Abstract
The presence of a nucleus and other membrane-bounded intracellular compartments is the defining feature of eukaryotic cells. Endosymbiosis accounts for the origins of mitochondria and plastids, but the evolutionary ancestry of the remaining cellular compartments is incompletely documented. Resolving the evolutionary history of organelle-identity encoding proteins within the endomembrane system is a necessity for unravelling the origins and diversification of the endogenously derived organelles. Comparative genomics reveals events after the last eukaryotic common ancestor (LECA), but resolution of events prior to LECA, and a full account of the intracellular compartments present in LECA, has proved elusive. We have devised and exploited a new phylogenetic strategy to reconstruct the history of the Rab GTPases, a key family of endomembrane-specificity proteins. Strikingly, we infer a remarkably sophisticated organellar composition for LECA, which we predict possessed as many as 23 Rab GTPases. This repertoire is significantly greater than that present in many modern organisms and unexpectedly indicates a major role for secondary loss in the evolutionary diversification of the endomembrane system. We have identified two Rab paralogues of unknown function but wide distribution, and thus presumably ancient nature; RabTitan and RTW. Furthermore, we show that many Rab paralogues emerged relatively suddenly during early metazoan evolution, which is in stark contrast to the lack of significant Rab family expansions at the onset of most other major eukaryotic groups. Finally, we reconstruct higher-order ancestral clades of Rabs primarily linked with endocytic and exocytic process, suggesting the presence of primordial Rabs associated with the establishment of those pathways and giving the deepest glimpse to date into pre-LECA history of the endomembrane system.
Key words: Evolution, Membrane traffic, Phylogenetics
Introduction
Intracellular compartmentalization is a major evolutionary transition, and a defining feature of essentially all eukaryotic cells (Cavalier-Smith, 2002; Stanier, 1970), representing a major advance in cellular complexity. The organelles comprising the endomembrane system arose by autogenous evolution, i.e. from pre-existing components and/or structures within ancestral (prokaryotic-like) organisms (Dacks and Field, 2007), differentiating them from the endosymbiotic mitochondrion and plastids (Embley and Martin, 2006; Keeling, 2010). The endomembrane system consists of many discrete, interconnected compartments with distinct protein and lipid compositions, morphologies and functions that enable the uptake (endocytosis) and export (exocytosis) of macromolecules, particles and other metabolites. Numerous pathological conditions are associated with defects in endomembrane activity (Huizing et al., 2008; Olkkonen and Ikonen, 2006).
Maintaining this organellar system requires mechanisms for targeting specific molecules to individual organelles and is, in part, achieved by co-operative action of multiple paralogue-rich protein families, including SNAREs, vesicle coat complexes and – importantly – Rab GTPases (Cai et al., 2007; Stenmark, 2009; Südhof and Rothman, 2009). Rab orthologues conserve, to a rather remarkable degree, their functions and intracellular locations between highly divergent species, underpinning their exploitation as valuable markers for intracellular compartments (Brighouse et al., 2010; Stenmark, 2009; Woollard and Moore, 2008). The presence of paralogue-containing protein families at the core of membrane trafficking and organellar definition suggests a common origin for many intracellular transport steps, and also a rationale explaining the evolutionary plasticity facilitating the generation of new compartments. Recently we proposed a model for organelle evolution whereby gene duplication and co-evolution of multiple specificity-encoding proteins drives increased organellar complexity, and enabled a single primordial endomembrane compartment to differentiate into an array of non-endosymbiotic organelles as present in modern cells (Dacks and Field, 2007; Dacks et al., 2009; Dacks et al., 2008). This model implied that reconstruction of the evolutionary history of an endomembrane specificity-encoding protein, for example Rab GTPases, would also reveal the evolutionary relationships between the endomembrane organelles.
Rabs are vital players (Dacks and Field, 2007; Elias, 2010), and possibly even principal drivers, of endomembrane evolution (Gurkan et al., 2007). However, previous explorations of Rab protein evolution focused on either limited taxa (e.g. Bright et al., 2010; Pereira-Leal, 2008; Pereira-Leal and Seabra, 2001) or restricted Rab paralogue diversity (e.g. Elias et al., 2009; Mackiewicz and Wyroba, 2009). Systematic reconstructions deduced that the last eukaryotic common ancestor (LECA) possessed up to 14 ancient Rab paralogues, but only 8–10 of these were robustly reconstructed by phylogenetics (Bright et al., 2010; Pereira-Leal, 2008; Pereira-Leal and Seabra, 2001). Most unicellular eukaryotes possess approximately 10–20 distinct Rabs, but several have many more (Bright et al., 2010; Carlton et al., 2007; Saito-Nakano et al., 2005), whereas multicellular organisms can possess over 60 (Pereira-Leal and Seabra, 2001; Rutherford and Moore, 2002). The precise biological implications of an increased Rab repertoire remain unclear. Furthermore, a comprehensive Rab phylogeny remains elusive, with the consequence that understanding the extent and timing of Rab family innovations, and by inference the frequency of lineage-specific trafficking pathways, in most eukaryotic lineages is lacking. This also prevents accurate reconstruction of the LECA. Finally, lack of definition of deep relationships between Rab proteins hinders development of a model for early endomembrane system evolution prior to the LECA, and hence determination of the earliest events in eukaryogenesis.
A phylogeny for the Rab GTPases directly addresses three fundamental evolutionary cell biology questions: (1) what was the intracellular transport complexity in the LECA; (2) how has transport complexity evolved post-LECA; and (3) what steps led to this complexity pre-LECA? Here, we solve two confounding problems for addressing these questions: data quality and phylogenetic resolution. Utilizing recently generated genomic and transcriptomic data we compiled a curated, annotated and taxonomically broad Rab dataset. Furthermore, we describe a novel phylogenetic workflow, ScrollSaw, which provides increased resolution between Rab clades, and reconstructs the backbone of the Rab phylogenetic tree with unprecedented clarity.
Results
Dataset construction
A comprehensive database of manually curated Rab sequences was assembled from a combination of complete genomic sequences and EST survey data. This database comprises 1453 sequences from 55 organisms selected so that the known eukaryotic phylogenetic diversity is encompassed as broadly as possible, but also minimizing redundancy and overemphasis on specific lineages (supplementary material Table S1, Fig. S1). Because Rabs are traditionally considered as a distinct family within the Ras superfamily, we included sequences giving higher BLASTp scores to known Rabs than to members of the other GTPases. We also retained two additional Rab-like subfamilies [RTW (RABL2) and IFT27 (RABL4)] lacking the typical C-terminal geranylgeranyl modification signal. Ran, which is involved in multiple activities at the nucleus, was included as a potential outgroup.
Traditional analysis based on selected taxa
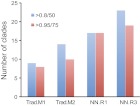
Attempts to analyse this entire dataset, or subsets encompassing all sequences from a cohort of species representing phylogenetically diverse lineages, yielded little resolution. Because we wished to reconstruct ancestral Rab clades, i.e. those representing paralogues present in the LECA, two datasets were constructed, each containing all Rab sequences from a representative from each of the presumably monophyletic eukaryotic supergroups, either Opisthokonta, Excavata, Amoebozoa, Archaeplastida, Chromalveolata and Rhizaria (Trad.M1) or Opisthokonta, Excavata, Amoebozoa, Archaeplastida, SAR and CCTH (Trad.M2) after accommodating recent taxonomic controversies [see Walker et al., 2011 and references therein (Walker et al., 2011)]. Using criteria whereby an ancestral Rab clade must contain representatives from at least three eukaryotic supergroups, analyses of these datasets provided some resolution, suggesting between eight and 14 Rab subfamilies in the LECA (Fig. 1; supplementary material Fig. S2, Fig. S3, Table S2), and were consistent with earlier reconstructions (Bright et al., 2010; Pereira-Leal, 2008; Pereira-Leal and Seabra, 2001). However, phylogenetic analyses of both the Trad.M1 and Trad.M2 datasets suffer severe organismal sampling bias and left placement of a great many Rab sequences unresolved.
Fig. 1.
Increased phylogenetic resolution using ScrollSaw versus a traditional (Trad) approach. Two test datasets (Trad.M1 and Trad.M2 or NN.R1 and NN.R3) were analysed in each instance, and the numbers of clades reconstructed by each analysis are given. Blue bars are moderate stringency, and red bars high stringency cutoff for assigning confidence to a clade. Statistical support is given for MrBayes (posterior probability) or ML (bootstrap) as indicated.
ScrollSaw, a new phylogenetic approach, provides increased resolution for the Rab family
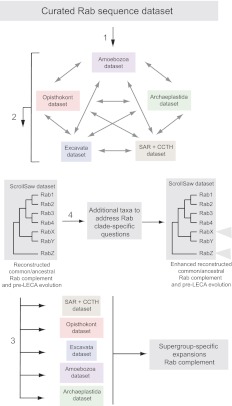
The Rab protein family has undergone extensive duplications and differential divergence rates and now contains many paralogues, necessitating a methodology that distinguishes slowly evolving Rabs from lineage-specific and divergent ones. We devised a phylogenetic strategy that mitigates major informational limitations arising from the data structure (i.e. a low number of informative positions per taxa) and the evolutionary mode of the Rab family. Briefly, this new approach, ScrollSaw, divides the dataset by established taxonomic criteria, and relies on a series of inter-subset comparisons to re-assemble evolution of the overall protein family (Fig. 2).
Fig. 2.
The ScrollSaw workflow. A dataset, here curated predicted Rab protein sequences, is subdivided on taxonomic grounds (1). All pairwise comparisons are made between the relationships for each data subset to identify the minimal distance pairs (2), and the resultant data used to reconstruct a ScrollSaw dataset, which now includes representatives across the whole sampled taxonomic range (3). This analysis allows reconstruction of supergroup-specific expansions and reconstruction of the LECA. Additional taxa are introduced into the analysis to address specific issues (4) that could not be resolved adequately using the main data subset (3).
We reasoned that distinguishing slowly evolving Rab paralogues from lineage-specific divergent Rabs, and limiting phylogenetic analyses to the former, would allow elucidation of at least backbone relationships within the Rab family. We assembled five non-overlapping Rab sequence sets, each restricted to a single supergroup (supplementary material Table S1), and calculated pair-wise maximum likelihood distances for all ten dataset pairs. We then determined those pairs of sequences that exhibited the lowest mutual distances in between-supergroup comparisons with each pair consisting of sequences from two different supergroups. Such pairs are expected to represent the least divergent orthologous representatives of the respective Rab lineage within a given supergroup. Conversely, a lineage-specific divergent paralogue should be excluded, as it would lack a homologue in another supergroup with which it would exhibit reciprocally minimal distance (supplementary material Fig. S4). By relying on the minimal reciprocal distances, the approach may also overlook ancient Rab paralogues with very rapidly evolving sequences. However, as ten separate between-supergroup comparisons were performed and every pair of sequences with mutually minimal distances was investigated (Fig. 2), we consider it highly unlikely that a cryptic ancient Rab paralogue would have failed to be identified.
An initial tree inferred from the resulting dataset (NN.R1) revealed 17 strongly supported ancestral clades and several receiving moderate to low statistical support (Fig. 1; supplementary material Fig. S5, Table S2). Re-inspection revealed additional features. For instance, the Rab24-related clade possessed a deep, strongly supported division into two subclades, raising the possibility that it comprises multiple paralogues predating eukaryotic radiation. Analysis of a Rab24-specific dataset (supplementary material Fig. S6) revealed that these subclades indeed represent distinct ancestral paralogues, one typified by Rab24 and the other by Rab20. Additionally, although Rab1 and Rab14 were not reconstructed as monophyletic clades by all methods, we operated on the hypothesis of monophyly for further analyses, which was validated as described below. The clades reconstructed in these analyses not only suggest the presence of these Rab subfamilies in the LECA, but also identify putative supergroup-specific losses. To confirm these losses, datasets of each supergroup, along with representatives of the putatively absent clades were constructed and analysed (supplementary material Figs S7–S16). This identified several additional candidate representatives for Rabs originally deduced as lost by specific supergroups.
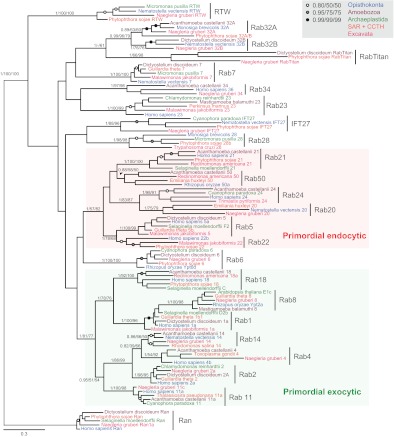
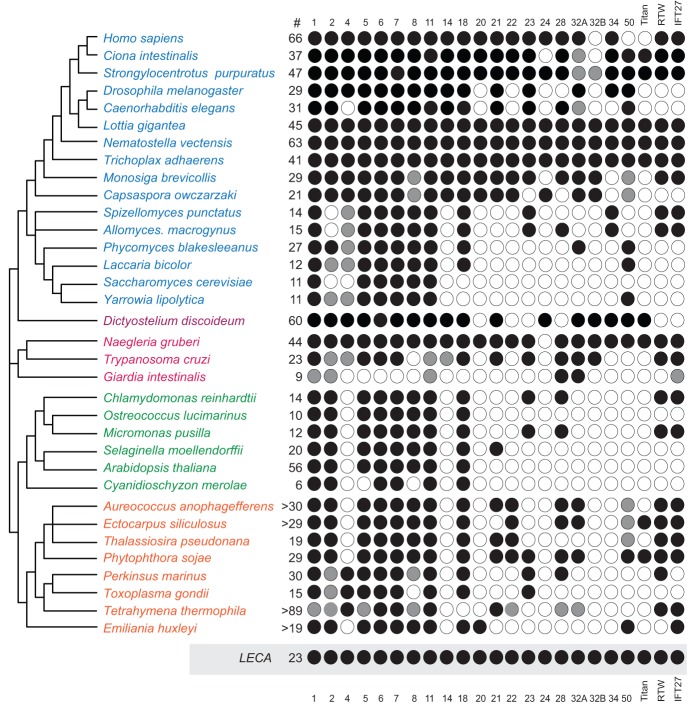
Analysis of a second dataset (NN.R3) comprising the single least divergent representative of each putatively ancestral Rab clade produced a highly resolved phylogeny (Fig. 3) and yielded several key findings. Defining an ancestral Rab clade as containing sequences from at least three supergroups, and supported by >0.95 posterior probability (PP) and >75% bootstrap (BP) support by either ML method, we reconstructed the LECA as possessing Ran, Rabs 2, 4, 5, 6, 7, 8, 11, 18, 20, 21, 23, 24, 28 and 34, two Rab-like paralogues, IFT27 and RTW, two previously undetected ancient subfamilies within the Rab32 clade, i.e. Rab32A and 32B, and a new subfamily, designated here as RabTitan because of its early origin and the large size of its members (generally much longer than canonical Rabs). Using less conservative criteria (0.8PP and 50% BP) allowed inclusion of Rabs 14, 22 and another new subfamily, here named Rab50 for convenience. Rab1 is reconstructed as a paraphyletic group from which Rab8 emerges, but because both Rab groups are broadly conserved among diverse eukaryotes, they can be categorized as separate ancient Rab subfamilies. Thus the LECA had a minimum of 19 distinct Rab and Rab-related proteins, and potentially as many as 23 (Fig. 1), representing a strikingly complex repertoire, which is notably larger than many extant unicellular organisms (Fig. 4).
Fig. 3.
Evolutionary relationships of core Rab clades predicted to be present in the LECA. Phylogenetic tree for the Rab clades predicted to have been present in the last eukaryotic common ancestor following ScrollSaw. Clades are indicated by vertical bars, and putative functional groupings for primordial endocytic and exocytic Rabs are shaded. Ran is used as an outgroup and the best MrBayes tree topology is shown. Individual leaves are colour-coded according to supergroup, and statistical support is indicated by the raw values for important nodes defining Rab clades, or are iconized as indicated (upper right). Supergroup divisions are sensu Adl et al. (Adl et al., 2005), except the SAR + CCTH supergroup, which contains the stramenopiles, alveolates and Rhizaria together with the cryptophyte-centrohelid-telonemid-haptophyte grouping (Burki et al., 2009).
Fig. 4.
Rab representation for select eukaryotes. Individual Rab clades, inferred as present in the LECA, are shown as columns. Taxa are shown as rows, with the hypothetical LECA as the lowest row (grey box). A schematic phylogeny for the taxa is drawn on the left and derived from Walker et al. and references therein (Walker et al., 2011). The total number of Rabs found in each genome is also indicated on the right of the taxon labels, and by a hash. Here, and in Fig. 5, black circles indicate at least one member of the clade has been identified with phylogenetic support (>0.80/50/50 MrBayes/PhyML/RaxML) and grey circles indicate naming based on BLAST results. Taxa are colour-coded by supergroup as in Fig. 3.
Previously unrecognized ancient Rabs, lineage-specific complexity and ancient relationships
The newly identified RabTitan is an ancient Rab subfamily containing a C-terminal extension, which in some representatives also includes an SH2 domain (supplementary material Fig. S17). Re-analysis of the above dataset, but with all putative RabTitan orthologues, including those from species that had not been systematically investigated above, revealed clear orthologues restricted to the SAR+CCTH, Amoebozoa and Excavata supergroups (supplementary material Fig. S18, Table S1). However, some metazoan genes also clustered with RabTitan (supplementary material Fig. S18), albeit with moderate support, implying a potential presence also in the opisthokonts.
Remarkably, ScrollSaw allowed both reconstruction of deep evolutionary events and determination of the Rab complements of modern eukaryotes. For example, there is an abundance of evolutionarily novel Rab paralogues in the stem lineage of Metazoa (Figs 5, 6), potentially correlated with increased trafficking complexity and/or multicellularity, as suggested previously (Pereira-Leal and Seabra, 2001). Consequently, we reconstructed Rab complements for several crucial eukaryotic phylogenetic nodes (Fig. 6). We found few expansions in the stem lineages of Fungi, Amoebozoa, Excavata, Stramenopiles, Alveolata or Archaeplastida (supplementary material Figs S7–S16). Notably, we failed to see equivalent expansions in independently arisen multicellular lineages within these groups (including in the embryophytes) suggesting that significant Rab family expansions are not driven by multicellularity per se.
Fig. 5.
Evolutionarily novel Rab clades present in Holozoa and Metazoa. For the data shown here and in Fig. 4, either positive identification by phylogenetics or lack of identification of at least one homologue of each Rab clade in each organism by either BLAST or ScrollSaw, was attained in 93% of the cases, with positive assignment by BLAST alone being necessary less than 7% of the time. Data are based on the phylogenetic reconstructions in Fig. 3 and supplementary material Fig. S6 and Fig. S18.
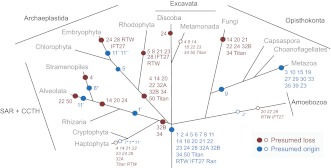
Fig. 6.
The birth and death of Rab subfamilies in eukaryote supergroups. Points of presumed origin (blue) and loss (magenta) are shown as circles overlaid onto a schematic taxonomy of the eukaryotes. Evidence for losses and origins is based on the data in supplementary material Figs S7–S16. Closed circles indicate that two full genomes or more support the predicted event, and open circles with italic text are events where support is derived from only one completed genome plus EST data. Rabs are indicated by numbers except for RTW, IFT27, Titan and Ran; primes indicate that the ancestral clade has divided to generate a new clade but one that is clearly derived from the ancestral Rab clade.
Most significantly, well-supported relationships between many Rab paralogues were reconstructed for the first time (Fig. 3). Robust higher-order clades encompassing Rab 2, 4, 14, and Rab 1, 8, 18 were found, which consistently group with Rab 11 in a major super-clade. Another major super-clade containing Rab 5, 20, 21, 22, 24 and 50 was also reconstructed with high support values. These reconstructions provide resolution of more than half of the deduced ancestral Rab subfamilies, a significant advance in our understanding of Rab evolution pre-LECA.
Discussion
Our analyses of a curated, taxonomically broad Rab protein sequence dataset yielded unprecedented resolution of Rab phylogenetic relationships. This significant improvement is the result of a simple, novel and general approach, here named ‘ScrollSaw’. ScrollSaw improves phylogenetic resolution by concentrating on the minimally derived representatives of paralogues that are conserved between distant taxonomic supergroups to provide reconstruction across the entire taxon range, here all eukaryotes.
ScrollSaw is preferable to the ‘traditional’ strategy of using all genes from a representative taxon for each supergroup for theoretical and empirical reasons. First, analysing the entirety of the dataset avoids taxon bias, as a priori selecting taxa to best represent a particular group is frequently necessary in the traditional approach for computational tractability, and is clearly subjective. The inherent problems are evident in the inconsistencies in the clade reconstruction between the two ‘traditional’ datasets, which were each anticipated to behave well in phylogenetic analysis (Fig. 1), but with Trad.M1 reconstructing eight and Trad.M2 14 ancestral clades. Second, ScrollSaw does not rely exclusively on characterized query sequences and so escapes the constraints of searching only for orthologues of proteins studied in model systems such as animals and fungi. Rather, because ScrollSaw provides resolution of paralogous gene families, it facilitates identification of previously unknown Rab innovation across the range of taxa studied. Ancient families, including Rab20, 32A, 32B, 34 or RabTitan were not anticipated and/or are absent from the well-characterized model organisms of mammalian cells or fungi. Perhaps most significantly, ScrollSaw was designed primarily for analysis of highly paralogue-rich gene families, and therefore is potentially applicable to any large dataset, and across any taxonomic range. With massive sequence datasets increasingly common, the application of ScrollSaw to other large paralagous families with only restricted regions of informative sequence, e.g. kinases, proteases or myosins, should provide a powerful method for gaining analytical insights into these data.
The improved resolution of the Rab dataset revealed several major insights into evolution of the ancient eukaryotic cell. The first is that the LECA possessed up to 23 Rab paralogues, although this number might fractionally decrease depending on the position of the eukaryotic root, for which there is currently no strong consensus (Roger and Simpson, 2009). Nonetheless, the number of widely conserved Rab paralogues revealed by the present analysis is one third or more higher than that reported previously (Bright et al., 2010; Pereira-Leal, 2008; Pereira-Leal and Seabra, 2001), and also does not take into account potential multiple subfamily members that might have also been present.
The second insight relates to deduced details of cellular complexity in the LECA. A consensus is emerging from comparative genomics that the LECA was a highly sophisticated and complex organism, further supported by the recent description of the Naegleria gruberi genome, which revealed unprecedented levels of metabolic and cellular flexibility and complexity in a unicellular free-living organism (Fritz-Laylin et al., 2010; Koonin, 2010). We extend this paradigm by deducing the presence of both the core endocytic and exocytic pathways in LECA, together with several additional and less well-characterized pathways. Extrapolating from the functions of Rab paralogues as experimentally defined (Lumb et al., 2011; Stenmark, 2009; Woollard and Moore, 2008), the LECA possessed multiple Rab proteins mediating anterograde transport or regulation at the ER (Rab1 and 8). This indicates the presence of potentially multiple anterograde routes, and also implies the presence of active autophagic systems. Rab5, 21 and 22, which each mediate comparatively early endocytic events, suggest a rather complex endosomal network containing multiple sorting and recycling steps. Furthermore, Rab proteins involved in late endosomal and/or lysosomal trafficking (Rab7, 2 and 32), retrograde transport through the Golgi complex (Rab2 and 6) and the endosomal recycling and exocytic system (Rab4, 11) clearly indicate that bidirectional movement of molecules through the endomembrane system was firmly established in the LECA. Finally, the presence of IFT27, Rab23, 8 and 11, all suggest multiple transport pathways integrating the endomembrane system and the flagellum. The detection of several ancestral, widely distributed Rabs with no known function [e.g. Rab20 and 50, RabTitan, RTW (RABL2)] suggests that there remain many fundamental aspects of Rab biology that are yet to be described.
In addition to identifying those ancient Rab paralogues that emerged prior to the LECA, or at least before the diversification of most eukaryotic supergroups, we also uncovered a great many expansions and secondary losses. The significance of lineage-specific expansions in the Rab family has been previously acknowledged by phylogenetic analyses in various taxa (e.g. Lal et al., 2005; Rutherford and Moore, 2002; Saito-Nakano et al., 2005; Saito-Nakano et al., 2010). Our aim was not to provide a full description of all Rab duplication events that have occurred, but rather to reconstruct the Rab complement at important nodes of the eukaryotic phylogeny, and hence estimate the extent of innovation at the establishment of the major eukaryotic groups. Our reconstructions reveal the stem lineage of multicellular animals (Metazoa) as a particularly prominent ‘hotspot’ of Rab evolution, with possibly 11 new paralogues added to the ancestral Rab family (Fig. 6), representing a 50% expansion of the complement inherited from unicellular metazoan ancestors. Because no equivalent expansions are seen for stem lineages of other multicellular taxa, i.e. embryophytes, a subset of fungi and brown algae, we posit that metazoan multicellularity is uniquely intertwined with a sophisticated endomembrane system. Indeed, some paralogues that have been well characterized clearly evolved to mediate various specialized exocytic and endocytic processes responsible for intercellular communication through an array of signalling molecules including hormones, morphogenetic factors and neurotransmitters, e.g. Rab3 and Rab27 (Fukuda et al., 2000).
In further contrast, few, if any, expansions in the Rab family could be inferred for the ancestors of most major non-metazoan eukaryotic clades (Fig. 6), indicating that fundamental evolutionary transitions are not necessarily coupled to major modifications of the endomembrane system. Our analysis, however, is limited to current genome sequence availability, which is somewhat restricted for several supergroups. Improved genome sampling and ScrollSaw will make it possible to uncover additional paralogues and define the ultimate phylogenetic origins of many lineage-specific Rab proteins.
Unlike paralagous expansion, secondary loss has not been fully appreciated as a significant force in sculpting the Rab protein family and, by extension, the membrane-trafficking system. Strikingly, the LECA appears to have possessed at least as large a Rab complement as many living species and rather more than in numerous experimentally important fungal and other unicellular organisms (Fig. 4). Arguably, an intermittent phylogenetic distribution for several Rab subfamilies could be explained by dissemination of more recently established, lineage-specific paralogues to distant lineages through horizontal gene transfer (HGT). Although we cannot exclude HGT as contributing to Rab evolution, this would require unparsimonious extensive gene transfer, and at multiple taxonomic levels. Robust evidence for this is lacking. Hence we conclude that the Rab family is shaped by the balance of sculpting by loss of ancient paralogues together with elaboration by lineage-specific and subfamily-specific expansion.
Our findings are consistent with a very recent analysis of Rab diversity across eukaryotes by Diekmann et al. (Diekmann et al., 2011). In agreement with our findings, they observed frequent and uneven expansions and secondary loss of Rab complements in various eukaryotic lineages, interpreted as a complex Rab complement in the LECA and an unappreciated role of secondary loss. Because their and our analytical approaches differ substantially, we found more ancestral Rab families than Diekmann et al., but, nonetheless, the overall conclusions are similar and the datasets substantially agree. The data are also congruent with analyses on additional aspects of the trafficking-specificity machinery. The adaptin complexes appear to be ancient but subject to sporadic loss, with the newly discovered adaptin 5 being the most prominent example [(Hirst et al., 2011), inter alia]. Similarly analyses of SNARE proteins found examples of both reduction (Ayong et al., 2007; Elias et al., 2008) and expansion (Dacks and Doolittle, 2002; Kissmehl et al., 2007; Kloepper et al., 2007; Sanderfoot, 2007).
Perhaps the greatest advance here over previous analyses is resolution of higher-order clades among ancestral Rab paralogues. We can conclude that Rab1 and 8, Rab20 and 24, and Rab32A and 32B are closely related paralogous pairs. Even more significantly, our analysis resolved two remarkable Rab super-clades, one comprising paralogues primarily implicated in anterograde trafficking (Rab1, 8, 18, 2, 4, 14 and 11), and the other including paralogues governing endocytosis (Rab5, 21 and 22) and, perhaps, autophagy (Rab24).
The LECA was clearly a highly complex cell, but because of the lack of resolution between organelle- and pathway-specific paralogues, the emergence of this complexity has appeared to be difficult to explain. We previously proposed that resolving the evolutionary history of specificity-encoding factors would suggest an order for endomembrane organelle evolution (Dacks and Field, 2007). Recent work (Hirst et al., 2011) has provided some insight into the steps pre-LECA of the evolution of the adaptin complexes. However, these complexes are restricted to a subset of trafficking organelles: the Rab proteins are found across the membrane-trafficking system. From present data we propose an expansion to this model, whereby in a protein family with as extensive an ancestral complement as the Rabs, the resolved order might better reflect the innovation of pathways, rather than organelles per se. Thus, we suggest that the Rab ancestors of the two super-clades functioned, respectively, as regulators of exocytic and endocytic processes, associated with a simple primordial endomembrane system, and importantly that these were established prior to the genesis of at least some individual compartments. Subsequent duplications within the primordial endocytic and exocytic clades finally giving rise to multiple (seven or six, respectively) paralogues in the LECA drove further diversification and sophistication of endomembrane compartments and trafficking pathways.
Several Rab paralogues phylogenetically excluded from these two super-clades are associated with the late endosomes and/or lysosomes (Rab7 and 28) or lysosome-derived compartments (Rab32), whereas others (Rab23, IFT27) mediate transport events to or within the flagellum. Speculatively, the placement of Rab7 outside of the primordial endocytic clade might reflect a separate origin of this pathway from that of the phagosomal pathway. The integration of these paralogues, along with other Rabs not yet functionally characterized, and indeed other GTPases (Arf/Sar), and trafficking factors such as the SNAREs and proto-coatomer-derived complexes will be crucial to develop a more complete evolutionary view of the eukaryotic cell.
What remains to be achieved to enable an even finer picture of early Rab evolution? First, several Rab paralogues present in the LECA are uncharacterized in any detail, and functional information on the other paralogues is limited to a few eukaryotic model organisms. The investigation of Rab function in representatives for each eukaryotic supergroup will continue to provide invaluable information and render important cell biological context to the evolutionary reconstructions (Agop-Nersesian et al., 2009; Bright et al., 2010; Field and Carrington, 2004; Nakada-Tsukui et al., 2010; Rutherford and Moore, 2002). Second, the relationship between many ancestral Rab paralogues remains unresolved, even utilizing minimally derived Rab sequences, and awaits further advances in phylogenetic methodology. The final essential piece is the true position of the Rab phylogeny root: our use of Ran sequences as an outgroup (Fig. 3) is arbitrary (Colicelli, 2004). This last point is at the same time challenging and exciting and, given the integration of these GTPases in diverse cellular systems, once achieved it should prove illuminating not only for evolution of the membrane-trafficking system, but for the entire eukaryotic cell.
In conclusion, we present comprehensive evidence for ongoing sculpting within the Rab family, with unexpected ancient complexity and with paralogues destroyed, and to a lesser extent created, at all levels of the evolutionary process, i.e. comparatively proximal to the emergence of the modern supergroups and also in the more recent emergence of the individual taxonomic groups. Importantly, this pattern is seen in all supergroups, suggesting that the Rab protein family provides a potent force for endomembrane and cellular evolution across the entire range of Eukaryota.
Materials and Methods
Assembling the sequence dataset
Rab homologues from 55 species representing as many major eukaryotic lineages as possible were identified with BLASTp and tBLASTn searches (Altschul et al., 1997) against appropriate sequence databases; the source of sequences for each species is provided in supplementary material Table S1. For the purpose of this study, we worked further with sequences showing closer similarity to known Rabs than to members of other GTPase families [Ras, Rho, Miro, Rjl, RABL3 (Lip1), RABL5, Tem1 (Spg1), Roco, Arf, etc.]. We also excluded some highly divergent Rab-like sequences that were difficult to align, but we kept sequences of the RAN family, considered as a potential outgroup for phylogenetic analyses, and of two Rab-like families, RTW (RABL2) and IFT27 (RABL4), which differ from typical Rabs by the lack of a hypervariable C-terminal tail with a cysteine geranylgeranylation motif. We deliberately omitted some species (Trichomonas vaginalis, Paramecium tetraurelia, Entamoeba histolytica, microsporidians) that exhibit rather divergent and/or extremely expanded families of Rab sequences. Nonetheless, other representatives of the respective lineages are included in the dataset (i.e. Giardia lamblia, Trimastix pyriformis, Tetrahymena thermophila, Dictyostelium discoideum and various fungi) ensuring representation of the relevant taxonomic groups. Existing protein predictions were carefully verified and corrected whenever necessary.
Standard phylogenetic analyses
Sequences were initially aligned using ClustalX (Thompson et al., 1997) and the alignment was extensively edited manually, guided by solved structures of multiple diverse Rabs available from the Protein Data Bank (http://www.pdb.org/pdb/). Poorly conserved N- and C-terminal regions were excluded and a few highly variable internal regions were masked in the final ‘master’ alignment used for phylogenetic analyses (supplementary material Fig. S1). The various sub-datasets are available upon request. Different subsets of the aligned sequences were used to infer trees using two different implementations of maximum likelihood methods [RAxML v7.0.0 (Stamatakis, 2006) and PhyML v2.44 (Guindon and Gascuel, 2003)]. Bayesian inference was implemented in MrBayes v3.2 (Ronquist and Huelsenbeck, 2003), generally with 5×106 MCMC generations. In the case of the Rab32 analysis only 1×106 generations were needed to obtain convergence, whereas in several other datasets, analysis was run up to 18×106 generations in order for convergence to be achieved, as measured by a splits frequency below 0.1 being reached. Posterior probability values were obtained with burnin values determined by removing trees either prior to a graphically determined plateau of –LnL values or graphically or prior to the convergence generation, which ever was most conservative. Substitution models employed for inferring the trees were selected using ProtTest v1.3 (Abascal et al., 2005).
The ScrollSaw method
Five subsets of Rab sequences were assembled, each comprising sequences from a set of species representing one presumably monophyletic eukaryotic supergroup (Opisthokonta, Amoebozoa, Excavata, Archaeplastida and SAR+CCTH). The supergroup-specific datasets were combined in all possible pairwise combinations (10 in total) and for each paired dataset genetic distances between the sequences were inferred with the maximum likelihood method implemented in Tree-Puzzle 5.2 (Schmidt et al., 2002) and using the WAG+γ+I substitution model. Each of the resulting ten distance matrices were analysed to identify sequence pairs, each sequence from a different supergroup, that have mutually minimal distances among all distances to sequences from the opposite supergroup. Given the scale of our analysis this was performed using a script written in the R package (available upon request). After pooling the sequences from all these pairs and removing redundancies, trees were inferred using all three methods employed in this study (MrBayes, RAxML, PhyML). These trees were compared and dissected to define orthologous relationships among the sequences. Ancestral clades were reconstructed as supported by 0.95PP and at least 75% bootstrap support in one ML method. Similar criteria were applied when probing taxon-specific datasets with least diverged representatives of ancestral Rab paralogues (supplementary material Figs S7–S16). To resolve actual orthologous relationships among Rab24-related (supplementary material Fig. S6) and Rab32-related sequences (supplementary material Fig. S19), additional targeted analyses were conducted with the standard phylogenetic methods.
Supplementary Material
Acknowledgements
We thank the DOE Joint Genome Institute, BCM Human Genome Sequencing Center and the Broad Institute for generating and releasing prior to publication some of the draft genome assemblies and annotations exploited in this study. The authors are grateful to the following for discussions on the manuscript: John Archibald, Ryan McKay, James Kaufman and Michael Rout. We thank Jiri Neustupa (Charles University, Prague) for providing a script for analysing distance matrices.
Footnotes
Funding
The research was supported by the Czech Science Foundation [grant number P305/10/0205 to M.E.]; the Institute of Environmental Technologies [project registration number CZ.1.05/2.1.00/03.0100 to M.E.]; a Natural Sciences and Engineering Research Council of Canada Discovery Grant [grant number RPGIN 372638-09 to J.B.D.]; Alberta Innovates Technology Futures [grant number NFAO201000076 to J.B.D.]; and a Wellcome Trust program grant [grant number 082813 to M.C.F.]. Deposited in PMC for release after 6 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.101378/-/DC1
References
- Abascal F., Zardoya R., Posada D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104-2105 [DOI] [PubMed] [Google Scholar]
- Adl S. M., Simpson A. G., Farmer M. A., Andersen R. A., Anderson O. R., Barta J. R., Bowser S. S., Brugerolle G., Fensome R. A., Fredericq S., et al. (2005). The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52, 399-451 [DOI] [PubMed] [Google Scholar]
- Agop-Nersesian C., Naissant B., Ben Rached F., Rauch M., Kretzschmar A., Thiberge S., Menard R., Ferguson D. J., Meissner M., Langsley G. (2009). Rab11A-controlled assembly of the inner membrane complex is required for completion of apicomplexan cytokinesis. PLoS Pathog. 5, e1000270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayong L., Pagnotti G., Tobon A. B., Chakrabarti D. (2007). Identification of Plasmodium falciparum family of SNAREs. Mol. Biochem. Parasitol. 152, 113-122 [DOI] [PubMed] [Google Scholar]
- Brighouse A., Dacks J. B., Field M. C. (2010). Rab protein evolution and the history of the eukaryotic endomembrane system. Cell. Mol. Life Sci. 67, 3449-3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright L. J., Kambesis N., Nelson S. B., Jeong B., Turkewitz A. P. (2010). Comprehensive analysis reveals dynamic and evolutionary plasticity of Rab GTPases and membrane traffic in Tetrahymena thermophila. PLoS Genet. 6, e1001155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F., Inagaki Y., Bråte J., Archibald J. M., Keeling P. J., Cavalier-Smith T., Sakaguchi M., Hashimoto T., Horak A., Kumar S., et al. (2009). Large-scale phylogenomic analyses reveal that two enigmatic protist lineages, Telonemia and Centroheliozoa, are related to photosynthetic chromalveolates. Genome Biol. Evol. 1, 231-238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H., Reinisch K., Ferro-Novick S. (2007). Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev. Cell 12, 671-682 [DOI] [PubMed] [Google Scholar]
- Carlton J. M., Hirt R. P., Silva J. C., Delcher A. L., Schatz M., Zhao Q., Wortman J. R., Bidwell S. L., Alsmark U. C., Besteiro S., et al. (2007). Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315, 207-212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. (2002). The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int. J. Syst. Evol. Microbiol. 52, 297-354 [DOI] [PubMed] [Google Scholar]
- Colicelli J. (2004). Human RAS superfamily proteins and related GTPases. Sci. STKE 2004, re13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacks J. B., Doolittle W. F. (2002). Novel syntaxin gene sequences from Giardia, Trypanosoma and algae: implications for the ancient evolution of the eukaryotic endomembrane system. J. Cell Sci. 115, 1635-1642 [DOI] [PubMed] [Google Scholar]
- Dacks J. B., Field M. C. (2007). Evolution of the eukaryotic membrane-trafficking system: origin, tempo and mode. J. Cell Sci. 120, 2977-2985 [DOI] [PubMed] [Google Scholar]
- Dacks J. B., Poon P. P., Field M. C. (2008). Phylogeny of endocytic components yields insight into the process of nonendosymbiotic organelle evolution. Proc. Natl. Acad. Sci. USA 105, 588-593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacks J. B., Peden A. A., Field M. C. (2009). Evolution of specificity in the eukaryotic endomembrane system. Int. J. Biochem. Cell Biol. 41, 330-340 [DOI] [PubMed] [Google Scholar]
- Diekmann Y., Seixas E., Gouw M., Tavares-Cadete F., Seabra M. C., Pereira-Leal J. B. (2011). Thousands of rab GTPases for the cell biologist. PLOS Comput. Biol. 7, e1002217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias M. (2010). Patterns and processes in the evolution of the eukaryotic endomembrane system. Mol. Membr. Biol. 27, 469-489 [DOI] [PubMed] [Google Scholar]
- Elias E. V., Quiroga R., Gottig N., Nakanishi H., Nash T. E., Neiman A., Lujan H. D. (2008). Characterization of SNAREs determines the absence of a typical Golgi apparatus in the ancient eukaryote Giardia lamblia. J. Biol. Chem. 283, 35996-36010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias M., Patron N. J., Keeling P. J. (2009). The RAB family GTPase Rab1A from Plasmodium falciparum defines a unique paralog shared by chromalveolates and Rhizaria. J. Eukaryot. Microbiol. 56, 348-356 [DOI] [PubMed] [Google Scholar]
- Embley T. M., Martin W. (2006). Eukaryotic evolution, changes and challenges. Nature 440, 623-630 [DOI] [PubMed] [Google Scholar]
- Field M. C., Carrington M. (2004). Intracellular membrane transport systems in Trypanosoma brucei. Traffic 5, 905-913 [DOI] [PubMed] [Google Scholar]
- Fritz-Laylin L. K., Prochnik S. E., Ginger M. L., Dacks J. B., Carpenter M. L., Field M. C., Kuo A., Paredez A., Chapman J., Pham J., et al. (2010). The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 140, 631-642 [DOI] [PubMed] [Google Scholar]
- Fukuda R., McNew J. A., Weber T., Parlati F., Engel T., Nickel W., Rothman J. E., Söllner T. H. (2000). Functional architecture of an intracellular membrane t-SNARE. Nature 407, 198-202 [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696-704 [DOI] [PubMed] [Google Scholar]
- Gurkan C., Koulov A. V., Balch W. E. (2007). An evolutionary perspective on eukaryotic membrane trafficking. Adv. Exp. Med. Biol. 607, 73-83 [DOI] [PubMed] [Google Scholar]
- Hirst J., Barlow L. D., Francisco G. C., Sahlender D. A., Seaman M. N., Dacks J. B., Robinson M. S. (2011). The fifth adaptor protein complex. PLoS Biol. 9, e1001170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizing M., Helip-Wooley A., Westbroek W., Gunay-Aygun M., Gahl W. A. (2008). Disorders of lysosome-related organelle biogenesis: clinical and molecular genetics. Annu. Rev. Genomics Hum. Genet. 9, 359-386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling P. J. (2010). The endosymbiotic origin, diversification and fate of plastids. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 729-748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissmehl R., Schilde C., Wassmer T., Danzer C., Nuehse K., Lutter K., Plattner H. (2007). Molecular identification of 26 syntaxin genes and their assignment to the different trafficking pathways in Paramecium. Traffic 8, 523-542 [DOI] [PubMed] [Google Scholar]
- Kloepper T. H., Kienle C. N., Fasshauer D. (2007). An elaborate classification of SNARE proteins sheds light on the conservation of the eukaryotic endomembrane system. Mol. Biol. Cell 18, 3463-3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V. (2010). Preview. The incredible expanding ancestor of eukaryotes. Cell 140, 606-608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal K., Field M. C., Carlton J. M., Warwicker J., Hirt R. P. (2005). Identification of a very large Rab GTPase family in the parasitic protozoan Trichomonas vaginalis. Mol. Biochem. Parasitol. 143, 226-235 [DOI] [PubMed] [Google Scholar]
- Letunic I., Doerks T., Bork P. (2009). SMART 6: recent updates and new developments. Nucleic Acids Res. 37, D229-D232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumb J. H., Leung K. F., Dubois K. N., Field M. C. (2011). Rab28 function in trypanosomes: interactions with retromer and ESCRT pathways. J. Cell Sci. 124, 3771-3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackiewicz P., Wyroba E. (2009). Phylogeny and evolution of Rab7 and Rab9 proteins. BMC Evol. Biol. 9, 101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada-Tsukui K., Saito-Nakano Y., Husain A., Nozaki T. (2010). Conservation and function of Rab small GTPases in Entamoeba: annotation of E. invadens Rab and its use for the understanding of Entamoeba biology. Exp. Parasitol. 126, 337-347 [DOI] [PubMed] [Google Scholar]
- Olkkonen V. M., Ikonen E. (2006). When intracellular logistics fails - genetic defects in membrane trafficking. J. Cell Sci. 119, 5031-5045 [DOI] [PubMed] [Google Scholar]
- Pereira-Leal J. B. (2008). The Ypt/Rab family and the evolution of trafficking in fungi. Traffic 9, 27-38 [DOI] [PubMed] [Google Scholar]
- Pereira-Leal J. B., Seabra M. C. (2001). Evolution of the Rab family of small GTP-binding proteins. J. Mol. Biol. 313, 889-901 [DOI] [PubMed] [Google Scholar]
- Roger A. J., Simpson A. G. (2009). Evolution: revisiting the root of the eukaryote tree. Curr. Biol. 19, R165-R167 [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572-1574 [DOI] [PubMed] [Google Scholar]
- Rutherford S., Moore I. (2002). The Arabidopsis Rab GTPase family: another enigma variation. Curr. Opin. Plant Biol. 5, 518-528 [DOI] [PubMed] [Google Scholar]
- Saito-Nakano Y., Loftus B. J., Hall N., Nozaki T. (2005). The diversity of rab GTPases in Entamoeba histolytica. Exp. Parasitol. 110, 244-252 [DOI] [PubMed] [Google Scholar]
- Saito-Nakano Y., Nakahara T., Nakano K., Nozaki T., Numata O. (2010). Marked amplification and diversification of products of ras genes from rat brain, Rab GTPases, in the ciliates Tetrahymena thermophila and Paramecium tetraurelia. J. Eukaryot. Microbiol. 57, 389-399 [DOI] [PubMed] [Google Scholar]
- Sanderfoot A. (2007). Increases in the number of SNARE genes parallels the rise of multicellularity among the green plants. Plant Physiol. 144, 6-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H. A., Strimmer K., Vingron M., von Haeseler A. (2002). TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18, 502-504 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688-2690 [DOI] [PubMed] [Google Scholar]
- Stanier R. (1970). Some aspects of the biology of cells and their possible evolutionary significance. In Organization and Control in Prokaryotic and Eukaryotic Cells (ed. Charles H., Knight B.), pp. 1-38 Cambridge, UK: Cambridge University Press; [Google Scholar]
- Stenmark H. (2009). Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513-525 [DOI] [PubMed] [Google Scholar]
- Südhof T. C., Rothman J. E. (2009). Membrane fusion: grappling with SNARE and SM proteins. Science 323, 474-477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876-4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G., Dorrell R. G., Schlacht A., Dacks J. B. (2011). Eukaryotic systematics: a user's guide for cell biologists and parasitologists. Parasitology 138, 1638-1663 [DOI] [PubMed] [Google Scholar]
- Woollard A. A., Moore I. (2008). The functions of Rab GTPases in plant membrane traffic. Curr. Opin. Plant Biol. 11, 610-619 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.