Abstract
Escherichia coli chemotaxis serves as a paradigm for the way living cells respond and adapt to changes in their environment. The chemotactic response has been characterized at the level of individual flagellar motors and in populations of swimming cells. However, it has not been previously possible to quantify accurately the adaptive response of a single, multiflagellated cell. Here, we use our recently developed optical trapping technique to characterize the swimming behavior of individual bacteria as they respond to sudden changes in the chemical environment. We follow the adaptation kinetics of E. coli to varying magnitudes of step-up and step-down changes in concentration of chemoattractant. We quantify two features of adaptation and how they vary with stimulus strength: abruptness (the degree to which return to prestimulus behavior occurs within a small number of run/tumble events) and overshoot (the degree of excessive response before the return to prestimulus behavior). We also characterize the asymmetry between step-up and step-down responses, observed at the single-cell level. Our findings provide clues to an improved understanding of chemotactic adaptation.
Keywords: bacterial chemotaxis, optical tweezers, single cell studies
Many species of bacteria swim by rotating helical filaments (flagella) driven by bidirectional rotary motors (1). In the peritrichously flagellated Escherichia coli, counterclockwise (CCW) rotation induces formation of a flagellar bundle that propels the cell forward in a “run.” Clockwise (CW) rotating flagella break from the bundle and cause the cell to change swimming direction abruptly in a “tumble” (2–4). A swimming cell alternates stochastically between running and tumbling, exploring its surroundings in a random walk (5). The flagellar motor’s rotational bias, and thus the cell’s swimming behavior, is modulated by the chemicals in the surrounding environment. Exposure to attractants causes cells to tumble less frequently whereas depletion of attractant causes them to tumble more, leading to net migration toward favorable environments (6). This chemotactic response is governed by a protein network that is well-characterized (7, 8) (SI Discussion). One hallmark of this network is its ability to adapt to a wide range of chemical environments. When cells are exposed to a sudden change in environment, they respond by temporarily changing their swimming behavior (tumbling more or less frequently), but then return over time to their prestimulus swimming state (9, 10). Chemotactic adaptation is believed to be exact (11), allowing cells to maintain a high sensitivity to their environment over a wide range of background chemoeffector concentrations.
Various techniques have been used to study the chemotactic response and adaptation in E. coli. By following populations of swimming cells, the relation between adaptation time and stimulus strength, as well as the robustness of exact adaptation, have been studied (10–13). Detailed features of the flagellar motor’s response to chemical stimuli of various forms have been characterized by tethering individual cells to the surface of a microscope slide and monitoring the motor’s rotational direction (9, 14–16). More recently, a Förster resonance energy transfer (FRET) reporter system was used to probe the activity of the essential kinase (CheA) in the chemotactic protein network from a population of cells (17–19). Despite providing invaluable information, the approaches above suffer significant limitations. Population measurements average over cell-to-cell variability, potentially masking important features of the single-cell response (12). At the other end, single-motor measurements may not accurately reflect the swimming behavior of the cell because this behavior arises from the collective state of multiple flagellar motors (3, 4).
In this study, we utilize our recently developed optical tweezers technique (20) to apply controlled step-up and step-down chemical stimuli to individual swimming E. coli cells and to monitor their chemotactic adaptation. The acquisition of trajectories from many individual cells, for a long duration (> 10 min) at high temporal resolution (approximately 100 Hz) allowed us to characterize adaptation kinetics at an unprecedented level of detail. In particular, we quantified two features of adaptation: (i) abruptness (the degree to which return to prestimulus behavior occurs within a small number of run/tumble events) and (ii) overshoot (the degree of excessive response before the return to prestimulus behavior). Though abruptness and overshoot have been previously reported in the literature (9, 16, 21–23), they have not yet been characterized in detail. Here, we quantify both features in response to both step-up and step-down stimuli across a broad range of stimulus strengths. We also characterize the striking asymmetry in the cell’s response to step-up and step-down stimuli. We suggest how our findings provide clues to an improved understanding of chemotactic adaptation.
Results
Optical Trapping Enables Following Adaptation Kinetics in Individual Swimming Cells.
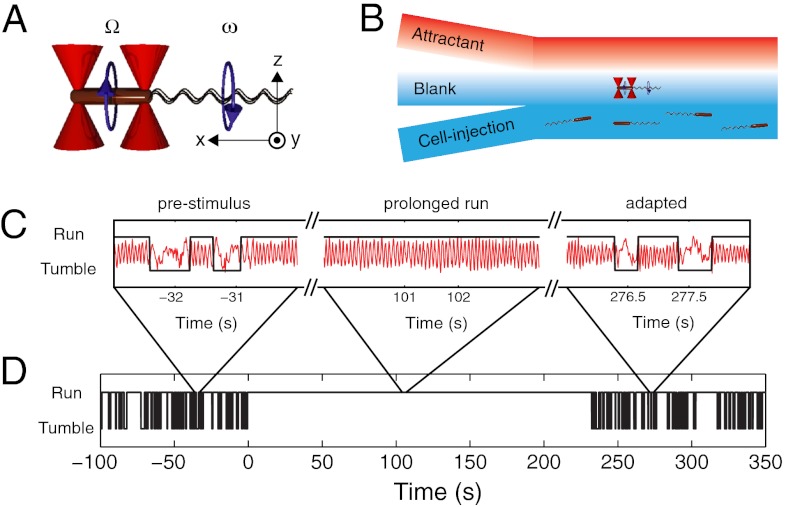
To enable precise long-term measurement of a single cell’s swimming behavior, we used our optical trapping assay (20) (Fig. 1). Briefly, two optical traps were used to hold a single cell by the two ends of its body and to orient it horizontally in the imaging plane (the xy plane in Fig. 1A). Movement of the trapped cell was monitored by imaging the trap light scattered by the cell onto position-sensitive photodiodes. For a freely swimming cell, rotation of the flagellar bundle causes the body to counter-rotate about its long axis during a run (24). During a tumble, the cell body moves erratically as it changes direction (3). For an optically trapped cell, both flagellar bundle and cell body are free to rotate even though the cell is immobilized in space. Thus, runs and tumbles manifest themselves in the trap as oscillatory and erratic signals from rotation and random motion of the cell body, respectively (20) (Fig. 1C). We have previously shown that trapped cells exhibit a normal swimming phenotype as reflected by the measured run/tumble statistics (20).
Fig. 1.
Measuring the chemotactic response of individual bacteria. (A) Schematic representation of an E. coli bacterium (brown cylinder) held by two optical traps (red cones). The cell body counter-rotates (Ω) in a direction opposite to the flagellar bundle’s rotation (ω). Experimental coordinates are also shown. (B) Schematic of the laminar-flow chamber used to establish chemical gradients. To apply a chemical stimulus, the trapped bacterium is moved perpendicularly to the flow direction, in or out of the channel containing chemoattractant. (C) Representative cell-body rotation signals from a trapped cell before stimulus (Left), during adaptation (Middle), and after adaptation is complete (Right). Runs and tumbles (black line) are distinguished by using an automated routine. (D) A long-term run/tumble binary time trace obtained from the same cell. Stimulus was applied at t = 0. For experimental details see SI Materials and Methods.
To deliver chemical stimuli to the optically trapped cells, we created a chemical attractant concentration profile in a laminar flow chamber. In this chamber, three separate streams containing different solutions merged into a central channel: a “cell-injection” stream, a “blank” stream, and an “attractant” stream (Fig. 1B). The cross-section of the flow chamber (100 μm × 1000 μm per inlet) and the flow speed (70 μm/s) ensured that fluid flow was laminar with minimal mixing, creating well-defined boundaries in the chemical profile along the direction perpendicular to the flow (the change from 10% to 90% of the maximum concentration occurred within approximately 300 μm) (SI Materials and Methods and Fig. S1 A–C). In a typical experiment, a swimming cell was captured from the cell-injection stream of the flow chamber containing many cells and oriented along the flow direction using the optical traps. By moving the flow chamber using a motorized translation stage, the trapped cell was then positioned into the blank stream containing trap motility buffer (TMB) (see Materials and Methods). Following measurement of the steady-state swimming behavior for up to 5 min, the trapped cell was moved rapidly to the attractant stream containing the chemoattractant l-aspartate (5, 11, 14–16, 25) and monitored for at least an additional 7 min. Because the trapped cells were moved at a speed of 100 μm/s, they experienced a chemical stimulus in the form of a step up in attractant concentration over a span of approximately 3 s (Fig. S1). A unique aspect of our technique is the ability to apply step-down stimuli simply by moving cells in the reverse manner. Step down is difficult to achieve in free-swimming assays, where flow cannot be used to remove chemoeffectors without flushing cells away.
Fig. 1C shows three short segments out of an approximately 5-min measurement (Fig. 1D) of a trapped cell undergoing a step up in l-aspartate concentration, from 0 μM to 100 μM. In the first segment the cell was in its steady-state and exhibited alternating periods of oscillatory and erratic signals corresponding to runs and tumbles. In the next segment the cell underwent a prolonged oscillation (a long run) in response to the applied chemical stimulus. The last segment shows the cell after it adapted to the new level of attractant concentration, where switching between oscillatory and erratic signals resumed. The trap signal was converted to a binary time series of runs and tumbles (Fig. 1D) as described previously (20) (SI Materials and Methods).
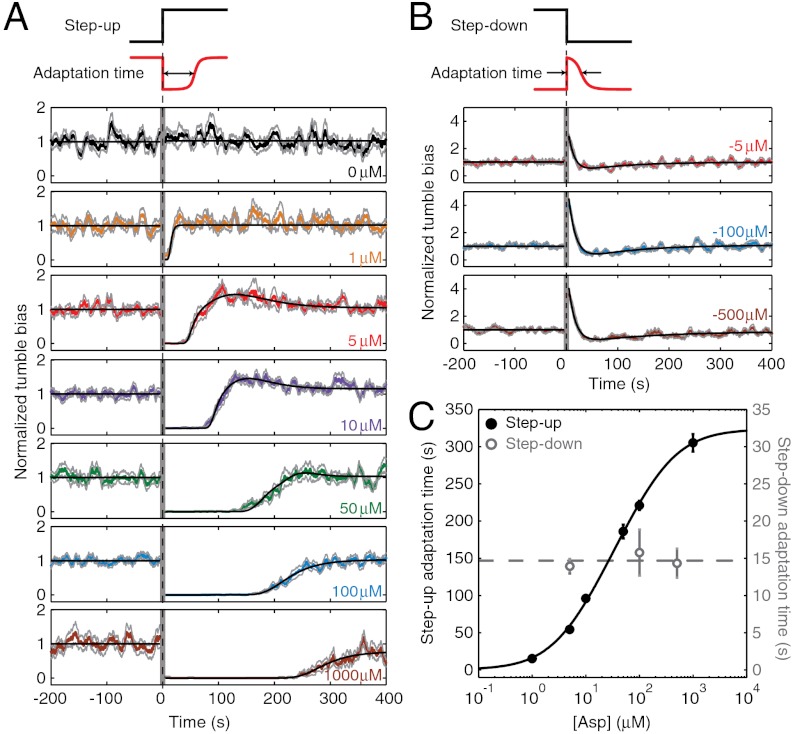
Using the methods described above, we characterized the adaptation of individual cells (E. coli strain RP437, wild type for chemotaxis) (26) in response to a step up in l-aspartate concentration of varying strength (0 μM to 1–1,000 μM) (Fig. 2). From each single-cell binary time series, we determined the adaptation response by calculating the tumble bias in a running 10-s time window (Fig. S2). When the adaptation response was averaged over many individual cells, the average response curve showed a gradual adaptation time course (Fig. 2A), similar to that observed in previous studies (11, 14, 17, 19, 25). For each stimulus, we also determined the average adaptation time, defined as the time elapsed between the application of the stimulus and the recovery of tumble bias to 50% of its prestimulus average value. The dependence of adaptation time on attractant concentration (Fig. 2C) exhibits the Michealis–Menten-like behavior reported in earlier studies (9, 10). The curve is also in quantitative agreement with a recent theoretical model for the chemotactic network (27). In the long term, after the transient response to the step-up stimulus, cells achieved a steady-state tumble bias. For each stimulus, we determined the exactness in adaptation, defined as the ratio of post- to prestimulus steady-state tumble biases. For the majority of stimulus strengths assayed, adaptation was exact within our experimental error (Fig. S3), as expected from previous studies (11, 16, 19). [For the highest step up in l-aspartate concentration (1000 μM), we did observe a decrease in steady-state tumble bias poststimulus, though we attribute this to limited observation time]. This agreement with previous experimental and theoretical results on chemotactic adaptation, taken together with our previous report regarding the free-swimming behavior of trapped cells (20), demonstrates that optically trapped cells exhibit a normal behavior in all aspects of motility, including chemotactic response.
Fig. 2.
Population-averaged response to step-up and step-down stimuli. (A) The population-averaged response to a step up in attractant concentration, delivered at t = 0. Individual tumble bias traces were normalized by the mean prestimulus tumble bias before averaging across the population. Solid colored lines designate the averaged response at different stimulus levels [changing from 0 μM to 0 (control), 1, 5, 10, 50, 100, and 1,000 μM of l-aspartate, color coded in black, orange, red, purple, green, blue, and brown, respectively). Light gray lines denote one standard error above and below the mean. Black lines describe fit to a theoretical model of the chemotaxis network, with an added overshoot feature. The vertical gray band near t = 0 corresponds to the time when cells were moved along the chemical gradient and data was not recorded. The number of different cells (each cell was stimulated only once) included at each stimulus level: n = 10, 13, 22, 26, 20, 39, and 14, from top to bottom. (B) Same as A, for step-down stimuli (changing from 5, 100, and 500 μM to 0 μM of l-aspartate, color coded in red, blue, and brown, respectively). In this case, the theoretical fit (black lines) after stimulus is the sum of two exponentials. The number of different cells (each cell was stimulated only once) included at each stimulus level: n = 13, 15, and 14, from top to bottom. (C) The average adaptation time as a function of stimulus strength. Adaptation time in step-up experiments (solid black circles) was the time at which the model fit recovered to half the pre-stimulus tumble bias. Error bars are standard errors obtained from bootstrapping. The solid black line is a fit to a receptor free-energy model (27). The dashed gray line is the mean of the three step-down data points (open gray circles). See SI Materials and Methods for more details of data analysis and modeling.
Individual Cells Exhibit a Stimulus-Dependent Abruptness of Adaptation.
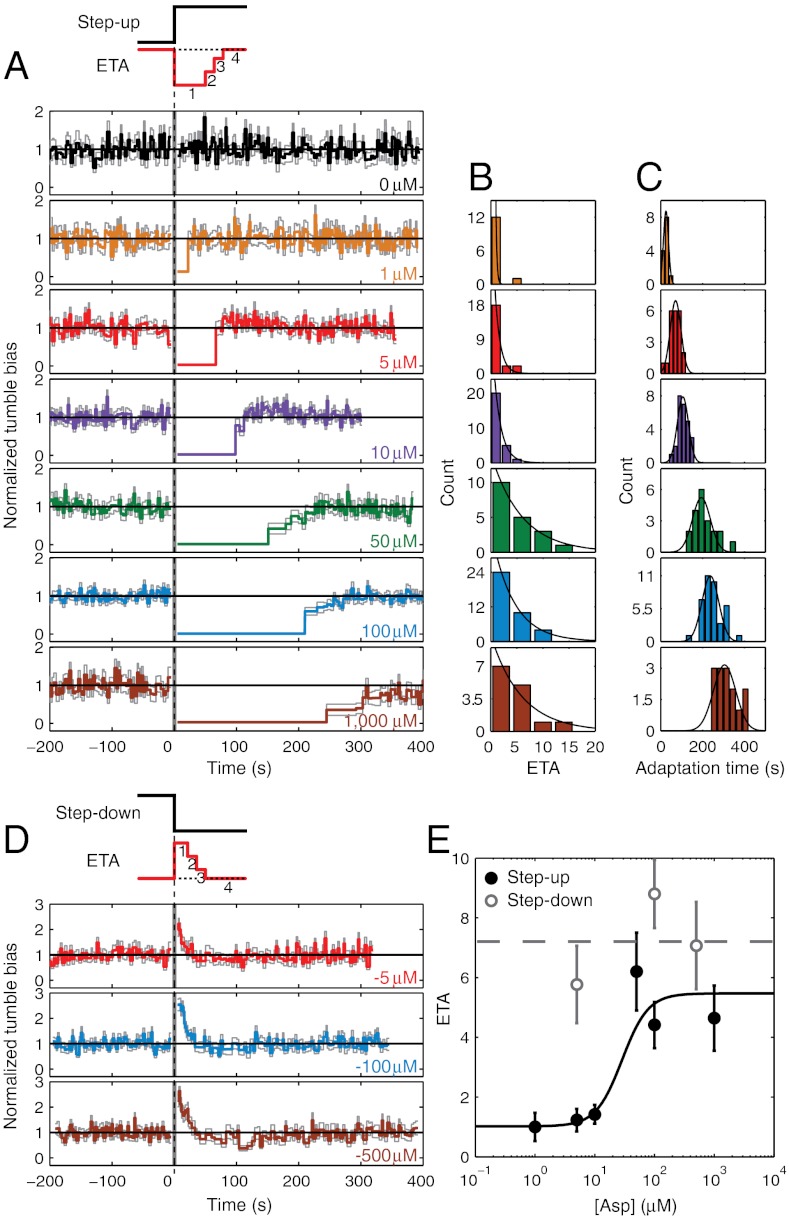
The average response curves in Fig. 2A, though useful when comparing our results to previous studies, mask important features of adaptation kinetics at the single-cell level. As seen from the binary time traces (Fig. S4), individual cells exhibit large cell-to-cell variations in adaptation times because of the stochastic nature of the underlying network reactions as well as variability in the chemotaxis network protein numbers (12). Thus, the population-averaged traces in Fig. 2A smooth over cell-to-cell differences in adaptation kinetics. In order to elucidate the “typical” behavior of the individual cell, we analyzed our data using a recently introduced scheme (16) in which individual traces are indexed by “events”—run and tumble pairs—rather than time (Materials and Methods and Fig. S5). Fig. 3A displays the result of averaging individual traces according to run/tumble event number, aligned relative to the delivery of the stimulus (i.e., run/tumble events were enumerated from the time the stimulus was applied). The ordinate represents the mean tumble bias, and the abscissa, the mean duration of the i- th run/tumble pair averaged across the cell population. This averaging scheme is not subject to stochastic variability in run or tumble duration, and thus better captures the typical adaptation kinetics of individual cells (16).
Fig. 3.
Abruptness of adaptation to step-up and step-down stimuli. (A) The event-based average response to a step up in attractant concentration, delivered at t = 0. Individual tumble-bias traces were normalized by the mean prestimulus tumble bias. Color notations as in Fig. 2. The vertical gray band near t = 0 corresponds to the time when cells were moved along the chemical gradient and data was not recorded. The same raw data as in Fig. 2A were used in this analysis. (B) Histograms of the number of run/tumble pair ETA from individual cells. Black line is a fit to an exponential. (C) Histograms of adaptation time from individual cells. Black line is a fit to a Gaussian. (D) Same as A, for a step-down stimulus. The same raw data as in Fig. 2B were used in this analysis. (E) The average number of ETA as a function of stimulus strength for step up (black solid circles, values obtained from C) and step down (open gray circles). Error bars designate standard error of the mean. The solid black line is a fit to a sigmoidal function. The dashed gray line is the mean of the three step-down data points. See SI Materials and Methods for details of the event-based analysis.
In comparison to the population-averaged response curves in Fig. 2A, the corresponding event-averaged curves in Fig. 3A reveal the abruptness with which individual cells adapt. The predominant adaptive response to a step-increase in attractant consisted of a single, long run/tumble event (specifically, a single long run; tumble duration did not change significantly) (Fig. S6 A and B), after which the cell’s swimming returned to its prestimulus behavior. Abrupt adaptation kinetics at the level of individual motors were reported many years ago for the case of saturating stimuli (9) and, more recently, for small stimuli (16). However, a detailed characterization of this feature in individual swimming cells, over a wide range of chemical stimulus strengths, has not been conducted. To quantify adaptation abruptness in individual cells we determined the number of run/tumble pair “events to adaptation” (ETA) for individual cells at each stimulus level (Materials and Methods and Fig. S7). Histograms of single-cell ETAs are shown in Fig. 3B, and are well described by exponential distributions. The event-based analysis was also used to define an adaptation time for individual cells, determined by summing the durations of all run/tumble pair events leading up to adaptation. Fig. 3C shows the corresponding histograms of single-cell adaptation times at each stimulus level, with fits to normal distributions. We note that the single-cell–based estimates of adaptation times are in good agreement with the population-based estimates (Fig. 2C and Fig. S8), as expected. Numerical simulations support the idea that cell-to-cell variability in adaptation time arises from protein-number fluctuations in the chemotaxis network (Fig. S4B and SI Materials and Methods). Consistent with this idea, we found the variability in adaptation times within the same cell to be smaller than the cell-to-cell variability, in agreement with previous reports (9, 12). The coefficient of variation in adaptation times in individual cells that underwent two consecutive stimulations was 0.056 ± 0.013 (mean ± SEM, 9 cells), whereas the coefficient of variation in adaptation times between pairs of cells was 0.189 ± 0.006 (mean ± SEM, 496 pairs) for similar stimulation strengths.
Examining the ETA reveals an unexpected feature of adaptation: The abruptness in an individual cell’s adaptive response depends on the stimulus strength. At low stimulus levels (up to 10 μM), the majority of individual cells adapted within one event. In contrast, for higher stimulus levels (50–1,000 μM), event durations were typically longer than their steady-state value for several events following the stimulus. In Fig. 3E, the average ETAs (extracted from the histograms in Fig. 3B) are plotted against the stimulus strength. The average ETA exhibits an almost stepwise increase from one to approximately six as the stimulus level exceeds 50 μM. ETAs obtained from the population-averaged event-based adaptation curves in Fig. 3A exhibit similar behavior (Fig. S8). Below we discuss possible explanations for the stimulus-dependent abruptness of adaptation exhibited by individual cells.
Individual Cells Exhibit an Overshoot Response.
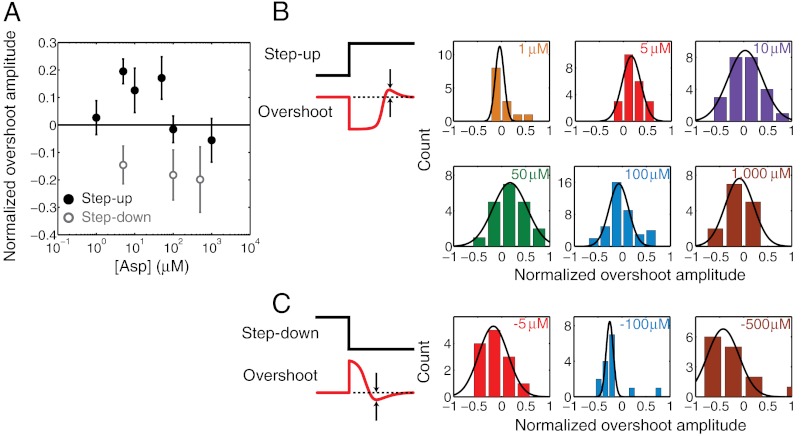
After the application of a step-up stimulus and the resulting long run/tumble event(s) discussed above, many cell traces exhibited an overshoot, during which the tumble bias exceeded the prestimulus steady-state. The tumble bias eventually returned to the prestimulus value. This feature was observed in population-averaged traces (Fig. 2A) and event-averaged traces (Fig. 3A) alike, and quantified at the single-cell level (Fig. S7 and SI Materials and Methods). Fig. 4A displays the average amplitude of the overshoot—defined as the fractional excess tumble bias over the poststimulus steady-state—for different stimulus strengths. Interestingly, the overshoot amplitude exhibited a nonmonotonic dependence on stimulus strength, negligible at our lowest (1 μM) and highest (1 mM) stimulus strengths but peaking to a value of approximately 20% at intermediate (5–50 μM) strengths. An overshoot response of individual motors was reported many years ago (9) but is absent from later studies of chemotactic adaptation (11, 14, 19) (see Discussion). Fig. 4B displays the corresponding single-cell histograms of the overshoot.
Fig. 4.
Overshoot response to step-up and step-down stimuli. (A) The overshoot amplitude, normalized by the steady-state tumble bias and averaged over individual cells, is plotted as a function of the step up (solid black circles) and step down (gray open circles) stimulus. Error bars designate standard error of the mean. (B) Histograms of single-cell overshoot amplitudes in response to varying magnitudes of step-up stimuli. (C) Same as B, for step-down stimuli. Black lines are fits to a Gaussian. Color notations and sample sizes at each stimulus level are the same as in Fig. 3. See SI Materials and Methods for details of the overshoot calculation.
Adaptation Kinetics Show Asymmetry in Response to a Step Up Versus a Step Down.
In addition to the above measurements, we also quantified the response of individual cells to a step down in l-aspartate concentration. In agreement with recent reports (19), the chemotactic response was not merely a mirror image of that seen for a step-up stimulus; distinctly different adaptation kinetics were observed in the two cases (compare Fig. 2 A and B). We characterized this asymmetry between step-up and step-down responses in detail. Cells adapted to step-down stimuli in much shorter times (6). Whereas adaptation times for the step-up stimuli ranged from approximately 15 s to over 4 min in the range of concentration jumps tested, adaptation times for the step-down stimuli saturated at approximately 15 s and showed little variation over two orders of magnitude change in the step-down concentration jumps (Fig. 2C).
Analysis of individual cell traces (Fig. 3D) revealed additional differences. In contrast to adaptation to step-up stimuli, the average number of run/tumble events before adaptation to a step down was consistently high (approximately seven) and was largely independent of stimulus strength (Fig. 3E). This behavior is explained by the fact that run/tumble events were significantly shorter during step-down stimuli compared to events during a step up (Fig. S6 C and D) and the adaptation time was uniform (approximately 15 s) across the range of stimulus strengths. Finally, adaptation traces exhibited significant overshoot (approximately 20%) at all stimulus strengths tested (Fig. 4 A and C). Overshoot was noticeable in the population-averaged traces as well (Fig. 2B). Below we discuss possible explanations for the observed asymmetry between step-up and step-down responses in individual cells.
Discussion
Our ability to perturb the chemotaxis network through step-up and step-down changes in chemoattractant concentration and to measure the response of the individual cell reveals features of the adaptation kinetics that are masked in population-averaged measurements. Specifically, the precise, long-term characterization of chemotactic adaptation allowed us to quantify two features of adaptation kinetics in individual cells, and how they vary as a function of the stimulus strength.
A first such feature is the abrupt adaptation to a step up in attractant concentration. Upon stimulation, swimming cells entered a run of extended duration, but returned to their steady-state behavior after a small number of run/tumble event pairs (Fig. 3A). As seen from the histograms of ETA values in Fig. 3B, the most common response shown by cells at all stimulus levels was a single, prolonged run. The average number of events until adaptation ranged between one and six, tending to increase with stimulus strength (Fig. 3E). In contrast, adaptation to a step down in attractant concentration involved a larger number of run/tumble event pairs, exhibiting no clear dependence on the stimulus strength (Fig. 3E).
In the literature, chemotactic adaptation is typically described as a gradual process (6, 11, 14, 17, 19, 25). It is important to note, however, that these studies all involved averaging over multiple cells. As noted above, such averaging masks important features of single-cell adaptation kinetics because of the asynchrony in adaption between different cells (compare Figs. 2A and 3A). In studies where the response of individual flagellar motors was examined, the motors were described to undergo abrupt switches in behavior during the course of adaptation (9, 16), but this abruptness was not characterized in detail. Our measurements extend these findings by quantifying the level of abruptness as a function of stimulus strength, and by moving from the level of single flagellar motors to the (physiologically relevant) whole-cell swimming behavior.
What is the source of abrupt adaptation, and what makes the abruptness stimulus-dependent? As noted above, abruptness is already observed at the single-motor level (16). Therefore, the source of abruptness cannot be in the transition from the individual motors to the whole-cell behavior. Rather, the source must lie upstream in the cascade of interactions. Prima facie, a natural candidate to consider is the switch-like dependence of the flagellar rotational state on CheY-P level (the signaling molecule that controls the rotational bias of the motor) (28, 29). However, a simple theoretical argument shows that the cooperativity exhibited by the motor (30) has little to no effect on the abruptness of adaptation (Fig. S9A and SI Discussion). This claim is further supported by numerical simulations (Fig. S10 and SI Materials and Methods). Instead, we believe the evidence points towards a role for strongly interacting receptor clusters in creating the stimulus-dependent adaptation abruptness observed in our experiments. Several experiments on receptor interaction and dynamics in addition to our own theoretical analysis of the chemotactic network support this view (SI Discussion). An alternate mechanism for abruptness may be the recently discovered remodeling of FliM, a component of the flagellar motor to which CheY-P binds, during adaptation (31). Initial theoretical analysis based on this new result demonstrates that remodeling may lead to more abrupt adaptation, but it is unclear how it would explain the dependence of abruptness on stimulus strength (SI Discussion and Figs. S9B and S10D).
A second feature quantified in this work is the degree of overshoot in chemotactic response to step-up and step-down stimuli. Overshoot was evident at intermediate values of step-up stimulus (5–50 μM), and at all values of step-down stimulus (Fig. 4A). To the best of our knowledge, there is only one previous experimental account of chemotactic overshoot, in tethered cells (9). Curiously, other studies of chemotactic adaptation did not report an overshoot, instead describing a monotonous temporal response (6, 11, 14, 17, 19, 25). We speculate that, at least in some of these studies, the reason for failing to detect an overshoot was the insufficient duration of the experiments or an insufficient temporal resolution. The specific choice of stimulus level is also critical for the degree of overshoot (Fig. 4A), making this feature quite easy to miss. The potential mechanisms for an overshoot may provide additional arguments as to why it has not been widely observed. In general, an overshoot response may occur whenever different components of the network adapt at different rates. A recent theoretical model of the chemotaxis network (23) postulates that the overshoot response is caused by the differences in methylation kinetics between different types of receptors. Interestingly, one of the studies mentioned above (19) was conducted using a mutant strain that only expressed a single receptor type, potentially explaining why an overshoot was not reported. FliM remodeling (31) may be another potential candidate mechanism for overshoot, if the remodeling kinetics differ from those of CheA adaptation (SI Discussion and Fig. S10D). This mechanism would account for a lack of overshoot observed in studies probing the chemotaxis network upstream of the flagellar motor (e.g., at the level of CheA). Further work will be necessary to determine the source of the overshoot.
Lastly, we observed several differences in the adaptation response to step-down stimuli compared to the response to a step up. First, we observed that cells adapted much more rapidly to step-down stimuli (Fig. 2B), in agreement with the existing literature (6, 9, 19). It is generally believed that this difference arises from up-regulation of CheB activity by CheA during the step-down response (19, 32). Second, we observed that adaption times for step-down stimuli were independent of stimulus strength. This result conforms to measurements of swimming and tethered cells, in which short adaptation times (< 15 s) were consistently observed for high step-down stimulus strength (6, 9). However, it is in marked contrast with FRET measurements of CheA activity, which revealed longer CheA adaptation times that increased with higher step-down stimulus strength (19). The discrepancy between the FRET study and our own suggests that the source for stimulus independence may lie downstream from CheA in the chemotaxis network, possibly at the level of the flagellar motor or further downstream, at the mapping between individual motors and the whole-cell swimming behavior. Third, the step-down response exhibited a clear overshoot at all stimulus strengths tested, in contrast to step up (Fig. 4A). Because the overshoot arises from two components of the network adapting at widely different rates, we hypothesize that the overshoot may persist over a wider range of stimuli (because CheB kinetics are consistently faster for step down compared to those of CheR for step up). Finally, at all stimulus levels tested, the ETAs for step-down adaptation were consistently larger than for step up (Fig. 4D). In contrast, a prior single-motor study (9) reported only one prolonged CW event prior to adaptation to a step-down stimulus. We speculate that this discrepancy may reflect the differences between single-motor and whole-cell swimming behavior. As the CW bias increases, it is known that CW intervals in single motors increase in duration, whereas tumble durations in swimming cells remain relatively constant (29). Our data is consistent with this picture: Tumble durations remain short throughout the course of step-down adaptation (Fig. S6). Further measurements will be necessary to resolve these many interesting questions.
This last point motivates the need for investigating chemotaxis at the whole-cell level. The cell’s swimming phenotype is the complex outcome of multiple flagellar motors acting together. On one hand, individual motors are independent entities that switch stochastically between CCW and CW rotation (33–35). On the other hand, motors exhibit some synchrony because of common control via diffusive CheY-P fields within the cell (36) and, potentially, because of hydrodynamic interactions outside the cell (37). The mapping that connects the conformational state of individual flagella to the cell swimming state (3, 4) has remained elusive, largely due to the lack of quantitative experimental results. Recent theoretical studies predict that cells with different numbers of flagella should exhibit different swimming behavior (38, 39). Our data may provide important clues as to the way multiple motors collectively produce the swimming behavior of a cell. As discussed above, certain features of adaptation kinetics observed at the whole-cell level are reproduced at the single-motor level (and upstream in the network) while others are not. These discrepancies may reflect differences in the way the cell enters into the tumbling state, when the flagellar bundle is disrupted, and exits from that state, when the bundle reforms.
In summary, our ability to elucidate the mechanisms governing bacterial chemotaxis is driven first and foremost by our experimental ability to characterize the system’s behavior with increasing resolution and precision (17, 20, 30, 40). Here, by following the chemotactic response of individual multiflagellated cells in unprecedented detail, we were able to quantify fine kinetic features of adaptation, providing unique insights into the workings of the chemotactic network. Further technological enhancements to our ability to quantify cellular parameters during the chemotactic response will provide additional findings and allow us to further refine our understanding of the chemotaxis system. A promising future direction is the addition of a high spatial resolution fluorescence imaging module to our trapping device, which will allow us to follow in real time the spatiotemporal dynamics of key cellular players—such as flagella, membrane receptors, and intracellular proteins—as the chemotactic response is taking place.
Materials and Methods
An overnight culture of E. coli strain RP437 (wild type for chemotaxis) (26) was diluted 100-fold into 1 ml tryptone broth and grown at 30 °C for 4.5 h (OD600 approximately 0.5). Chemotaxis experiments were conducted in “trap motility buffer” (TMB). TMB contained 70 mM NaCl, 0.1 mM methionine, 100 mM Tris-Cl, 2% (wt/vol) glucose, and an oxygen-scavenging system (80 μg ml-1 glucose oxidase and 13 μg ml-1 catalase). A detailed description of the optical tweezers and flow cell design can be found elsewhere (20).
Determination of runs and tumbles from the optical trap signal was done as described previously (20). The run/tumble binary time traces were subsequently analyzed using two different methods. In the first method, the tumble bias was determined from a 10-second moving window by calculating the fraction of time the cell spent tumbling within the window (Fig. S2). Adaptation parameters were obtained from the average of a population of cells that experienced the same stimulus. In the second method, the run/tumble binary time traces were analyzed by pairing each run with its subsequent tumble (16). For each run/tumble pair event we determined a corresponding tumble bias value [tumble duration/(run duration + tumble duration)] and a duration (run duration + tumble duration). Adaptation parameters were then obtained from individual cell traces. Population averages were also obtained from the event-based analysis. Here, the average tumble bias and duration were determined for each run/tumble event, enumerated relative to the application of the stimulus, across the cell population. Detailed descriptions of the experimental setup and the data-analysis methods are provided in SI Materials and Methods.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Chris Rao for advice. We thank members of the Chemla, Golding, Rao, and Ha Laboratories for providing help with experiments. The work was supported by US National Science Foundation Grant 082265 (Physics Frontiers Center: Center for the Physics of Living Cells). Work in the Chemla Laboratory is supported by US National Science Foundation Grant MCB 09-52442 (CAREER), the Burroughs Wellcome Fund–Career Awards at the Scientific Interface (CASI), and the Alfred P. Sloane Research Fellowship. Work in the Golding Laboratory is supported by National Institutes of Health Grant R01GM082837, Human Frontier Science Program Grant RGY 70/2008, and Welch Foundation Grant Q-1759.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120218109/-/DCSupplemental.
References
- 1.Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 2.Silverman M, Simon M. Flagellar rotation and the mechanism of bacterial motility. Nature. 1974;249:73–74. doi: 10.1038/249073a0. [DOI] [PubMed] [Google Scholar]
- 3.Turner L, Ryu WS, Berg HC. Real-time imaging of fluorescent flagellar filaments. J Bacteriol. 2000;182:2793–2801. doi: 10.1128/jb.182.10.2793-2801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darnton NC, Turner L, Rojevsky S, Berg HC. On torque and tumbling in swimming Escherichia coli. J Bacteriol. 2007;189:1756–1764. doi: 10.1128/JB.01501-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berg HC, Brown DA. Chemotaxis in Escherichia coli analyzed by 3-dimensional tracking. Nature. 1972;239:500–504. doi: 10.1038/239500a0. [DOI] [PubMed] [Google Scholar]
- 6.Macnab RM, Koshland DE., Jr The gradient-sensing mechanism in bacterial chemotaxis. Proc Natl Acad Sci USA. 1972;69:2509–2512. doi: 10.1073/pnas.69.9.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker MD, Wolanin PM, Stock JB. Signal transduction in bacterial chemotaxis. BioEssays. 2006;28:9–22. doi: 10.1002/bies.20343. [DOI] [PubMed] [Google Scholar]
- 8.Wadhams GH, Armitage JP. Making sense of it all: Bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 9.Berg HC, Tedesco PM. Transient response to chemotactic stimuli in Escherichia coli. Proc Natl Acad Sci USA. 1975;72:3235–3239. doi: 10.1073/pnas.72.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spudich JL, Koshland DE., Jr Quantitation of the sensory response in bacterial chemotaxis. Proc Natl Acad Sci USA. 1975;72:710–713. doi: 10.1073/pnas.72.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alon U, Surette MG, Barkai N, Leibler S. Robustness in bacterial chemotaxis. Nature. 1999;397:168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- 12.Spudich JL, Koshland DE., Jr Non-genetic individuality: Chance in the single cell. Nature. 1976;262:467–471. doi: 10.1038/262467a0. [DOI] [PubMed] [Google Scholar]
- 13.Barkai N, Leibler S. Robustness in simple biochemical networks. Nature. 1997;387:913–917. doi: 10.1038/43199. [DOI] [PubMed] [Google Scholar]
- 14.Segall JE, Block SM, Berg HC. Temporal comparisons in bacterial chemotaxis. Proc Natl Acad Sci USA. 1986;83:8987–8991. doi: 10.1073/pnas.83.23.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Block SM, Segall JE, Berg HC. Adaptation kinetics in bacterial chemotaxis. J Bacteriol. 1983;154:312–323. doi: 10.1128/jb.154.1.312-323.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park H, et al. Interdependence of behavioural variability and response to small stimuli in bacteria. Nature. 2010;468:819–823. doi: 10.1038/nature09551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sourjik V, Berg HC. Receptor sensitivity in bacterial chemotaxis. Proc Natl Acad Sci USA. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimizu TS, Tu Y, Berg HC. A modular gradient–sensing network for chemotaxis in Escherichia coli revealed by responses to time-varying stimuli. Mol Syst Biol. 2010;6:382. doi: 10.1038/msb.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meir Y, Jakovljevic V, Oleksiuk O, Sourjik V, Wingreen NS. Precision and kinetics of adaptation in bacterial chemotaxis. Biophys J. 2010;99:2766–2774. doi: 10.1016/j.bpj.2010.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Min TL, et al. High-resolution, long-term characterization of bacterial motility using optical tweezers. Nat Methods. 2009;6:831–835. doi: 10.1038/nmeth.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanders DA, Koshland DE., Jr Receptor interactions through phosphorylation and methylation pathways in bacterial chemotaxis. Proc Natl Acad Sci USA. 1988;85:8425–8429. doi: 10.1073/pnas.85.22.8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen CH, Sourjik V, Wingreen NS. A dynamic-signaling-team model for chemotaxis receptors in Escherichia coli. Proc Natl Acad Sci USA. 2010;107:17170–17175. doi: 10.1073/pnas.1005017107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lan G, Schulmeister S, Sourjik V, Tu Y. Adapt locally and act globally: Strategy to maintain high chemoreceptor sensitivity in complex environments. Mol Syst Biol. 2011;7:475. doi: 10.1038/msb.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chattopadhyay S, Moldovan R, Yeung C, Wu XL. Swimming efficiency of bacterium Escherichia coli. Proc Natl Acad Sci USA. 2006;103:13712–13717. doi: 10.1073/pnas.0602043103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jasuja R, Keyoung J, Reid GP, Trentham DR, Khan S. Chemotactic responses of Escherichia coli to small jumps of photoreleased L-aspartate. Biophys J. 1999;76:1706–1719. doi: 10.1016/S0006-3495(99)77329-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parkinson JS, Houts SE. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keymer JE, Endres RG, Skoge M, Meir Y, Wingreen NS. Chemosensing in Escherichia coli: Two regimes of two-state receptors. Proc Natl Acad Sci USA. 2006;103:1786–1791. doi: 10.1073/pnas.0507438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scharf BE, Fahrner KA, Turner L, Berg HC. Control of direction of flagellar rotation in bacterial chemotaxis. Proc Natl Acad Sci USA. 1998;95:201–206. doi: 10.1073/pnas.95.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alon U, et al. Response regulator output in bacterial chemotaxis. EMBO J. 1998;17:4238–4248. doi: 10.1093/emboj/17.15.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cluzel P, Surette M, Leibler S. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science. 2000;287:1652–1655. doi: 10.1126/science.287.5458.1652. [DOI] [PubMed] [Google Scholar]
- 31.Yuan J, Branch RW, Hosu BG, Berg HC. Adaptation at the output of the chemotaxis signalling pathway. Nature. 2012;484:233–236. doi: 10.1038/nature10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anand GS, Goudreau PN, Stock AM. Activation of methylesterase CheB: Evidence of a dual role for the regulatory domain. Biochemistry. 1998;37:14038–14047. doi: 10.1021/bi980865d. [DOI] [PubMed] [Google Scholar]
- 33.Macnab RM, Han DP. Asynchronous switching of flagellar motors on a single bacterial-cell. Cell. 1983;32:109–117. doi: 10.1016/0092-8674(83)90501-9. [DOI] [PubMed] [Google Scholar]
- 34.Ishihara A, Segall JE, Block SM, Berg HC. Coordination of flagella on filamentous cells of Escherichia-coli. J Bacteriol. 1983;155:228–237. doi: 10.1128/jb.155.1.228-237.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korobkova EA, Emonet T, Park H, Cluzel P. Hidden stochastic nature of a single bacterial motor. Phys Rev Lett. 2006;96:058105. doi: 10.1103/PhysRevLett.96.058105. [DOI] [PubMed] [Google Scholar]
- 36.Terasawa S, et al. Coordinated reversal of flagellar motors on a single Escherichia coli cell. Biophys J. 2011;100:2193–2200. doi: 10.1016/j.bpj.2011.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reichert M, Stark H. Synchronization of rotating helices by hydrodynamic interactions. Eur Phys J E Soft Matter. 2005;17:493–500. doi: 10.1140/epje/i2004-10152-7. [DOI] [PubMed] [Google Scholar]
- 38.Vladimirov N, Lebiedz D, Sourjik V. Predicted auxiliary navigation mechanism of peritrichously flagellated chemotactic bacteria. PLoS Comput Biol. 2010;6:e1000717. doi: 10.1371/journal.pcbi.1000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sneddon MW, Pontius W, Emonet T. Stochastic coordination of multiple actuators reduces latency and improves chemotactic response in bacteria. Proc Natl Acad Sci USA. 2011;109:805–810. doi: 10.1073/pnas.1113706109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altindal T, Chattopadhyay S, Wu XL. Bacterial chemotaxis in an optical trap. PLoS One. 2011;6:e18231. doi: 10.1371/journal.pone.0018231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.