Abstract
The transcriptional coactivator PPAR gamma coactivator 1 α (PGC-1α) is a key regulator of metabolic processes such as mitochondrial biogenesis and respiration in muscle and gluconeogenesis in liver. Reduced levels of PGC-1α in humans have been associated with type II diabetes. PGC-1α contains a negative regulatory domain that attenuates its transcriptional activity. This negative regulation is removed by phosphorylation of PGC-1α by p38 MAPK, an important kinase downstream of cytokine signaling in muscle and β-adrenergic signaling in brown fat. We describe here the identification of p160 myb binding protein (p160MBP) as a repressor of PGC-1α. The binding and repression of PGC-1α by p160MBP is disrupted by p38 MAPK phosphorylation of PGC-1α. Adenoviral expression of p160MBP in myoblasts strongly reduces PGC-1α's ability to stimulate mitochondrial respiration and the expression of the genes of the electron transport system. This repression does not require removal of PGC-1α from chromatin, suggesting that p160MBP is or recruits a direct transcriptional suppressor. Overall, these data indicate that p160MBP is a powerful negative regulator of PGC-1α function and provide a molecular mechanism for the activation of PGC-1α by p38 MAPK. The discovery of p160MBP as a PGC-1α regulator has important implications for the understanding of energy balance and diabetes.
Keywords: PGC-1α, Mybbp1a, mitochondria, repressor, p38 MAPK
Most biological programs involving alterations in gene expression are regulated by changes in amounts or activities of DNA-binding transcription factors. However, recent evidence has shown that transcriptional coactivators can be important targets for physiologic regulation. PPAR gamma coactivator 1 α (PGC-1α) has emerged as a key regulator of several aspects of mammalian energy metabolism. It was originally cloned as a binding partner for PPARγ (Puigserver et al. 1998), but interacts with and coactivates many transcription factors, such as nuclear respiratory factor 1 (NRF-1), myocyte-enhancing factor-2C (MEF2C), and most members of the nuclear hormone receptor family (for review, see Puigserver and Spiegelman 2003).
PGC-1α activates a broad program of adaptive thermogenesis characteristic of brown fat cells when expressed in white fat cells (Puigserver et al. 1998; Tiraby et al. 2003). This is characterized by mitochondrial biogenesis and an induction of the uncoupling protein 1, resulting in elevated respiration. PGC-1α expression in muscle cells leads to elevated respiration and oxidative phosphorylation with increases in the expression of mitochondrial genes and the glucose transporter GLUT4 (Wu et al. 1999; Michael et al. 2001; St-Pierre et al. 2003). Importantly, expression of PGC-1α in skeletal muscle in vivo via a transgene triggers the conversion of type IIb muscle fibers to type IIa and type I fibers, as evidenced by mitochondrial biogenesis and expression of myofibrillar proteins characteristic of slow-twitch muscle (Lin et al. 2002; Handschin et al. 2003). Most recently, decreases in skeletal muscle PGC-1α have been associated with human type 2 diabetes and those at risk for the development of diabetes (Mootha et al. 2003; Patti et al. 2003). PGC-1α is also associated with increased oxidative capacity in heart. It is dramatically induced at birth when the heart has an increased requirement for mitochondrial respiration (Lehman et al. 2000). Moreover, PGC-1α stimulates fatty acid oxidation in heart, which provides fuel for the respiratory chain (Vega et al. 2000).
PGC-1α is induced in liver in response to fasting, and activates key genes of the fasting response, including the genes of the gluconeogenic pathway and β-oxidation of fatty acids (Herzig et al. 2001; Yoon et al. 2001; Puigserver et al. 2003; Rhee et al. 2003). This is of critical importance in diabetes because dysregulated gluconeogenesis is a key component of the hyperglycemia characteristic of both type I and type II diabetes.
In addition to these physiological changes in PGC-1α gene expression, this coactivator is also controlled at the posttranslational level. PGC-1α contains a negative regulatory region between amino acids 170 and 350, which reduces the function of the transcriptional activation domain located between amino acids 1 and 170 (Puigserver et al. 1999). This transcriptional suppression can be relieved via phosphorylation by p38 MAPK at residues Thr 262, Ser 265, and Thr 298 of murine PGC-1α, all within the negative regulatory domain. These sites are likely to be physiologically relevant, as signaling upstream of p38 MAPK is activated during certain states of increased respiration and metabolic rates. For example, p38 MAPK is activated downstream of inflammatory cytokine stimulation in muscle (Knutti et al. 2001; Puigserver et al. 2001) and β3-adrenergic stimulation in brown adipocytes (Cao et al. 2001), correlating with states of cachexia and cold exposure, respectively.
PGC-1α that has been phosphorylated by p38 MAPK is both more stable and has augmented transcriptional activity relative to unphosphorylated protein (Puigserver et al. 2001). The increased stability of the p38 MAPK phosphorylated protein does not, however, fully explain the observed increase in transcriptional coactivation. A mutant PGC-1α with alanine substitutions at the phosphorylation sites has similarly increased stability, but is much less active in stimulating mitochondrial gene expression and oxygen consumption (Puigserver et al. 2001). Hence, the p38 MAPK-mediated phosphorylation is able to activate PGC-1α function through at least two distinct mechanisms.
We describe here the identification of p160 myb binding protein (Mybbp1a, herein called p160MBP) as a repressor of PGC-1α that interacts with PGC-1α's negative regulatory domain and reduces its transcriptional and biological activities. Moreover, p160MBP binding is disrupted by p38 MAPK phosphorylation of PGC-1α, providing a mechanistic explanation for the positive regulation of this coactivator by p38 MAPK.
Results
p160MBP interacts with the regulatory domain of PGC-1α and inhibits its transcriptional activity
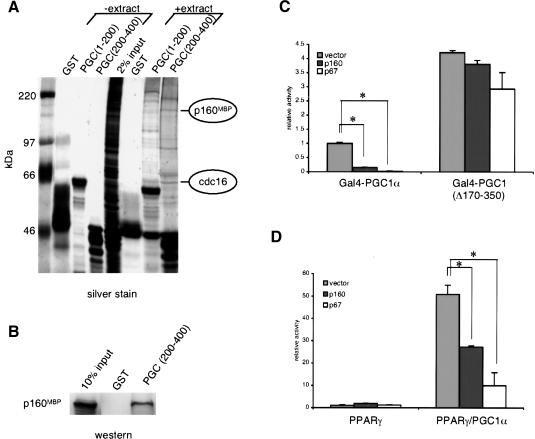
To investigate the mechanism of regulation of PGC-1α through its suppressor domain, we performed affinity chromatography using a GST-fusion protein containing amino acids 200-400 of PGC-1α and nuclear extracts from C2C12 myoblasts. The parent GST protein and GST fused to amino acids 1-200 of PGC-1α [GSTPGC1(1-200)] were used as controls. After binding and washing, the GST proteins and associated proteins were eluted with free glutathione, separated by SDS-PAGE, and silver stained. A unique constellation of proteins interacted with GST-PGC1(200-400) compared with GST-PGC1(1-200) or the control GST protein (Fig. 1A).
Figure 1.
p160MBP interacts with GST-PGC1(200-400) and suppresses PGC-1α-mediated transcription. C2C12 muscle cell nuclear extracts were incubated with bacterially expressed GST fusion proteins on glutathione sepharose beads. PGC(1-200) is GST-PGC1α(1-200) and PGC(200-400) is GST-PGC1α(200-400). The interacting proteins were run on an SDS-PAGE gel and (A) silver stained or (B) Western blotted with antibodies against p160MBP. (C) p160MBP or p67MBP was cotransfected with Gal4-PGC1α or Gal4-PGC1(Δ170-350) and a UAS-luciferase reporter in HIB1B cells. Luciferase activity was measured after 24 h. (Asterisks) P < 0.001, paired t-test. (D) PPARγ and RXRα with or without PGC-1α were cotransfected with a DR-1-luciferase reporter into HIB1B cells. p160MBP or p67MBP were also added and luciferase levels were measured after 24 h. All luciferase assays were normalized for transfection efficiencies using β-galactosidase. (Asterisks) P < 0.001, paired t-test.
Proteins binding to GST-PGC1(200-400) were excised from SDS-PAGE gels and identified by a combination of peptide mass fingerprinting using matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF) mass spectrometry (MS), and mass spectrometric sequencing using NanoES triple-quadruple MS/MS (Winkler et al. 2002). Several proteins containing hnRNP or splicing motifs were among those found to interact with GSTPGC1(200-400), highlighting PGC-1α's role in RNA processing (Supplemental Table 1). We also purified two components of the anaphase-promoting complex, tsg24 and cdc16. cdc16 is indicated in Figure 1A and will be examined in later experiments. Our attention is focused on another interacting protein, p160MBP, because it was previously identified as a negative regulator of a transcription factor, c-myb (Tavner et al. 1998). To confirm this identification, we used antibodies against p160MBP in Western blots of the proteins purified with the GST fusions. As shown in Figure 1B, p160MBP indeed interacts strongly with GST-PGC1(200-400) compared with GST alone.
p160MBP has been previously shown to be expressed ubiquitously and is posttranslationally processed in some cells to create an N-terminal fragment of 67 kD, p67MBP (Tavner et al. 1998). Previous work showed that p160MBP did not alter c-myb's transcriptional activity in transfection assays, whereas p67MBP reduced c-myb transcription by 80%. We cloned mouse p160MBP and p67MBP separately into mammalian expression vectors and tested them on PGC-1α.
HIB1B brown fat cells were cotransfected with a fusion protein connecting the yeast Gal4 DNA binding domain (Gal4-DBD) with full-length PGC-1α, along with an appropriate Gal4-UAS-luciferase reporter, and p160MBP or p67MBP. Luciferase assays were conducted 24 h later and revealed that both p160MBP and p67MBP dramatically repressed Gal4-PGC1α transcriptional activity, by 80% and 96%, respectively (Fig. 1C). In contrast, a version of PGC-1α that has lost its negative regulatory region, Gal4-PGC1(Δ170-350), was not significantly affected by either p160MBP or p67MBP expression. These data demonstrate that both p160MBP and p67MBP have inhibitory effects on PGC-1α activity and that these effects are dependent on the regulatory region of PGC-1α to which these factors bind.
Wild-type PGC-1α does not bind to DNA directly, but rather functions as a coactivator of transcription factors. To study the effects of p160MBP and p67MBP on PGC-1α in a more native context, we cotransfected vectors expressing these proteins with PGC-1α and one of its transcription factor targets, PPARγ. A multimerized PPARγ response element (DR-1) linked to a luciferase reporter gene was used to measure transcriptional activity. Although p160MBP and p67MBP had minimal effects on PPARγ alone, they both significantly diminished PGC-1α's coactivation of PPARγ (Fig. 1D). These results indicate that both p160MBP and p67MBP are able to repress PGC-1α's trancriptional coactivation in transient transfection.
Both N- and C-terminal domains of p160MBP interact with PGC-1α
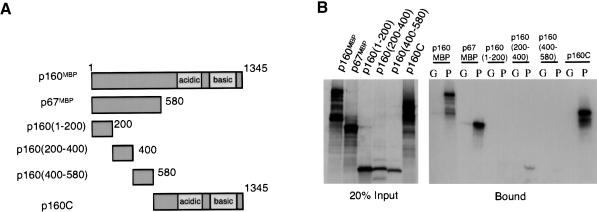
A series of deletions of p160MBP were constructed to determine which regions of this protein are responsible for the interaction with PGC-1α. Given the repressive effects of p67MBP on PGC-1α, it was expected that the N terminus of this molecule would interact with PGC-1α. Therefore, in addition to the p67MBP protein, this N-terminal region was also divided into three portions, amino acids 1-200, 200-400, and 400-580. An expression vector for the remainder of p160MBP, which is not encompassed in p67MBP, was also generated and is termed p160C (Fig. 2A).
Figure 2.
PGC-1α binds to amino acids 200-400 and to the C terminus of p160MBP. (A) Summary of p160MBP deletion constructs used in this experiment. (B) GST, labeled G, or GST-PGC1(200-400), labeled P, were incubated with in vitro translated 35S-p160MBP deletions for 1 h. Interacting proteins were run on an SDS-PAGE gel and visualized by autoradiography.
When in vitro translated portions of p160MBP were incubated with GST-PGC1(200-400), it was clear that PGC-1α interacted with p160MBP through both the N-terminal and C-terminal halves of p160MBP (Fig. 2B). Within the subdomains of the N terminus of p160MBP, the only segment that showed detectable binding to PGC-1α was p160(200-400) (Fig. 2B). It is interesting that p160MBP is predicted to contain two leucine zipper motifs, one at aa 307 to 335, and another at 900 to 928 (Tavner et al. 1998). Both of these lie within segments that bind to PGC-1α, and it is possible that they play a role in this interaction. In fact, p160MBP interacts with leucine-rich regions of c-myb and c-jun, and mutations of specific leucines on PGC-1α disrupt p160MBP binding (discussed in Fig. 3). Neither p160C nor any of the smaller fragments of p67MBP were able to suppress the transcriptional activity of Gal4-PGC1α (data not shown), suggesting that some of the key functional domains of p160MBP lie outside of the PGC-1α interaction region.
Figure 3.
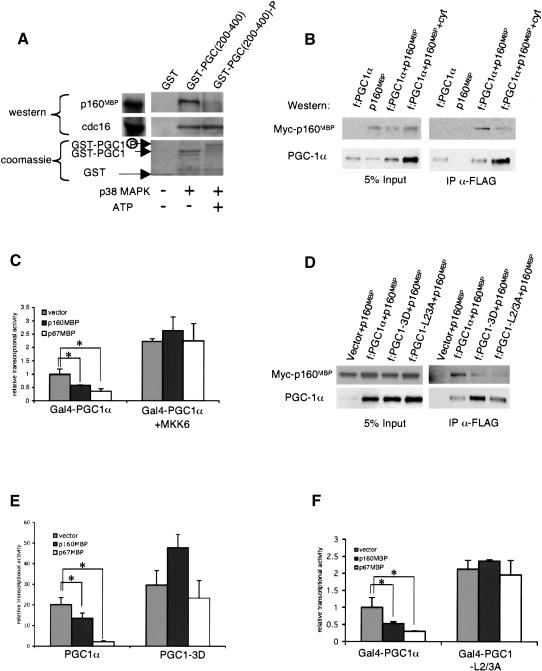
p160MBP's interaction with PGC-1α is modulated by p38 MAPK phosphorylation. (A) GST-PGC1(200-400) was phosphorylated in vitro by activated p38 MAPK in the presence of ATP [GST-PGC1(200-400)-P] or mock phosphorylated in the absence of ATP [GST-PGC1(200-400)]. The GST proteins were then incubated with C2C12 nuclear extracts and interacting proteins were visualized by Western blot by using antibodies against p160MBP or against cdc16 (Santa Cruz Biotechnology). (B) Flag-tagged PGC-1α (f:PGC1α) was cotransfected with myc-tagged p160MBP (myc-p160MBP) in BOSC cells and after 24 h treated with the cytokines IL-1α, IL-1β, and TNFα overnight in DMEM + 0.5% BSA. Whole-cell extracts were immunoprecipitated with M2 anti-Flag antibodies (Sigma) and Western blots were conducted with antibodies against myc or PGC-1α (Santa Cruz Biotechnology). The left panel is a Western blot of 5% of the whole-cell extract before immunoprecipitation and the right panel is a Western blot of the immunoprecipitated proteins. (C) Gal4-PGC1α was cotransfected in C2C12 cells with a UAS-luciferase reporter and a vector expressing MKK6, the upstream activator of p38 MAPK. Medium was changed to DMEM + 0.5% BSA after 24 h, and luciferase activity was measured after 48 h. (Asterisks) P < 0.05, paired t-test. (D) Myc-p160MBP was cotransfected with f:PGC1α, f:PGC1α with the phosphorylation sites mutated to aspartic acid (f:PGC1-3D), or f:PGC1α with selected leucines at L2 and L3 mutated to alanine (f:PGC1-L2/3A). Extracts were immunoprecipitated with anti-Flag antibodies as described earlier. (E) PGC-1α or PGC1-3D were cotransfected with PPARγ, RXR, and DR-1 luciferase reporter in HIB1B cells. Cells were harvested after 24 h and fold activation over PPARγ was graphed. (Asterisks) P < 0.05, paired t-test. (F) Gal4-PGC1 or Gal4-PGC1-L2/3A were cotransfected with a UAS-TATA-luciferase reporter in C2C12 cells, and luciferase activity was measured after 24 h. (Asterisks) P < 0.05, paired t-test.
p38 MAPK phosphorylation of PGC-1α blocks its interaction with p160MBP
p38 MAPK directly phosphorylates amino acids Thr 262, Ser 265, and Thr 298 in the repressor region of PGC-1α and leads to a protein that is both more stable and more transcriptionally active (Knutti et al. 2001; Puigserver et al. 2001). The p38 MAPK sites are highly conserved across species and are downstream of both cytokine and β-adrenergic signaling cascades (Tracey et al. 1987; Cao et al. 2001). We therefore investigated the possibility that phosphorylation of PGC-1α could interfere with p160MBP binding to the negative regulatory region of PGC-1α. p38 MAPK was used to phosphorylate GSTPGC1(200-400) in vitro under conditions in which the vast majority of the protein becomes phosphorylated and has reduced mobility in SDS-PAGE [GST-PGC(200-400)P] (Fig. 3A). Identical protein that was incubated with p38 MAPK in the absence of ATP was used as a control [GST-PGC1(200-400)]. As seen in Figure 3A, GST-PGC1 (200-400)-P bound much less p160MBP from C2C12 nuclear extracts than the mock phosphorylated GST-PGC1 (200-400) protein (Fig. 3A). In contrast, cdc16 binds equally well to PGC-1α with or without phosphorylation. These data indicate that p38 MAPK phosphorylation of PGC-1α specifically inhibits its binding to p160MBP.
We also asked whether phosphorylation of PGC-1α disrupts its interaction with p160MBP in cells. Flag-tagged PGC-1α (f:PGC1α) and myc-tagged p160MBP were cotransfected into BOSC cells and treated with the cytokines interleukin-1α (IL-1α), interleukin-1β (IL-1β), and tumor necrosis factor α (TNFα) to induce p38 MAPK-mediated phosphorylation of PGC-1α (Puigserver et al. 2001). PGC-1α was then immunoprecipitated and the complex was run on SDS-PAGE and analyzed by Western blotting. Despite the fact that cytokines increased the amount of PGC-1α protein, as expected, less myc-tagged p160MBP was immunoprecipitated with PGC-1α when cells were treated with cytokines (Fig. 3B).
We next examined the effects of p38 MAPK on PGC-1α and p160MBP by measuring transcriptional activity in cells. MKK6, the upstream activator of p38 MAPK, was used to stimulate phosphorylation of Gal4-PGC1α in C2C12 myoblasts. As shown previously (Knutti et al. 2001; Puigserver et al. 2001) and in Figure 3C, MKK6 activates the transcriptional activity of Gal4-PGC1α. However, whereas p160MBP and p67MBP suppress the transcriptional activity of Gal4-PGC1α, MKK6 completely relieves this suppression.
To confirm that the p38 MAPK modulation of the PGC-1α and p160MBP interaction is a consequence of the direct action of p38 MAPK on PGC-1α, we created a mutant of PGC-1α that replaces the phosphorylated residues with aspartic acids (PGC1-3D). The aspartate residues are used to mimic a phosphorylated version of PGC-1α. There may also be additional effects of MKK6 on other components of the PGC-1α pathway, but the PGC1-3D mutant allows us to test the portion of the MKK6 activation that directly targets PGC-1α protein itself. As shown in Figure 3D, the mutated Flag-tagged PGC-1α (f:PGC1-3D) coimmunoprecipitated significantly less myc-tagged p160MBP when cotransfected in BOSC cells. This mutant is more transcriptionally active than wild-type PGC-1α, coactivating PPARγ by 30-fold as compared with wild-type PGC-1α's 20-fold on a DR-1 luciferase reporter (Fig. 3E). When p160MBP and p67MBP were coexpressed with PGC-1α or PGC1-3D, PGC1-3D was resistant to repression by p160MBP and p67MBP. Together, these data indicate that the positive effects of p38 MAPK on PGC-1α activity occur largely via direct modulation of PGC-1α to diminish p160MBP binding.
It has previously been shown that a titratable repressor interacts with PGC-1α and is dependent on the leucine motifs LXXLL (L2), beginning at amino acid 147, and LLXXL (L3), starting at amino acid 210 (Knutti et al. 2001). Mutation of the leucine motifs to AXXLL and AAXXL, respectively, relieves the suppressive effects of the regulatory domain of PGC-1α. Figure 3D shows that a PGC-1α containing mutations in the leucine motifs (f:PGC1-L2/3A) was unable to coimmunoprecipitate p160MBP. p160MBP and p67MBP, when transfected into C2C12 cells, failed to suppress the transcriptional activity of a version of Gal4-PGC1α containing the leucine mutations (Gal4-PGC1-L2/3A; Fig. 3F). Because mutation of either the L2 or L3 motif alone was not sufficient to completely abolish p160MBP repression, we conclude that both the L2 and L3 motifs contribute to p160MBP's interaction with full-length PGC-1α (data not shown). Despite the fact that the leucine motifs are 50 amino acids away from the phosphorylation sites, Gal4-PGC1-L2/3A no longer undergoes phosphorylation-induced increases in transcription (Knutti et al. 2001). This is consistent with the data indicating that p160MBP binding is regulated by PGC-1α phosphorylation. If p160MBP cannot bind to the Gal4-PGC1-L2/3A mutant, phosphorylation of this mutant would not be expected to result in increased activity via p160MBP dissociation.
Adenoviral expression of p160MBP decreases PGC-1α-mediated respiration and mitochondrial gene expression
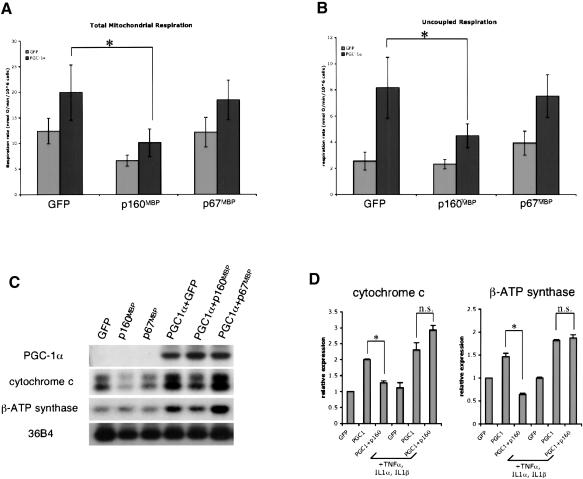
Adenoviral vectors for p160MBP and p67MBP were generated to investigate their activity on physiological functions of PGC-1α. These viruses were used in conjunction with adenoviral-PGC1α to infect C2C12 muscle cells, which were then harvested after 48 h for respiration studies and RNA analyses. As shown in Figure 4, A and C, and has been shown previously, expression of PGC-1α causes a rise in total mitochondrial oxygen consumption and the expression of genes of the mitochondrial electron transport system. Coexpression of p160MBP reversed the effects of PGC-1α, causing a complete elimination of PGC-1α-driven mitochondrial oxygen consumption. This was accompanied by a reduction in the target genes cytochrome c and β-ATP synthase. There was also a slight decrease in the respiration of cells infected with adenoviral GFP and p160MBP, although this did not reach statistical significance. The small change in basal respiration may be due to the repression of the endogenous PGC-1α in C2C12 myoblasts, or to an as yet undescribed activity of p160MBP on respiration. Interestingly, p67MBP did not alter oxygen consumption or PGC-1α target gene expression in these experiments despite the data shown earlier indicating that p67MBP is a more potent repressor of PGC-1α in transient transfections. Western blots confirmed that both adenoviral p160MBP and p67MBP were expressed efficiently in these experiments, with p67MBP expressed at slightly higher levels than p160MBP (data not shown). Thus, it appears that full-length p160MBP protein is necessary for suppressive function on endogenous PGC-1α target genes. This suggests that the C terminus of p160MBP is important for repression on chromatinized, endogenous DNA, but it is not required on plasmid DNA.
Figure 4.
p160MBP suppresses PGC-1α's biological effects in muscle cells. C2C12 myoblasts were coinfected with adenoviral PGC1α and p160MBP. Adenoviral GFP was used in the control infections. After 48 h, (A) total mitochondrial oxygen consumption and (B) uncoupled respiration were measured in live muscle cells. Uncoupled respiration represents the fraction of mitochondrial respiration that is not coupled to ATP production and was attained through addition of the ATP-synthase inhibitor oligomycin. (Asterisks) P < 0.05, repeat ANOVA, a posteriori Tukey test. (C) Same as in A and B, except instead of using cells for oxygen consumption assays, RNA was harvested and Northern blots were performed. (D) C2C12 myoblasts were coinfected with adenoviral PGC1α and p160MBP in serum-free media. TNFα, IL-1α, and IL-1β were added at the time of infection and cells were harvested after 24 h. Real-time PCR was used to quantitate RNA levels. (Asterisks) P < 0.05, paired t-test; (n.s.) not statistically significant, paired t-test.
We also measured the effect of p160MBP on the uncoupled fraction of mitochondrial respiration by adding the drug oligomycin to block ATP synthase. PGC-1α has been shown to have more potent effects on induction of mitochondrial leak than on induction of total oxygen consumption (St-Pierre et al. 2003). Figure 4B shows that uncoupled respiration is robustly induced by PGC-1α, and p160MBP represses the PGC-1α-driven proton leak by >60%. p160MBP's repression of proton leak is specific for PGC-1α-infected cells as compared with cells infected with GFP. Together, these experiments indicate a role for p160MBP in the suppression of PGC-1α's regulation of mitochondrial respiration and gene expression.
We next added cytokines and tested whether p160MBP would still be able to suppress PGC-1α induction of target genes. Because cytokines lead to phosphorylation of PGC-1α, p160MBP should no longer suppress PGC-1α activity. C2C12 myotubes were infected with adenoviral PGC-1α and p160MBP, with or without cytokines, in the absence of serum. Because of the toxicity associated with the cytokines and serum starvation, RNA was harvested after only 24 h, and subjected to real-time PCR. Although p160MBP suppressed cytochrome c and βATP synthase expression in the absence of cytokines, addition of a cocktail of TNFα, IL-1α, and IL-1β completely eliminated this suppression (Fig. 4D). This further supports a model in which p160MBP's action on PGC-1α is inhibited by PGC-1α phosphorylation.
p160MBP has intrinsic transcriptional repressive activity
There are several possible mechanisms, not mutually exclusive, for how p160MBP might mediate repression of PGC-1α function. Our primary hypotheses were based on previous work indicating that cytokines altered both the protein stability and transcriptional activity of PGC-1α. We studied this in two different cell types relevant to the biology of PGC-1α: brown fat and muscle. We first looked at PGC-1α protein levels in the brown fat HIB1B cells cotransfected with p160MBP. p160MBP caused a dramatic decrease in the amount of full-length PGC-1α protein (Fig. 5A), and a similar decrease was seen with a truncated version of PGC-1α containing only the first 400 amino acids. In contrast, a mutant of PGC-1α containing only the first 180 amino acids, which does not bind to p160MBP, was not affected. The triple aspartic acid mutant, PGC1-3D, which also does not bind significantly to p160MBP, was similarly unchanged. Although we were unable to ascertain effects of p160MBP on PGC-1α RNA because of the low transfection, the fact that all of these versions of PGC-1α are expressed from the same vector strongly suggests that the differences seen in protein level represent a change in protein stability rather than a change in transcript expression from the CMV promoter.
Figure 5.
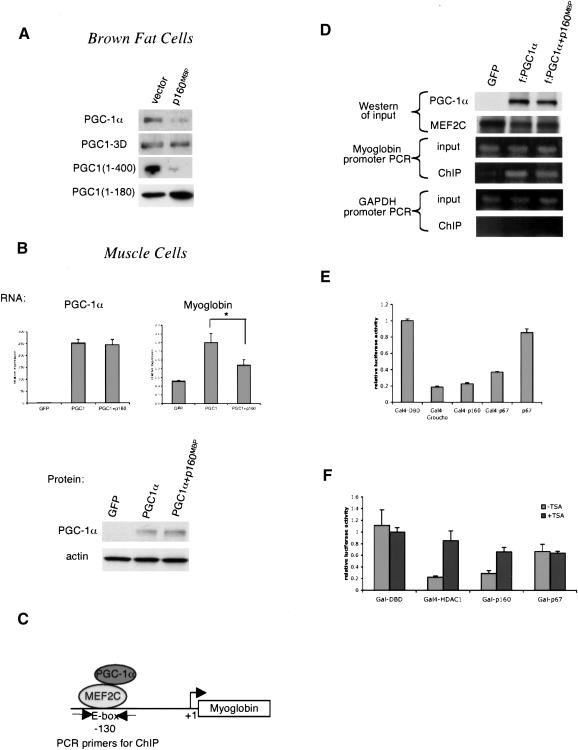
p160MBP does not alter PGC-1α protein level or recruitment to DNA. (A) HIB1B brown fat cells were cotransfected with p160MBP and various versions of PGC-1α. Cell extracts were Western blotted for PGC-1α. (B) C2C12 myotubes were coinfected with adenoviral MEF2C, PGC-1α, and p160MBP and harvested for RNA and protein. RNA was subjected to real-time PCR analysis to determine the amounts of PGC-1α and myoglobin transcripts. Protein was blotted with antibodies against PGC-1α or the loading control actin. (Asterisk) P < 0.05, paired t-test. (C) Diagram of PGC-1α interaction with MEF2C on the myoglobin promoter; sites for PCR primers for chromatin immunoprecipitation assays are indicated. (D) C2C12 myotubes were coinfected with adenoviral MEF2C, Flag-tagged PGC-1α, and p160MBP. Chromatin immunoprecipitations were carried out using M2 anti-Flag antibodies. The input and immunoprecipitated DNA were used as templates for PCR with primers flanking the MEF2C site at -130 on the myoglobin promoter. Primers amplifying a region of the GAPDH gene were used as a negative control. (E) Gal4 fusion protein constructs were transfected into C2C12 myoblasts with a UAS-tk-luciferase reporter. The cells were harvested after 24 h and luciferase activity was measured. (F) Similar to E, except 100 μg/mL trichostatin A (TSA) was added after 12 h.
We also investigated p160MBP-mediated PGC-1α repression in C2C12 muscle cells, the cell type in which the oxygen consumption assays and target gene studies were conducted. In contrast to the data shown earlier with HIB1B, we did not see a clear effect of p160MBP on PGC-1α protein amount in C2C12 (Fig. 5B). To investigate whether repression occurs through the direct action of p160MBP on the promoter of PGC-1α target genes, we used a well-characterized PGC-1α target gene in muscle cells, myoglobin. PGC-1α coactivates MEF2C on a well-defined site on the myoglobin promoter (diagram in Fig. 5C; Yan et al. 2001; Handschin et al. 2003).
C2C12 myotubes were infected with MEF2C, PGC-1α, and p160MBP. RNA analyses and Western blots were conducted in parallel to determine the amount of PGC-1α RNA and protein in the presence of p160MBP. Real-time PCR analysis revealed that p160MBP represses PGC-1α-induced activation of myoglobin by almost 60% without altering the amount of PGC-1α mRNA or protein (Fig. 5B). We therefore conclude that, in C2C12 myotubes, p160MBP is working through a mechanism other than alteration of PGC-1α protein stability.
We next used chromatin immunoprecipitations to ask whether PGC-1α remained bound to the myoglobin promoter under the influence of p160MBP. C2C12 myotubes were infected with MEF2C, Flag-tagged PGC-1α, and p160MBP. Protein and DNA were cross-linked and chromatin was sheared before immunoprecipitation using anti-Flag antibodies. PCR with primers flanking the MEF2C site of the myoglobin promoter was then used to detect if PGC-1α was bound to the myoglobin promoter. Figure 5D shows that PGC-1α is recruited to the myoglobin MEF2C site, but that p160MBP does not alter this recruitment. Thus, PGC-1α remains bound to the myoglobin promoter even in the presence of p160MBP's suppression of this target gene. Western blots for PGC-1α and MEF2C and a PCR of the MEF2C site from input DNA prior to the immunoprecipitation serve as controls. A region of the GAPDH gene was also used for PCR from the input and immunoprecipitated DNA, and confirmed that PGC-1α specifically immunoprecipitated the myoglobin promoter, but not GAPDH.
Because p160MBP does not displace PGC-1α from the promoter of a target gene, we hypothesized that p160MBP itself might contain inherent transcriptional repressive activity. Hence, the mere recruitment of p160MBP to PGC-1α would serve to lower transcription. To study p160MBP's intrinsic repressive activity, we fused p160MBP to Gal4's DNA binding domain and tested it on a UAS-tk-luciferase reporter. The tk promoter has significant basal transcriptional activity and therefore is useful for testing transcriptional repressors. We used Gal4-Groucho, a known transcriptional repressor, as a positive control for this assay. Gal4-Groucho expression results in an 80% reduction in transcription. Strikingly, Gal4-p160MBP and Gal4-p67MBP were capable of repressing the UAS-tk-luciferase reporter to nearly the same extent as Gal4-Groucho (Fig. 5E). Expression of a version of p67MBP that was not fused to the Gal4 DNA binding domain had no effect, confirming that recruitment to the promoter is essential for this repression. These data are consistent with a model in which p160MBP binds to PGC-1α on the DNA and directly represses its transcriptional activity.
A common mechanism of transcriptional repression is through deacetylation of histones. We used the histone deacetylase inhibitor trichostatin A (TSA) to investigate the possibility that p160MBP possesses intrinsic histone deacetylase activity or recruits a histone deacetylase. In a transient transfection in C2C12 myoblasts, Gal4-p160MBP's repressive activities were diminished by the addition of TSA (Fig. 5F). This indicates that at least part of the suppressive activity of p160MBP is dependent on histone deacetylases. It is notable that TSA had no effect on Gal-p67MBP, highlighting the functional difference in the properties of these two molecules. Gal4-HDAC1 was used as a positive control. These studies provide insight into the mechanism of p160MBP function, specifically suggesting the involvement of histone deacetylases.
Discussion
Although most biological regulation at the transcriptional level studied to date is attributed to alterations in amounts or activities of DNA-binding transcription factors, it is now clear that major control can be elicited through regulation of coactivator proteins. The first example of this was through the identification of OcaB, a B-cell selective coactivator of octomer binding proteins (Luo and Roeder 1995). Perhaps the most dramatic example of this, however, has been the coactivator PGC-1α, which is a major regulator of metabolic functions in many tissues. PGC-1α activates mitochondrial biogenesis and respiration in both brown fat and skeletal muscle, and is induced in the cold in both tissues (Puigserver et al. 1998; Wu et al. 1999). PGC-1α is also expressed at high levels in skeletal muscle rich in type I fibers and can regulate muscle fiber type switching (Lin et al. 2002). PGC-1α is induced in the liver on fasting, where it activates mitochondrial β-oxidation of fatty acids and gluconeogenesis (Herzig et al. 2001; Yoon et al. 2001).
Although PGC-1α is greatly regulated at the transcriptional level, one mechanism has been described that indicates that important post-translational control of this coactivator also occurs. p38 MAPK directly phosphorylates sites in the suppressor domain of PGC-1α, and greatly enhances its transcriptional activity (Puigserver et al. 2001). Although we originally described this p38 MAPK-mediated modification as a control point in the well-established role of cytokines in the activation of energy expenditure in vivo, recent data have shown that p38 MAPK is also activated downstream of cyclic AMP signaling in brown fat (Cao et al. 2001). Because cyclic AMP and CREB have been shown to be major players in the transcriptional control of PGC-1α expression in many tissues, this raises the possibility that the p38 MAPK activation of PGC-1α may be part of a coordinated program to increase the function of this coactivator at several levels simultaneously. Despite the data pointing to the importance of p38 MAPK modulation of PGC-1α as a key control point, virtually nothing has been known about how these phosphorylation sites might exert their effects.
The data presented here describe p160MBP as a repressor of PGC-1α function. p160MBP has been relatively uncharacterized to date. It was originally cloned as a repressor of c-myb, and its truncated form, p67MBP, was found to repress c-myb-mediated transcription (Tavner et al. 1998). p160MBP also interacts with the transactivation domain of the aromatic hydrocarbon receptor. In this context, p160MBP activates transcription in transient transfection (Jones et al. 2002). Thus far, the mechanism of p160MBP-mediated activation or repression has been unknown.
Several lines of evidence support the notion that modulation of PGC-1α function by p38 MAPK is mediated, in significant measure, by p160MBP. First, p160MBP was isolated in an unbiased biochemical purification as binding to the region of PGC-1α, aa 200-400, which is associated with transcriptional suppression. Second, p38 MAPK-mediated phosphorylation of PGC-1α in vitro greatly reduces the binding of p160MBP. Third, p160MBP reduces the transcriptional activity and biological effects of PGC-1α, and in all cases, p38 MAPK-mediated phosphorylation reduces or completely blocks all of the effects of p160MBP. In addition, we have found that the binding of p160MBP to PGC-1α is reduced by mutation of leucines at motifs LXXLL (L2), which begins at amino acid 147, and LLXXL (L3), which begins at amino acid 210. These mutations have been previously suggested by Kralli and colleagues to block the binding of a putative titratable repressor of PGC-1α (Knutti et al. 2001).
The discovery of molecular repressors of PGC-1α is important because all of the experiments conducted with PGC-1α to date have been gain-of-function experiments. A repressor could provide a valuable tool for studying the effects of decreasing PGC-1α activity. A recent publication has suggested that the docking of the orphan nuclear receptor estrogen-related receptor α (ERRα) may depress PGC-1α function through the same negative regulatory domain. However, the docking of ERRα is not modulated by p38 MAPK-mediated phosphorylation, a modification known to reverse the function of the repressor domain (Ichida et al. 2002).
Our earlier work had indicated that the p38 MAPK-mediated phosphorylation worked to suppress PGC-1α function in at least two distinct ways: via changes in protein stability and via changes in transcriptional function that could not be explained simply through the changes in protein stability (Puigserver et al. 2001). This was shown most clearly in a mutant of PGC-1α that had alanine substitutions (3A) at all three of the MAPK phosphorylation sites. This protein was stabilized to a comparable extent as the fully phosphorylated PGC-1α, but was far less active transcriptionally and biologically than the phosphorylated protein. The data presented here indicate that p160MBP can dock on PGC-1α and regulate its transcriptional activity and biological functions, notably in mitochondrial respiration and expression of genes of the electron transport system. This suppression seems to occur without modulation of PGC-1α protein levels, at least in skeletal muscle, or removal of PGC-1α from chromatin. As shown in chromatin immunoprecipitation assays (Fig. 5D), PGC-1α remains bound to the myoglobin promoter at the crucial MEF2 binding site even under the negative regulation of p160MBP. This suggests a model whereby p160MBP is recruited to PGC-1α targeted promoters through the suppressor domain of this coactivator (Fig. 6). That p160MBP has inherent transcriptional suppression activity is shown by the fact that this molecule can suppress transcription even when covalently linked to the DNA-binding domain of Gal4. Addition of the histone deacetylase inhibitor trichostatin A partially reverses this repression, suggesting the involvement of histone deacetylase activity in p160MBP-mediated transcriptional repression.
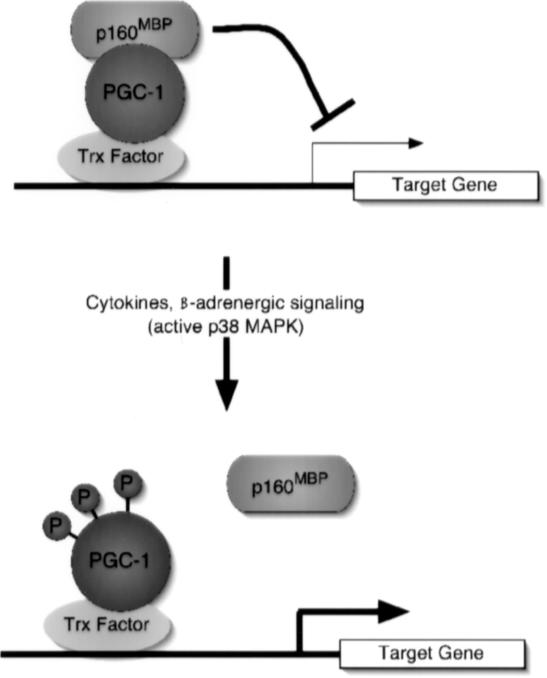
Figure 6.
Schematic representation of p160MBP repression of PGC-1α on DNA; regulation by p38 MAPK signaling events. p160MBP binds to PGC-1α and represses transcription of PGC-1α target genes. On cytokine or β-adrenergic signaling, p38 MAPK becomes active and phosphorylates PGC-1α. The phosphates disrupt p160MBP binding to PGC-1α, leading to increased activation of PGC-1α target genes.
The role of p160MBP in controlling the stability of PGC-1α is not completely clear at present. In some cell types, such as HIB1B, coexpression of p160MBP reduces the amount of PGC-1α protein, and this reduction is dependent on the p160MBP binding domain of PGC-1α. On the other hand, we have not been able to show an effect on PGC-1α protein stability in other cell types, such as C2C12 myotubes. These data suggest that p160MBP can control the amount of PGC-1α protein, probably via stability, but that there may be additional molecules involved in this process. The purification of certain subunits of the anaphase promoting complex with the suppressor domain of PGC-1α certainly suggests the possible involvement of the protein-destruction apparatus in this process.
The mechanisms illustrated here—namely, the p160MBP docking on PGC-1α in a p38 MAPK-regulatable fashion—has therapeutic implications. Particularly in muscle, PGC-1α has been shown to stimulate glucose uptake as well as mitochondrial biogenesis and respiration, all associated with increased energy expenditure. In fact, recent work from two groups has shown a decrease in expression of PGC-1α and its target genes in the muscle of type 2 diabetics and prediabetics, individuals with no disease, but with a high likelihood of developing diabetes (Mootha et al. 2003; Patti et al. 2003). These studies have suggested that even a mild to moderate increase in PGC-1α function might benefit these patients. The data here suggest that inhibition of the docking of p160MBP on PGC-1α would increase the biological effects of this coactivator. Although inhibition of protein-protein interactions by a small molecule might not be easy, the fact that this docking event is apparently structured so that it is disrupted by a limited number of phosphorylations indicates that the relevant protein surfaces controlling this binding may not be large. Such an inhibitor could prove useful in the treatment of type II diabetes.
Materials and methods
Preparation of GST fusion proteins and nuclear extracts
Bacterially produced GST-PGC1α fusion proteins were generated by inducing BL21 cells containing GST-PGC1α plasmids with 1 mM IPTG for 3 h. The cells were then sonicated and centrifuged to remove cell debris. The supernatant was incubated with glutathione-sepharose beads to purify the GST protein. The beads were then washed in PBS and stored at 4°C. To generate phosphorylated GST-PGC1α, we carried out an in vitro phosphorylation reaction using activated p38α MAPK (Upstate Biotechnology). Nuclear extracts were prepared from C2C12 myoblasts using the Dignam protocol (Dignam et al. 1983). Cells were incubated on ice with hypotonic Solution A (10 mM HEPES-KOH at pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 0.5% NP-40, protease inhibitors) for 5 min, then vortexed and spun at 1000g for 5 min to pellet the nuclei. The nuclei were resuspended in Solution B (20 mM HEPES-KOH at pH 7.9, 400 mM NaCl, 1.5 mM MgCl2, 0.5 mM DTT, 0.2 mM EDTA, 15% glycerol, protease inhibitors) and shaken, not stirred, at 4°C for 20 min to extract nuclear proteins. Samples were spun at 16,000g for 10 min and supernatant was stored at -80°C.
Affinity purification with PGC-1α regulatory domain
C2C12 nuclear extracts were diluted in three volumes of dilution buffer (20 mM HEPES-KOH at pH 7.9, 1.5 mM MgCl2, 1 mM DTT, 0.2 mM EDTA, 15% glycerol, protease inhibitors) and precleared on glutathione sepharose beads for 1 h at 4°C. The extract was then incubated with the GST-PGC1α fusion proteins overnight at 4°C. After washing in HEGN (20 mM HEPES at pH 7.6, 0.15 M KCl, 0.1 mM EDTA, 10% glycerol, 0.1% NP-40, 1 mM DTT, protease inhibitors), the GST proteins were eluted with 20 mM glutathione (20 mM glutathione, 20 mM HEPES at pH 7.6, 0.1 M KCl, 0.1 mM EDTA, 10% glycerol, 0.025% NP-40, 1 mM DTT, protease inhibitors), TCA precipitated, and run on an SDS-PAGE gel. The gels were either subjected to silver staining or Western blotting as indicated.
Protein identification by mass spectrometry
Gel-resolved proteins were digested with trypsin, partially fractionated, and the resulting peptide mixtures analyzed by matrix-assisted laser-desorption/ionization reflectron time-of-flight (MALDI-reTOF) MS (Reflex III; BRUKER Daltonics), as described (Erdjument-Bromage et al. 1998); and also using an electrospray ionization triple quadrupole MS/MS instrument (API300; ABI/MDS SCIEX) modified with an ultrafine ionization source (Geromanos et al. 2000). Selected precursor or fragment ion masses from the MALDI-TOF MS or NanoES-MS/MS spectra were taken to search a nonredundant protein database, as described (Winkler et al. 2002). MS/MS spectra also were inspected for y′′ ion series to compare with the computer-generated fragment ion series of the predicted tryptic peptides.
GST purifications of 35S in vitro-translated proteins
GST proteins were incubated with 35S-labeled in vitro translated proteins (TnT kit, Promega) in binding buffer containing 100 mM HEPES (pH 7.7), 375 mM KCl, 0.5 mM EDTA, 12.5 mM MgCl2, 0.25% NP-40, and 50% glycerol supplemented with 0.5% milk for 1 h at room temperature. The beads were washed three times in binding buffer without milk before being resuspended in SDS loading buffer and run on SDS-PAGE gels.
Mammalian cell culture and transfections/infections
C2C12, HIB1B, and BOSC cells were grown in Dulbecco's Modified Eagle's Medium supplemented with either 10% cosmic calf serum or 10% fetal bovine serum. For transcription assays, cells were transfected with luciferase reporter plasmids as well as Gal4 fusions or PPARγ and p160MBP or p67MBP as indicated. Trichostatin A (Sigma) was added to cells at 100 μg/mL for 12 h as indicated. After 1 d, cells were lysed, tested for luciferase activity, and normalized to β-galactosidase activity for transfection efficiency. To visualize protein, we generated whole-cell extracts by incubating the cells in lysis buffer (20 mM HEPESKOH at pH 7.9, 125 mM NaCl, 0.1% NP-40, 1 mM EDTA, 1 mM DTT, protease inhibitors), freeze-thawing in liquid nitrogen, and centrifuging to remove the debris. Immunoprecipitations were carried out in M2 lysis buffer (50 mM Tris-HCl at pH 7.8, 137 mM NaCl, 10 mM NaF, 1 mM EDTA, 1% Triton-X-100, 0.2% sarkosyl, 1 mM DTT, 10% glycerol, fresh proteinase, phosphatase inhibitors) for 4 h and washed three times in M2 lysis buffer. For Northern blots, cells were harvested in Trizol reagent (Invitrogen) for subsequent RNA purification. For cytokine treatment, C2C12 cells were incubated in culture medium containing 0.5% BSA in place of serum. Human TNFα (10 ng/mL) and human IL-1α and IL-1β (2 ng/mL) were added as indicated for 12 h for coimmunoprecipitations and 24 h for target gene experiments. For adenoviral experiments, adenoviruses were generated in 293 cells and infection rates were monitored using GFP fluorescence.
Oxygen consumption assays
Oxygen consumption measurements were carried out as described previously (Wu et al. 1999; St-Pierre et al. 2003). Basically, cells were isolated by washing with PBS and trypsinizing for 5 min. DMEM with 10% FBS was added to step the reaction and the cells were resuspended and spun twice at 1000 rpm for 5 min. Finally, cells were resuspended in DPBS supplemented with 25 mM glucose, 1 mM pyruvate, and 2% BSA.
Plasmids
p160MBP and its deletion constructs were cloned into the mammalian expression vector pcDNA3.1-Myc-His (Invitrogen) between EcoRV and HindIII. Nuclear localization signals were added to p160MBP deletions 1-200, 200-400, and 400-580. Gal4-p160/p67 constructs were cloned into pCMX between EcoRI and EcoRV. Point mutants were generated using the Quikchange site-directed mutagenesis kit (Stratagene). Adenop160/p67 constructs were cloned into pAdTrack-CMV between EcoRV and HindIII before being recombined into pAdEasy.
Chromatin immunoprecipitation assay
C2C12 myotubes were infected with adenoviral MEF2C, Flag-tagged PGC1α, and p160MBP. Cells were harvested after 2 d and chromatin immunoprecipitation was conducted by using the Upstate Biotechnology Kit (Cat#17-295) with M2 anti-Flag antibodies. Input DNA and immunoprecipitated DNA were subjected to PCR analysis with the primers 5′-AAGTCCAGACA GTGACCTGGCTG-3′ and 5′-TGGGTGCCACATGCCCTTC CTGC-3′, which flank the MEF2C site at nucleotide -130 on the myoglobin promoter (Yan et al. 2001), and the primers 5′-GTCGTGGAGTCTACTGGTGTC-3′ and 5′-CTAAGCAGTT GGTGGTGCAGG-3′, which flank a region of the GAPDH gene.
Acknowledgments
We thank Dr. Tom Gonda, Dr. Phil Hieter, Dr. Anthony Hollenberg, Dr. Sanders Williams, Dr. Jeff Parvin, Dr. Guillaume Adelmant, Dr. Zhidan Wu, and Bryan Sun for reagents. We thank Dr. Christoph Rachez, Stuart Levine, Dr. Stefanie Hauser, and members of the Spiegelman Lab, especially Paul Tarr, Dr. Geoffrey Girnun, Dr. Wenli Yang, and Lindsay Rohas, for thoughtful discussion and technical assistance. This work was supported by NIH grant DK54477 (to B.M.S.) and NCI Cancer Center Support Grant P30CA08748 (to P.T.). M.F. was supported by NIH Training Grant 5T32EY07110.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1152204.
Supplemental material is available at http://www.genesdev.org.
References
- Cao W., Medvedev, A.V., Daniel, K.W., and Collins, S. 2001. beta-Adrenergic activation of p38 MAP kinase in adipocytes: cAMP induction of the uncoupling protein 1 (UCP1) gene requires p38 MAP kinase. J. Biol. Chem. 276: 27077-27082. [DOI] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz, R.M., and Roeder, R.G. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acid Res. 11: 1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdjument-Bromage H., Lui, M., Lacomis, L., Grewal, A., Annan, R.S., McNulty, D.E., Carr, S.A., and Tempst, P. 1998. Examination of micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J. Chromatogr. A 826: 167-181. [DOI] [PubMed] [Google Scholar]
- Geromanos S., Freckleton, G., and Tempst, P. 2000. Tuning of an electrospray ionization source for maximum peptide-ion transmission into a mass spectrometer. Anal. Chem. 72: 777-790. [DOI] [PubMed] [Google Scholar]
- Handschin C., Rhee, J., Lin, J., Tarr, P.T., and Spiegelman, B.M. 2003. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc. Natl. Acad. Sci. 100: 7111-7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig S., Long, F., Jhala, U.S., Hedrick, S., Quinn, R., Bauer, A., Rudolph, D., Schutz, G., Yoon, C., Puigserver, P., et al. 2001. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 413: 179-183. [DOI] [PubMed] [Google Scholar]
- Ichida M., Nemoto, S., and Finkel, T. 2002. Identification of a specific molecular repressor of the peroxisome proliferator-activated receptor gamma Coactivator-1 alpha (PGC-1alpha). J. Biol. Chem. 277: 50991-50995. [DOI] [PubMed] [Google Scholar]
- Jones L.C., Okino, S.T., Gonda, T.J., and Whitlock Jr., J.P. 2002. Myb-binding protein 1a augments AhR-dependent gene expression. J. Biol. Chem. 277: 22515-22519. [DOI] [PubMed] [Google Scholar]
- Knutti D., Kressler, D., and Kralli, A. 2001. Regulation of the transcriptional coactivator PGC-1 via MAPK-sensitive interaction with a repressor. Proc. Natl. Acad. Sci. 98: 9713-9718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman J.J., Barger, P.M., Kovacs, A., Saffitz, J.E., Medeiros, D.M., and Kelly, D.P. 2000. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J. Clin. Invest. 106: 847-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Wu, H., Tarr, P.T., Zhang, C.Y., Wu, Z., Boss, O., Michael, L.F., Puigserver, P., Isotani, E., Olson, E.N., et al. 2002. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature 418: 797-801. [DOI] [PubMed] [Google Scholar]
- Luo Y. and Roeder, R.G. 1995. Cloning, functional characterization, and mechanism of action of the B-cell-specific transcriptional coactivator OCA-B. Mol. Cell. Biol. 15: 4115-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael L.F., Wu, Z., Cheatham, R.B., Puigserver, P., Adelmant, G., Lehman, J.J., Kelly, D.P., and Spiegelman, B.M. 2001. Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. Proc. Natl. Acad. Sci. 98: 3820-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha V.K., Lindgren, C.M., Eriksson, K.F., Subramanian, A., Sihag, S., Lehar, J., Puigserver, P., Carlsson, E., Ridderstrale, M., Laurila, E., et al. 2003. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately down-regulated in human diabetes. Nat. Genet. 34: 267-273. [DOI] [PubMed] [Google Scholar]
- Patti M.E., Butte, A.J., Crunkhorn, S., Cusi, K., Berria, R., Kashyap, S., Miyazaki, Y., Kohane, I., Costello, M., Saccone, R., et al. 2003. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc. Natl. Acad. Sci. 100: 8466-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P. and Spiegelman, B.M. 2003. Peroxisome proliferator-activated receptor-gamma coactivator 1alpha (PGC-1alpha): Transcriptional coactivator and metabolic regulator. Endocr. Rev. 24: 78-90. [DOI] [PubMed] [Google Scholar]
- Puigserver P., Wu, Z., Park, C.W., Graves, R., Wright, M., and Spiegelman, B.M. 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92: 829-839. [DOI] [PubMed] [Google Scholar]
- Puigserver P., Adelmant, G., Wu, Z., Fan, M., Xu, J., O'Malley, B., and Spiegelman, B.M. 1999. Activation of PPARgamma coactivator-1 through transcription factor docking. Science 286: 1368-1371. [DOI] [PubMed] [Google Scholar]
- Puigserver P., Rhee, J., Lin, J., Wu, Z., Yoon, J.C., Zhang, C.Y., Krauss, S., Mootha, V.K., Lowell, B.B., and Spiegelman, B.M. 2001. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARgamma coactivator-1. Mol. Cell 8: 971-982. [DOI] [PubMed] [Google Scholar]
- Puigserver P., Rhee, J., Donovan, J., Walkey, C.J., Yoon, J.C., Oriente, F., Kitamura, Y., Altomonte, J., Dong, H., Accili, D., et al. 2003. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature 423: 550-555. [DOI] [PubMed] [Google Scholar]
- Rhee J., Inoue, Y., Yoon, J.C., Puigserver, P., Fan, M., Gonzalez, F.J., and Spiegelman, B.M. 2003. Regulation of hepatic fasting response by PPARgamma coactivator-1alpha (PGC-1): Requirement for hepatocyte nuclear factor 4alpha in gluconeogenesis. Proc. Natl. Acad. Sci. 100: 4012-4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre J., Lin, J., Krauss, S., Tarr, P.T., Yang, R., Newgard, C.B., and Spiegelman, B.M. 2003. Bioenergetic analysis of PGC-1alpha and PGC-1beta in muscle cells. J. Biol. Chem. 278: 26597-26603. [DOI] [PubMed] [Google Scholar]
- Tavner F.J., Simpson, R., Tashiro, S., Favier, D., Jenkins, N.A., Gilbert, D.J., Copeland, N.G., Macmillan, E.M., Lutwyche, J., Keough, R.A., et al. 1998. Molecular cloning reveals that the p160 Myb-binding protein is a novel, predominantly nucleolar protein which may play a role in transactivation by Myb. Mol. Cell. Biol. 18: 989-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiraby C., Tavernier, G., Lefort, C., Larrouy, D., Bouillaud, F., Ricquier, D., and Langin, D. 2003. Acquirement of brown fat cell features by human white adipocytes. J. Biol. Chem. 278: 33370-33376. [DOI] [PubMed] [Google Scholar]
- Tracey K.J., Fong, Y., Hesse, D.G., Manogue, K.R., Lee, A.T., Kuo, G.C., Lowry, S.F., and Cerami, A. 1987. Anti-cachectin/TNF monoclonal antibodies prevent septic shock during lethal bacteraemia. Nature 330: 662-664. [DOI] [PubMed] [Google Scholar]
- Vega R.B., Huss, J.M., and Kelly, D.P. 2000. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol. Cell. Biol. 20: 1868-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler G.S., Lacomis, L., Philip, J., Erdjument-Bromage, H., Svejstrup, J.Q., and Tempst, P. 2002. Isolation and mass spectrometry of transcription factor complexes. Methods 26: 260-269. [DOI] [PubMed] [Google Scholar]
- Wu Z., Puigserver, P., Andersson, U., Zhang, C., Adelmant, G., Mootha, V., Troy, A., Cinti, S., Lowell, B., Scarpulla, R.C., et al. 1999. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98: 115-124. [DOI] [PubMed] [Google Scholar]
- Yan Z., Serrano, A.L., Schiaffino, S., Bassel-Duby, R., and Williams, R.S. 2001. Regulatory elements governing transcription in specialized myofiber subtypes. J. Biol. Chem. 276: 17361-17366. [DOI] [PubMed] [Google Scholar]
- Yoon J.C., Puigserver, P., Chen, G., Donovan, J., Wu, Z., Rhee, J., Adelmant, G., Stafford, J., Kahn, C.R., Granner, D.K., et al. 2001. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413: 131-138. [DOI] [PubMed] [Google Scholar]