 12 species. For the comparison of each morphological parameter between green and white sectors, Student’s t-test was used with Stacle2 (OMS publishing Inc., Saitama, Japan) after checking the equal variance of the two groups. If the assumption of equal variance was not met, t-test with unequal variance was used. For the comparison of signal intensity of mitochondrial or plastid DNA between the sectors, a general linear model was used with JMP 8.0 (SAS Institute, Cary, NC, USA). In this analysis, the identity of plant individual (#1–3), the sector (green or white) from which the sample was taken, and the signal intensity of nuclear DNA (measured in the dilution series of 75, 150, and 300 ng/dot or 50, 100, and 200 ng/dot) were designated as independent variables. The signal intensity of either mitochondrial or plastid DNA in the same dilution series was designated as a dependent variable. Nuclear DNA signal was included because a positive correlation was expected between signal intensities of nuclear DNA and mitochondrial or plastid DNA in the dilution series. The individual identity was treated as a random factor, using the EMS (expected value of mean squares) method as implemented in JMP. Normality of the residuals was checked for each analysis, but the data were not transformed because a significant deviation from the normal distribution was detected in only one out of 24 (mitochondrial or plastid DNA in 12 species) independent analyses.
Results
Morphological characterization of variegated leaves
Variegated leaves from the plants used in this study, which include both dicotyledonous and monocotyledonous plants, are shown in Fig. 1. The variegation patterns are roughly classified into three types — central, random, or marginal — in which white sectors/stripes are observed predominantly around the central region of the leaves, randomly, or near leaf margins, respectively. H. canariensis, classified as random type in Fig. 1, may represent an intermediate type between random and marginal types; although their leaf margins were not always white and their white sectors appeared to distribute rather randomly, their white sectors usually included leaf margins and thus appeared to originate from marginal region.
Fig. 1.
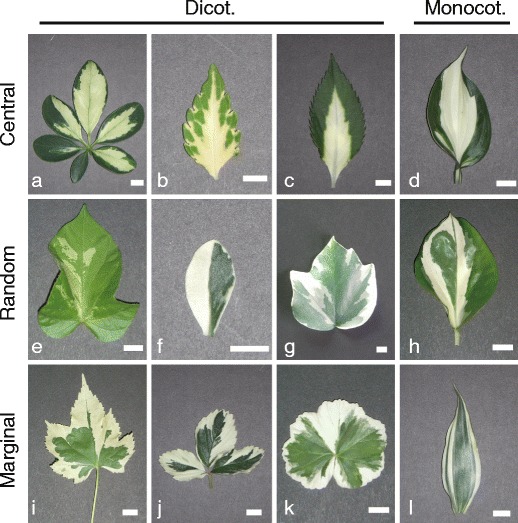
Representative images of variegated leaves used in this study. a Schefflera arboricola; b Solenostemon scutellarioides; c Impatiens hybrids; d Hosta sp.; e Ipomea nil; f Felicia amelloides; g Hedera canariensis; h Epipremnum aureum; i Abutilon variegatum; j Fragaria × ananassa; k Pelargonium zonale; l Dracaena sanderiana. Dicot. dicotyledonous plants, Monocot. monocotyledonous plants. Variegation patterns (Central, Random, and Marginal types) are shown to the left. Scale bars represent 1 cm
In white sectors of these leaves, all, or part of mesophyll cell layers lacked green chloroplast (Fig. 2). The white sectors of A. variegatum (Fig. 2(i2)) were an exceptional case; they completely lacked any mesophyll cell layer. Instead, their abaxial epidermal cells contained chloroplasts and appeared green (Fig. 2(i2), inset). In white sectors of S. arboricola (Fig. 2(a2)) and I. nil (Fig. 2(e2)), loss of chloroplasts was essentially restricted to palisade cell layers only: the spongy cell layers usually retained green chloroplasts. In white sectors of the other nine plant species, both palisade and spongy cell layers generally lacked chloroplasts. However, H. canariensis, E. aureum, and P. zonale leaves had pale green regions in addition to green and white regions (see Fig. 1g, h, k), where chloroplasts were absent only in either of the palisade or spongy cell layers (data not shown). As shown in H. canariensis and E. aureum (Fig. 2(g2, h2)), white sectors sometimes contained islands of green mesophyll cells with chloroplasts within chloroplast-free cell populations.
Fig. 2.
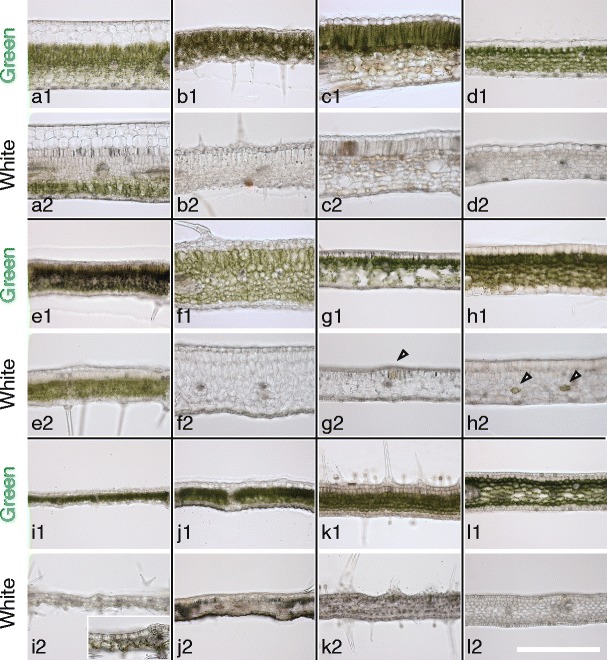
Cross sections of variegated leaves used in this study. a1/a2 Schefflera arboricola b1/b2 Solenostemon scutellarioides; c1/c2 Impatiens hybrids; d1/d2 Hosta sp.; e1/e2 Ipomea nil; f1/f2 Felicia amelloides; g1/g2 Hedera canariensis; h1/h2 Epipremnum aureum; i1/i2 Abutilon variegatum; j1/j2 Fragaria × ananassa; k1/k2 Pelargonium zonale; l1/l2 Dracaena sanderiana. Cross sections of green (a1, b1, c1, d1, e1, f1, g1, h1, i1, j1, k1, l1) and white (a2, b2, c2, d2, e2, f2, g2, h2, i2, j2, k2, l2) sectors are shown. Arrowheads in g2 and h2 show green cells in white sectors. Inset in i2 is a higher-magnification image showing the green color of abaxial epidermal cells. Scale bar in l2 represents 500 μm
The difference between green and white sectors was further characterized by measuring chlorophyll content (Fig. 3). “White” sectors of S. arboricola and I. nil, whose spongy tissue remained green (see Fig. 2(a2, e2)), contained considerable amounts of chlorophyll (Fig. 3a, e). White sectors of other plant species contained only trace amounts of chlorophyll (Fig. 3b–d, f–l). These results indicated that loss or reduction of green color in the white sectors of the variegated leaves used in this study did not result from an optical effect (such as random reflection of light), but was indeed caused by loss of functional chloroplasts. The small amount of chlorophyll found in the white sectors of A. variegatum that lack mesophyll cells (Fig. 3i) might have come from the green abaxial epidermal cells (see Fig. 2(i2)). In other cases, chlorophyll in the white sectors might have come from green cells that were sparsely distributed within the white mesophyll cell population (see Fig. 2(g2, h2)), guard cells of stomata, or small plastids possibly present in the apparently white mesophyll cells.
Fig. 3.
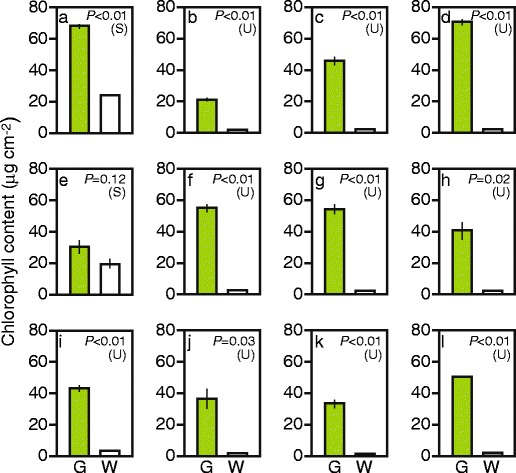
Chlorophyll contents of green and white sectors. a Schefflera arboricola; b Solenostemon scutellarioides; c Impatiens hybrids; d Hosta sp.; e Ipomea nil; f Felicia amelloides; g Hedera canariensis; h Epipremnum aureum; i Abutilon variegatum; j Fragaria × ananassa; k Pelargonium zonale; l Dracaena sanderiana. G green sector, W white sector. Each bar represents mean ± SE (n = 3). Results of Student’s t-test (S) or t-test with unequal variance (U) are also shown
Tables 1 and 2 summarize the morphological parameters of green and white sectors. In leaves with marginal type variegation, white sectors were thinner than green sectors; this difference was probably influenced by the position of the white sectors within a leaf (Table 1). Nonetheless, in most of the plant species examined here (including marginal type plants), the mesophyll cell density per leaf area, which was calculated by multiplying the “number of mesophyll cell layers” by the square of “density of palisade cells in the paradermal direction”, was not significantly different between green and white sectors (Table 2).
Table 1.
Comparison of the morphological parameters of green and white sectors of variegated leaves used in this study (1): variegation pattern and leaf thickness
| Species (genus) | Variegation pattern | Leaf thickness (μm) | |||||
|---|---|---|---|---|---|---|---|
| Organ level | Tissue level | Green | White | W/G ratio | t-Test | ||
| t | P | ||||||
| Schefflera | C | P | 428 ± 14 | 424 ± 18 | 0.99 | 0.18 | 0.87 |
| Solenostemon | C | P+S | 215 ± 9 | 233 ± 5 | 1.08 | 0.17 | 0.17 |
| Impatiens | C | P+S | 380 ±20 | 382 ± 14 | 1.01 | −0.10 | 0.92 |
| Hosta | C | P+S | 232 ± 8 | 217 ± 10 | 0.94 | 1.22 | 0.29 |
| Ipomea | R | P | 212 ± 4 | 206 ± 5 | 0.97 | 0.96 | 0.39 |
| Felicia | R | P+S | 382 ± 23 | 429 ± 11 | 1.12 | −1.80 | 0.15 |
| Hedera | R/M | P, S, P+S | 227 ± 3 | 202 ± 5 | 0.89 | 4.32 | 0.01 |
| Epipremnum | R | P, S, P+S | 305 ± 5 | 257 ± 23 | 0.84 | 2.08 | 0.11 |
| Abutilon | M | (w/o P, S) | 92 ± 3 | 67 ± 2 | 0.73 | 6.98 | <0.01 |
| Fragaria | M | P+S | 157 ± 4 | 141 ± 3 | 0.90 | 3.28 | 0.03 |
| Pelargonium | M | P, S, P+S | 194 ± 7 | 149 ± 8 | 0.77 | 4.02 | 0.02 |
| Dracaena | M | P+S | 236 ± 4 | 201 ± 6 | 0.85 | 4.99 | <0.01 |
For leaf thickness in green and white sectors, each datum represents an average ± standard error (n = 3). The results of Student’s t-tests are also shown.
W/G ratio, ratio of the average values for green and white sectors; C central type, R random type, M marginal type, P pallisade tissue, S spongy tissue
Table 2.
Comparison of the morphological parameters of green and white sectors of variegated leaves used in this study (2): mesophyll cell density
| Species (genus) | Number of mesophyll cell layer (cells) | Paradermal pallisade cell density (cells mm−1) | Mesophyll cell density index (104 cell equivalents mm−2) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Green | White | W/G ratio | t-Test | Green | White | W/G ratio | t-Test | Green | White | W/G ratio | t-Test | |||||||
| t | P | Type | t | P | Type | t | P | Type | ||||||||||
| Schefflera | 8.7 ± 0.2 | 9.2 ± 0.6 | 1.06 | −0.94 | 0.40 | S | 51 ± 2 | 52 ± 3 | 1.02 | −0.52 | 0.63 | S | 2.3 ± 0.3 | 2.5 ± 0.3 | 1.09 | −0.29 | 0.78 | S |
| Solenostemon | 4.1 ± 0.1 | 5.0 ± 0.2 | 1.22 | −4.00 | 0.02 | S | 39 ± 2 | 31 ± 1 | 0.79 | 2.63 | 0.03 | S | 0.6 ± 0.1 | 0.5 ± 0.1 | 0.79 | 1.30 | 0.26 | S |
| Impatiens | 7.9 ± 0.2 | 9.4 ± 0.1 | 1.19 | −6.26 | <0.01 | S | 42 ± 1 | 43 ± 2 | 1.02 | −0.20 | 0.85 | S | 1.4 ± 0.1 | 1.7 ± 0.2 | 1.21 | −1.18 | 0.30 | S |
| Hosta | 6.1 ± 0.1 | 6.4 ± 0.1 | 1.05 | −2.12 | 0.10 | S | 34 ± 3 | 34 ± 3 | 1.00 | −0.07 | 0.94 | S | 0.7 ± 0.2 | 0.8 ± 0.1 | 1.06 | −0.16 | 0.88 | S |
| Ipomea | 6.7 ± 0.4 | 6.7 ± 0.2 | 1.00 | 0.00 | 1.00 | S | 84 ± 3 | 88 ± 3 | 1.05 | −0.95 | 0.37 | S | 4.7 ± 0.5 | 5.1 ± 0.3 | 1.09 | −0.78 | 0.48 | S |
| Felicia | 6.9 ± 0.3 | 6.8 ± 0.2 | 0.99 | 0.30 | 0.78 | S | 29 ± 2 | 34 ± 1 | 1.17 | −1.73 | 0.11 | S | 0.6 ± 0.1 | 0.8 ± 0.0 | 1.28 | −1.27 | 0.27 | S |
| Hedera | 6.2 ± 0.1 | 6.3 ± 0.3 | 1.02 | −0.32 | 0.77 | S | 34 ± 1 | 47 ± 1 | 1.38 | −8.46 | <0.01 | S | 0.9 ± 0.1 | 1.4 ± 0.1 | 1.65 | −3.41 | 0.03 | S |
| Epipremnum | 6.6 ± 0.3 | 5.0 ± 0.2 | 0.76 | 4.43 | 0.01 | S | 42 ± 2 | 37 ± 1 | 0.88 | 1.65 | 0.15 | U | 1.2 ± 0.2 | 0.7 ± 0.0 | 0.58 | 1.99 | 0.18 | U |
| Abutilon | 2.8 ± 0.3 | 0.0 ± 0.0 | 0.00 | – | a | – | 90 ± 4 | 0 ± 0 | 0.00 | – | a | – | 2.3 ± 0.5 | 0.0 ± 0.0 | 0.00 | – | a | – |
| Fragaria | 2.7 ± 0.2 | 3.1 ± 0.1 | 1.15 | −2.00 | 0.12 | S | 94 ± 4 | 87 ± 2 | 0.93 | 1.51 | 0.16 | S | 2.4 ± 0.3 | 2.4 ± 0.1 | 1.00 | −0.04 | 0.97 | S |
| Pelargonium | 4.1 ± 0.1 | 3.8 ± 0.1 | 0.93 | 2.12 | 0.10 | S | 43 ± 1 | 46 ± 2 | 1.07 | −1.42 | 0.21 | U | 0.8 ± 0.1 | 0.8 ± 0.1 | 1.05 | −0.50 | 0.64 | S |
| Dracaena | 6.4 ± 0.1 | 6.3 ± 0.5 | 0.98 | 0.21 | 0.84 | S | 32 ± 1 | 40 ± 2 | 1.25 | −4.26 | <0.01 | S | 0.7 ± 0.0 | 1.0 ± 0.1 | 1.54 | −2.77 | 0.05 | S |
Each datum represents an average ± standard error (n = 3). The results of t-test and type of t-test used (S, Student’s t-test; U, t-test with unequal variance) are also shown.
W/G ratio, ratio of the average values for green and white sectors
aNot subjected to statistical analysis due to lack of variance, but apparently different
Comparison of plastid and mitochondrial DNA levels between green and white sectors
We then performed semi-quantitative Southern dot-blot hybridization to compare organelle DNA levels between green and white sectors. Total DNA was extracted from green and white sectors from individual leaves and blotted in a dilution series. Three sets of blots with identical contents were prepared, and each was hybridized with probes specific to mitochondrial, plastid, or nuclear DNA sequences. Intensities of hybridization signals were measured and subjected to subsequent analyses.
When relative signal intensities for plastid DNA were plotted against relative signal intensities for nuclear DNA (Fig. 4A), the data points for white sectors appeared to be up-shifted when compared to those for green sectors in eight species (e.g., Fig. 4A(c, l)). In these eight species, the ratio of plastid DNA to nuclear DNA should be higher in the white sectors than in the green sectors. In several other species, however, distribution of the data points for green and white sectors appeared nearly identical (e.g., Fig 4A(b, d, i)). Moreover, in Fragaria × ananassa (Fig. 4A(j)), the data points for white sectors appeared to be down-shifted relative to those for green sectors, suggesting that plastid DNA levels were lower in white sectors than in green sectors. These results, together with the results of statistical analyses (Table 3), demonstrated that plastid DNA levels in white sectors might be either increased (eight species), unaffected (three species), or decreased (one species, Fragaria × ananassa), relative to those in green sectors.
Fig. 4.
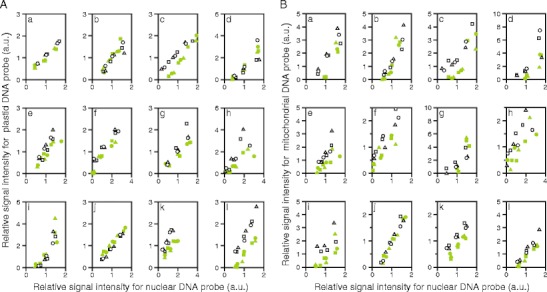
Comparison of organelle DNA levels in green and white sectors. A Comparison of plastid DNA level. B Comparison of mitochondrial DNA level. Total DNAs were extracted from green and white sectors of the same leaves and dot-blotted to nylon membrane as dilution series (75, 150, and 300 ng/dot or 50, 100, and 200 ng/dot). Three sheets of membrane with identical contents were prepared, each of which was hybridized with a single probe that was specific to a plastid, a mitochondrial, or a nuclear DNA sequence. For each plant species, hybridization experiments were repeated three times using three different sets of DNA samples. Intensities of hybridization signals were measured and transformed to relative values, where, for each plant species, each probe and each white/green pair, the average of the signal intensities from green and white sector samples at middle dose (100 or 150 ng/dot) was arbitrarily set at 1. Finally, the relative signal intensities for plastid DNA probe (A) and mitochondrial DNA probe (B) were plotted against the relative signal intensities for nuclear DNA probe. Green and white symbols indicate data from green and white sectors, respectively. Different figures of symbols (circles, triangles, and squares) indicate results from different set of leaves. Data with extremely low signals (i.e., those without linear dose–signal relationships) were excluded from the analyses. a Schefflera arboricola; b Solenostemon scutellarioides; c Impatiens hybrids; d Hosta sp.; e Ipomea nil; f Felicia amelloides; g Hedera canariensis; h Epipremnum aureum; i Abutilon variegatum; j Fragaria × ananassa; k Pelargonium zonale; l Dracaena sanderiana. For results of statistical analyses, see Table 3
Table 3.
Comparison of plastid and mitochondrial DNA levels between green and white sectors
| Species (genus) | Plastid DNA level | Mitochondrial DNA level | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Individual ID | Relative signal intensity for nuclear DNA | White or green | n | Individual ID | Relative signal intensity for nuclear DNA | White or green | |||||||
| F | P | F | P | F | P | F | P | F | P | F | P | |||
| Schefflera | 18 | 0.38 | 0.69 | 436.00 | <0.01 | 15.69 | <0.01 | 15 | 2.63 | 0.12 | 112.46 | <0.01 | 25.20 | <0.01 |
| Solenostemon | 18 | 0.74 | 0.49 | 95.21 | <0.01 | 2.07 | 0.17 | 18 | 1.81 | 0.20 | 109.52 | <0.01 | 0.05 | 0.83 |
| Impatiens | 18 | 0.05 | 0.95 | 224.10 | <0.01 | 47.62 | <0.01 | 18 | 1.26 | 0.32 | 75.03 | <0.01 | 22.55 | <0.01 |
| Hosta | 18 | 1.95 | 0.18 | 98.18 | <0.01 | 0.25 | 0.62 | 14 | 1.71 | 0.23 | 32.32 | <0.01 | 9.87 | 0.01 |
| Ipomea | 17 | 0.69 | 0.52 | 62.49 | <0.01 | 11.79 | <0.01 | 17 | 2.12 | 0.16 | 23.44 | <0.01 | 19.17 | <0.01 |
| Felicia | 18 | 0.17 | 0.85 | 231.41 | <0.01 | 22.79 | <0.01 | 18 | 1.05 | 0.38 | 79.07 | <0.01 | 14.72 | <0.01 |
| Hedera | 18 | 0.14 | 0.87 | 89.34 | <0.01 | 8.45 | 0.01 | 14 | 0.11 | 0.90 | 13.48 | 0.01 | 0.01 | 0.91 |
| Epipremnum | 18 | 0.92 | 0.42 | 32.45 | <0.01 | 9.25 | <0.01 | 18 | 0.46 | 0.64 | 33.88 | <0.01 | 7.42 | 0.02 |
| Abutilon | 18 | 1.80 | 0.20 | 66.30 | <0.01 | 0.88 | 0.36 | 18 | 5.02 | 0.02 | 32.92 | <0.01 | 18.91 | <0.01 |
| Fragaria | 18 | 0.20 | 0.82 | 752.57 | <0.01 | 34.80 | <0.01 | 18 | 2.72 | 0.10 | 587.60 | <0.01 | 4.44 | 0.06 |
| Pelargonium | 18 | 0.12 | 0.89 | 54.62 | <0.01 | 40.32 | <0.01 | 18 | 0.35 | 0.71 | 228.22 | <0.01 | 97.85 | <0.01 |
| Dracaena | 18 | 0.29 | 0.75 | 182.32 | <0.01 | 113.26 | <0.01 | 18 | 1.05 | 0.38 | 185.16 | <0.01 | 24.26 | <0.01 |
Data shown in Fig. 4 was analyzed using a general linear model. For details, see "Materials and methods"
When relative signal intensities for mitochondrial DNA were plotted against relative signal intensities for nuclear DNA (Fig. 4B), the data points for white sectors appeared to be up-shifted when compared to those for green sectors, in most of the plant species examined. The results shown in Fig. 4, together with the results of statistical analyses (Table 3) revealed that mitochondrial DNA level was significantly higher in white sectors than in green sectors in nine out of 12 species examined, and the difference between green and white sectors of Fragaria × ananassa (Fig. 4B(j)) was marginally significant (P = 0.06). Moreover, there was no species with mitochondrial DNA level that were significantly lower in white sectors than in green sectors. Thus, we concluded that there was a general trend in which mitochondrial DNA levels were higher in white sectors than in green sectors in various plant species with variegated leaves.
The hybridization results were subjected to another type of analysis, which was more quantitative, but was not suitable for statistical analysis; the process and results of this analysis are shown in Figs. S1 and S2. The intensities of respective hybridization signals were plotted against the amount of blotted DNA, and regression lines were drawn for respective samples. Then, for each probe and each sample, the amounts of total DNA necessary to generate an appropriate, arbitrarily set level of signal intensity were estimated, and the relative concentrations of organelle and nuclear DNAs within the sample DNA solutions were calculated. Then, the white/green ratios of DNA concentrations were calculated for each probe and each green-white set of the samples. Finally, the white/green ratios of organelle DNAs were corrected by dividing them with the white/green ratio of nuclear DNA (Fig. S1).
The results of this analysis demonstrated that there were no clear trends for changes in plastid DNA level accompanying white sector formation (Fig. S2A). In eight out of 12 species, average white/green ratio for plastid DNA level was higher than 1. Among the eight species, however, only one (P. zonale) species exhibited a white/green ratio higher than 2. In the remaining four species, white/green ratio for plastid DNA level did not exceed 1. In spite of the loss of functional chloroplasts in white sectors, extensive reduction of plastid DNA level there was not observed for any of the plant species examined.
In contrast to the case with plastid DNA level, mitochondrial DNA level exhibited a clear trend; mitochondrial DNA levels were higher in white sectors than in green sectors (Fig. S2B). Among the 12 species examined, 11 species exhibited average white/green ratio higher than 1; specifically, three species (I. nil, Impatiens hybrids, and A. variegatum) exhibited white/green ratio higher than 2. There was no species that exhibited the average white/green ratio lower than 1.
Photosynthesis and dark respiration in green and white sectors
The results of measurement of O2 evolution/consumption with a liquid-phase O2 electrode clearly demonstrated that gross photosynthetic rates were lower in white sectors than in green sectors (Fig. 5A). Among the 12 species examined, I. nil, whose white sectors retained chloroplasts in spongy cell layers, showed relatively high photosynthetic rate in white sectors, and the difference between green and white sectors was not statistically significant (Fig. 5A(e)). In the remaining 11 species, photosynthetic rates were significantly lower in white sectors than in green sectors, demonstrating chloroplast dysfunction in white sectors.
Fig. 5.
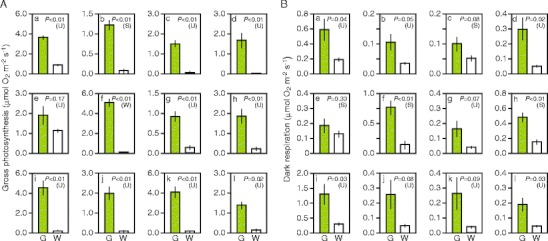
Gross photosynthesis and dark respiration rates of green and white sectors. A Gross photosynthesis rate. B Dark respiration rate. The rates of O2 consumption in the dark and O2 generation under illumination were measured with a liquid-phase O2 electrode. a Schefflera arboricola; b Solenostemon scutellarioides; c Impatiens hybrids; d Hosta sp.; e Ipomea nil; f Felicia amelloides; g Hedera canariensis; h Epipremnum aureum; i Abutilon variegatum; j Fragaria × ananassa; k Pelargonium zonale; l Dracaena sanderiana. G green sector, W white sector. Each bar represents mean ± SE (n = 6; six leaves from three plants). Results of Student’s t-test (S) or t-test with unequal variance (U) are also shown
White sectors tended to exhibit dark respiration rates lower than those in green sectors (Fig. 5B). Among the 12 species examined, seven species showed significantly lower dark respiration rates in white sectors. Even in the remaining five species, average values for dark respiration rates in white sectors ranged from 12% to 68% of the values for green sectors. In four out of the five species (i.e., Impatiens hybrid, H. canariensis, Fragaria × ananassa, and P. zonale; Fig. 5B(c, g, j, k)), the difference between green and white sectors was marginally significant (0.05 <P <0.10). Thus, increased mitochondrial DNA levels in white sectors did not lead to high dark respiration rates.
Discussion
Chloroplast dysfunction is accompanied by elevated mitochondrial DNA level
Of the 12 species examined in this study, only one species (Solenostemon scutellarioides) did not exhibit signs of increased mitochondrial DNA levels in white sectors; nine species had mitochondrial DNA levels that were significantly higher in white sectors than in green sectors (Fig. 4B, Table 3), and two of the remaining species (H. canariensis and Fragaria × ananassa) had average white/green ratio (ratio of mitochondrial DNA content in white sectors versus green sectors) higher than 1 (Fig. S2B). This general trend of increased mitochondrial DNA level in white sectors might be attributable to the loss of functional chloroplasts, which was the only factor demonstrably common among white sectors of all the plant species examined here.
In Sol. scutellarioides, which exhibited no signs of increased mitochondrial DNA level in white sectors, the “white” sectors in very young leaves appeared pale green and then became whiter during leaf maturation (data not shown), indicating that complete loss of functional chloroplast occurred relatively late in leaf development. In other species, which exhibited some signs of elevated mitochondrial DNA levels in white sectors, white sectors in young leaves appeared whiter than those in young Sol. scutellarioides leaves. Thus, the timing of chloroplast dysfunction might be an important factor in inducing elevation of mitochondrial DNA level in white sectors.
Our results are basically consistent with the conclusion drawn from the analyses on albostrians barley by Hedtke et al. (1999) that loss of functional chloroplasts leads to increases in mitochondrial DNA level. Moreover, our results further demonstrated that mitochondrial DNA level was elevated not only in albino leaves that were fully heterotrophic, but also in white sectors of variegated leaves that grow photo-autotrophically as a whole, indicating that neither mutations that causes albino phenotype nor heterotrophic culture conditions for albino plants are the cause for increased mitochondrial DNA level observed in white cells. However, the extent of elevation of mitochondrial DNA level in white sectors (1.1- to 2.7-fold; Fig. S2B) was smaller than that reported for albino leaves of albostrians barley (2.5- to 3.0-fold; Hedtke et al. 1999). Thus, complete loss of the photosynthetic cells within the leaves and/or the growth condition of the whole organ/plant may affect the extent to which mitochondrial DNA levels are elevated.
Mitochondria in white sectors may be arrested at early developmental stages
The higher mitochondrial DNA levels may result from either increased replication or reduced degradation of mitochondrial DNA. Plastids and mitochondria utilize common DNA polymerase encoded on nuclear genome (Elo et al. 2003; Christiansen et al. 2005; Ono et al. 2007; Mori et al. 2005), and they share a common intracellular nucleotide pool for replication of their DNAs. If abnormal development of chloroplasts in the white sector cells results in less active incorporation of organelle DNA polymerase/nucleotide molecules into the plastids, mitochondrial DNA level may become higher by using the surplus polymerase/nucleotides. In this case, the plastid DNA level is expected to be lower in white sectors than in green sectors. However, preferential replication of mitochondrial DNA over plastid DNA seems unlikely, because plastid DNA level was not drastically lower in white sectors than in green sectors in any of the plant species examined (Table 3).
Sakamoto et al. (2009) found that the plastids in the white sectors of nine variegated plant species contained large nucleoids (DNA–protein complexes) similar to those observed in immature chloroplasts, and they suggested that the plastids in the white sectors were arrested at an early stage of chloroplast development. Their suggestion prompted us to hypothesize that the mitochondria in the white sector might also be arrested at an early developmental stage. During normal leaf development, the level of mitochondrial DNA (Fujie et al. 1994; Topping and Leaver 1990) declines while chloroplasts develop and become photosynthetically competent. The declines in the organelle DNA levels during normal leaf development and senescence are probably attributable to the onset of DNA degrading activity within the organelles, as demonstrated for chloroplasts (Sodmergen et al. 1991). If the onset of DNA degradation requires normal development and/or function of chloroplasts as a cue, mitochondrial DNA degradation may not occur in the white sector cells, resulting in higher mitochondrial DNA levels in white sectors (where degradation does not occur) than in green sectors (where the degradation process is turned on correctly). This hypothesis can explain the reason why “white” sectors of Sol. scutellarioides, which were initially pale green and later became whiter, exhibited mitochondrial DNA level similar to that in green sectors; in Sol. scutellarioides, partial chloroplast development in “white” sectors during early leaf development might have activated the organelle DNA degradation process. To examine this possibility, future studies should focus on changes in the status of mitochondrial nucleoids, mitochondrial DNA levels and nucleolytic activities in green and white sectors throughout the course of leaf development.
Increased mitochondrial DNA levels in white sectors did not lead to high respiration rates
In the present study, we carefully dissected green and white sectors, and compared their O2 consumption rates individually using liquid-phase O2 electrode. There was a strong trend; specifically, dark respiration rates were lower in white sectors than in green sectors (Fig. 5B). Yoshioka et al. (2009) suggested that dark respiration rate was the same in green and white sectors in variegated Hedera helix leaves, based on measurements of O2 consumption rates of leaf discs with different green/white ratios by using gas-phase O2 electrode. The reason for the difference between this and previous studies should be examined, but the results of the two reports are common in that white sectors do not exhibit higher respiration rate than green sectors.
Our measurements and comparisons of dark respiration rates were made on basis of unit leaf area (Fig. 5). However, dark respiration rates in white sectors should be lower than those in green sectors even if the comparisons were based on cell numbers. In nine out of 12 plant species examined, the mesophyll cell density indices (per leaf area) in the white sectors were nearly equal to, or even higher than, those in the green sectors (Table 2); thus, the difference between green and white sectors should become larger when comparison are based on cell number, rather than leaf area. Among the remaining three species, A. variegatum should be considered separately because its white sectors lack mesophyll cells. In Sol. scutellarioides and E. aureum, the mesophyll cell density indices in white sectors were reduced to 79% and 58% (Table 2), while dark respiration rates per leaf area in white sectors were 36% and 33% (Fig. 5), of the values in green sectors, respectively. Thus, dark respiration rates should be still lower in white sectors than in green sectors of these plant species even when comparison are based on cell numbers.
Clearly, increased mitochondrial DNA in white sectors did not lead to enhanced respiration. Because we measured respiration rates without any stimulation for respiratory O2 consumption, the measured values might be different from maximum respiratory potentials, to which mitochondrial DNA level might be correlated. However, it is likely that potential for respiratory electron transport is similar between green and white sectors, because COX II protein (a constituent of respiratory chain) accumulated to similar levels in green and white sectors of variegated I. nil leaves (Katsumata et al., unpublished result). We also observed that not only mitochondrial DNA level but also mitochondrial transcript (coxI mRNA) level was higher in white sectors than in green sectors of I. nil, suggesting that the effects of increased mitochondrial gene dosage in white sectors are cancelled at translational and/or post-translational levels and thus the equal accumulation of respiratory proteins and absence of enhanced respiration result.
We further suppose that physiological interaction between plastids (chloroplasts) and mitochondria may be the basis for the lower dark respiration rates in white sectors. It is well known that sun-leaves exhibit higher respiration rates as compared to shade-leaves, probably due to a greater demand for respiratory energy (to maintain cells with greater protein content and/or to export photosynthetic products from the leaf) and higher carbohydrate supply (Lambers et al. 1998). Conversely, the low energy demand and limited carbohydrate supply in white sector cells (due to loss of functional chloroplasts) may result in lower dark respiration rates in white sectors, in spite of their relatively high mitochondrial DNA level (and equal accumulation of respiratory chain proteins).
Conclusion and working hypothesis for further analyses
Our results demonstrated two strong trends, i.e., increased mitochondrial DNA levels and decreased respiration rates in white sectors of variegated leaves from various plant species. Based on these results, we propose that chloroplast dysfunction leads to increases in mitochondrial DNA level, probably because of the absence of the activation of mechanism(s) to degrade mitochondrial DNA, which is(are) usually turned on early during normal development of leaf cells. We also propose, based on the low dark respiration rates in white sectors, that respiration rate is not determined by genetic factors (e.g., mitochondrial DNA level), but is largely determined by physiological factors (e.g., energy demand and/or supply of respiratory substrates). Then, two questions arise. (1) What is the signal for the presence or absence of functional chloroplasts and triggering mitochondrial DNA degradation? (2) Is there any connection between the “absence of mitochondrial DNA degradation” and “low dark respiration” in the white sectors?
One possible explanation is that reactive oxygen species (ROS) may play a central role in both triggering mitochondrial DNA degradation and connecting respiration rate and mitochondrial DNA content. Although the major site of ROS generation in the illuminated mesophyll cells is the chloroplasts (Foyer and Noctor 2003), mitochondria also generate ROS during the process of respiratory electron transfer. During the process of normal leaf development, the rate of ROS generation in the leaf mesophyll cells should rise in both chloroplasts and mitochondria, according to the start of photosynthesis and concomitant activation of respiration, respectively. During the development of variegated leaves, ROS generation should increase similarly in green sectors, whereas the rate of ROS generation should remain lower in white sectors. Indeed, selective accumulation of ROS in green sectors has been reported in variegated leaves of Arabidopsis var2 mutant (Yoshida et al. 2008; Miura et al. 2010). Among the ROS generated in chloroplasts and mitochondria, the latter appears a more likely candidate for the trigger of mitochondrial DNA degradation, because it may readily damage mitochondrial DNA and thereby induce DNA degrading mechanism. Moreover, the observation that mitochondrial transcript level was elevated in norflurazon-treated leaves, where excess ROS generation in the chloroplasts should occur (Hedtke et al. 1999), suggest that high ROS generation in the chloroplasts may not result in mitochondrial DNA degradation. Thus, we tentatively propose that the low ROS generation in mitochondria due to low respiration rates, which ultimately result from the absence of photosynthetically functional chloroplasts, is the cause for the absence of mitochondrial DNA degradation in white-sector cells, resulting in the higher mitochondrial DNA level in white sectors than in green sectors. In this scenario, the higher mitochondrial DNA level in the white sectors is the result of lower respiration activity, which is completely different from our initial expectation that high mitochondrial DNA level in white sectors may lead to higher respiration rate there.
Variegated plants are useful resources to study the effects of the developmental and/or physiological status of plastids on mitochondria. Further analyses to examine the effects of chloroplast function on the genetic/physiological status of mitochondria, as well as those to explore the possible relationship between the changes in the affected mitochondria, are now in progress, using several representative variegated plants.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Procedure for the determination of organelle DNA levels in white sectors relative to those in green sectors. A, An example of the process used to determine relative intracellular plastid DNA level. Total cellular DNAs extracted from white and green sectors were blotted onto nylon membrane as dilution series (50, 100, and 200 ng/dot in this case), and hybridized with probes specific to plastid DNA (pt-DNA, left) or nuclear DNA (nuc-DNA, right) (upper panels). Intensities of hybridization signals for plastid DNA were measured and plotted against the amounts of blotted DNA, and regression line was drawn for each of samples. Amounts of total cellular DNA necessary to give hybridization signal with an arbitrarily set intensity (e.g., 110 in this case; selected so as to cross the both of the regression lines near middle or upper data points) was determined for green and white sector samples (186 and 110 ng, respectively), and the ratio (white/green) of plastid DNA concentrations in the sample DNA solutions was calculated (1.69). The ratio (white/green) of nuclear DNA concentrations in the same sample DNA solutions was calculated in a similar way (e.g., 0.89). Finally, the ratio (white/green) of intracellular plastid DNA levels was found by dividing the former ratio by the latter ratio (1.69/0.89 = 1.90). B, An example of the process to determine relative intracellular mitochondrial DNA level. The process is essentially the same as that described above, except that the mitochondrial-DNA specific probe was used instead of plastid-DNA specific probe. Results for Pelargonium zonale are shown as examples in A and B. For each species and each probe, hybridization was repeated three times using three different sets of DNA samples extracted from green and white sectors. (PDF 2349 kb)
Intracellular organelle DNA levels of green and white sectors. A, Intracellular plastid DNA level. B, Intracellular mitochondrial DNA level. a Schefflera arboricola; b Solenostemon scutellarioides; c Impatiens hybrids; d Hosta sp.; e Ipomea nil; f Felicia amelloides; g Hedera canariensis; h Epipremnum aureum; i Abutilon variegatum; j Fragaria × ananassa; k Pelargonium zonale; l Dracaena sanderiana. G green sector, W white sector. The values for green sectors (green bars) were always 1 due to the principal of the calculation (see legend to Fig. S1). White bars represent mean ± SE (n = 3) for the white sectors (PDF 329 kb)
Acknowledgements
The authors thank Miss Y. Sawai for her critical reading of the manuscript. This work was partly supported by Grants-in-Aid for Scientific Research on Priority Areas (No. 16085208 to A.S.) from the Japan Society for the Promotion of Science.
Conflict of interest
The authors declare that they have no conflict of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Christiansen AC, Lyznik A, Mohammed S, Elowsky CG, Elo A, Yule R, Mackenzie SA. Dual-domain, dual-targeting organellar protein presequences in Arabidopsis can use non-AUG start codons. Plant Cell. 2005;17:2805–2816. doi: 10.1105/tpc.105.035287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elo A, Lyznik A, Gonzalez ZO, Kachman SD, Mackenzie SA. Nuclear genes that encode mitochondrial proteins for DNA and RNA metabolism are clustered in the Arabidopsis genome. Plant Cell. 2003;15:1619–1631. doi: 10.1105/tpc.010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan PM, Whelan J, Millar AH, Zang Q, Smith MK, Wiskich JT, Day DA. Differential expression of the multigene family encoding the soybean mitochondrial alternative oxidase. Plant Physiol. 1997;114:455–466. doi: 10.1104/pp.114.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. Redox sensing and signalling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol Plant. 2003;119:355–364. doi: 10.1034/j.1399-3054.2003.00223.x. [DOI] [Google Scholar]
- Fujie M, Kuroiwa H, Kawano S, Mutoh S, Kuroiwa T. Behavior of organelles and their nucleoids in the shoot apical meristem during leaf development in Arabidopsis thaliana L. Planta. 1994;194:395–405. doi: 10.1007/BF00197541. [DOI] [PubMed] [Google Scholar]
- Hedtke B, Wagner I, Börner T, Hess WR. Inter-organellar crosstalk in higher plants: impaired chloroplast development affects mitochondrial gene and transcript levels. Plant J. 1999;19:635–643. doi: 10.1046/j.1365-313x.1999.00554.x. [DOI] [PubMed] [Google Scholar]
- Jarvis P. Intracellular signalling: the language of the chloroplast. Curr Biol. 2003;13:R314–R316. doi: 10.1016/S0960-9822(03)00235-5. [DOI] [PubMed] [Google Scholar]
- Kato Y, Miura E, Matsushima R, Sakamoto W. White leaf sectors in yellow variegated2 are formed by viable cells with undifferentiated plastids. Plant Physiol. 2007;144:952–960. doi: 10.1104/pp.107.099002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk JTO, Tilney-Bassett RAE. The plastids: their chemistry, structure, growth, and inheritance. Amsterdam: Elsevier/North-Holland; 1978. [Google Scholar]
- Lambers H, Chapin III FS, Pons TL (1998) Photosynthesis, Respiration, and Long-distance Transport. In: Plant physiological ecology. Springer, New York, pp 10–153
- Miura E, Kato Y, Sakamoto W. Reactive oxygen species derived from impaired quality control of photosystem II are irrelevant to plasma-membrane NADPH oxidases. Plant Signal Behav. 2010;5:264–266. doi: 10.4161/psb.5.3.10604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y, Kimura S, Saotome A, Kasai N, Sakaguchi N, Uchimiya Y, Ishibashi T, Yamamoto T, Chiku H, Sakaguchi K. Plastid DNA polymerases from higher plants, Arabidopsis thaliana. Biochem Biophys Res Commun. 2005;334:43–50. doi: 10.1016/j.bbrc.2005.06.052. [DOI] [PubMed] [Google Scholar]
- Noctor G, De Paepe R, Foyer CH. Mitochondrial redox biology and homeostasis in plants. Trends Plant Sci. 2007;12:125–134. doi: 10.1016/j.tplants.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Ogren WL. Photorespiration: pathways, regulation, and modification. Annu Rev Plant Physiol. 1984;35:415–442. doi: 10.1146/annurev.pp.35.060184.002215. [DOI] [Google Scholar]
- Ono Y, Sakai A, Takechi K, Takio S, Takusagawa M, Takano H. NtPolI-like 1 and NtPolI-like 2, bacterial DNA polymerase I homologues isolated from BY-2 cultured tobacco cells, encode DNA polymerases engaged in DNA replication in both plastids and mitochondria. Plant Cell Physiol. 2007;48:1679–1692. doi: 10.1093/pcp/pcm140. [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson A, Friedelman PE. Determination of accurate extraction and simultaneously equation for assaying chlorophyll a and b extracted with different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta. 1989;975:384–394. doi: 10.1016/S0005-2728(89)80347-0. [DOI] [Google Scholar]
- Sakai A, Suzuki T, Miyazawa Y, Kawano S, Nagata T, Kuroiwa T. Comparative analysis of plastid gene expression in tobacco chloroplasts and proplastids: relationship between transcription and transcript accumulation. Plant Cell Physiol. 1998;39:581–589. doi: 10.1093/oxfordjournals.pcp.a029408. [DOI] [Google Scholar]
- Sakamoto W, Uno Y, Zhang Q, Miura E, Kato Y, Sodmergen (2009) Arrested differentiation of proplastids into chloroplasts in variegated leaves characterized by plastid ultrastructure and nucleoid morphology. Plant Cell Physiol 50:2069–2083 [DOI] [PubMed]
- Sodmergen KS, Tano S, Kuroiwa T. Degradation of chloroplast DNA in second leaves of rice (Oryza sativa) before leaf yellowing. Protoplasma. 1991;160:89–98. doi: 10.1007/BF01539960. [DOI] [Google Scholar]
- Suzuki T, Kawano S, Sakai A, Fujie M, Kuroiwa H, Nakamura H, Kuroiwa T. Preferential mitochondrial and plastid DNA synthesis before multiple cell divisions in Nicotiana tabacum. J Cell Sci. 1992;103:831–837. [Google Scholar]
- Topping JF, Leaver CJ. Mitochondrial gene expression during wheat leaf development. Planta. 1990;182:399–407. doi: 10.1007/BF02411391. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Terashima I, Noguchi K. Up-regulation of mitochondrial alternative oxidase concomitant with chloroplast over-reduction by excess light. Plant Cell Physiol. 2007;48:606–614. doi: 10.1093/pcp/pcm033. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Watanabe C, Kato Y, Sakamoto W, Noguchi K. Influence of photo-oxidative stress on mitochondrial alternative oxidase capacity and respiratory properties: a case study with Arabidopsis yellow variegated 2. Plant Cell Physiol. 2008;49:592–603. doi: 10.1093/pcp/pcn031. [DOI] [PubMed] [Google Scholar]
- Yoshioka N, Imanishi Y, Yasuda K, Sakai A. Effects of chloroplast dysfunction in a subpopulation of leaf mesophyll cells on photosynthetic and respiratory activities of a whole leaf: a study using variegated leaves of Hedera helix L. Plant Morphol. 2009;21:87–91. doi: 10.5685/plmorphol.21.87. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Procedure for the determination of organelle DNA levels in white sectors relative to those in green sectors. A, An example of the process used to determine relative intracellular plastid DNA level. Total cellular DNAs extracted from white and green sectors were blotted onto nylon membrane as dilution series (50, 100, and 200 ng/dot in this case), and hybridized with probes specific to plastid DNA (pt-DNA, left) or nuclear DNA (nuc-DNA, right) (upper panels). Intensities of hybridization signals for plastid DNA were measured and plotted against the amounts of blotted DNA, and regression line was drawn for each of samples. Amounts of total cellular DNA necessary to give hybridization signal with an arbitrarily set intensity (e.g., 110 in this case; selected so as to cross the both of the regression lines near middle or upper data points) was determined for green and white sector samples (186 and 110 ng, respectively), and the ratio (white/green) of plastid DNA concentrations in the sample DNA solutions was calculated (1.69). The ratio (white/green) of nuclear DNA concentrations in the same sample DNA solutions was calculated in a similar way (e.g., 0.89). Finally, the ratio (white/green) of intracellular plastid DNA levels was found by dividing the former ratio by the latter ratio (1.69/0.89 = 1.90). B, An example of the process to determine relative intracellular mitochondrial DNA level. The process is essentially the same as that described above, except that the mitochondrial-DNA specific probe was used instead of plastid-DNA specific probe. Results for Pelargonium zonale are shown as examples in A and B. For each species and each probe, hybridization was repeated three times using three different sets of DNA samples extracted from green and white sectors. (PDF 2349 kb)
Intracellular organelle DNA levels of green and white sectors. A, Intracellular plastid DNA level. B, Intracellular mitochondrial DNA level. a Schefflera arboricola; b Solenostemon scutellarioides; c Impatiens hybrids; d Hosta sp.; e Ipomea nil; f Felicia amelloides; g Hedera canariensis; h Epipremnum aureum; i Abutilon variegatum; j Fragaria × ananassa; k Pelargonium zonale; l Dracaena sanderiana. G green sector, W white sector. The values for green sectors (green bars) were always 1 due to the principal of the calculation (see legend to Fig. S1). White bars represent mean ± SE (n = 3) for the white sectors (PDF 329 kb)


