Abstract
Sialic acid-recognizing Ig-like lectins (Siglecs) are signaling receptors that modulate immune responses, and are targeted for interactions by certain pathogens. We describe two primate Siglecs that were rendered nonfunctional by single genetic events during hominin evolution after our common ancestor with the chimpanzee. SIGLEC13 was deleted by an Alu-mediated recombination event, and a single base pair deletion disrupted the ORF of SIGLEC17. Siglec-13 is expressed on chimpanzee monocytes, innate immune cells that react to bacteria. The human SIGLEC17P pseudogene mRNA is still expressed at high levels in human natural killer cells, which bridge innate and adaptive immune responses. As both resulting pseudogenes are homozygous in all human populations, we resurrected the originally encoded proteins and examined their functions. Chimpanzee Siglec-13 and the resurrected human Siglec-17 recruit a signaling adapter and bind sialic acids. Expression of either Siglec in innate immune cells alters inflammatory cytokine secretion in response to Toll-like receptor-4 stimulation. Both Siglecs can also be engaged by two potentially lethal sialylated bacterial pathogens of newborns and infants, agents with a potential impact on reproductive fitness. Neanderthal and Denisovan genomes show human-like sequences at both loci, corroborating estimates that the initial pseudogenization events occurred in the common ancestral population of these hominins. Both loci also show limited polymorphic diversity, suggesting selection forces predating the origin of modern humans. Taken together, these data suggest that genetic elimination of Siglec-13 and/or Siglec-17 represents signatures of infectious and/or other inflammatory selective processes contributing to population restrictions during hominin origins.
Sialic acids (Sias) are monosaccharides typically found at the outermost ends of complex glycan chains that decorate all vertebrate cell surfaces (1, 2). Sias are essential for embryonic development (3) and mediate important intrinsic organismal functions (1, 2). However, given their location and density, Sia-bearing glycans are also targets for recognition by many pathogen-binding proteins and toxins (1, 4, 5). Adding complexity to these opposing evolutionary selection forces, many important bacterial pathogens have evolved convergent mechanisms for molecular mimicry of host Sias (4, 6, 7). For all these reasons, both Sias and Sia-recognizing proteins are rapidly evolving in some taxa. Current data suggest that humans are an extreme example, with Sia-related genes representing a “hotspot” in human evolution (5). Of less than 70 human genes known to be involved in Sia biology, more than 10 have been documented to exhibit human-specific changes relative to the chimpanzee, our closest evolutionary cousins (5). Most of these human-specific genetic changes are in Sia-recognizing Ig-like lectin (SIGLEC) genes (5).
SIGLEC genes encode a family of transmembrane receptors that bind Sia-containing ligands via their amino-terminal extracellular Ig-like domains and modulate cellular responses via cytosolic signaling motifs (7–10). The CD33-related subset of Siglecs (CD33rSiglecs) is rapidly evolving within vertebrates (7–10). The most likely reason is that CD33rSiglecs are prominently expressed on innate immune cells, and modulate responses to pathogens. In this regard, Siglec-3 and Siglecs-5 to -11 in humans seem to recognize sialylated ligands as “self-associated molecular patterns” (11), limiting unwanted reactivity against other cells in the same organism (7–10). However, certain immune-modulating bacterial pathogens carry out molecular mimicry of sialylated CD33rSiglec ligands (12), dampening host innate immune cell responses and facilitating infection (13). In one instance, a human bacterial pathogen evolved a more stable protein–protein interaction with an inhibitory CD33rSiglec to suppress innate immunity (14).
Evolution has also generated CD33rSiglecs with opposing activatory potential (7, 15, 16), transmitting positive signals to immune cells via recruitment of the immunoreceptor tyrosine-based activation motif (ITAM)-containing DAP12 adaptor protein (17). Some activatory Siglecs pair with inhibitory ones (15, 16), supporting the notion that they represent a host evolutionary response to pathogen mimicry and engagement of inhibitory CD33rSiglecs (7). However, low avidity engagement of activatory Siglecs can mediate paradoxical inhibitory responses (ITAMi) (18, 19).
Two genomic loci encoding ITAM-containing primate Siglecs (SIGLEC14 and SIGLEC16) are polymorphic, with their common alleles being nonfunctional (15, 16, 20). These polymorphisms exist in African populations, indicating that they likely originated before the migration of modern humans out of Africa about 60,000–70,000 y ago. Thus, functional and pseudogene alleles of SIGLEC genes can be maintained in populations over long periods. This finding likely reflects ongoing selection forces involving the need to maintain innate immune self-recognition and control damaging inflammatory responses, all against a backdrop of potential pathogen subversion of these mechanisms (7, 8, 20). An evolutionary balancing act is also supported by the high frequency of human-specific pathogens that carry out molecular mimicry of Sias through convergent evolutionary mechanisms (4, 6).
Current genetic, archeological, skeletal, and radioactive dating evidence indicates that all modern humans are derived from a population with an effective size of 10,000 or fewer that originated in Africa ∼100,000–200,000 y ago (21–25) and later spread across the planet, replacing other hominin species and having limited interbreeding with our closest extinct cousins, the Neanderthals (26) and Denisovans (27). Even smaller estimates have been made of the effective population size of Neanderthals (28). The reasons for the small effective population sizes are unknown, and host–pathogen interactions have not been previously considered as contributors. Here, we report two genomic inactivation events in SIGLEC genes that became fixed before the emergence of modern humans in Africa. Our evidence indicates that these genes may have been inactivated in the hominin lineage, because they can be engaged by pathogens associated with life-threatening invasive infections in newborns and infants. It is also possible that the immunomodulatory capacity of these genes was costly in contexts beyond bacterial engagement of Siglecs, such as toxic inflammatory effects on ancestral hominin immune cells.
Results and Discussion
Single Alu-Mediated Deletion Event Inactivated SIGLEC13 in the Hominin Lineage.
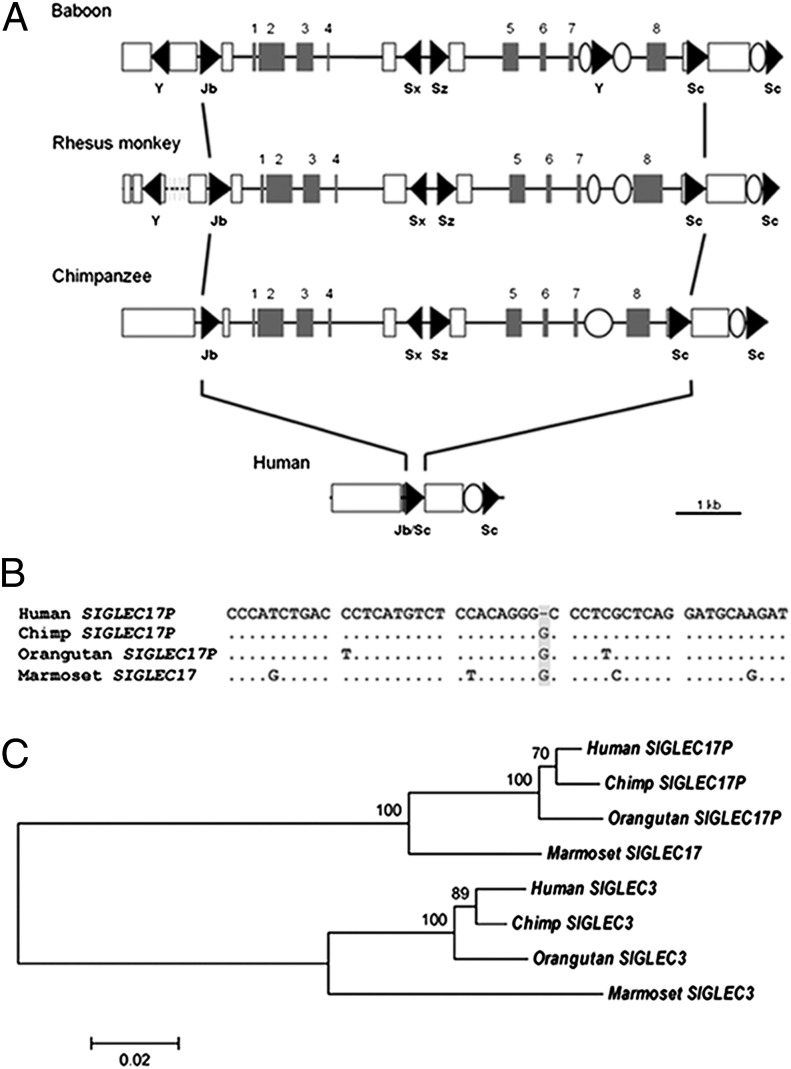
Analysis of genomic BAC clones indicated that the primate SIGLEC13 gene was missing from the human genome but present in chimpanzees and baboons (29). To understand events accounting for this apparently human-specific deletion, we analyzed the genomic region encompassing the SIGLEC13 locus in the currently available sequence builds of the human, chimpanzee, baboon, and rhesus genomes. RepeatMasker software identified several repetitive elements in this region in all of these species. These elements include Alu elements, which are primate-specific short interspersed elements (30). In the chimpanzee genome, five Alu elements are located in an ∼10-kb genomic region containing the SIGLEC13 locus (Fig. 1A). One Alu element belonging to the AluJb family is located upstream of the SIGLEC13 locus, one AluSx and one AluSz elements are within the locus, and two AluSc elements are found downstream. These elements are also found in orthologous regions in the baboon and rhesus monkey genomes (Fig. 1A). In the human genome, we found just one composite Alu element (AluJb/Sc) occupying the region of ∼7 kb that contains the SIGLEC13 locus in chimpanzee, rhesus monkey, and baboon (Fig. 1A). The ∼7-kb region deleted in the human genome is sandwiched between two ancestral AluJb and AluSc elements (Fig. 1A). Thus, a single Alu-mediated recombination event was the likely mechanism for human-specific deletion of SIGLEC13, leaving a single fused Alu element in the human genome.
Fig. 1.
Inactivation of two SIGLEC genes during hominin evolution. (A) Comparison of genomic structure surrounding the SIGLEC13 locus among humans and other primates. Coding regions are represented by shaded boxes, and Alu elements are represented by triangles. Names of Alu subfamilies are shown. Open boxes and ellipses indicate LINE (long interspersed element) and LTR (long terminal repeat) elements, respectively. The dotted line indicates a sequence gap. (B) DNA sequence alignment using Clustal W in MEGA4 shows the human-specific loss of G in SIGLEC17P (highlighted in gray). (C) Reconstructed neighbor-joining (NJ) tree of SIGLEC17P and SIGLEC3 among primates. Bootstrap values of 1,000 replicates are shown on internal branches. MEGA4 was used for NJ tree reconstruction and bootstrap analysis.
Human-Specific Mutational Events Functionally Altered and then, Pseudogenized Primate SIGLEC17.
We previously described the human SIGLECP3 locus with an inactivating deletion in the predicted ORF (29). We now note that the rest of the predicted ORF remains intact. We found more than one cDNA clone predicting a full-length transcript derived from SIGLECP3 (for example, BC041072 in SI Appendix, Fig. S1). In addition to the single nucleotide deletion, the remnant human SIGLECP3 coding region harbors a human-unique missense mutation of the codon encoding an Arg residue that would have been involved in Sia recognition when the ORF was intact (SI Appendix, Fig. S2A). Taken together with evidence for Sia-binding properties of the resurrected protein when the Arg codon is restored (see below) and evidence for an intact marmoset ortholog (SI Appendix, Fig. S2A), we redesignate the original SIGLECP3 locus as primate SIGLEC17 and the corresponding human pseudogene as SIGLEC17P.
Phylogenetic sequence comparisons showed that the 1-bp deletion in SIGLEC17P is human-specific (Fig. 1B). A BLAST query using the human sequence identified an orthologous gene with a predicted intact ORF encoding Siglec-17 in the marmoset genome, with 93% DNA sequence similarity and 89% predicted protein sequence similarity (SI Appendix, Fig. S2A). Analysis of the RT-PCR–derived orthologous segment from chimpanzee peripheral blood mononuclear cell mRNA confirmed that, although the 1-bp deletion is human-specific, independent pseudogenization events have occurred in the chimpanzee as well as orangutan genomes. The homologous region containing the SIGLEC17P locus seems to be completely deleted in the rhesus and baboon genomes (29). Thus, although well-conserved in primate evolution from New World monkeys to ancestral hominins, the SIGLEC17 gene has also undergone independent deletion or pseudogenization events in multiple primate taxa, with a distinct event in the hominin lineage.
Additional BLAST analyses showed that SIGLEC17 is most closely related to SIGLEC3 (encoding Siglec-3/CD33), with the two loci evolving as paralogs in primates (Fig. 1C). The predicted V-set and C2-set domains of the resurrected human SIGLEC17 and human SIGLEC3 genes share 66% DNA sequence similarity (SI Appendix, Fig. S2B). The human pseudogene SIGLECP6 also has homology to human SIGLEC17P. A phylogenetic tree of primate SIGLEC3, SIGLECP6, and SIGLEC17P gave no evidence for a gene conversion during primate evolution among these loci.
Inactivation Events of SIGLEC13 and SIGLEC17 Are Human-Universal.
The Alu-mediated SIGLEC13 deletion and the SIGLEC17 frame-shift mutation were homozygous in 28 HapMap human samples representing a worldwide population (11 Yoruba Africans, 9 Japanese and Chinese, and 8 North Europeans). These events were not found in genomes from 19 chimpanzees (samples provided by Pascal Gagneux, University of California, San Diego, CA) or 6 orangutans (samples from the Coriell repository), ruling out an ancestral hominid polymorphism. Notably, except for the universal frame-shift mutation, the rest of the SIGLEC17 ORF was intact in all 28 humans. The HapMap sample results left open the possibility that rare functional alleles might persist in some African populations. However, both mutations were found to be in a homozygous state in ∼230 ethnically and geographically diverse Africans comprising many major African population groups (SI Appendix, Table S1) (31). This finding is in contrast to SIGLEC14 and SIGLEC16, which show polymorphic segregating pseudogenization in all human populations (15, 16).
As these pseudogenization events of SIGLEC13 and SIGLEC17 are both human-unique and -universal compared with other primates, we decided to explore the functional implications by studying the chimpanzee ortholog of SIGLEC13 and the functionally resurrected form of human SIGLEC17.
Siglec-13 Is Selectively Expressed on Chimpanzee Monocytes.
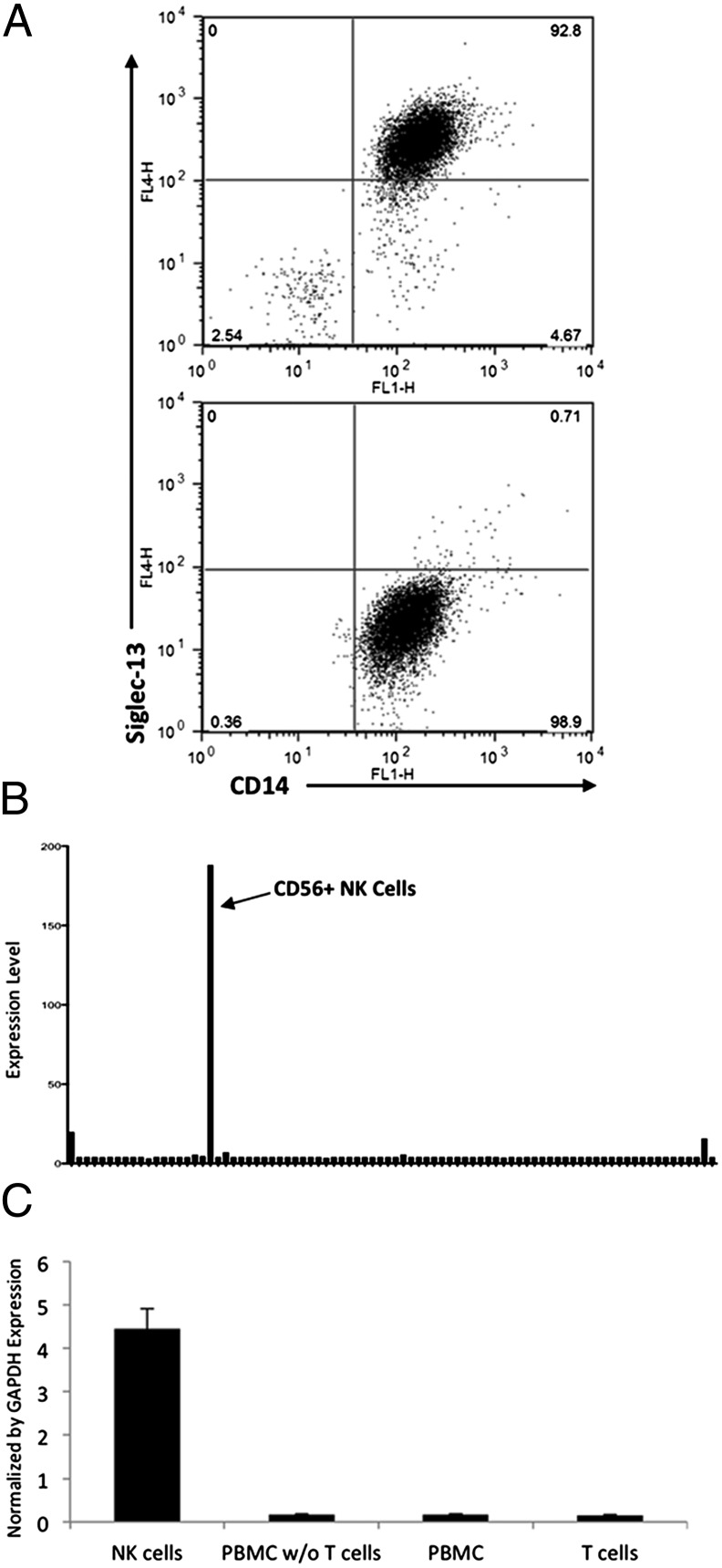
A monoclonal antibody against the recombinant soluble extracellular domains of chimpanzee Siglec-13 (SI Appendix) was used for flow cytometry analysis of chimpanzee peripheral blood leukocytes, showing expression predominantly on monocytes (Fig. 2A). As expected from the human SIGLEC13 deletion, there was no specific antibody binding to human leukocytes. Thus, earlier hominin ancestors likely expressed Siglec-13 on monocytes, a cell type participating in innate immune defenses against bacterial pathogens.
Fig. 2.
Cell-type specific expression of Siglec-13 and -17. (A) Chimpanzee and human peripheral blood mononuclear cells (PBMCs) were labeled with monoclonal anti–Siglec-13 mouse IgG. Colabeling for CD14 (a marker for PBMCs) showed specific expression in chimpanzee (Upper), but not in human (Lower) monocytes. (B) Transcript expression profile of human SIGLEC17P in 84 human tissues and cell lines acquired from BioGPS with probe gnf1h07492_at. The list of tissues and cell types studied can be found in the SI Appendix. (C) Quantitative RT-PCR for SIGLEC17P expression in human NK cells. Human T cells, whole-blood PBMCs, and whole-blood PBMCs depleted of T cells were used as controls. Results were normalized to GAPDH expression.
Human SIGLEC17P Message Is Selectively Expressed in Natural Killer Cells.
Because the SIGLEC17 ORF is interrupted by independent events in the human and chimpanzee genomes, we could not study chimpanzee cells to understand the original function of this protein. Gene Atlas analysis showed prominent expression of SIGLEC17P message almost exclusively in natural killer (NK) cells (Fig. 2B). This finding was confirmed by quantitative RT-PCR in human peripheral blood mononuclear cells, with the strongest signal in isolated NK cells (Fig. 2C). Thus, hominin ancestors likely expressed Siglec-17 on NK cells, a cell type that can activate macrophages to mount antibacterial functions and bridge the innate and adaptive immune responses.
Chimpanzee Siglec-13 and Functionally Resurrected Human Siglec-17 Bind Sias.
Recombinant soluble chimpanzee Siglec-13-Fc and human Siglec-17-Fc with a functionally restored ORF were prepared by fusing sequences encoding their first two extracellular Ig-like domains with coding regions for the Fc portion of human IgG1. Both Siglec-Fcs bound to a sialoglycan microarray, and binding was lost when the essential arginine residue was absent (SI Appendix, Fig. S3). These data show that these genes originally encoded functional Siglecs in hominin ancestors.
Both Proteins Interact with the DAP12 Signaling Adaptor Protein.
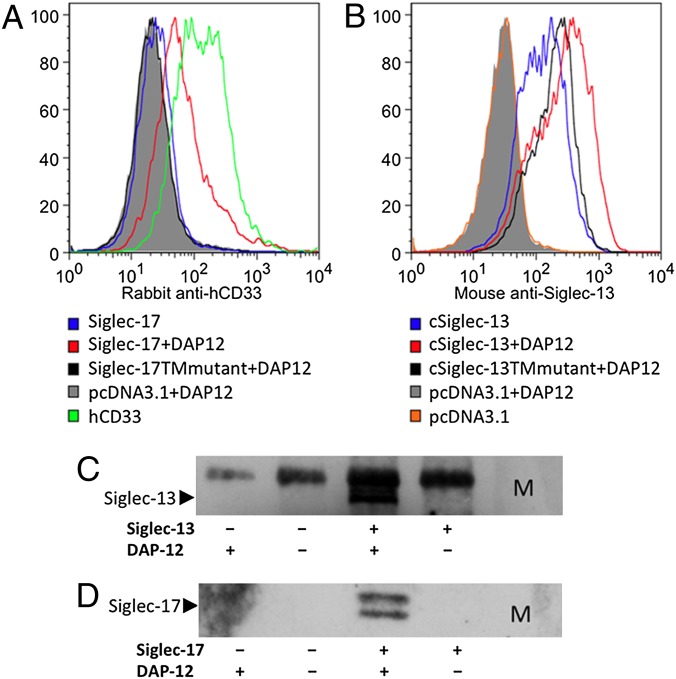
The ORFs of chimpanzee SIGLEC13 and resurrected human SIGLEC17 each encode a transmembrane protein with a single positively charged residue within the membrane-spanning region. Other immune cell proteins with this feature (including human Siglec-14 and -16) can associate with DNAX-activating protein of 12 kDa (DAP12) (17), which has a corresponding negatively charged residue within the membrane-spanning region (15, 16). Transient transfection of a FLAG-tagged DAP12 cDNA into 293T cells along with SIGLEC13 or SIGLEC17 cDNAs was required for optimal surface expression of both molecules (Fig. 3 A and B). Consistent with this finding, a DAP12–FLAG-tagged protein could coimmunoprecipitate with Siglec-13 or -17 (Fig. 3 C and D). Mutation of the single positively charged residue within the transmembrane domain of each Siglec to a neutral alanine residue diminished the expression of both proteins. Thus, the positively charged residue is involved in surface expression of both SIGLEC13 and SIGLEC17 genes (Fig. 3 A and B).
Fig. 3.
Importance of DAP12 for optimal surface expression of Siglec-13 and -17. (A) 293T cells were transiently transfected with a Siglec-17 cDNA in pcDNA3.1 with or without cotransfection with FLAG-tagged DAP12. Human CD33-transfected cells were used positive controls for detection by a rabbit anti-human CD33 antibody, which partially cross-reacts with human Siglec-17. Cells cotransfected with pcDNA3.1 and DAP12 were used as a negative control. Fluorescence was measured after staining with rabbit anti-human CD33 and then Alexa Fluor 647 donkey anti-rabbit IgG. The Siglec-17TMmutant in pcDNA3.1 was made from Siglec-17–pcDNA3.1 by introducing a K253A mutation. Cotransfected pIRES2-EGFP (Clontech) was used to gate positively transfected cells. (B) The 293T cells were transiently transfected with a Siglec-13 cDNA in pcDNA3.1 with or without cotransfection with FLAG-tagged DAP12. Controls were as in A. Fluorescence was measured after staining with mouse anti–Siglec-13 and then Alexa Fluor 647 goat anti-mouse IgG. The cSiglec-13TMmutant in pcDNA3.1 was made from cSiglec-13–pcDNA3.1 by introducing a K352A mutation. Cotransfected pIRES2-EGFP (Clontech) was used to gate positively transfected cells. (C and D) 293T cells transiently transfected with cDNAs for chimpanzee Siglec-13 (C) or human Siglec-17 (D) with/without DAP12 were lysed. M2 agarose beads were used to pull down FLAG tagged DAP12. Mouse anti-Siglec-13 or rabbit anti-human CD33 (which cross reacts with Siglc-17) were used in Western blots followed by HRP conjugated secondary antibodies, as shown in C and D, respectively. The upper band on the coimmunoprecipitation is nonspecific because of the use of M2 beads carrying a mouse antibody (C). M indicates All Blue protein standard (BIO-RAD).
Functional Analysis of Chimpanzee Siglec-13 or Resurrected Human Siglec-17.
To examine signaling events mediated by chimpanzee Siglec-13 and resurrected Siglec-17, we used RAW246.7 macrophages that intrinsically expresses DAP12 (32). Semistably transfected RAW264.7 cells with Siglec-13 or -17 cDNAs in pcDNA3.1 were acquired after 3 wk of selection in 1.5 mg/mL G418. Expression of the Siglecs in the transfected cells was confirmed by flow cytometry or RT-PCR (SI Appendix, Fig. S4). As shown in SI Appendix, Fig. S5 A and B, semistably transfected cells with Siglec-13 or -17 showed increased intracellular TNF levels after low-dose LPS stimulation [Toll-like receptor 4 (TLR4) activation] compared with mock-transfected cells. With Siglec-13 transfection, untreated cells sometimes displayed constitutive TNF production, which was further boosted by LPS (SI Appendix, Fig. S5A). Thus, Siglec-13 and -17 likely mediated signaling through DAP12 in ancestral monocytes and NK cells, modulating cytokine secretion.
Specific Interactions of Siglec-13 and -17 with Sialylated Bacterial Pathogens.
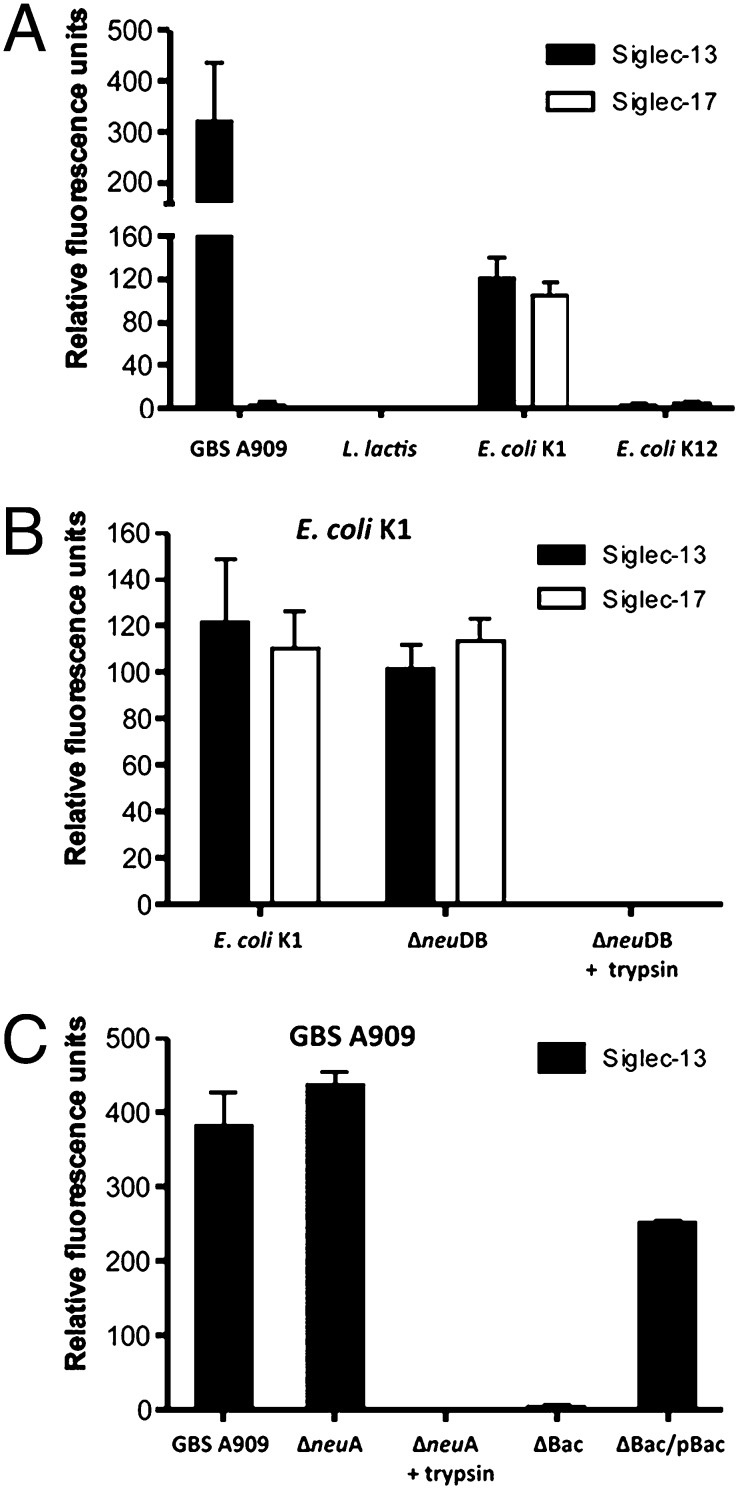
Some other CD33-related Siglecs are known to recognize certain pathogenic bacteria (13, 14) [e.g., Group B Streptococcus (GBS), a Gram-positive bacterial pathogen that expresses Sias and causes invasive infections in human newborn infants (33)]. GBS engagement of Siglecs can involve the Sias (13) and/or a specific cell surface-anchored protein, the β-protein (14). Because Siglec-13 and -17 could respond to a bacterial product (LPS) (SI Appendix, Fig. S5), we hypothesized that they would also interact with certain important sialylated human pathogens. Indeed, the extracellular domain of Siglec-13 bound to sialylated GBS A909 but not the nonpathogenic Gram-positive bacteria Lactococcus lactis (Fig. 4A). Extracellular domains of both Siglec-13 and -17 also bound to the sialylated Gram-negative pathogen Escherichia coli K1 (another leading cause of sepsis and meningitis in human newborns) but not nonpathogenic E. coli K-12 (Fig. 4A). Interestingly, although some Sia-dependent binding is observed, these interactions prominently involved trypsin-sensitive protein–protein interactions (Fig. 4 and SI Appendix, Fig. S6). With GBS, analysis of an isogenic bacterial mutant identified the likely binding partner for Siglec-13 as the GBS β-protein (Fig. 4C), an interaction previously shown to suppress human leukocyte responses through Siglec-5 (14).
Fig. 4.
Chimpanzee Siglec-13 or resurrected Siglec-17 interacts selectively with bacterial pathogens. (A) Chimpanzee Siglec-13-Fc or resurrected human Siglec-17-Fc (with Arg) chimeras were immobilized to ELISA wells by protein A, and binding of FITC-labeled sialylated strains GBS A909 (serotype Ia) or E. coli K1 RS218 (StrR) was studied. Negative controls were L. lactis and nonencapsulated laboratory E. coli K-12 strain DH5α. (B) Binding of E. coli K1 strain RS218 (StrR), isogenic Sia-deficient E. coli K1 ΔneuDB, or E. coli K1 ΔneuDB pretreated with trypsin was studied as in A. (C) Binding of GBS A909 serotype Ia, isogenic Sia-deficient GBS ΔneuA, or GBSΔneuA pretreated with trypsin was studied as in A. StrainsΔBac (lacking β-protein) or the plasmid complemented mutant ΔBac + pBac were also studied. All values are means from three independent experiments ± SD.
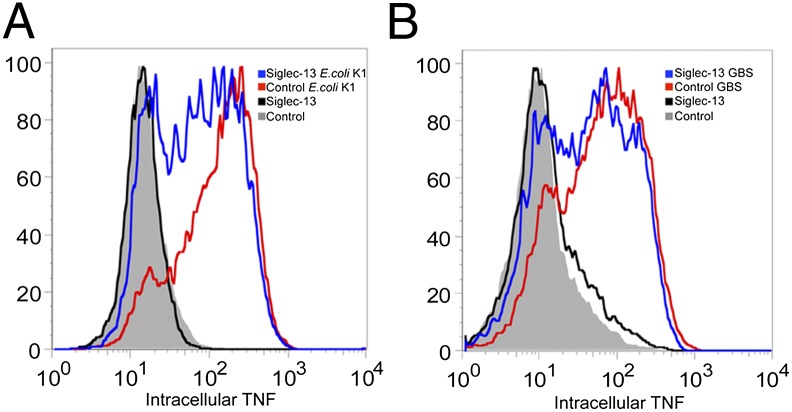
Reduced Intracellular TNF in Siglec-13–Transfected RAW 264.7 Cells in Response to the Bacterial Infection.
Semistably transfected RAW264.7 cells with Siglec-13 cDNA in pcDNA3.1 were acquired as mentioned above. Cells were infected with E. coli K1 or GBS A909 for 1 h at low multiplicity of infection (MOI; 0.6 and 0.1, respectively), and the level of intracellular TNF was then measured by an APC (Allophycocyanin) rat anti-mouse TNF antibody for all of the infected cells. Interestingly, compared with the mock control, the Siglec-13–transfected cells showed reduced intracellular TNF (Fig. 5 A and B). In this regard, it is known from work on other DAP12-interacting proteins that engagement of corresponding receptors can mediate either activating responses (through classical ITAM-Syk kinase signaling) or paradoxical inhibitory responses [ITAMi through Src kinase and Src homology phosphatase (SHP-1)] (18, 19). Thus, pathogenic bacteria might have been taking advantage of the cell surface-expressed Siglec-13 in ancestral hominins to dampen host innate immune cell responses and facilitate infection. Similar studies with Siglec-17 were not possible, because semistable expression in these cells resulted in markedly retarded growth.
Fig. 5.
Reduced intracellular TNF of Siglec-13–transfected RAW264.7 cells in response to bacterial pathogen infection; 500,000 cells semistably transfected with Siglec-13 or vector only and selected with G418 for 3 wk were seeded in a 12-well plate. The next day, 2 h before infection, cells were washed three times with HBSS, and regular culture medium without G418 added. Cells were infected with bacterial pathogens at an appropriate MOI for 1 h at 37 °C. The BD Fixation and Permeabilization Solution Kit (555028) with APC rat anti-mouse TNF-α was used to detect the intracellular TNF level using the recommended protocol. Cells without bacterial infection were used as controls. Cells transfected with pcDNA3.1 vector were used as a control. (A) Cells infected by E. coli K1 RS218 (StrR) at MOI = 0.6. (B) Cells infected by GBS A909 (serotype Ia) at MOI = 0.1.
Attempting to further recreate ancestral interactions of cells expressing Siglec-13 and -17 with human pathogenic bacteria, we tried to generate stable macrophage cell lines expressing them. However, compared with control-transfected cells, those expressing Siglec-13 or -17 grew very slowly in culture, and we could not generate long-term stable lines in either mouse RAW264.7 or human THP-1 macrophage cell lines. Similar difficulties were encountered with making stable transfectants of Siglec-17 in the human NK cell line NK-92. Apparently, sustained expression of these molecules is toxic to these cell types. Given the expression of Siglec-13 on chimpanzee monocytes, we assume that ancestral hominin macrophages were able to tolerate expression of this Siglec. Regardless, it is reasonable to suggest that Siglec-13–positive monocytes and/or Siglec-17–positive NK cells on ancestral innate immune cells may have influenced interactions with pathogenic bacteria and/or have had toxic effects.
Timing of SIGLEC Pseudogenization Events During Hominin Evolution.
The two hominin pseudogenization events occurred after a common ancestor with the chimpanzee, but predated the common origin of modern humans. Fresh analysis of genomic sequence data from a newly obtained well-preserved Neanderthal sample from Monti Lessini (MLS3) (SI Appendix, see ancient DNA analysis) showed that the pseudogenized allele of SIGLEC17 was already present before the population divergence of Neanderthals and modern humans (between 270,000 and 440,000 y ago) (26). The same genotype was noted in published Neanderthal genomic sequences. Whether SIGLEC13 was also deleted from the Neanderthal genome could not be determined with certainty. However, we found no evidence for the chimpanzee version of the gene in the Neanderthal sample analyzed here (SI Appendix) or the published Neanderthal genome sequence. The recently published Denisovan genome sequence (27) also showed evidence of the modern human versions of SIGLEC17P and the possible deletion of SIGLEC13.
Taken together, the data indicate that both the SIGLEC13 deletion allele and the modern human SIGLEC17P allele were already present in the common ancestral populations of Neanderthals, Denisovans, and humans. However, other similar SIGLEC genes (SIGLEC14 and SIGLEC16) show polymorphic pseudogenization in modern humans, with moderate frequency persistence of the intact functional allele in all human populations (15, 16). Initial pseudogenization of SIGLEC16 was estimated to occur at least 3 Mya (20). Given the small population of Neanderthals and Denisovans analyzed and the limited quality of these data, we cannot be certain regarding the timing of fixation of the SIGLEC13 and SIGLEC17 pseudogenes in hominin populations. Nevertheless, we hypothesize that expression of these genes became detrimental to survival under the selective pressure of pathogenic bacteria that were able to bind to the Siglecs and subvert their homeostatic immune functions. Another (not mutually exclusive) selection pressure for Siglec elimination could have been toxic overactivation of the immune system. To seek evidence for these hypotheses, we looked for residual signatures of selection surrounding these loci.
Genomic Evidence for Selection at the SIGLEC13 and SIGLEC17 Loci.
When positive directional selection involves specific genes or pseudogenes, the genomic regions encompassing such loci can show limited variation in the time period immediately after (34, 35) depending on the type of selection and whether the allele has reached fixation. This kind of signature of selection will be eroded over time by additional random mutations and/or recombination. The deepest time at which such signatures can still be confidently detected is thought to be 5,000–10,000 generations or about 100,000–200,000 y (34, 35). This depth of time happens to be similar to the depth of time proposed for the origin of modern humans (21, 22, 25). Thus, it is impossible to conclusively prove a classical “selective sweep” before the origin of modern humans ∼200,000 y ago. However, we decided to look for any residual evidence of selection surrounding these two loci compared with data for adjacent genes.
We, indeed, found low π- (nucleotide diversity) and θ- (nucleotide polymorphism) values for SIGLEC17P and the SIGLEC13 flanking regions relative to adjacent loci such as SIGLEC8 and SIGLEC10. Using GENECONV, we did not find any evidence that either locus was involved in gene conversions that may explain our observations. Furthermore, the estimated H values of Fay and Wu of human SIGLEC13 flanking region and SIGLEC17 are significantly negative, and the DH and DHEW tests (36, 37) show significant results for SIGLEC17P (SI Appendix, Table S4). However, not all of the statistics significantly rejected neutral evolution of two loci in our coalescent analysis (SI Appendix, Table S4). Notably, relatively low polymorphisms were also observed in SIGLEC7, which is ∼10 kb away from SIGLEC17P. Like SIGLEC17P (SI Appendix, Table S4), SIGLEC7 also showed significant deviation from neutrality in some statistical tests. However, SIGLEC9, another adjacent locus at a comparable distance, showed neither low polymorphisms (SI Appendix, Table S2) nor statistical significance in any of the tests. Thus, SIGLEC7 could not be the locus of selection. In addition, an HKA (Hudson–Kreitman–Aguadé) test (38) between SIGLEC17P and SIGLEC7 loci indicated the significantly lower polymorphism in SIGLEC17P compared with SIGLEC7 with the chimpanzee sequence as an outgroup (P < 0.05), supporting the notion that SIGLEC17P might be the center of the proposed ancient selective event.
Interestingly, the resurrected Siglec-17 also shows a low nonsynonymous/synonymous rate ratio (dN/dS) that is similar to most other currently functional Siglecs, suggesting that it was subject to purifying selection before its pseudogenization (SI Appendix, Fig. S7). Notably, with the exception of the human-universal single base pair deletion, the SIGLEC17 ORF remains conserved in all 28 HapMap humans studied, suggesting inadequate time for accumulation of other random mutations. We, of course, cannot rule out the possibility that this pseudogene is still undergoing purifying selection because of some other unknown function (39) (e.g., as a small RNAi-altering gene expression (40).
Although no single test can be conclusive at this depth of evolutionary time, our collective data indicate a residual signature of ancient selection forces acting on both of these loci, which must have predated the common origin of modern humans. However, such signatures should have faded in 100,000–200,000 y (34, 35). Thus, it is reasonable to speculate that positive selection on these pseudogenization events may have been involved in population bottlenecks close to the origin of modern humans.
Dating the Selection on the Inactivation of SIGLEC13 and SIGLEC17.
The hominin-specific events inactivating SIGLEC13 and SIGLEC17 likely occurred first in the common ancestral populations of modern humans, Neanderthal, and Denisovans. In keeping with this finding, coalescence analysis estimated the time of the most recent common ancestor of identified haplotypes at 800,000–900,000 for the SIGLEC13 deletion locus and SIGLEC17P (SI Appendix). At first glance, this timing may seem at odds with aforementioned signals of selection at both loci, which should have been completely erased over such long periods of time. However, we can suggest a scenario consistent with all of the data—that some active alleles of these genes persisted in the common ancestral population of modern humans, Neanderthal, and Denisovans until selection eliminated them at some point close to the common origin of modern humans. To seek additional evidence for this possibility, we calculated dates for selection acting on these events (SI Appendix). Approximate times were calculated as ∼105,000 y for SIGLEC17P and ∼46,000 y for SIGLEC13. These numbers are approximations, and the method does not allow error estimations. Because both pseudogenization events are universal to all modern human populations, they must actually date back to at least the common origin of modern humans ∼100,000–200,000 y ago. Regardless of exact timing, these data provide support for selection acting on these pseudogenes close to the common origin of modern humans.
Bacterial Pathogens as Selective Agents in Hominin Evolution?
Independent of exactly when fixation of these pseudogenes occurred in ancestral hominins, the question arises as to what selective forces were involved. Alteration of innate immune defense against invasive human neonatal pathogens such as GBS and E. coli K1 would exert a powerful selection pressure on reproductive success, and other prevalent microbial pathogens could also have subverted Siglec signaling to promote infection. Thus, in addition to other extant theories about the origin of modern humans, interactions of infectious agents with the innate immune system of early humans should be examined as a source of potential selection. After all, infectious agents are already widely recognized as selective agents to explain human polymorphisms (e.g., the role of malaria in selecting for the sickle cell hemoglobin trait). Alternative (but not mutually exclusive) hypotheses include Siglec-mediated overactivation of immune cells and/or changes in commensal–host interactions, which could have compromised reproductive success or the ability to care for the young. Additional studies are needed to further investigate the theoretical role of bacterial selection pressure during human origins. Strong selection by pathogens could result in severe population restrictions and bottlenecks, such as seen in hominin origins. If so, we may need to consider the possibility of an “infectious origin” of modern humans.
Materials and Methods
DNA samples and population groups are described in Results. PCR, RT-PCR, sequencing details, and details of flow cytometry for detection of Siglecs or intracellular TNF expression are in SI Appendix. Cells transfected with expression constructs for Siglec-13 or -17 with or without DAP12–FLAG were analyzed by coimmunoprecipitation and Western blotting (SI Appendix). Details of array fabrication and binding assays, transfection of Siglecs in mouse Raw264.7 cells, molecular and cellular assays, and human population genetic analysis are in SI Appendix. Interaction of Siglec-Fcs with bacteria was studied as described (33) with minor modifications (SI Appendix).
Supplementary Material
Acknowledgments
We thank Pascal Gagneux for valuable comments. This work was funded by grants listed in the SI Appendix.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
2A complete list of the National Institutes of Health Intramural Sequencing Center Comparative Sequencing Program Staff can be found in the SI Appendix.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119459109/-/DCSupplemental.
References
- 1.Schauer R. Sialic acids as regulators of molecular and cellular interactions. Curr Opin Struct Biol. 2009;19:507–514. doi: 10.1016/j.sbi.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen X, Varki A. Advances in the biology and chemistry of sialic acids. ACS Chem Biol. 2010;5:163–176. doi: 10.1021/cb900266r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwarzkopf M, et al. Sialylation is essential for early development in mice. Proc Natl Acad Sci USA. 2002;99:5267–5270. doi: 10.1073/pnas.072066199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Severi E, Hood DW, Thomas GH. Sialic acid utilization by bacterial pathogens. Microbiology. 2007;153:2817–2822. doi: 10.1099/mic.0.2007/009480-0. [DOI] [PubMed] [Google Scholar]
- 5.Varki A. Colloquium paper: Uniquely human evolution of sialic acid genetics and biology. Proc Natl Acad Sci USA. 2010;107(Suppl 2):8939–8946. doi: 10.1073/pnas.0914634107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis AL, et al. Innovations in host and microbial sialic acid biosynthesis revealed by phylogenomic prediction of nonulosonic acid structure. Proc Natl Acad Sci USA. 2009;106:13552–13557. doi: 10.1073/pnas.0902431106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao H, Crocker PR. Evolution of CD33-related siglecs: Regulating host immune functions and escaping pathogen exploitation? Immunology. 2011;132:18–26. doi: 10.1111/j.1365-2567.2010.03368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varki A, Angata T. Siglecs—the major subfamily of I-type lectins. Glycobiology. 2006;16:1R–27R. doi: 10.1093/glycob/cwj008. [DOI] [PubMed] [Google Scholar]
- 9.von Gunten S, Bochner BS. Basic and clinical immunology of Siglecs. Ann N Y Acad Sci. 2008;1143:61–82. doi: 10.1196/annals.1443.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez PH, Schnaar RL. Gangliosides in cell recognition and membrane protein regulation. Curr Opin Struct Biol. 2009;19:549–557. doi: 10.1016/j.sbi.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varki A. Since there are PAMPs and DAMPs, there must be SAMPs? Glycan “self-associated molecular patterns” dampen innate immunity, but pathogens can mimic them. Glycobiology. 2011;21:1121–1124. doi: 10.1093/glycob/cwr087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones C, Virji M, Crocker PR. Recognition of sialylated meningococcal lipopolysaccharide by siglecs expressed on myeloid cells leads to enhanced bacterial uptake. Mol Microbiol. 2003;49:1213–1225. doi: 10.1046/j.1365-2958.2003.03634.x. [DOI] [PubMed] [Google Scholar]
- 13.Carlin AF, et al. Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response. Blood. 2009;113:3333–3336. doi: 10.1182/blood-2008-11-187302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carlin AF, et al. Group B Streptococcus suppression of phagocyte functions by protein-mediated engagement of human Siglec-5. J Exp Med. 2009;206:1691–1699. doi: 10.1084/jem.20090691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angata T, Hayakawa T, Yamanaka M, Varki A, Nakamura M. Discovery of Siglec-14, a novel sialic acid receptor undergoing concerted evolution with Siglec-5 in primates. FASEB J. 2006;20:1964–1973. doi: 10.1096/fj.06-5800com. [DOI] [PubMed] [Google Scholar]
- 16.Cao H, et al. SIGLEC16 encodes a DAP12-associated receptor expressed in macrophages that evolved from its inhibitory counterpart SIGLEC11 and has functional and non-functional alleles in humans. Eur J Immunol. 2008;38:2303–2315. doi: 10.1002/eji.200738078. [DOI] [PubMed] [Google Scholar]
- 17.Lanier LL. DAP10- and DAP12-associated receptors in innate immunity. Immunol Rev. 2009;227:150–160. doi: 10.1111/j.1600-065X.2008.00720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turnbull IR, Colonna M. Activating and inhibitory functions of DAP12. Nat Rev Immunol. 2007;7:155–161. doi: 10.1038/nri2014. [DOI] [PubMed] [Google Scholar]
- 19.Pfirsch-Maisonnas S, et al. Inhibitory ITAM signaling traps activating receptors with the phosphatase SHP-1 to form polarized “inhibisome” clusters. Sci Signal. 2011;4:ra24. doi: 10.1126/scisignal.2001309. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, et al. Evolution of Siglec-11 and Siglec-16 genes in hominins. Mol Biol Evol. 2012 doi: 10.1093/molbev/mss077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell MC, Tishkoff SA. African genetic diversity: Implications for human demographic history, modern human origins, and complex disease mapping. Annu Rev Genomics Hum Genet. 2008;9:403–433. doi: 10.1146/annurev.genom.9.081307.164258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobs Z, et al. Ages for the Middle Stone Age of southern Africa: Implications for human behavior and dispersal. Science. 2008;322:733–735. doi: 10.1126/science.1162219. [DOI] [PubMed] [Google Scholar]
- 23.Gunz P, et al. Early modern human diversity suggests subdivided population structure and a complex out-of-Africa scenario. Proc Natl Acad Sci USA. 2009;106:6094–6098. doi: 10.1073/pnas.0808160106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White TD. Human origins and evolution: Cold Spring Harbor, deja vu. Cold Spring Harb Symp Quant Biol. 2009;74:335–344. doi: 10.1101/sqb.2009.74.016. [DOI] [PubMed] [Google Scholar]
- 25.Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reich D, et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Briggs AW, et al. Targeted retrieval and analysis of five Neandertal mtDNA genomes. Science. 2009;325:318–321. doi: 10.1126/science.1174462. [DOI] [PubMed] [Google Scholar]
- 29.Angata T, Margulies EH, Green ED, Varki A. Large-scale sequencing of the CD33-related Siglec gene cluster in five mammalian species reveals rapid evolution by multiple mechanisms. Proc Natl Acad Sci USA. 2004;101:13251–13256. doi: 10.1073/pnas.0404833101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmid CW. Does SINE evolution preclude Alu function? Nucleic Acids Res. 1998;26:4541–4550. doi: 10.1093/nar/26.20.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tishkoff SA, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Humphrey MB, et al. TREM2, a DAP12-associated receptor, regulates osteoclast differentiation and function. J Bone Miner Res. 2006;21:237–245. doi: 10.1359/JBMR.051016. [DOI] [PubMed] [Google Scholar]
- 33.Carlin AF, Lewis AL, Varki A, Nizet V. Group B streptococcal capsular sialic acids interact with siglecs (immunoglobulin-like lectins) on human leukocytes. J Bacteriol. 2007;189:1231–1237. doi: 10.1128/JB.01155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Przeworski M. Estimating the time since the fixation of a beneficial allele. Genetics. 2003;164:1667–1676. doi: 10.1093/genetics/164.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sabeti PC, et al. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 36.Zeng K, Fu YX, Shi S, Wu CI. Statistical tests for detecting positive selection by utilizing high-frequency variants. Genetics. 2006;174:1431–1439. doi: 10.1534/genetics.106.061432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng K, Shi S, Wu CI. Compound tests for the detection of hitchhiking under positive selection. Mol Biol Evol. 2007;24:1898–1908. doi: 10.1093/molbev/msm119. [DOI] [PubMed] [Google Scholar]
- 38.Hudson RR, Kreitman M, Aguadé M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Balakirev ES, Ayala FJ. Pseudogenes: Are they “junk” or functional DNA? Annu Rev Genet. 2003;37:123–151. doi: 10.1146/annurev.genet.37.040103.103949. [DOI] [PubMed] [Google Scholar]
- 40.Wen YZ, et al. Pseudogene-derived small interference RNAs regulate gene expression in African Trypanosoma brucei. Proc Natl Acad Sci USA. 2011;108:8345–8350. doi: 10.1073/pnas.1103894108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.