Abstract
Our knowledge of the O-glycoproteome [N-acetylgalactosamine (GalNAc) type] is highly limited. The O-glycoproteome is differentially regulated in cells by dynamic expression of a subset of 20 polypeptide GalNAc-transferases (GalNAc-Ts), and methods to identify important functions of individual GalNAc-Ts are largely unavailable. We recently introduced SimpleCells, i.e., human cell lines made deficient in O-glycan extension by zinc finger nuclease targeting of a key gene in O-glycan elongation (Cosmc), which allows for proteome-wide discovery of O-glycoproteins. Here we have extended the SimpleCell concept to include proteome-wide discovery of unique functions of individual GalNAc-Ts. We used the GalNAc-T2 isoform implicated in dyslipidemia and the human HepG2 liver cell line to demonstrate unique functions of this isoform. We confirm that GalNAc-T2–directed site-specific O-glycosylation inhibits proprotein activation of the lipase inhibitor ANGPTL3 in HepG2 cells and further identify eight O-glycoproteins exclusively glycosylated by T2 of which one, ApoC-III, is implicated in dyslipidemia. Our study supports an essential role for GalNAc-T2 in lipid metabolism, provides serum biomarkers for GalNAc-T2 enzyme function, and validates the use of GALNT gene targeting with SimpleCells for broad discovery of disease-causing deficiencies in O-glycosylation. The presented glycoengineering strategy opens the way for proteome-wide discovery of functions of GalNAc-T isoforms and their role in congenital diseases and disorders.
Keywords: apolipoproteins, angiopoietin-like proteins, genetic engineering, glycoproteins
The GALNT2 gene involved in O-glycosylation has been associated with aberrant serum levels of triglyceride (TG) and high-density lipoprotein cholesterol (HDL-C) by several genome-wide association studies (GWAS) (1, 2). A direct functional role of this gene was obtained by transient knockdown and overexpression of galnt2 in mouse liver, which resulted in increased and lowered plasma HDL-C, respectively (3). GALNT2 is a member of the largest family of homologous glycosyltransferase isoforms (up to 20) catalyzing the same glycosidic linkage (GalNAcα1-O-Ser/Thr) and initiating protein O-glycosylation (4, 5). These isoforms have overlapping but distinct peptide substrate specificities and identifying essential unique functions for individual GalNAc-T isoforms has been notoriously difficult. In search of putative functions of GALNT2 in lipid metabolism, we previously screened a number of known and predicted O-glycoproteins with roles in lipid metabolism for O-glycosylation by the encoded GalNAc-T2 enzyme and identified ANGPTL3 (6). We found that site-specific O-glycosylation of a Thr residue adjacent to the proprotein convertase (PC) processing site in ANGPTL3 that activates this lipase inhibitor was performed only by the GalNAc-T2 isoform. More recently, a heterozygous mutation in GALNT2 resulting in slightly reduced kinetic properties of the encoded GalNAc-T2 enzyme was found in two probands with elevated HDL and reduced TG (7). Serum ApoC-III in these individuals also showed a slight increase in the nonglycosylated ApoC-III variant.
GalNAc-type (mucin-type) O-glycosylation is arguably the most abundant and complex form of protein glycosylation, and yet the least understood with respect to site of attachments in proteins (5, 8). Site-specific O-glycosylation is an important regulator of protein function, with the most recently discovered function being coregulation of proprotein convertase processing of proteins (6, 9). Compared with other types of protein glycosylation, GalNAc-type O-glycosylation is unique with its 20 distinct isoenzymes (5, 8). This allows for an incomparable level of cell- and protein-specific, as well as dynamic, regulation of site-directed glycosylation, which can be essential for development and health (8–10). However, technical limitations hamper our understanding of the O-glycoproteome and functions of site-specific O-glycosylation.
We recently developed an O-glycoproteomic strategy to identify O-glycosylation sites using zinc finger nuclease (ZFN) glycoengineered human cell lines (11). ZFN targeting of the O-glycan elongation pathway results in stable “SimpleCell” lines with homogenous truncated O-glycosylation with GalNAcα (Tn) or NeuAcα2–6GalNAcα (STn) O-glycans. This method allows for straightforward isolation and identification of GalNAc O-glycopeptides from total cell lysates or secretions using lectin chromatography and nanoflow liquid chromatography tandem mass spectrometry (nLC/MS/MS) with electron transfer dissociation (ETD) for glycan site specification (Fig. 1A). Using this strategy, we have identified hundreds of unique O-glycoproteins and O-glycan sites.
Fig. 1.
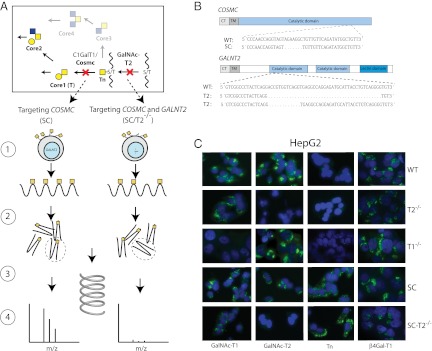
Probing isoform-specific functions of GalNAc-Ts by ZFN gene targeting glycosyltransferases. (A) Depiction of the strategy for identification of differential GalNAc-T isoform-specific O-glycoproteomes. (Section 1) ZFN targeting of COSMC in HepG2 (SC) generates cell lines with homogenous and truncated O-glycosylation (GalNAc), and subsequent targeting of GALNT2 (SC/T2−/−) eliminates GalNAc-T2 nonredundant O-glycosylation allowing for comparative analysis of O-glycoproteomes of isogenic cell lines with and without GALNT2. (Section 2) Proteins from total cell lysates or cell culture supernatant from HepG2-SC or SC/T2−/− are digested by trypsin. (Section 3) GalNAc-glycopeptides are isolated and separated by lectin weak affinity chromatography. (Section 4) O-glycosylation sites are identified by nLC/MS/MS and comparison between the two cell lines produce candidates for GalNAc-T2–specific contribution. (B) Depiction of Cosmc and GalNAc-T2 protein domains and targeted DNA sequences where introduced mutations are shown. (C) Immunofluorescence staining of isogenic HepG2 cell lines as indicated.
Here, we have extended the strategy to include differential analysis of the function of a single GalNAc-T isoform, GalNAc-T2, in the human HepG2 liver cell line. We generated isogenic cell models with and without GalNAc-T2 and identified nonredundant functions of this isoform in HepG2, including glycosylation of ApoC-III and ANGPTL3. The presented strategy now opens the way for proteome-wide discovery of site-specific O-glycosylation events controlled by distinct GalNAc-T isoforms.
Results
Generation of a HepG2 SimpleCell Line Deficient in GALNT2.
We used the epithelial hepatocellular carcinoma cell line HepG2 to sequentially knock out COSMC and GALNT2. We first targeted COSMC [HepG2-SC (clones 2G4 and 3B5)] followed by GALNT2 [HepG2-SC/T2−/− (clones 2G4-1F3 and 2G4-3G11)]. We also targeted GALNT2 in wild-type HepG2 cells [HepG2-T2−/− (clones 2C4 and 2G2)] (Fig. 1A). As controls, we generated HepG2 cell lines with knockout of the other common GalNAc-T expressed in the liver, GALNT1 [HepG2-SC/T1−/− (clones 2G4-2G9 and 2G4-2E11) and HepG2-T1−/− (clones 1F5 and 1F3)]. Furthermore, to confirm that effects observed in GALNT2 knockout clones were specific, we reintroduced GALNT2 under control of the AAVS1 promoter at the AAV1 safe harbor site using ZFN targeting in HepG2-T2−/− [HepG2-T2−/−/T2+/+ (clone 2C4-4B10)]. The O-glycan structures produced in HepG2 are mainly ST (Neu5Acα2–6Galβ1–3GalNAcα1-O-Ser/Thr) and very little if any STn (Neu5Acα2–6GalNAcα1-O-Ser/Thr), so COSMC knockout cells were selected by anti-Tn reactivity. DNA from selected clones was sequenced to confirm mutations (Fig. 1B). We also monitored glycosyltransferase expression in HepG2 wild-type and knockout clones with our panel of monoclonal antibodies to GalNAc-Ts (T1, T2, T3, T4, T6, T11, T12, and T14), sialyltransferases (ST3Gal-I and ST6GalNAc-I), and a galactosyltransferase (β4Gal-T1), and observed no changes in expression except for loss of GalNAc-T2 (Fig. 1C). Thus, no evidence of compensatory regulation was found.
Using Differential O-Glycoproteomics to Define Isoform-Specific Functions of GalNAc-Ts.
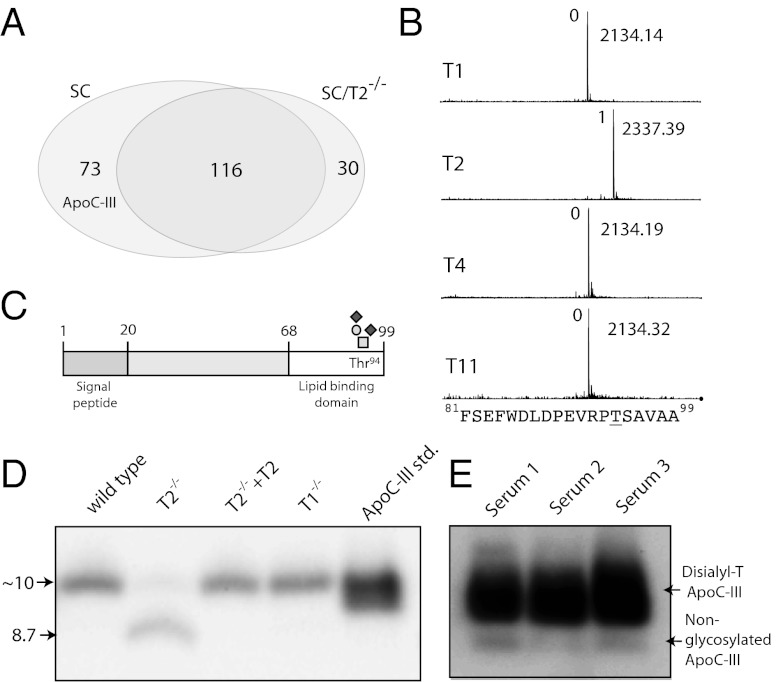
We used our previously reported SimpleCell glycoproteomics strategy to isolate and sequence GalNAc glycopeptides from tryptic digests by lectin weak-affinity chromatography (LWAC) on immobilized Vicia villosa agglutinin (VVA) lectin followed by nLC/MS/MS analysis (11). The protocol for analysis of secreted glycoproteins was modified to include an initial enrichment step by chromatography on a short VVA column to reduce volume and irrelevant protein content before trypsin digestion. We identified 219 O-glycosylation sites in total, of which 73 were found only in HepG2-SC, whereas 30 were identified only in HepG2-SC/T2−/− (Fig. 2A and Table S1). Compared with our previous study (11), which included in total three SimpleCell and two wild-type cell lines, the HepG2 SimpleCells yielded an additional 81 O-glycopeptides, identifying 45 additional O-glycoproteins. The secretome contained more glycopeptides derived from predicted secreted (∼75%) compared with predicted intracellular or membrane-bound proteins (∼25%), whereas the opposite was found for the total lysate (∼35% compared with ∼65%), as assessed by ontology terms of annotated proteins in UniProt.
Fig. 2.
GalNAc-T2 controls O-glycosylation of ApoC-III in HepG2 cells. (A) Venn diagram illustrating distribution of identified O-glycoproteins. ApoC-III was identified in HepG2-SC and SC/T2−/− secretomes and therefore selected for analysis by GalNAc-T enzyme assays. (B) In vitro glycosylation of an ApoC-III peptide substrate as indicated with GalNAc-T1, T2, T4, and T11 monitored by MALDI-TOF analysis (2-h time point shown). Masses of peptides and glycopeptides are shown with the predicted number of incorporated GalNAc residues indicated Left of the peaks. (C) Schematic drawing of ApoC-III indicating the position of O-glycosylated Thr94. (D) Immunoprecipitation of ApoC-III from cell culture media of HepG2 wild-type cells, HepG2-T2−/−, HepG2-T2−/−/T2+/+, HepG2-T1−/−, and 50 ng ApoC-III standard from human serum. SDS/PAGE Western blot with anti–ApoC-III. (E) SDS/PAGE Western blot of 1 μL human serum from three healthy blood donors with anti–ApoC-III. Migration of glycosylated and nonglycosylated ApoC-III is indicated with arrows.
The present proteomic analysis, although not comprehensive, was more than sufficient to quickly identify a significant number of candidate GalNAc-T isoform-specific O-glycan sites. Thus, the 73 sites identified only in HepG2-SC and not HepG2-SC/T2−/− are clearly potential isoform-specific substrates for GalNAc-T2 in HepG2. To further explore these sites, we synthesized representative peptide substrates for these sites and tested in vitro glycosylation using the most common GalNAc-Ts expressed in HepG2 (Table 1 and Fig. S1).
Table 1.
Summary of O-glycopeptides identified in HepG2-SC cells and in vitro GalNAc-T enzyme analysis
| Protein name | Sequence† | T1‡ | T2 | T4 | T11 | Sites in T2+§ | Sites in T2− | |
| 16 | AFP | 122 HNCFLAHKKPTPASIPLFQV 141 | −¶ | 1* | − | − | 1 | 0 |
| AHSG | 337 RKTRTVVQPSVGAAAGPVVP 356 | − | 1* | NA | NA | 1 | 0 | |
| APP | 361 PVKLPTTAASTPDAVDK 377 | − | 1 | 1 | − | 1 | 0 | |
| APOC2 | 36 FLTQVKESLSSYWESAKTAA 55 | − | − | − | − | 1 | 0 | |
| APOC3 | 81 FSEFWDLDPEVRPTSAVAA 99 | − | 1* | − | − | 1 | 0 | |
| CRLF1 | 324 AGIWSEWSHPTAASTPRSER 343 | 2–3 | 1–2 | − | − | 1 | 0 | |
| DKK1 | 50 GGAAGHPGSAVSAAPGILYP 69 | 1 | 1 | 1 | 1 | 1 | 0 | |
| IGFBP6 | 117 PKESKPQAGTARPQDVNRRD 136 | 1 | 1 | − | − | 1 | 0 | |
| ITIH2 | 591 QLLAERSLAPTAAAKRRITR 610 | − | 1* | − | − | 1 | 0 | |
| LRP1 I | 3723 GDGTDEEDCEPPTAHTTHCK 3742 | − | − | − | − | 1 | 0 | |
| MATN3 | 43 GPGGSPGRRPSPAAPDGAPA 62 | − | 1* | − | − | 2 | 0 | |
| NPTXR | 72 PALPGAPAASAHPLPPGPLF 91 | − | 1* | − | − | 2 | 0 | |
| OS9 | 521 VVPKKPPPSPQPTEEDPEHR 540 | − | 1–2* | − | − | 1 | 0 | |
| RELN | 174 AQQLCEQGAPTDVTVHPHLA 193 | 1 | 1 | 1 | 1 | 1 | 0 | |
| ROBO1 | 650 DPVKTQDVLPTSQGVDHKQV 669 | − | 1* | − | − | 1 | 0 | |
| STC2 | 241 EAGHHLPEPSSRETGRGAKG 260 | − | − | − | − | 1 | 0 | |
| 10 | APOE | 301 VQAAVGTSAAPVPSDNH 317 | − | 1–2 | 1–2 | − | 2 | 2 |
| DLK1 | 251 SPQQVTRLPSGYGLAYRLTP 270 | 1 | − | NA | 1 | 1 | 1 | |
| GPC6 | 365 RPYNPEERPTTAAGTSLDRL 384 | 1 | 1 | NA | − | 1 | 1 | |
| LRP1 II | 3527 PKEECDERTCEPYQFRCKNN 3546 | − | − | − | − | 1 | 1 | |
| NID1 | 304 GLEDVGTTPFSYKALRRGGA 323 | 1 | 1 | NA | − | 2 | 2 | |
| NUCB1 | 32 GAPNKEETPATESPDTGLYY 51 | 1 | − | 1 | − | 1 | 1 | |
| PLTP | 476 RPADVRASTAPTPSTAAV 493 | 2–3 | 1–2 | NA | NA | 3 | 3 | |
| SDC2 | 91 LNIQNKIPAQTKSPEETDKE 110 | 1 | − | NA | − | 1 | 1 | |
| SDC4 | 90 PERAGSGSQVPTEPKKLEEN 109 | − | − | − | − | 1 | 1 | |
| ST6GalI | 66 SSTQDPHRGRQTLGSLRGLA 85 | 1–3 | − | 1 | − | 3 | 3 | |
| 5 | APOA2 | 45 GKDLMEKVKSPELQAEAKSY 64 | − | − | − | − | 0 | 1 |
| APOF | 262 DQKDANISQPETTKEGLRAI 281 | 1 | 1 | − | − | 0 | 1 | |
| CX3CL1 | 188 VPPVSTAATWQSSAPHQPGP 207 | 1–2 | 1–2 | 1–2 | − | 0 | 4 | |
| IGFBP1 | 159 DGSKALHVTNIKKWKEPCRI 178 | − | − | − | − | 0 | 1 | |
| PRAP1 | 64 PVQKPKLLTTEEKPRGQGRG 83 | − | − | − | − | 0 | 2 |
NA, not analyzed.
*T2-specific sites.
†Sequence of synthetic peptide designed to cover identified O-glycosylation site(s) (underlined).
‡In vitro glycosylation of synthetic peptides using GalNAc-T1, -T2, -T4, and -T11.
§Number of sites identified in glycopeptides from HepG2 SC (T2+) or HepG2 SC/GALNT2−/− (T2−). 0 (glycopeptide not identified), 1, 2, or 3 [1, 2, or 3 site(s) identified].
¶Number of GalNAc residues added to the peptide substrate. (-) no GalNAc added.
Among the peptide substrates identified only in HepG2-SC and not in HepG2-SC/T2−/− (16 in total), 8 peptides were glycosylated exclusively by GalNAc-T2, 5 were glycosylated by T2 and several other isoforms, and 3 were not glycosylated by any isoform tested as summarized in Table 1. Among peptide substrates identified in both SC and SC/T2−/− (10 in total), 8 were glycosylated by several isoforms, and 2 were not glycosylated by any isoform tested. We also included a selection of glycopeptides identified only in SC/T2−/− (5 in total). Among the corresponding synthetic peptides, two were glycosylated by several isoforms and three were not glycosylated in vitro. These results demonstrate nonredundant isoform-specific functions of GalNAc-T2 and provide documentation for a good correlation between in vitro (test tube) and ex vivo enzyme specificity determinations as previously discussed (5, 12, 13).
Among the identified O-glycoproteins several are linked to HDL and/or TG metabolism; however only one of these, ApoC-III, was exclusively glycosylated by GalNAc-T2. ApoC-III has previously been identified as a serum biomarker of deficiency in O-glycan sialylation (14), and a recent study has provided evidence for slightly altered efficiency of O-glycosylation of ApoC-III in individuals with a heterozygous missense mutation in GALNT2, which results in a twofold increase in the apparent Km of the encoded enzyme (7). We also identified glycopeptides in ApoC-II and the LDL-receptor related protein-1 (LRP1), but the peptides used for in vitro glycosylation did not serve as substrate for any of the tested GalNAc-T isoforms. Because we were unable to identify the actual site of glycosylation, it is possible that the design of the peptide prevents in vitro glycosylation.
A GalNAc-T2–specific site was identified in α-2-HS glycoprotein (AHSG) just C-terminal to the connecting peptide processing site RKTR↓KVVQPS346V. Furthermore, we identified an additional unique O-glycosylation site in the central region of amyloid βA4 (APP) (PTTAAST371PD, Ser370, or Thr371) specifically glycosylated by GalNAc-T2, whereby we increase the number of identified sites of O-glycosylation in APP to nine (11, 15, 16). Thr529 in OS9, a lectin required for endoplasmic reticulum-associated degradation, and Thr660 of ROBO1, a potential serum biomarker for hepatocellular carcinomas, were also specific substrates for GalNAc-T2 (17).
GalNAc-T2 Controls O-Glycosylation of ApoC-III in HepG2 Liver Cells.
ApoC-III is a known serum O-glycoprotein with one sialylated Core1 (T) O-glycan attached to Thr94 in the C-terminal lipid-binding domain (18). Analysis of secreted ApoC-III from HepG2 wild-type and HepG2-T2−/− showed that ApoC-III secreted from HepG2-T2−/− migrated ∼1 kDa lower than the secreted form from wild-type cells (Fig. 2D). Moreover, when we reintroduced GalNAc-T2 into HepG2-T2−/− cells, the migration was restored, unambiguously establishing that GalNAc-T2 is required for O-glycosylation of ApoC-III in HepG2 cells. Knockout of another GalNAc-T isoform, GalNAc-T1 (HepG2-T1−/−), did not affect migration of ApoC-III. These experiments also lend support for the validity of the strategy.
ANGPTL3 Is Not Glycosylated and Processed in HepG2-T2−/−.
ANGPTL3 is a specific substrate of GalNAc-T2 and site-specific O-glycosylation by GalNAc-T2 modulates PC processing and activation (6). We did not identify glycopeptides from ANGPTL3 in HepG2-SC, and interestingly ANGPTL3 was also not identified in a recent deep proteomic analysis of HepG2 cells (19). This may be due to the low amounts of ANGPTL3 normally produced in HepG2 (less than 5 ng/mL). Nevertheless, we were able to immunoprecipitate ANGPTL3 from the medium of HepG2 and HepG2-T2−/− cells and show, in agreement with our previous studies, that ANGPTL3 was intact in wild-type cells but processed in the T2-deficient cells (Fig. 3). This unequivocally confirms that GalNAc-T2–directed site-specific O-glycosylation inhibits cleavage and activation of this important lipase inhibitor and further illustrates utility of isogenic wild-type cells with deficiencies in individual GalNAc-T isoforms.
Fig. 3.
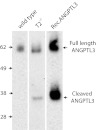
Increased PC processing of ANGPTL3 in HepG2-T2−/− cells. (A) Immunoprecipitation of ANGPTL3 from HepG2 wild-type and T2−/− cells induced with 5 μM T0901317 for 48 h. SDS/PAGE Western blot with anti-ANGPTL3. Control lane includes 20 ng recombinant nonglycosylated ANGPTL3. Full-length ANGPTL3 migrates to ∼62 kDa and cleaved N-terminal ANGPTL3 migrates to ∼38 kDa as indicated by arrows.
Discussion
The O-glycoproteome, and in particular the contribution of individual polypeptide GalNAc-Ts to its makeup, has long remained elusive. The GalNAc-type O-glycoproteome is the most complex regulated glycoproteome with the 20 GalNAc-Ts controlling sites of O-glycan attachments on proteins (5). It is our hypothesis that differential regulation of the expression of GalNAc-Ts in cells provides for a variable and dynamic O-glycoproteome, which has specific biological impact for protein functions such as serving as coregulator of PC processing (20). The major obstacle to be overcome to gain insight into this glycoproteome is the technical difficulty in identifying and monitoring site-specific O-glycosylation on a proteome-wide basis. Our previously introduced SimpleCell strategy has enabled proteome-wide discovery of O-glycoproteins with sites of O-glycan attachments (11), and as shown here, combining the SimpleCell strategy with targeted knockout and knockin of individual GalNAc-Ts, now allows for broad ex vivo discovery of GalNAc-T isoform-specific functions. The strategy was applied to the GALNT2 isoform implicated in lipid metabolism, and a number of unique functions of this enzyme were identified. Among these was ApoC-III, which is an inhibitor of LPL-mediated hydrolysis of triglycerides in very (V)LDL and chylomicrons (21, 22), providing another potential link between GALNT2 and dysregulated HDL and TG in addition to ANGPTL3 (6). A number of GALNT genes have been associated by GWAS and other studies with diseases and specific biological functions (5), but methods to uncover specific molecular mechanisms for these interesting associations have not existed. The ZFN gene targeting strategy presented here should be widely applicable to all GalNAc-Ts, and it provides a major leap in our understanding of site-specific functions of O-glycosylation.
Our strategy involves isogenic cell lines at two levels: (i) GalNAc-T knockout/knockin in SimpleCells to define GalNAc-T isoform nonredundant contributions to the glycoproteome; and (ii) parallel knockout/knockin in corresponding wild-type cells to decipher nonredundant biological functions. The isogenic SimpleCell lines with and without GalNAc-Ts are thus primarily used to define sites of O-glycosylation, but these cells may also provide clues to biological functions if the initial GalNAc O-glycosylation without O-glycan extension is sufficient to conserve the effect of glycosylation on target proteins. In many cases, however, extended, branched, and/or terminally modified O-glycan structures are required for the biological functions (23), in which case comparative studies in isogenic wild-type cells with and without specific GalNAc-Ts are needed. The isogenic cell systems are stable and can be used for comprehensive proteome, transcriptome, and more in-depth cell biology analyses to identify biological pathways affected by loss of a single GalNAc-T. This is an important feature considering a number of exciting, albeit often conflicting, proposed roles of GalNAc-Ts.
Thus, specific GALNTs have been implicated in TGFβ signaling (24), regulation of sensitivity to TNF-related apoptosis-inducing ligand (TRAIL)-induced apoptosis in colonic cancer cells lines (25), regulation of cell surface expression of MUC1 in breast cancer (26), and function of fibronectin in the epithelial–mesenchymal transition (EMT) process (27, 28). Although these studies are intriguing, they mainly rely on RNA silencing strategies, which have inherent problems with glycosyltransferases, where complete and specific knockout is required. Furthermore, the specific glycosylation events underlying the biological functions were not elucidated. Introducing ZFN engineered isogenic cell models and SimpleCells now allows for analysis of phenotypic and functional consequences of single GalNAc-T isoforms.
GALNTs appear to play driver roles in malignancies, and stable isogenic cancer cell models with deficiencies in GALNTs have the potential to uncover functions of site-specific O-glycosylation in carcinogenesis. The repertoire of enzyme isoforms expressed in cancer cells is markedly altered from the normal counterparts (29–34); however, the consequences of these changes for the O-glycoproteome and carcinogenesis remain unknown. The locus 9q22 including the GALNT12 gene is a susceptibility locus for colorectal cancer (35) and heterozygous germ line as well as somatic inactivating mutations have been identified in cancer patients (36). GALNT12 is thus a prime candidate for the proposed strategy.
We previously developed LWAC for isolation of Tn glycopeptides released from total cell lysates. Here we further developed a two-step VVA lectin strategy to isolate secreted GalNAc glycoproteins in culture medium with serum followed by digestion and isolation of GalNAc glycopeptides. This strategy provides a unique opportunity to identify secreted O-glycoproteins from different human cells that may have potential for serving as serum biomarkers such as ApoC-III. We did not identify ANGPTL3 in the proteomic strategy used, but the isogenic wild-type cell systems still allowed us to confirm that activation of this inhibitor of lipases that also affects HDL and TG is inhibited by GalNAc-T2 O-glycosylation (Fig. 3). We are currently developing strategies to improve the coverage of the GalNAc glycoproteome using different endoproteases and isoelectric focusing.
An important finding was a high degree of correlation between ex vivo substrate specificity of GalNAc-Ts and the substrate specificity determined by in vitro assays (Table 1). Although expected on the basis of past studies with a few select sequences (9, 37, 38), this hypothesis has been validated on a proteome-wide scale. GalNAc-Ts function in Golgi where their protein substrates are folded. It is thus not empirically logical that the substrate specificities of GalNAc-Ts would be characterized by a short linear sequence motif (5, 39). The two POFUTs initiating protein O-fucosylation have strict substrate specificities for the correctly folded EGF repeat domain and do not function with short synthetic peptides (40). This restricted specificity may be because O-fucosylation occurs in ER at the same site as folding, allowing for differential glycosylation of Notch and Thrombospondin EGF repeats by the two isoenzymes (41). The 20 GalNAc-Ts offer a much more complex scenario, and this may be one reason for topological separation from ER in contrast to the initiation step of most other types of protein glycosylation (5). Interestingly, however, Bard and colleagues have found that the GalNAc-Ts and the initiation step of O-glycosylation can be selectively relocated to ER by activation of Src (42). It is conceivable that this induces substantial changes in the O-glycoproteome and perhaps alterations in other O-glycoproteomes by competition in ER (8).
In summary, we present a unique strategy to identify nonredundant O-glycosylation controlled by individual polypeptide GalNAc-T isoforms and to uncover site-specific biological functions hereof. Given that there is increasingly more evidence, from GWAS and other studies, demonstrating important biological functions of the large GalNAc-T gene family, it is becoming even more urgent to uncover the underlying molecular mechanisms as well as identify biological pathways affected. The isogenic cell models presented opens the way for such studies and the strategies applied are generally applicable to other biological pathways.
Materials and Methods
ZFN Gene Targeting.
ZFN targeting constructs for COSMC and GALNT2 were custom produced (Sigma-Aldrich) with the following binding sites. Cutting sites are indicated in parentheses: COSMC 5′-CCCAACCAGGTAGT(AGAAGGCT)GTTGTTCAGATATGGCTGTT-3′; and GALNT2 5′-GTCGGCCCTACTCAGGAC(CGTGGT)CAGGTGAGGCCAGGAGAT-3′. For stable integration of GALNT2, a ZFN integration vector was designed for insertion into the adeno-associated virus integration site 1 (AAVS1) locus. Expression was driven by the endogenous AAVS1 promoter as previously described (43), by including a splice acceptor and a 2A “self-cleaving” peptide directly 5′ to the full GALNT2 ORF. A polyA signal sequence was appended 3′ to the ORF sequence, and all of the aforementioned sequences were flanked by left and right arms of homology from the pZDonor-AAVS1 puromycin vector (Sigma-Aldrich) at the 5′ and 3′ ends, respectively. Finally, the above synthetic gene was inserted into the pZDonor vector (Sigma-Aldrich) via Blpl–Pfol restriction enzyme digestion, thus generating the pAAVS1-Sig2a-GALNT2-ZFN integration (pAAVS1-T2) vector.
HepG2 cells (kind gift from Novo Nordisk) were transfected with 1 vial of mRNA (Sigma-Aldrich) or 5 μg of endotoxin free pAAVS1-T2 plasmid DNA for knockin experiments (mixed with 5 μL of pAAVS1 ZFN mRNA (Sigma-Aldrich) using nucleofection on an Amaxa Nucleofector (Lonza). Cells were cloned by limited dilution, trypsinized and fixed in ice-cold acetone on teflon-coated slides and stained with monoclonal antibodies (MAbs) to Tn (5F4), GalNAc-T2 (UH4 and 2E10), GalNAc-T1 (UH3), or C1GalT (5B6) (11). COSMC knockout clones (SimpleCells) were selected by reactivity with MAbs to Tn and loss of reactivity with MAb to the C1GalT1 enzyme, and clones were confirmed to have COSMC mutations using PCR and sequencing using primers 5′-AGGGAGGGATGATTTGGAAG-3′ and 5′-TTGTCAGAACCATTTGGAGGT-3′. GALNT2 and GALNT1 knockout clones were selected for loss of reactivity with MAbs to GalNAc-T2 and -T1, and mutations confirmed by PCR using primers GALNT2PZFN F/R (5′-CCATCCCAGTTGGTCAGTCT-3′/5′-CTGTGCTGAGCAGTCAGGAG-3′) or GALNT1PZFN F/R (5′-GAATAGTGCCAGGCCACACT-3′/5′-AAAGCAAACTTGGGAGGAAAT-3′) and sequencing. GALNT2 knockin clones were screened for reactivity with MAbs to GalNAc-T2. For immunocytochemistry, HepG2 cells were grown on sterile Diagnostic Imaging printed slides (Clearcell, Histolab) and fixed with acetone. Cells were then sequentially incubated with MAbs overnight and FITC-conjugated rabbit antibody to mouse Ig (Dako) for 45 min and mounted with ProLong Gold antifade reagent (DAPI) (Invitrogen). Fluorescence microscopy was performed using a Zeiss Axioskop 2 plus with an AxioCam MR3. Bit depth and pixel dimensions were 36 bit and 1,388 × 1,040 pixels, respectively.
LWAC Isolation of Tn O-Glycopeptides.
A total of 100 mL of cell culture supernatant (secretome) or 0.5 mL packed cells (total cell lysate) was harvested from HepG2 COSMC knockout and HepG2 COSMC/GALNT2 double knockout. Secretome: Cell culture supernatants were cleared by centrifugation (2,500 g for 10 min), dialyzed (molecular weight cutoff, 3,500 Da) twice against 5 L 1 mM Tris-HCl, pH 7.4, filtered (0.45 μm), diluted in 100 mL buffer A (10 mM Tris, pH 8, 150 mM NaCl, 1 mM CaCl2/MgCl2/MnCl2/ZnCl2, 1 M urea), and subjected to the first Vicia villosa agglutinin (VVA) lectin chromatography for enrichment of glycoproteins. The VVA agarose (Vector Laboratories) (0.8 mL in 2-mL syringe) was equilibrated in buffer A, the sample loaded twice followed by 10–20 column volumes (CV) wash in buffer A, and enriched Tn glycoproteins eluted with 2 × 1 mL 0.2 M GalNAc. The eluate was dialyzed against 2 × 5 L 50 mM ammonium bicarbonate, lyophilized, dissolved in 0.5 mL 50 mM ammonium bicarbonate, and further processed as cell lysates for digestion and isolation of Tn glycopeptides. Total cell lysates: Packed cells were lysed in 0.1% RapiGest (Waters) in 50 mM ammonium bicarbonate with a sonic probe and the solution cleared by centrifugation (1,000 g for 10 min). The cleared lysate and secretome samples were heated for 10 min at 80 °C, followed by reduction (5 mM DTT, 60 °C, 0.5 h) and alkylation (10 mM iodoacetamide, RT, 30 min), and digestion with trypsin (25 μg) (Roche) [37 °C, overnight (ON)]. Digests were treated with TFA (6 μL, 37 °C, 20 min), cleared by centrifugation, purified on C18 Sep-Pak (Waters), lyophilized, resuspended in 1 mL buffer A, and injected to a preequilibrated 2.6 m long VVA column (packed in PFA tubing 1/16 inch × 50 feet, flow 100 μL min−1) for isolation of glycopeptides (11). The column was washed with 0.4 M glucose in buffer A, eluted with 0.2 M GalNAc (2 CV) and 0.4 M GalNAc (1 CV), and glycopeptide fractions purified by Stage Tips (Thermo Scientific) for analysis. A detailed step-by-step protocol is published elsewhere (44).
nLC/MS/MS Analysis.
Liquid chromatography–tandem mass spectrometry was performed on a system composed of an EASY-nLC II (Thermo Fisher Scientific) interfaced via a nanoSpray Flex ion source to an LTQ-Orbitrap XL hybrid spectrometer (Thermo Fisher Scientific), equipped for both higher-energy C-trap dissociation (HCD)- and ETD-MS2 modes, enabling peptide sequence analysis without and with retention of glycan site-specific fragments, respectively. The conditions of LC analysis were essentially as described previously (11), except that the nLC was operated using a single analytical column set up (polar end-capped C18-silica; 10 cm length, 75 μm inner diameter and 3-μm particle size).
A data-dependent mass spectral acquisition routine, HCD triggering of subsequent ETD scan, was used for all runs. Briefly, a precursor MS1 scan (m/z 350–1,700) of intact peptides was acquired in the Orbitrap at a resolution setting of 30,000, followed by Orbitrap HCD-MS2 (m/z of 100–2,000) of the three most abundant multiply charged precursors above 5,000 counts in the MS1 spectrum; the appearance of a HexNAc fragment at m/z 204.086 (in practice a ± m/z 0.15 window was used) in the HCD-MS2 spectrum triggered a subsequent ETD-MS2 from the same precursor with a resolution setting of 15,000 (11).
Data Analysis.
Data processing was carried out using Proteome Discoverer 1.2 software (Thermo Fisher Scientific) as previously described (11), with only small changes in preprocessing and processing procedures (e.g., in the HexNAc subtraction routine for correctly interpreting HCD spectra, exact masses of 1×, 2×, 3×, and 4× HexNAc units were subtracted from the corresponding precursor ion mass, generating four distinct files, instead of only up to 3 HexNAc units). All candidate-matched glycopeptides associated with each protein were validated by inspection. The results obtained were accumulated in a single list (Table S1; raw data related to this paper can be downloaded from ProteomeCommons.org using the following tranche hash: JQoOtUklwXlG7eEiT9B9Avxb8yloxZj7dqeq62goVARNygxG4Pp8jnudiFZavgFT6OMpK6JkAqY4BlcsDKNhAJE1mGIAAAAAAABFyQ==).
Glycosyltransferase Assays.
All recombinant glycosyltransferases were expressed as soluble secreted truncated proteins in insect cells (45). Screening assays for GalNAc-T glycosylation of peptides (Schafer-N; NeoBioSci) were performed as product development assays in 25 μL of 25 mM cacodylic acid sodium, pH 7.4, 10 mM MnCl2, 0.25% Triton X-100, 1.5 mM UDP-GalNAc (Sigma-Aldrich), 10 μg of acceptor peptides, and 0.2–0.5 μg of purified enzyme.
Immunoprecipitation.
Protein A beads (cat. no. sc-2001, Santa Cruz) were washed in PBS and IP buffer (20 mM Tris-base, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, protease inhibitor mixture; Roche). Beads were preincubated with polyclonal rabbit antihuman ApoC-III (4 μg) (Abcam; ab21032) or rabbit antihuman ANGPTL3 1:2,000 (Alexis Biochemical; 210448) for 1.5 h at room temperature. The beads were washed and incubated with conditioned media harvested from HepG2 wild-type and mutant cells overnight at 4 °C. ANGPTL3 expression was induced with 5 μM T0901317 (LXR agonist) (Sigma-Aldrich) for 48 h before harvest of conditioned media. After a thorough wash, ApoC-III or ANGPTL3 was eluted in LDS loading buffer (NuPAGE; Invitrogen) and 5 mM DTT and beads were pelleted by centrifugation. Sample proteins were separated in 10 or 12% NuPAGE Novex Bis-Tris in MES running buffer and blotted onto nitrocellulose membranes. Membranes were blocked in 5% (wt/vol) dry milk for 40 min and incubated in primary antibodies diluted in Tris-buffered saline (TBS) plus 0.05% Tween 20 (polyclonal rabbit antihuman ApoC-III 1:3,000 or biotinylated sheep antihuman ANGPTL3 1:2,000; R&D Systems; BAF 3829) overnight at 4 °C. After wash, membranes were incubated in secondary antibodies (antirabbit-HRP 1:3,000 (Dako) and streptavidin-HRP (P0397) 1:3,000 and developed using ECL (Pierce).
Supplementary Material
Acknowledgments
This work was supported by Kirsten and Freddy Johansens Fond, A. P. Møller og Hustru Chastine Mc-Kinney Møllers Fond til Almene Formaal, the Carlsberg Foundation, the Novo Nordisk Foundation, the Danish Research Councils, a program of excellence from the University of Copenhagen, National Institutes of Health Grant (U01CA128437), and the Danish National Research Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See companion article on page 9672.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203563109/-/DCSupplemental.
References
- 1.Willer CJ, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kathiresan S, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White T, et al. Purification and cDNA cloning of a human UDP-N-acetyl-α-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase. J Biol Chem. 1995;270:24156–24165. doi: 10.1074/jbc.270.41.24156. [DOI] [PubMed] [Google Scholar]
- 5.Bennett EP, et al. Control of mucin-type O-glycosylation: A classification of the polypeptide GalNAc-transferase gene family. Glycobiology. 2011 doi: 10.1093/glycob/cwr182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schjoldager KT, et al. O-glycosylation modulates proprotein convertase activation of angiopoietin-like protein 3: Possible role of polypeptide GalNAc-transferase-2 in regulation of concentrations of plasma lipids. J Biol Chem. 2010;285:36293–36303. doi: 10.1074/jbc.M110.156950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holleboom AG, et al. Heterozygosity for a loss-of-function mutation in GALNT2 improves plasma triglyceride clearance in man. Cell Metab. 2011;14:811–818. doi: 10.1016/j.cmet.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gill DJ, Clausen H, Bard F. Location, location, location: New insights into O-GalNAc protein glycosylation. Trends Cell Biol. 2011;21:149–158. doi: 10.1016/j.tcb.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Kato K, et al. Polypeptide GalNAc-transferase T3 and familial tumoral calcinosis. Secretion of fibroblast growth factor 23 requires O-glycosylation. J Biol Chem. 2006;281:18370–18377. doi: 10.1074/jbc.M602469200. [DOI] [PubMed] [Google Scholar]
- 10.Schwientek T, et al. Functional conservation of subfamilies of putative UDP-N-acetylgalactosamine:polypeptide N-acetylgalactosaminyltransferases in Drosophila, Caenorhabditis elegans, and mammals. One subfamily composed of l(2)35Aa is essential in Drosophila. J Biol Chem. 2002;277:22623–22638. doi: 10.1074/jbc.M202684200. [DOI] [PubMed] [Google Scholar]
- 11.Steentoft C, et al. Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat Methods. 2011;8:977–982. doi: 10.1038/nmeth.1731. [DOI] [PubMed] [Google Scholar]
- 12.Hassan H, Bennett EP, Mandel U, Hollingsworth MA, Clausen H. In: Carbohydrates in Chemistry and Biology: A Comprehension Handbook. Ernst B, Hart GW, Sinay P, editors. New York: Wiley; 2000. pp. 273–292. [Google Scholar]
- 13.DeFrees S, et al. GlycoPEGylation of recombinant therapeutic proteins produced in Escherichia coli. Glycobiology. 2006;16:833–843. doi: 10.1093/glycob/cwl004. [DOI] [PubMed] [Google Scholar]
- 14.Wopereis S, et al. Apolipoprotein C-III isofocusing in the diagnosis of genetic defects in O-glycan biosynthesis. Clin Chem. 2003;49:1839–1845. doi: 10.1373/clinchem.2003.022541. [DOI] [PubMed] [Google Scholar]
- 15.Perdivara I, et al. Elucidation of O-glycosylation structures of the beta-amyloid precursor protein by liquid chromatography-mass spectrometry using electron transfer dissociation and collision induced dissociation. J Proteome Res. 2009;8:631–642. doi: 10.1021/pr800758g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halim A, et al. Site-specific characterization of threonine, serine, and tyrosine glycosylations of amyloid precursor protein/amyloid β-peptides in human cerebrospinal fluid. Proc Natl Acad Sci USA. 2011;108:11848–11853. doi: 10.1073/pnas.1102664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito H, et al. Identification of ROBO1 as a novel hepatocellular carcinoma antigen and a potential therapeutic and diagnostic target. Clin Cancer Res. 2006;12:3257–3264. doi: 10.1158/1078-0432.CCR-05-2787. [DOI] [PubMed] [Google Scholar]
- 18.Brewer HB, Jr, Shulman R, Herbert P, Ronan R, Wehrly K. The complete amino acid sequence of alanine apolipoprotein (apoC-3), and apolipoprotein from human plasma very low density lipoproteins. J Biol Chem. 1974;249:4975–4984. [PubMed] [Google Scholar]
- 19.Geiger T, Wehner A, Schaab C, Cox J, Mann M. Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.014050. M111.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gram Schjoldager KT, et al. A systematic study of site-specific GalNAc-type O-glycosylation modulating proprotein convertase processing. J Biol Chem. 2011;286:40122–40132. doi: 10.1074/jbc.M111.287912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jong MC, et al. Apolipoprotein C-III deficiency accelerates triglyceride hydrolysis by lipoprotein lipase in wild-type and apoE knockout mice. J Lipid Res. 2001;42:1578–1585. [PubMed] [Google Scholar]
- 22.Jong MC, Hofker MH, Havekes LM. Role of ApoCs in lipoprotein metabolism: Functional differences between ApoC1, ApoC2, and ApoC3. Arterioscler Thromb Vasc Biol. 1999;19:472–484. doi: 10.1161/01.atv.19.3.472. [DOI] [PubMed] [Google Scholar]
- 23.Liu W, et al. Identification of N-terminal residues on P-selectin glycoprotein ligand-1 required for binding to P-selectin. J Biol Chem. 1998;273:7078–7087. doi: 10.1074/jbc.273.12.7078. [DOI] [PubMed] [Google Scholar]
- 24.Herr P, Korniychuk G, Yamamoto Y, Grubisic K, Oelgeschläger M. Regulation of TGF-(beta) signalling by N-acetylgalactosaminyltransferase-like 1. Development. 2008;135:1813–1822. doi: 10.1242/dev.019323. [DOI] [PubMed] [Google Scholar]
- 25.Wagner KW, et al. Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat Med. 2007;13:1070–1077. doi: 10.1038/nm1627. [DOI] [PubMed] [Google Scholar]
- 26.Park JH, et al. Critical roles of mucin 1 glycosylation by transactivated polypeptide N-acetylgalactosaminyltransferase 6 in mammary carcinogenesis. Cancer Res. 2010;70:2759–2769. doi: 10.1158/0008-5472.CAN-09-3911. [DOI] [PubMed] [Google Scholar]
- 27.Park JH, Katagiri T, Chung S, Kijima K, Nakamura Y. Polypeptide N-acetylgalactosaminyltransferase 6 disrupts mammary acinar morphogenesis through O-glycosylation of fibronectin. Neoplasia. 2011;13:320–326. doi: 10.1593/neo.101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freire-de-Lima L, et al. Involvement of O-glycosylation defining oncofetal fibronectin in epithelial-mesenchymal transition process. Proc Natl Acad Sci USA. 2011;108:17690–17695. doi: 10.1073/pnas.1115191108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brooks SA, Carter TM, Bennett EP, Clausen H, Mandel U. Immunolocalisation of members of the polypeptide N-acetylgalactosaminyl transferase (ppGalNAc-T) family is consistent with biologically relevant altered cell surface glycosylation in breast cancer. Acta Histochem. 2007;109:273–284. doi: 10.1016/j.acthis.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 30.Rajpert-De Meyts E, et al. Changes in the profile of simple mucin-type O-glycans and polypeptide GalNAc-transferases in human testis and testicular neoplasms are associated with germ cell maturation and tumour differentiation. Virchows Arch. 2007;451:805–814. doi: 10.1007/s00428-007-0478-4. [DOI] [PubMed] [Google Scholar]
- 31.Mandel U, et al. Expression of polypeptide GalNAc-transferases in stratified epithelia and squamous cell carcinomas: Immunohistological evaluation using monoclonal antibodies to three members of the GalNAc-transferase family. Glycobiology. 1999;9:43–52. doi: 10.1093/glycob/9.1.43. [DOI] [PubMed] [Google Scholar]
- 32.Gomes J, et al. Expression of UDP-N-acetyl-D-galactosamine: Polypeptide N-acetylgalactosaminyltransferase-6 in gastric mucosa, intestinal metaplasia, and gastric carcinoma. J Histochem Cytochem. 2009;57:79–86. doi: 10.1369/jhc.2008.952283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berois N, et al. UDP-N-acetyl-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase-6 as a new immunohistochemical breast cancer marker. J Histochem Cytochem. 2006;54:317–328. doi: 10.1369/jhc.5A6783.2005. [DOI] [PubMed] [Google Scholar]
- 34.Sutherlin ME, et al. Expression of three UDP-N-acetyl-α-D-galactosamine:polypeptide GalNAc N-acetylgalactosaminyltransferases in adenocarcinoma cell lines. Cancer Res. 1997;57:4744–4748. [PubMed] [Google Scholar]
- 35.Wiesner GL, et al. A subset of familial colorectal neoplasia kindreds linked to chromosome 9q22.2-31.2. Proc Natl Acad Sci USA. 2003;100:12961–12965. doi: 10.1073/pnas.2132286100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guda K, et al. Inactivating germ-line and somatic mutations in polypeptide N-acetylgalactosaminyltransferase 12 in human colon cancers. Proc Natl Acad Sci USA. 2009;106:12921–12925. doi: 10.1073/pnas.0901454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bennett EP, Hassan H, Clausen H. cDNA cloning and expression of a novel human UDP-N-acetyl-α-D-galactosamine. Polypeptide N-acetylgalactosaminyltransferase, GalNAc-t3. J Biol Chem. 1996;271:17006–17012. doi: 10.1074/jbc.271.29.17006. [DOI] [PubMed] [Google Scholar]
- 38.Nehrke K, Hagen FK, Tabak LA. Isoform-specific O-glycosylation by murine UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase-T3, in vivo. Glycobiology. 1998;8:367–371. doi: 10.1093/glycob/8.4.367. [DOI] [PubMed] [Google Scholar]
- 39.Gerken TA, Raman J, Fritz TA, Jamison O. Identification of common and unique peptide substrate preferences for the UDP-GalNAc:polypeptide α-N-acetylgalactosaminyltransferases T1 and T2 derived from oriented random peptide substrates. J Biol Chem. 2006;281:32403–32416. doi: 10.1074/jbc.M605149200. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, et al. Modification of epidermal growth factor-like repeats with O-fucose. Molecular cloning and expression of a novel GDP-fucose protein O-fucosyltransferase. J Biol Chem. 2001;276:40338–40345. doi: 10.1074/jbc.M107849200. [DOI] [PubMed] [Google Scholar]
- 41.Shao L, Haltiwanger RS. O-fucose modifications of epidermal growth factor-like repeats and thrombospondin type 1 repeats: Unusual modifications in unusual places. Cell Mol Life Sci. 2003;60:241–250. doi: 10.1007/s000180300019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gill DJ, Chia J, Senewiratne J, Bard F. Regulation of O-glycosylation through Golgi-to-ER relocation of initiation enzymes. J Cell Biol. 2010;189:843–858. doi: 10.1083/jcb.201003055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeKelver RC, et al. Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res. 2010;20:1133–1142. doi: 10.1101/gr.106773.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steentoft C, Bennett EP, Clausen H. Glycoengineering of human cell lines using Zinc Finger nuclease gene targeting: SimpleCells with homogenous GalNAc O-glycosylation allow isolation of the O-glycoproteome by one-step lectin affinity chromatography. Methods Mol Biol. 2012 doi: 10.1007/978-1-62703-465-4_29. in press. [DOI] [PubMed] [Google Scholar]
- 45.Wandall HH, et al. Substrate specificities of three members of the human UDP-N-acetyl-α-D-galactosamine:Polypeptide N-acetylgalactosaminyltransferase family, GalNAc-T1, -T2, and -T3. J Biol Chem. 1997;272:23503–23514. doi: 10.1074/jbc.272.38.23503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.