Abstract
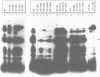
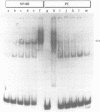
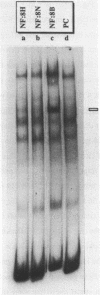
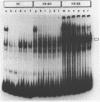
Control mechanisms of tRNA gene transcription were studied in vivo in Saccharomyces cerevisiae. In order to be able to monitor in vivo transcription products of an individual tRNA gene, a 'tester gene' was used which is readily transcribed in vivo in yeast but does not cross-hybridize with any cellular yeast tRNA. A series of insertion mutants were constructed, modifying thereby the immediate and further distant 5'-flanking region of the 'tester tRNA gene'. Small linker molecules of different length and different sequence were inserted at positions -3 and -56 on the non-coding strand. Resulting tRNA gene variants were transformed into yeast cells and in vivo synthesized products were monitored by primer extension analysis. From the experimental data we suggest that a few essential nucleotides within the flanking region are able to determine the in vivo transcription activity of the 'tester tRNA gene'. Our results are rationalized on a biochemical level by protein binding assays: At least one protein binds to the 5'-flanking region of the 'tester tRNA gene' and different protein complexes are sequestered on active or less active tRNA gene variants.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison D. S., Hall B. D. Effects of alterations in the 3' flanking sequence on in vivo and in vitro expression of the yeast SUP4-o tRNATyr gene. EMBO J. 1985 Oct;4(10):2657–2664. doi: 10.1002/j.1460-2075.1985.tb03984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold G. J., Gross H. J. Unrelated leader sequences can efficiently promote human tRNA gene transcription. Gene. 1987;51(2-3):237–246. doi: 10.1016/0378-1119(87)90312-x. [DOI] [PubMed] [Google Scholar]
- Burke D. J., Söll D. Functional analysis of fractionated Drosophila Kc cell tRNA gene transcription components. J Biol Chem. 1985 Jan 25;260(2):816–823. [PubMed] [Google Scholar]
- Camier S., Gabrielsen O., Baker R., Sentenac A. A split binding site for transcription factor tau on the tRNA3Glu gene. EMBO J. 1985 Feb;4(2):491–500. doi: 10.1002/j.1460-2075.1985.tb03655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M. F., Gerrard S. P., Cozzarelli N. R. Analysis of RNA polymerase III transcription complexes by gel filtration. J Biol Chem. 1986 Mar 25;261(9):4309–4317. [PubMed] [Google Scholar]
- Ciliberto G., Castagnoli L., Melton D. A., Cortese R. Promoter of a eukaryotic tRNAPro gene is composed of three noncontiguous regions. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1195–1199. doi: 10.1073/pnas.79.4.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco D., Sharp S., Söll D. Identification of regulatory sequences contained in the 5'-flanking region of Drosophila lysine tRNA2 genes. J Biol Chem. 1981 Dec 10;256(23):12424–12429. [PubMed] [Google Scholar]
- Dingermann T., Burke D. J., Sharp S., Schaack J., Söll D. The 5- flanking sequences of Drosophila tRNAArg genes control their in vitro transcription in a Drosophila cell extract. J Biol Chem. 1982 Dec 25;257(24):14738–14744. [PubMed] [Google Scholar]
- Dingermann T., Nerke K., Marschalek R. Influence of different 5'-flanking sequences of tRNA genes on their in vivo transcription efficiencies in Saccharomyces cerevisiae. Eur J Biochem. 1987 Dec 30;170(1-2):217–224. doi: 10.1111/j.1432-1033.1987.tb13689.x. [DOI] [PubMed] [Google Scholar]
- Dingermann T., Nerke K. Primer extension analysis of tRNA gene transcripts synthesized in vitro and in vivo. Anal Biochem. 1987 May 1;162(2):466–475. doi: 10.1016/0003-2697(87)90422-2. [DOI] [PubMed] [Google Scholar]
- Dunn T. M., Hahn S., Ogden S., Schleif R. F. An operator at -280 base pairs that is required for repression of araBAD operon promoter: addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5017–5020. doi: 10.1073/pnas.81.16.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman S. A., Engelke D. R., Geiduschek E. P. HeLa cell RNA polymerase III transcription factors. Functional characterization of a fraction identified by its activity in a second template rescue assay. J Biol Chem. 1984 Feb 10;259(3):1934–1943. [PubMed] [Google Scholar]
- Galli G., Hofstetter H., Birnstiel M. L. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature. 1981 Dec 17;294(5842):626–631. doi: 10.1038/294626a0. [DOI] [PubMed] [Google Scholar]
- Hipskind R. A., Clarkson S. G. 5'-flanking sequences that inhibit in vitro transcription of a xenopus laevis tRNA gene. Cell. 1983 Oct;34(3):881–890. doi: 10.1016/0092-8674(83)90545-7. [DOI] [PubMed] [Google Scholar]
- Koski R. A., Allison D. S., Worthington M., Hall B. D. An in vitro RNA polymerase III system from S. cerevisiae: effects of deletions and point mutations upon SUP4 gene transcription. Nucleic Acids Res. 1982 Dec 20;10(24):8127–8143. doi: 10.1093/nar/10.24.8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurjan J., Hall B. D., Gillam S., Smith M. Mutations at the yeast SUP4 tRNATyr locus: DNA sequence changes in mutants lacking suppressor activity. Cell. 1980 Jul;20(3):701–709. doi: 10.1016/0092-8674(80)90316-5. [DOI] [PubMed] [Google Scholar]
- Larson D., Bradford-Wilcox J., Young L. S., Sprague K. U. A short 5' flanking region containing conserved sequences is required for silkworm alanine tRNA gene activity. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3416–3420. doi: 10.1073/pnas.80.11.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassar A. B., Martin P. L., Roeder R. G. Transcription of class III genes: formation of preinitiation complexes. Science. 1983 Nov 18;222(4625):740–748. doi: 10.1126/science.6356356. [DOI] [PubMed] [Google Scholar]
- Lofquist A., Sharp S. The 5'-flanking sequences of Drosophila melanogaster tRNA5Asn genes differentially arrest RNA polymerase III. J Biol Chem. 1986 Nov 5;261(31):14600–14606. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Raymond G. J., Johnson J. D. The 5'-flanking sequence of yeast tRNA(Leu3) genes enhances the rate of transcription from stable pre-initiation complexes. Nucleic Acids Res. 1987 Dec 10;15(23):9881–9894. doi: 10.1093/nar/15.23.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond G. J., Johnson J. D. The role of non-coding DNA sequences in transcription and processing of a yeast tRNA. Nucleic Acids Res. 1983 Sep 10;11(17):5969–5988. doi: 10.1093/nar/11.17.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond K. C., Raymond G. J., Johnson J. D. In vivo modulation of yeast tRNA gene expression by 5'-flanking sequences. EMBO J. 1985 Oct;4(10):2649–2656. doi: 10.1002/j.1460-2075.1985.tb03983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaack J., Sharp S., Dingermann T., Burke D. J., Cooley L., Söll D. The extent of a eukaryotic tRNA gene. 5'- and 3'-flanking sequence dependence for transcription and stable complex formation. J Biol Chem. 1984 Feb 10;259(3):1461–1467. [PubMed] [Google Scholar]
- Schaack J., Sharp S., Dingermann T., Söll D. Transcription of eukaryotic tRNA genes in vitro. II. Formation of stable complexes. J Biol Chem. 1983 Feb 25;258(4):2447–2453. [PubMed] [Google Scholar]
- Sharp S. J., Schaack J., Cooley L., Burke D. J., Söll D. Structure and transcription of eukaryotic tRNA genes. CRC Crit Rev Biochem. 1985;19(2):107–144. doi: 10.3109/10409238509082541. [DOI] [PubMed] [Google Scholar]
- Sharp S., DeFranco D., Dingermann T., Farrell P., Söll D. Internal control regions for transcription of eukaryotic tRNA genes. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6657–6661. doi: 10.1073/pnas.78.11.6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp S., Dingermann T., Schaack J., DeFranco D., Söll D. Transcription of eukaryotic tRNA genes in vitro. I. Analysis of control regions using a competition assay. J Biol Chem. 1983 Feb 25;258(4):2440–2446. [PubMed] [Google Scholar]
- Shaw K. J., Olson M. V. Effects of altered 5'-flanking sequences on the in vivo expression of a Saccharomyces cerevisiae tRNATyr gene. Mol Cell Biol. 1984 Apr;4(4):657–665. doi: 10.1128/mcb.4.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague K. U., Larson D., Morton D. 5' flanking sequence signals are required for activity of silkworm alanine tRNA genes in homologous in vitro transcription systems. Cell. 1980 Nov;22(1 Pt 1):171–178. doi: 10.1016/0092-8674(80)90165-8. [DOI] [PubMed] [Google Scholar]
- Stillman D. J., Caspers P., Geiduschek E. P. Effects of temperature and single-stranded DNA on the interaction of an RNA polymerase III transcription factor with a tRNA gene. Cell. 1985 Feb;40(2):311–317. doi: 10.1016/0092-8674(85)90145-x. [DOI] [PubMed] [Google Scholar]
- Strauss F., Varshavsky A. A protein binds to a satellite DNA repeat at three specific sites that would be brought into mutual proximity by DNA folding in the nucleosome. Cell. 1984 Jul;37(3):889–901. doi: 10.1016/0092-8674(84)90424-0. [DOI] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Vigneron M., Matthes H., Wildeman A., Zenke M., Chambon P. Requirement of stereospecific alignments for initiation from the simian virus 40 early promoter. Nature. 1986 Jan 9;319(6049):121–126. doi: 10.1038/319121a0. [DOI] [PubMed] [Google Scholar]