Summary
Quantitative spatial distributions of ribosomes (S2-YFP) and RNA polymerase (β′-yGFP) in live E. coli are measured by superresolution fluorescence microscopy. In moderate growth conditions, Nucleoid-ribosome segregation is strong, and RNAP localizes to the nucleoid lobes. The mean copy numbers per cell are 4600 RNAPs and 55,000 ribosomes. Only 10–15% of the ribosomes lie within the densest part of the nucleoid lobes, and at most 4% of the RNAPs lie in the two ribosome-rich endcaps. The predominant observed diffusion coefficient of ribosomes is Dribo = 0.04 μm2/s, attributed to free mRNA being translated by one or more 70S ribosomes. We find no clear evidence of sub-diffusion, as would arise from tethering of ribosomes. The degree of DNA-ribosome segregation strongly suggests that in E. coli most translation occurs on free mRNA transcripts that have diffused into the ribosome-rich regions. Both RNAP and ribosome radial distributions extend to the cytoplasmic membrane, consistent with the transertion hypothesis. However, few if any RNAP copies lie near the membrane of the endcaps. This suggests that if transertion occurs, it exerts a direct radially expanding force on the nucleoid, but not a direct axially expanding force.
INTRODUCTION
The cytoplasm of bacterial cells exhibits remarkable spatial organization whose underlying causes are uncertain (Lewis, 2004). In E. coli, the chromosomal DNA is “condensed” into an irregularly shaped, central region called the nucleoid (Robinow & Kellenberger, 1994). The nucleoid does not fill the entire cytoplasmic volume, in spite of severe compaction of ~1.5 mm of DNA contour length within a 3 μm long × 1 μm diameter cell. The chromosomal origin oriC is anchored near the cell center until replication and segregation, when the two copies migrate to the quarter-cell positions (Nielsen et al., 2006). Certain proteins and lipids are known to concentrate in the polar or septal regions during specific phases of the cell cycle (Shapiro et al., 2009). There is recent evidence that specific mRNA transcripts carry information that targets them to specific cellular locations for translation (Nevo-Dinur et al., 2011).
In good growth conditions, multiple ribosomes are translating mRNA strands while they are being synthesized by RNA polymerase (“coupled transcription and translation”, or “co-transcriptional translation”) (Woldringh, 2002). Poly-ribosomes (“polysomes”) are necklaces of ribosomes serially translating the same mRNA copy (Brandt et al., 2009). Polysomes tethered to DNA via RNAP linkages were observed in early EM studies of E. coli extracts (Miller et al., 1970). Polysomes of compact, quasi-helical structure have recently been directly observed in cryoelectron tomography images (Brandt et al., 2009).
In some species, transcription and translation evidently occur predominantly in different sub-regions of the cytoplasm. In B. subtilis, the ribosomes are concentrated outside the nucleoid in ribosome-rich regions of the cytoplasm comprising the two polar end-caps, the space between nucleoid lobes, and a thin shell proximal to the cytoplasmic membrane (Lewis et al., 2000). Because some 80% of ribosomes are believed to be actively translating proteins in all growth conditions, the ribosome-rich regions evidently form a localized “protein factory” occupying some 50% of the cytoplasmic volume (Woldringh & Nanninga, 2006). Similarly, in E. coli early EM studies of thin slices of fixed cells found ribosomes concentrated near the periphery of the cell with the nucleoid avoiding the near vicinity of the cytoplasmic membrane (Robinow & Kellenberger, 1994). More recently, widefield immuno-fluorescence microscopy found the L7/L12 subunits of ribosomes to be strongly localized outside the nucleoid (Azam et al., 2000). In essential agreement with these data, a simple physical model of plectonemic DNA and hard-sphere ribosomes found that at sufficiently high DNA density, configurational entropy and excluded volume effects alone can cause strong nucleoid-ribosome segregation (Mondal et al., 2011). However, some species appear not to exhibit strong nucleoid-ribosome segregation. A recent study of Caulobacter crescentus found ribosomes and DNA to be spread throughout the cytoplasm (Llopis et al., 2010).
At least in B. subtilis and E. coli, the spatial separation of most of the chromosomal DNA from most of the ribosomes raises the question of the overall percentage of translation that could be coupled to transcription (Mascarenhas et al., 2001, Lewis et al., 2000). Early EM evidence from fixed E. coli cells indicates that RNA polymerase (RNAP) localizes near the boundary between the nucleoid and the cytoplasmic periphery (Durrenberger et al., 1988), but this has not been demonstrated in live cells. Comparison of the mean lifetime of mRNA (~5 min) with the mean time to transcribe a message (~20 s) suggests that most translation occurs on complete mRNA copies that have released from RNAP and diffused to the ribosome-rich regions (Llopis et al., 2010, Bernstein et al., 2002). Free messages coated with ribosomes and perhaps heat-shock proteins as well (El-Sharoud & Graumann, 2007) might avoid degradation by ribonucleases long enough to reach the ribosome-rich regions, where efficient polysomic translation could continue to protect them from damage.
The diffusion of free mRNA in live bacterial cells is not well characterized. In C. crescentus, a recent fluorescence in situ hybridization study found that mRNA strands did not diffuse significantly from the location where they were synthesized (Llopis et al., 2010). In sharp contrast, a recent study in E. coli found that free messages evidently diffuse to the specific regions of the cytoplasm where their protein products will be used, suggesting some type of location code within the message (Nevo-Dinur et al., 2011). A very long, 6000 kDa mRNA-MS2-GFP complex exhibited seemingly tethered diffusion with a short-time diffusion coefficient of 0.03 μm2-s−1 (Golding & Cox, 2004).
The new superresolution fluorescence microscopy methods (PALM, (Betzig et al., 2006) F-PALM, (Hess et al., 2006) and STORM (Rust et al., 2006)) are poised to provide a new level of quantitative information in live bacterial cells (Taniguchi et al., 2010, English et al., 2011, Lee et al., 2011, Wang et al., 2011). This includes protein copy numbers, spatial distributions with ~30-nm accuracy, and diffusion coefficients on sub-second time scales for a variety of key cytoplasmic components. Here we present a quantitative study of ribosomes in live E. coli labeled by the construct S2-YFP. Under moderate growth conditions, most cells exhibit two partially segregated nucleoid lobes and three “ribosome-rich regions”, the two endcaps and the region between nucleoid lobes. We find strong nucleoid-ribosome segregation. On average, only ~10–15% of the ribosomes are found within the dense regions of the nucleoid. The single-particle tracking measurements are consistent with free diffusion of 70S monomers or polysomes under effective confinement in the ribosome-rich regions, with Dribo ~ 0.04 μm2-s−1. This value is presumably describes the effective diffusion coefficient of free mRNA decorated with translating ribosomes. It follows that free mRNA born in the dense nucleoid region will find a ribosome-rich region in ~1 s or less. This is much shorter than typical mRNA degradation times of ~3–8 min (Bernstein et al., 2002). Finally, the diffusion data do not rule out coupled transcription and translation, but neither do we find any clear evidence of sub-diffusion of ribosomes, which would arise from tethering of ribosomes to DNA.
In a different strain of cells, superrresolution images of RNA polymerase are obtained from a β′-yGFP construct in live E. coli under the same growth conditions. yGFP is a YFP-like construct engineered from GFP (Nielsen et al., 2006). The spatial distribution of RNAP mimics that of DNA, as stained by the red dye DRAQ5. In moderate growth conditions, we do not find evidence for a high-density shell of RNAP surrounding the nucleoids. There are very few RNAP copies in the ribosome-rich regions.
Taken together, our results strongly suggest that at least in E. coli, most translation is not coupled with transcription (the “co-transcriptional translation” mechanism), counter to a view common in the literature. Instead, the data suggest that completed messages diffuse to find the ribosome-rich regions where the bulk of translation occurs. This is consistent with the fact that lifetime estimates of mRNA in live bacteria exceed typical times required for transcription of a message by a factor of 15 or so (Bernstein et al., 2002).
At the same time, we do observe a significant number of ribosome and RNAP copies lying within ~60 nm of the cytoplasmic membrane in close proximity to the dense nucleoid lobes. These copies are candidates for participation in “transertion”, which is the co-transcriptional translation of membrane proteins (Woldringh, 2002, Norris & Madsen, 1995). Transertion has been proposed as an “expanding” force on the nucleoids (Woldringh, 2002). Accordingly, we observe radial compaction of the nucleoids after treatment with rifampicin or chloramphenicol. However, the data do not support transertion of membrane proteins at the endcaps; there are essentially no RNAP copies close to the endcap sections of the cytoplasmic membrane. Thus transertion seemingly cannot provide the direct axial expanding force on the nucleoid that has been inferred from axial contraction of nucleoid lobes after chloramphenicol treatment.
Throughout this paper the x coordinate runs parallel to the long axis of the cell and the y coordinate runs perpendicular to x and along the short axis (Fig. 1B-inset).
Figure 1.
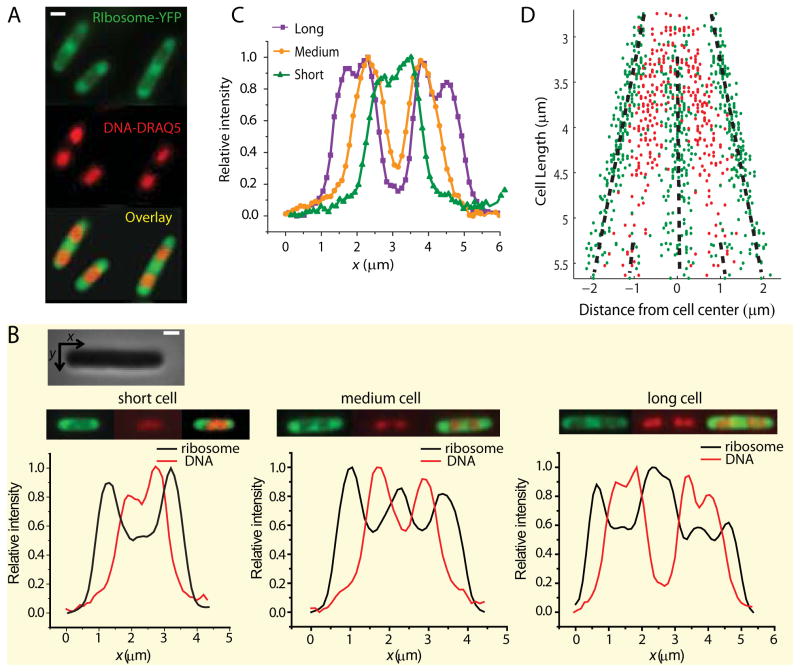
Widefield imaging of ribosomes (S2-YFP labels) and chromosomal DNA (DRAQ5 stain) for K-12 cells grown in EZRDM at 30°C. (A) Ribosome distribution (green) and DNA distribution (red) for three typical cells. The composite image shows the anti-correlation between the two distributions. (B) Axial intensity distributions in the ribosome and DNA channels for a short, medium, and long cell. Intensity is summed along y (short axis coordinate) at each x (long axis coordinate). The two channels are strongly anti-correlated. (Inset: Phase contrast image of a cell showing x (along the long axis) and y (along short axis) coordinates.) (C) DNA axial intensity distribution for a short, medium, and long cell plotted on same axes to show progressive segregation as the cell elongates. (D) The distance from the cell center of the local maxima in ribosome distribution (green dots) and DNA distribution (red dots) plotted for 286 cells of different cell length. The black dashed lines guide the eye. Scale bar = 1 μm.
RESULTS
Widefield co-imaging of ribosomes (S2-YFP labeling) and nucleoid (DRAQ5 staining)
For live cells growing in the complete, defined medium EZRDM at 30°C, Fig. 1A shows examples of widefield fluorescence images of ribosomes (labeled by the S2-YFP protein and observed in the yellow channel) and the chromosomal DNA (labeled by the permeable dye DRAQ5 and observed in the red channel). Ribosomes localize to the end caps and the central region between two segregated DNA lobes, the “ribosome-rich regions”. The DRAQ5-stained images show the chromosomal DNA segregated to varying degree into two nucleoid lobes. Axial intensity distributions (intensity summed along y at each axial position x) show that the degree of DNA segregation consistently increases from short to medium to long cells (Fig. 1C and 1D). As cells approach the division stage of the cycle, each of the two primary lobes begins to split into two sub-lobes (Fig. 1C).
The peak-to-valley intensity ratio in the S2-YFP axial intensity profiles is typically about 1.5:1.0, with ribosome peaks correlating with DNA valleys (Fig. 1B). However, the degree of ribosome segregation is underestimated from these axial fluorescence profiles obtained with the 1.49 NA objective. The S2-YFP images in Fig. 1A are in principle a superposition of broadly distributed cell autofluorescence; rapidly-diffusing, free S2-YFP or perhaps YFP copies; and a mixture of more slowly diffusing 30S subunits, 70S mono-ribosomes, and polysomes. Based on the diffusion evidence presented below, we believe the contribution of free S2-YFP and YFP is negligible. The background autofluorescence contributes only 2% of the yellow intensity, as judged by tests of wild-type cells grown and imaged under the same conditions. The widefield ribosome images are somewhat broadened by diffraction. However, the primary broadening is due to out-of-focus light collected by the 1.49 NA microscope objective, whose depth-of-focus parameter is only 500 nm (±250 nm about the focal point) (Hall, 2008). Widefield images of the ribosome distribution taken with a low numerical aperture objective (NA = 1.25, depth-of-focus parameter 660 nm) show stronger axial segregation (Fig. S1; peak-to-valley intensity ratios of 2:1 or even 3:1). The red DRAQ5 fluorescence images taken with the 1.49 NA objective are narrower than the S2-YFP images because the nucleoids are concentrated near the cell axis; there is much less out-of-focus light. Importantly, there is no clear evidence that the DNA distribution extends all the way to the tips of the cells, as seen by comparing the widefield DNA vs ribosome axial distributions (Fig. 1B).
The peaks in the axial profiles of ribosome-YFP intensity distributions are consistently anti-correlated to the peaks in the DNA-DRAQ5 intensity profile (Fig. 1B and 1D). The three typical cells in Fig. 1B were selected based on length: a short cell (2.8 μm tip-to-tip), a medium-length cell (3.2 μm), and a long cell (4.4 μm). For most of the short cells (2.6–3.0 μm tip-to-tip) the DNA localizes to the middle of the cell and the peak in DNA intensity appears near the center of the cell. In these short cells, ribosomes concentrate only at the end caps. For mid-length cells (3.0–4.5 μm tip-to-tip) DNA segregates into two lobes and the ribosome distribution has a third peak at mid-cell, the region between the segregated DNA lobes. Just before the cells divide, they are very long (4.5–5.5 μm tip-to-tip) and each DNA lobe begins to split into two sub-lobes (Fig. 1B at right). The ribosome distribution now has 5 peaks; even the minor DNA dips correspond to ribosome peaks. These same plots show the ribosome distribution filling both endcaps, whereas there is little or no DNA near the tips of the cell.
In Fig. 1D we plot the axial location relative to cell center of the peaks in the DNA and ribosome axial distributions from 286 cells of varying length. The locations of the peaks from each cell are displaced vertically according to total cell length, a rough measure of the phase of the cell cycle. As cells become longer, both ribosome and DNA peaks move symmetrically outward and remain segregated. Evidently the ribosome distribution responds faithfully to the DNA distribution, always avoiding the densest regions of DNA.
The radial distribution of S2-YFP intensity from the widefield images is significantly wider in the DNA-rich regions than in the ribosome-rich regions (data not shown). This is consistent with the presence of a shell of higher ribosome concentration near the cytoplasmic membrane and lower ribosome concentration near the axis of the cell where the DNA is densest (Mondal et al., 2011). This feature of the radial distribution of ribosomes becomes more obvious in the superresolution images presented below.
Widefield co-imaging of RNAP (β′-yGFP) and nucleoids (DRAQ5 staining)
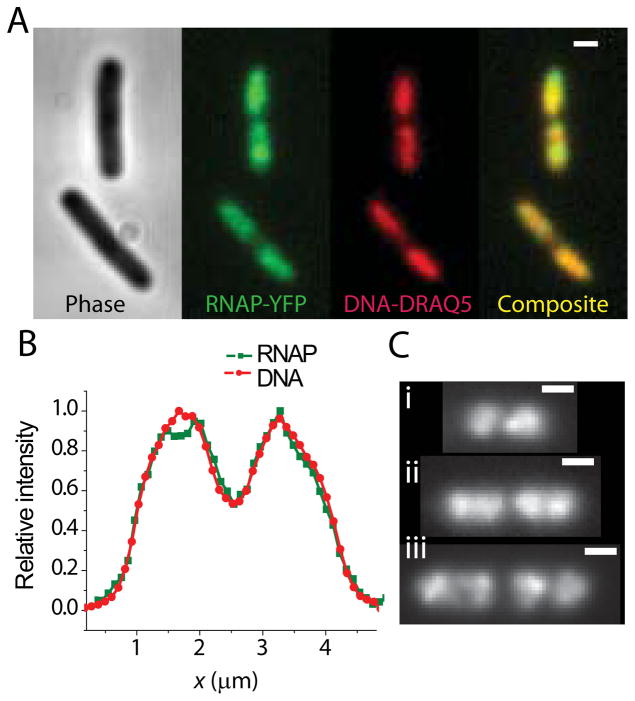
Figure 2A shows examples of widefield fluorescence images of RNAP (labeled by the β′-yGFP protein and observed in the yellow channel) and DRAQ5-labeled chromosomal DNA (observed in the red channel). As described before (Bratton et al., 2011, Cabrera et al., 2009), within each cell the RNAP distribution and the chromosomal DNA distribution are very similar (Fig. 2B). Like the DNA, the RNAP distribution typically exhibits two lobes; in the longest cells, each lobe has begun to sub-divide. The three examples in Figs. 2C show a typical short cell (2.6 μm tip-to-tip), mid-length cell (3.9 μm) and long cell (5.1 μm). The RNAP distribution follows the progressive segregation of DNA as the cell elongates. As noted elsewhere, the RNAP distribution also exhibits narrow, bright features that are absent in the DNA images (Cabrera et al., 2009, Bratton et al., 2011). These have been attributed to transcription foci, locations of particularly dense transcription activity (Cabrera & Jin, 2003, Lewis et al., 2000).
Figure 2.
Widefield imaging of RNA polymerase (β′-yGFP labels) and chromosomal DNA (DRAQ5 stain) for K-12 cells grown in EZRDM at 30°C. (A) Phase contrast image, RNAP image, and DNA image are shown for two different cells. The composite image shows RNAP co-localizes with DNA. Scale bar is 1 μm. (B) Axial intensity profile for RNAP-yGFP and DRAQ5-labeled DNA compared from a single cell. (C) RNAP images from three cells of length (i) 2.6 μm, (ii) 3.9 μm, and (iii) 5.1 μm. Scale bar is 1 μm. Progressive segregation of RNAP distribution is similar to DNA segregation shown in Fig. 1.
Superresolution ribosome spatial distribution
The superresolution images obtained here use the reversible photobleaching method (Biteen et al., 2008). Initially all the YFP copies in a cell are photobleached using 514 nm light. Subsequently, some 10–20% of the photobleached population spontaneously and gradually returns to the fluorescent state. The low density of these “revived” YFP copies enables single-molecule imaging. By plotting a point at the (x,y) coordinates of each location, we obtain a high-resolution spatial distribution averaged over the several-minute acquisition period. The dynamic localization accuracy is estimated to be σ~ 30 nm in each dimension x and y (Supporting Information). This superresolution method is impervious to background autofluorescence because almost no such fluorescence remains after the photobleaching step. In addition, use of the 1.49 NA objective plus the single-particle thresholding algorithm leads to significant sectioning of the cell in the x-y image. We are most sensitive to molecules in a horizontal, central slab of approximately 500–600 nm thickness (z in the range ±250–300 nm), which is substantially smaller than the 780 nm cell diameter (Bakshi et al., 2011). The top and bottom layers of the cytoplasm are not imaged (Fig. S3). Unlike the widefield images, the superresolution images obtained with the 1.49 NA objective are not broadened by out-of-focus light from the top and bottom of the cell; those molecules are rejected by the threshold criterion.
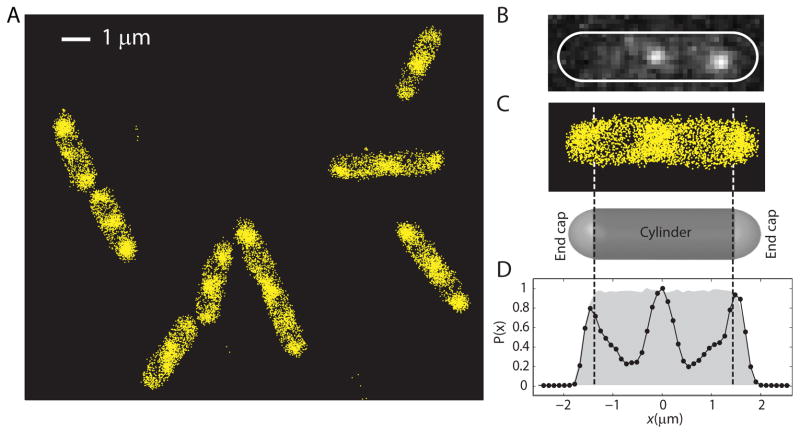
Ribosome superresolution images for several cells are shown in Fig. 3A. At the 33 Hz frame rate, we were able to study some 1,000–3,000 distinct S2-YFP copies in the typical, 3.5 μm long cell over a period of 2–3 min. The mean trajectory length under these imaging conditions is only 3 frames, so we typically obtain about 5,000–10,000 locations over the same period. The significant improvement in resolution and increased contrast between the DNA-rich region and the ribosome-rich region are immediately obvious. In Fig. 3C we display the superresolution image for a mid-length cell (3.6 μm tip-to-tip), which evidently has fairly strong nucleoid lobe separation. The histogram of Fig. 3D gives Nribo(x), the number of ribosomes detected within each axial bin at position x, with bin widths chosen at 100 nm to provide good signal-to-noise ratio. There are very few ribosomes within the nucleoid lobes. The peak-to-valley concentration ratio is at least 5:1 between the ribosome-rich regions and the densest part of the nucleoids. This may well be an underestimate due to the presence of a thin shell of ribosomes around each nucleoid lobe (Fig. S4B) and the irregular shape of the lobes themselves. This peak-to-valley ratio varies from cell to cell as illustrated in Fig. S2.
Figure 3.
Superresolution images of ribosomes (S2-YFP) within K-12 cells grown in EZRDM at 30°C. Each localization is plotted as a point at the calculated centroid position. (A) Nine representative cells. (B) Image of two single molecules within a cell prior to image filtering. Cell outline based on phase contrast image. (C) Expanded view of superresolution image of ribosomes in the same cell. A model spherocylinder is shown as a guide to the endcap positions. (D) Relative number of ribosomes at each axial position, with data along y at each x summed into 100 nm bins. The grey background shows the theoretical profile for a uniform distribution filling the model spherocylinder, taking account of measurement uncertainty and binning. Sectioning by the 1.49 NA objective does not affect the axial distribution significantly, as shown in Supporting Information (Fig. S3).
Evidence for the thin shell of ribosomes surrounding the nucleoid lobes is shown in Fig. S4B. Due to the sectioning by the 1.49 NA objective, the y-coordinate is a reasonable proxy for a radial coordinate. The y-axis profiles of the ribosome distribution limited to the regions containing the two nucleoid lobes for 12 cells are combined to make a smoother histogram. We often observe the axis of a nucleoid lobe to be off-center relative to the overall cell axis, meaning one edge of the thin ribosome shell is wider than the other. This causes broadening of features in the composite histogram. Nevertheless, the ribosome radial distribution clearly peaks away from the cell axis and extends all the way to the cytoplasmic membrane. As a control, we used cells expressing monomeric YFP from a plasmid to measure the y-distribution of free YFP molecules. The result is very similar to the theoretical distribution assuming YFP fills the cytoplasm uniformly and taking account of the sectioning by the 1.49 NA objective (Fig. S3).
Superresolution RNA polymerase spatial distribution
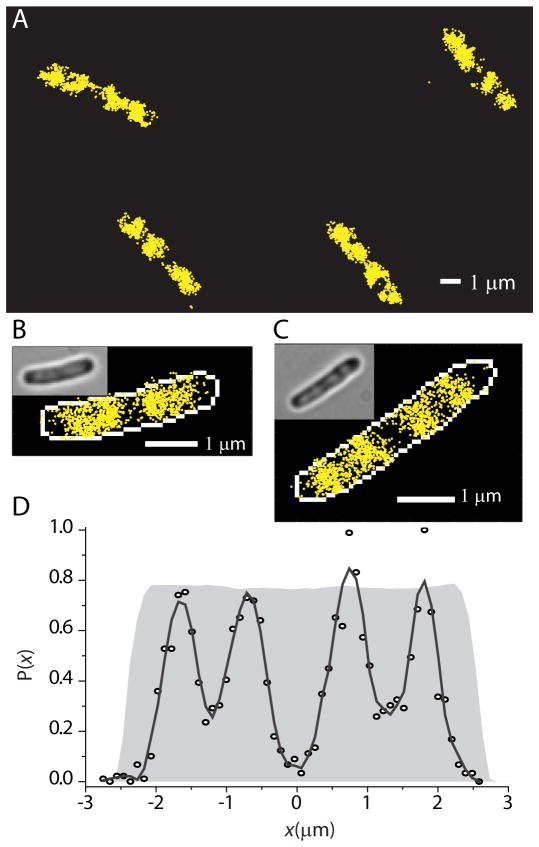
In the RNAP imaging experiments we obtain about 1000–3000 localizations of β′-yGFP from about 500 individual RNAP molecules. Fig. 4A shows images of four cells. The yellow dots are the centroid of the fluorescence image of single RNAP molecules in each camera frame. In Figs. 4B and 4C we show expanded superresolution images of the RNAP distribution in a medium-length cell (3.9 μm) and a long cell (5.2 μm tip-to-tip). The white outline is the mask created by edge detection from the white light image of the cell (Fig. S9). For the longer cell, the RNAP axial distribution (Fig. 4D) closely mimics the four-peaked DNA distribution common among the longest cells (Fig. 1B), but with enhanced spatial resolution. The endcap region and the region at mid-cell between the two pairs of DNA lobes are essentially devoid of RNAP. The number density of RNAP in the ribosome-rich region between nucleoid lobes is <10% of that in the densest part of the nucleoid lobes.
Figure 4.
Superresolution images of RNA polymerase (β′-yGFP) within K-12 cells grown in EZRDM at 30°C. (A) Four representative cells. (B) Expanded view of a medium-length cell (tip-to-tip length = 3.9 μm). (C) Expanded view of a long cell (tip-to-tip length = 5.2 μm). (D) Relative number of RNAP copies at each axial position x (100 nm bins). Solid line is the result of a Savitzky-Golay smoothing filter. The grey background shows the theoretical profile for a uniform distribution filling the model spherocylinder, as in Fig. 3. Note that RNAP avoids the endcaps.
Importantly, the y-coordinate profiles of the RNAP molecules, although noisy, clearly extend all the way to the cytoplasmic membrane (example in Fig. S4D). The presence of RNAP very near the cytoplasmic membrane suggests the presence of some DNA there as well, but only in the straight, cylindrical part of the cell. There is essentially no RNAP near the endcap portion of the cytoplasmic membrane (Fig. 4D). This has implications for the previously proposed transertion mechanism.
Copy numbers and number densities of ribosomes and RNAP
Because the S2-YFP gene replaces the normal S2 gene on the chromosome, all copies of S2 should carry the YFP label. We further assume that all S2-YFP copies are fluorescent and that essentially all S2 proteins are incorporated into 30S subunits. In Supporting Information, we show that original and “revived” YFP copies have the same fluorescence intensity under single-molecule imaging conditions (Fig. S16). We can then scale the total, pre-bleach YFP intensity to that of a single YFP molecule to estimate the copy numbers of ribosomes and RNAP in each cell (Taniguchi et al., 2010). For ribosomes (meaning the sum of 30S subunits and complete 70S ribosomes), the resulting estimated copy number ranges from ~30,000 to ~70,000 ribosomes per cell (42 cells, Fig. S6). The mean number of ribosomes per cell is ~55,000. The ribosome copy number increases monotonically with cell volume. The estimated copy number of β′-yGFP ranges from ~2000 per cell to ~10,000 per cell. The average number of β′-yGFP per cell is 4,600 (31 cells, Fig. S6). The number of β′-yGFP is correlated with cell volume.
To accurately determine cell geometry, we have also carried out superresolution imaging of free YFP expressed from a plasmid (Fig. S3, (Bakshi et al., 2011)). The cell radius R = 380 ± 20 nm is remarkably well conserved from cell to cell and is essentially independent of cell length. This enables calculation of the volume of each cell to about 10% accuracy. For each cell, the corrected fluorescence intensity and the known cell geometry then enables estimation of the average ribosome number density (i.e., the average concentration over the entire cytoplasmic volume) and the peak ribosome number density in the ribosome-rich regions. The average ribosome number density varies from 25,000 to 31,000 ribosomes/μm3 (Fig. S6) with a mean across cells of 27,000 ribosomes/μm3. The peak number density within ribosome-rich regions varies from 56,000 to 73,000 ribosomes/μm3 (Fig. S6) with a mean of 65,000 ribosomes/μm3. These values should prove useful in modeling ribosome diffusion (Ando & Skolnick, 2010). As shown by the axial distributions, the peak ribosome density is usually very similar in the central region and in the endcaps. From axial distributions such as those in Fig. 3D, we estimate that about 10–15% of the total ribosome population lies within the DNA-rich nucleoid lobes.
Similarly, we use the total RNAP-yGFP intensity and the known geometry to calculate the average number density of RNAP within the cytoplasm. This varies across cells from 1800 to 3500 RNAP/μm3, with a mean of 2200 RNAP/μm3. The peak RNAP number density within the nucleoid is difficult to estimate because the nucleoid lacks a sharply defined volume.
Clearly under these moderate growth conditions at 30°C, most of the 55,000 ribosomes and very few of the 4,600 RNAP molecules lie in the ribosome-rich regions of the cytoplasm. The cleanest quantitative estimates come from the endcaps, whose geometry is well defined; the central ribosome-rich region has poorly defined geometry. From composite data from five cells ranging in total length from 2.8 to 4.8 μm, we estimate that on average at most 180 copies (4%) of RNAP lie in the two endcaps combined. Essentially none of these few RNAP copies are proximal to the endcap membrane. The two endcap volumes contain some 17,000 ribosomes, either as 30S subunits or complete 70S ribosomes.
Diffusion of ribosomes
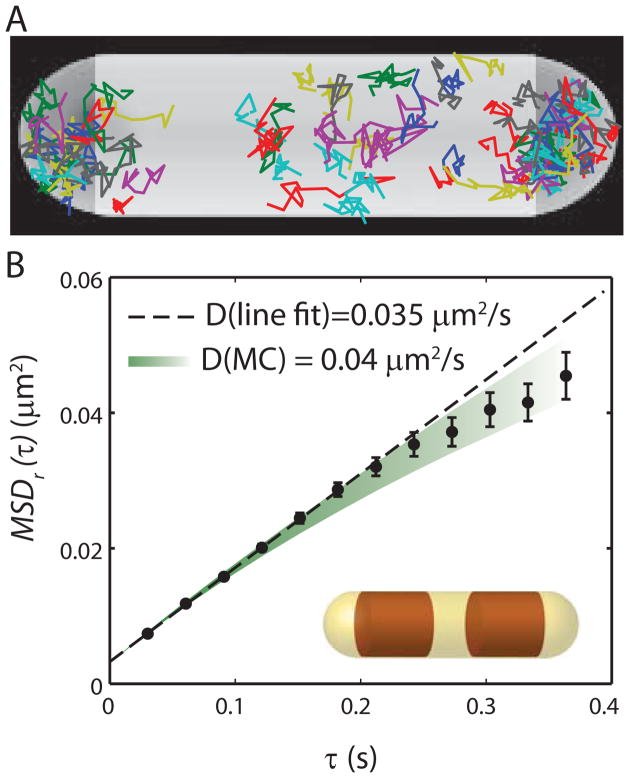
A detailed study of diffusion by tracking of single S2-YFP labels in two dimensions (using lower laser power) provides important clues to the nature of the species under study. The number of switched-on copies per cell was limited to 1–3 per frame, which usually enables accurate tracking of these slow-moving molecules. In practice, we were able to track 500–2000 molecules per cell over a period of 15–20 min. Reasonably good trajectories were obtained with 10 ms frames taken at 30 ms intervals. The distribution of trajectory lengths is roughly exponential, with a mean of 5–6 frames. In Fig. 5 we display those trajectories of 13 steps or longer for a single cell. There are typically ~50 such trajectories per cell. Trajectories longer than 13 steps are truncated at 13 for analysis. The trajectories are rather compact, and usually sample only one of the three ribosome-rich regions.
Figure 5.
Ribosome diffusion in untreated cells. (A) Trajectories of single ribosomes (S2-YFP labels) within one cell, plotted within a spherocylinder chosen to match the phase contrast image. (B) Mean-square displacement vs lag time, averaged over 53 single-molecule trajectories from the same cell, each consisting of 13 steps. Error estimates are ±1σof the MSD values from single molecules. Dashed line is a linear fit to first three data points, yielding a diffusion coefficient estimate of D = 0.035 μm2-s−1. Colored swath represents the range of theoretical MSDr(τ) plots (± one standard deviation of the mean) obtained from averaging fifty 13-step Monte Carlo simulated trajectories using D = 0.04 μm2-s−1, the best-fit value as judged by eye. The simulations were run in a spherocylinder within which two truncated cylinders (representing the segregated DNA nucleoid lobes) block ribosome diffusion, as shown in the inset.
For a particular cell, the mean-square displacement vs lag time, MSDr(τ), is computed as a running average (Fig. 5B). This is the average of over all molecules within one cell of the squared displacement from a starting location, with r = (x2 + y2)1/2. Three additional examples are given in Fig. S7. Cells exhibit negative curvature of MSDr(τ) after about five 30-ms steps. For free diffusion in an infinite volume, the mean-square displacement increases linearly in time. Finite volume in which to diffuse or sub-diffusion mechanisms (see below) could cause the MSD to rise more slowly than linearly in time.
We tested the effects of confinement in the ribosome-rich regions using Monte Carlo (random walk) simulations in a representative model geometry: a spherocylinder containing two impenetrable truncated cylinders to mimic the nucleoid lobes (Fig. 5B, inset). Initial particle positions were chosen randomly within the three regions available to ribosomes. The range of Monte Carlo outcomes for fixed D, each averaged over a number of trajectories similar to the amount of experimental data, is illustrated by the swath in Fig. 5. The simulations show that the curvature of MSDr(τ) is reasonably well explained by a model of free diffusion of ribosomes in a confined space. After testing the data for five cells against Monte Carlo simulations, we found that simply fitting a straight line to the slope of the first four points of MSDr(τ) underestimates D by a consistent factor of 1.15 ± 0.05. For consistency on 23 cells, we used the linear estimate with the correction factor to estimate D for each cell. The resulting distribution of D across cells is shown in Fig. S13. The mean diffusion coefficient is <D> = 0.04 ± 0.01 μm2/s (± one standard deviation). The dispersion is likely due to a combination of errors in D estimation and real cell-to-cell variation in ribosome diffusion, as observed for other species (Bratton et al., 2011).
The confined Monte Carlo simulations do not reproduce the curvature of the MSDr(τ) plots within the experimental uncertainty. This may be due to the oversimplified model of nucleoid geometry—in practice the edges of the nucleoid are “soft” and likely do not completely exclude ribosomes. Monte Carlo simulations using D = 0.04 μm2-s−1 curve towards an asymptote of ~0.1 μm2-s−1, but much of the curvature occurs on a time scale longer than 1 s (beyond our experimental range). We cannot completely rule out possible effects of sub-diffusion, defined here as a sub-linear increase in MSDr(τ) due to causes other than geometric confinement. Two possible sub-diffusion mechanisms are: (1) monosomes or ribosomes translating mRNA that remains tethered to DNA by co-transcriptional translation (Llopis et al., 2010, English et al., 2011) and (2) entanglement of free monosomes or polysomes due to long, trailing segments of unfolded nascent polypeptide chains. While the curvature in MSDr(τ) can be explained without recourse to sub-diffusion, the data would not be strongly affected by a 10% tethered component.
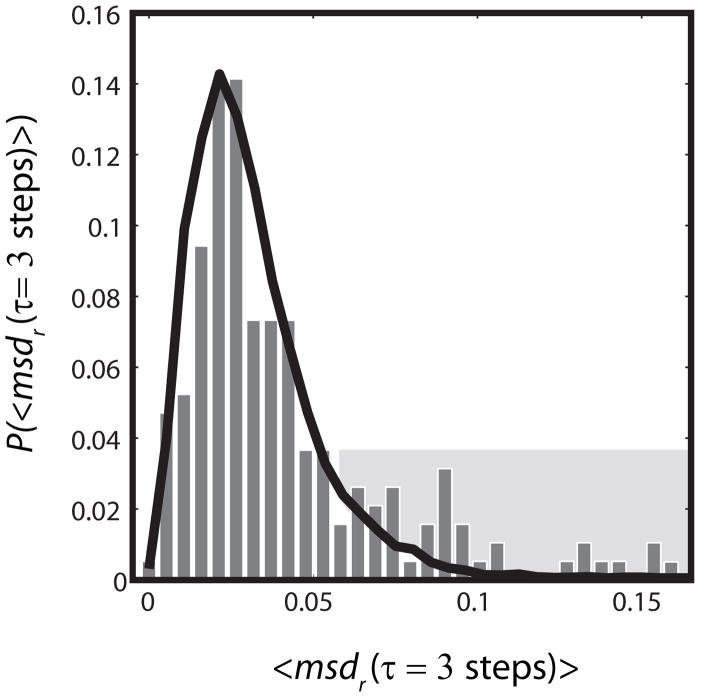
The near linearity of MSD plots does not preclude the possibility that different labeled copies have different diffusion coefficients, as would be expected for 30S subunits vs polysomes. We tested for such heterogeneity of the diffusion by examining the distribution of single-molecule mean-square displacements at fixed time lag τ. We call this single-molecule quantity msdr(τ); in contrast to MSDr(τ), it is not averaged over all trajectories obtained for each cell. In Fig. 6 we used all trajectories of 10 steps or longer to form the distribution of msdr(τ) within one particular cell. The experimental distribution is compared with that from analogous Monte Carlo simulations assuming free, homogeneous diffusion in the confined geometry of Fig. 5. The breadth of the model distribution illustrates how noisy short, diffusive trajectories are. When the simulation diffusion coefficient is chosen as D = 0.04 μm2-s−1 to match the narrow peak of the experimental distribution, the experimental amplitude of the peak and the long tail on the data are not well fit. When the diffusion coefficient is chosen as D = 0.05 μm2-s−1 to match the mean of the experimental data, the fit again fails. This indicates heterogeneity in the diffusive behavior, meaning at least two populations with diffusion coefficients sufficiently different that they are distinguishable over a lag time of 3 steps.
Figure 6.
Heterogeneity of ribosome diffusion. Experimental distribution (bars) of best-estimate single-ribosome diffusion coefficients calculated from the three-step mean-square displacement. Each trajectory of ten steps or longer in a single cell is truncated at 10 steps. For each single-molecule j, msdr,j at lag time τ= 3 steps = 30 ms is calculatedas a running average over all 10 steps. The single-molecule msd is estimated as: msdr,j = <r2(τ=3 steps)>j/4τ. The black line represents the distribution from a simulation with 10,000 trajectories of length 10 step, using a homogeneous diffusion coefficient of 0.04 μm2/s and normalized to match experiment. The geometric model includes two impenetrable cylinders to represent nucleoid lobes, as shown in Fig. 5. The peak and the shaded tail of the experimental distribution are not well fit by the model. See Fig. S8 for a two-component fit to composite data from three cells.
While the data almost surely contain contributions from many species with somewhat different diffusion coefficients, we have attempted to fit the data to the simplest heterogeneous model of only two non-exchanging populations. The fractional populations of the two components are f1 and f2 = 1 − f1 and the different diffusion coefficients D1 and D2 are independent of time over the period of measurement. In the present case, this might be slowly diffusing polysomes and faster diffusing monosomes or 30S subunits, for example. Composite data from three cells is reasonably well fit by a combination of two non-exchanging populations, ~80% of a slow component with D1 = 0.04 μm2-s−1 and ~20% of a faster component with D2 = 0.12 μm2-s−1, as shown in Fig. S8. Even under the assumption of only two components, this combination of parameters is by no means unique. The percentage of the slow component can be varied from about 65–85% while still achieving a reasonable fit. The two-population model is almost surely too simple, but the resulting estimates of diffusion coefficients and populations help us interpret the underlying nature of the diffusing species (Discussion).
Effects of rifampicin and chloramphenicol on distribution and diffusion of ribosomes
As is well known, rifampicin (Rif) halts transcription initiation. Chloramphenicol (Chl) halts translation, evidently by preventing release of ribosomes. Both drugs are known to significantly affect the morphology of the nucleoid (Cabrera et al., 2009). On long time scales after drug treatment, Rif expands the nucleoid, while Chl further compacts the nucleoid (Cabrera et al., 2009). We have measured the time-dependent effects of these drugs on nucleoid-ribosome segregation and on ribosome diffusion. On a time scale of 8 min after treatment, we find that both Rif and Chl contract the nucleoid, both radially and axially (data not shown).
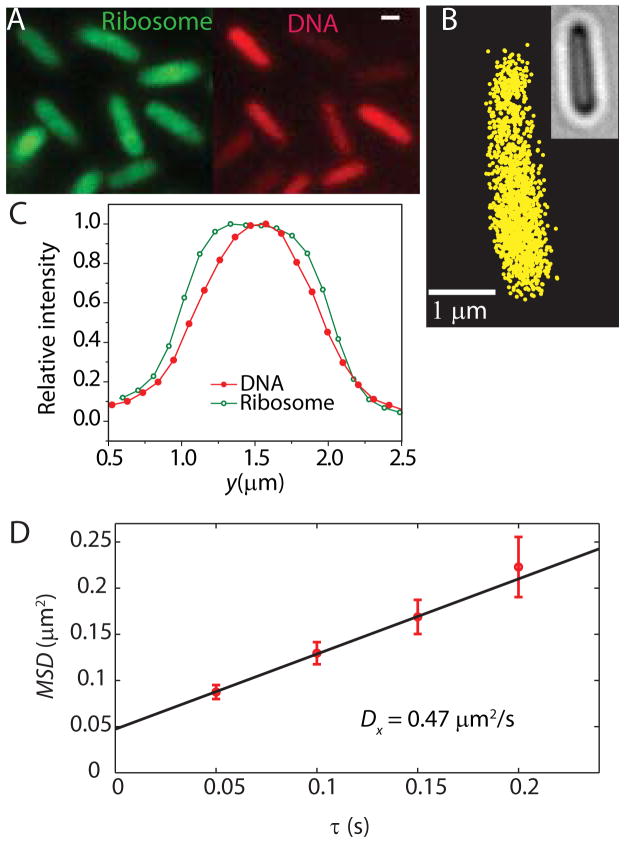
After 30 min of treatment with Rif at 200 μg/mL, the nucleoid has expanded axially, seemingly filling the cytoplasm uniformly (Fig. 7A). As measured by either widefield or superresolution microscopy, the ribosome distribution also expands (Fig. 7B). However, widefield intensity distributions along y show that the ribosome distribution remains somewhat broader than the DNA distribution (Fig. 7C). Evidently the DNA polymer continues to avoid the cytoplasmic boundary, in accord with a simple physical model of plectonemic DNA (Mondal et al., 2011). At the same time, the diffusion coefficient of the S2-YFP labels is enhanced about 10-fold. To minimize confinement effects, here we choose x as the analysis coordinate. An example of an MSDx(τ) plot is shown in Fig. 7D. The trajectories are short due to background introduced by Rif, which fluoresces weakly. In separate tests on wild-type cells, we have shown that the fluorescence quantum yield of Rif is too small to enable single-molecule detection. As estimated from the short-time slope, the diffusion coefficient after Rif treatment is <D30S> ~ 0.6 ± 0.2μm2/s (average over 9 cells). We tentatively attribute the much faster diffusion to 30S subunits.
Figure 7.
Effects of treatment with rifampicin. See text for details. (A) Widefield ribosome (S2-YFP) and DNA (DRAQ5) spatial distributions 30 min after rifampicin addition. Scale bar =1 μm. (B) Super-resolution image of ribosome distribution. See Fig. 3 for comparison with untreated cells. (C) Widefield intensity distributions for ribosomes and DNA along the short cell axis y. (D) MSDx(τ) plot for single-ribosome diffusion in one cell after rifampicin treatment. Linear fit yields the estimate D = 0.47 μm2-s−1.
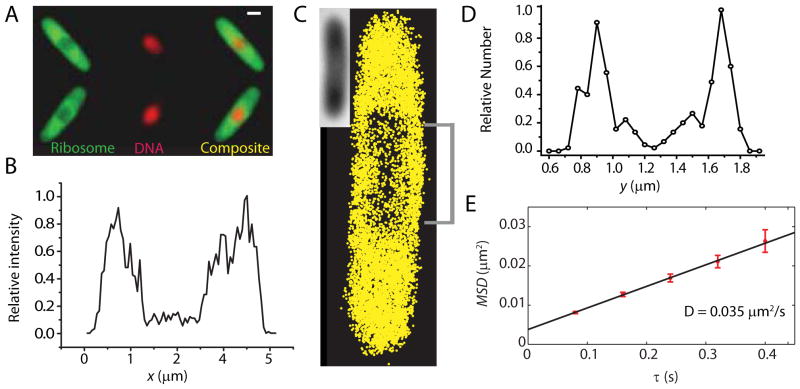
In sharp contrast, after 30 min of treatment with Chl at 200 μg/mL, the nucleoid has contracted both radially and axially, and very strong nucleoid-ribosome segregation is observed (Fig. 8A,C). The spatial distribution within a central 400-nm wide slice along the x-axis shows significantly greater ribosome-nucleoid segregation than in untreated cells, with peak-to-valley ratio of at least 10:1. From initial slopes of MSDr(τ) plots (Fig. 8E), we obtain <D> = 0.05 ± 0.02 μm2-s−1 (mean of 16 cells). Because Chl halts translation and does not permit release of the ribosomes, we tentatively attribute this value to a combination of ribosome monomers and polysomes similar to that in untreated cells.
Figure 8.
Effects of treatment with chloramphenicol. (A) Widefield ribosome (S2-YFP) and DNA (DRAQ5) spatial distributions 30 min after addition of chloramphenicol. Scale bar = 1 μm. (B) Axial ribosome profile from the super-resolution image shown in (C). Only the central 400 nm are included in the plot. (C) Superresolution image of ribosomes, 40 min after chloramphenicol treatment. (Inset = white light image of the cell). (D) Ribosome distribution along y, averaged over the 1 μm swath through the cell center as shown in (C). (E) MSDr(τ) plot from single-particle tracking of ribosomes 40 minutes after chloramphenicol treatment. Linear fit yields the estimate D = 0.035 μm2-s−1.
DISCUSSION
What S2-YFP-labeled species are located and tracked?
The first question is exactly what S2-YFP-containing species we are locating and tracking. Several time scales are important here. The maturation time of YFP in vitro is τmat ~ 10 min; this is limited by the lag time between translation and folding of the protein and the oxidation step that renders it fluorescent (Nagai et al., 2002). We assume a similar maturation time applies to the cytoplasm of E. coli (Sniegowski et al., 2005). The rise time for exponential growth of E. coli in EZRDM at 30°C is τg = 76 min. This base-e rise time is 1.44 times the ~54-min doubling time t1/2: τg = t1/2/ln2. As shown in the simple exponential growth model in Supplemental Information, the fraction of YFP that is mature is fmat = τg/(τmat + τg) ~ 0.9. The time for complete assembly of the 30S and 50S ribosomal subunits is τribo ~ 2–3 min (Lindahl, 1975).
Probably most S2-YFP copies are maturing to become fluorescent after incorporation into a 30S subunit. This means that some 90% of total YFP copies should be fluorescent and essentially all the fluorescent copies we detect are incorporated into 30S subunits. There is no evidence of the small, free proteins S2-YFP or YFP, which are expected to diffuse very rapidly in cytoplasm (Supporting Information). Based on the 55 kDa size of the S2-YFP construct, we estimate DS2-YFP ~ 5 μm2-s−1 (Bakshi et al., 2011), which is 100 times faster than the observed diffusion. If a significant fraction of the labels were free S2-YFP or YFP, the histogram of 3-step mean-square displacements (Fig. 6) would show a much longer tail. In Supplemental Information, we describe a rigorous test for rapidly diffusing molecules using faster frame times.
The species under study could be any combination of free 30S ribosomal subunits, translating 70S ribosome monomers, and polysomes (multiple 70S ribosomes simultaneously translating the same message). Classical data indicate that some 80% of ribosomes are translating at a given moment (Forschhamer & Lindahl, 1971) (Young & Bremer, 1976), meaning that most 30S subunits are incorporated into translating 70S ribosomes. Accordingly, the diffusion of the labels is heterogeneous with two resolvable components (Figs. 6 and S6): roughly 80% slow, with Dribo ~ 0.04 μm2-s−1 and roughly 20% faster, with D30S ~ 0.12 μm2-s−1. There is no obvious correlation of fast and slow diffusion with location in the different regions of space. Therefore, we infer that the faster component is due to free 30S subunits and the slower component is due to an undetermined mixture of ribosome monomers and polysomes. Because the 70S monosomes and polysomes are all translating mRNA, the slow component would then actually be attributed to diffusion of mRNA being translated by one or more ribosomes in live E. coli.
Data from the Rif-treated cells confirms this inference. In Rif-treated cells, D30S ~ 0.6 μm2-s−1, some 15 times faster than the major component and five times faster than the minor component in untreated cells. This remains ten times too slow to arise from free S2-YFP copies. Instead, we suggest that the Rif-treated cells contain predominantly 30S subunits. Treatment with Rif is known to halt transcription (Campbell et al., 2001). During the 30-min Rif treatment prior to observation, we expect all 70S ribosomes to complete translation of their messages and dissociate into free 30S and 50S subunits. Existing mRNA should degrade on a time scale of ~5 min. In the absence of new mRNA, the dominant YFP-labeled species should then be free 30S subunits. It is plausible that D30S ~ 0.6 μm2-s−1 in Rif-treated cells would be significantly faster than D30S ~ 0.12 μm2-s−1 in untreated cells because the 30S subunits are diffusing in a very different medium. The DNA meshwork has expanded, which would attenuate possible sieving effects on diffusion. The volume fraction occupied by ribosomal components (now 30S and 50S subunits) has roughly halved. As a result, crowding, sieving, and hydrodynamic effects should hinder diffusion significantly less for 30S subunits under Rif treatment than for 30S subunits in untreated cells.
A conceivable alternative explanation of our data posits that in untreated cells the faster component is due to 70S ribosome monomers translating mRNA and the slower component is dominated by polysomes. However, if that were so, we would have found no evidence of the ~20% 30S subunits expected in live E. coli (Forschhamer & Lindahl, 1971). Furthermore, we expect a smooth distribution of the number of ribosomes translating mRNA copies: ribosome monomers, pairs of ribosomes translating the same message, trios, and so forth up to larger polysomes. In all these cases, the diffusing entity is one or more ribosomes bound to a single mRNA message. Regardless of the details, the mean diffusion coefficient is expected to vary slowly with the number of ribosomes carried by a transcript. There is no reason for monomer diffusion to stand out from the other members of the distribution. In contrast, it seems sensible that diffusion of 30S might stand out from that of a smooth distribution of 70S monomers and polysomes. In addition to size and crowding considerations, free 30S is not bound to an mRNA message and is not trailing nascent polypeptide chains, both of which might impede diffusion of translating 70S ribosomes. We know little about the ribosome copy number per polysome.
Ribosome-nucleoid and protein-nucleoid segregation in bacterial cells
Our quantitative study of ribosome and DNA spatial distributions in live E. coli confirms previous reports of strong ribosome-DNA segregation in fixed cells. Early EM studies of thin slices of fixed E. coli cells by Robinow et al found ribosomes to be concentrated near the periphery of the cell and the nucleoid avoiding the near vicinity of the cytoplasmic membrane (Robinow & Kellenberger, 1994). From widefield immuno-fluorescence microscopy, the L7/L12 subunits of ribosomes were observed to localize outside the nucleoid (Azam et al., 2000). Our new live-cell results indicate that nucleoids of normal compaction strongly exclude ribosomes. We observe comparably strong ribosome-DNA segregation in the complete, chemically defined EZRDM, in minimal medium, and in the rich, undefined medium LB (Fig. S11).
Movies of widefield images with spatial resolution of ~250 nm show that strong nucleoid-ribosome segregation persists over the entire cell cycle. It is possible that the superresolution images acquired over several minutes underestimate the sharpness of the boundary between nucleoid and ribosome-rich regions due to time-dependent fluctuations in the underlying spatial distributions and also the possibility of complex, heterogeneous structure within dense nucleoid lobes on the scale of tens of nm. Regardless of these effects, the quantitative estimate of the fraction of ribosomes lying within the dense nucleoid regions in close proximity to DNA (10–15%) should be an accurate average over long periods of time.
In E. coli and in B. subtilis, we believe that strong exclusion of ribosomes from the dense portion of normal nucleoids determines the overall spatiobiology of transcription and translation. The present experimental results are consistent with a recent Monte Carlo simulation of ribosome-DNA segregation (Mondal et al., 2011). The single DNA polymer lobe is modeled as a branched, freely jointed chain with persistence length chosen to mimic that of plectonemic DNA. The ribosomes are modeled as hard spheres, and polysomes as freely-jointed chains of 10 hard spheres. Both DNA and ribosome concentrations were taken from E. coli content measurements. This simplified model predicts strong polysome-DNA segregation while accounting only for the effects of excluded volume and plectoneme chain conformational entropy (Fig. S5). With only one nucleoid lobe, the model finds ribosomes concentrated in the two endcaps and in a thin cylindrical shell surrounding the nucleoid, much as observed here (Figs. 1B and S2). The model does not include the transertion mechanism, which may anchor the DNA to the cell periphery by co-transcriptional translation of membranes proteins.
Two recent papers presented superresolution images of ribosomes in live E. coli under growth in minimal medium. Zhuang and co-workers (Wang et al., 2011) imaged S22-mEos2 and found ribosomes concentrated at the endcaps (their Fig. S3), much as observed here. Elf and coworkers show widefield images of S2-Dendra2 (English et al., 2011) that to our eye show strong ribosome segregation (their Fig. 3E). These copies of S2 were produced from the chromosome just like ours. However, the superresolution images of Dendra2-L25 (their Fig. 3D) seem to show less ribosome segregation. The initial slope of MSDr(τ) yields a diffusion coefficient of 0.5 μm2-s−1 for Dendra2-L25 single molecules (their Fig. 3A), which is much faster than our value of 0.04 μm2-s−1 using S2-YFP. The Dendra2-L25 was expressed from a leaky plasmid, not from the chromosome. Evidently these plasmid-expressed copies are distributed more uniformly and diffuse much more rapidly than our S2-YFP copies expressed from the chromosome. Further comparison studies are needed to resolve this discrepancy.
In C. crescentus, the evidence indicates that both ribosomes and DNA are dispersed throughout the cytoplasm (Llopis et al., 2010) in contrast to the results for E. coli and B. subtilis (Llopis et al., 2010, Nevo-Dinur et al., 2011). In C. crescentus, mRNA (possibly as a polysome) diffuses very little from the location of the gene from which it originated over some minutes (Llopis et al., 2010). It is conceivable that the density or morphology or local dynamics of DNA is substantially different in C. crescentus in a way that simultaneously enables ribosome-DNA mixing and also entangles polysomes within the DNA meshwork, greatly slowing their diffusion.
Ribosome and RNA polymerase copy numbers
We estimate that the mean copy number of S2-YFP per cell is ~55,000 for a modified K-12 strain under moderate growth conditions at 30°C. Based on heterogeneity in the distribution of single-molecule mean-square displacements (Figs. 6 and S8) and on earlier content studies (Dennis & Bremer, 1974) (Forschhamer & Lindahl, 1971), we estimate that some 80% are incorporated into complete 70S ribosomes and some 20% are in 30S subunits. As expected, the number of ribosomes per cell correlates with the cell volume (Fig. S6). Our estimate of mean copy number is about three times larger than the earlier value of 18,000 (interpolated between reported data for 40-min and 60-min doubling times) for E. coli B/r cells which were grown at 37°C (Dennis & Bremer, 1974). Part of the difference may be due to different temperatures and growth media. In addition, the mean cell volume in our study is almost two times larger than the mean volume reported for E. coli B/r cells. Thus the number of ribosomes per unit volume is more similar, ~27,000/μm3 in the K-12 strain vs ~18,000/μm3 in the B/r strain.
Roughly 85% of the ribosomes lie in the ribosome-rich regions. What is the biopolymer volume fraction in these regions? In the endcaps, whose geometry is well defined, the average density of ribosomes is ~55,000 ribosomes/μm3. Using an endcap radius of 400 nm, a ribosome radius of 10 nm, and assigning 30S half the volume of a 70S ribosome, we estimate that ~0.22 of the volume in the endcaps is occupied by 30S and 70S copies. The ribosome volume fraction is evidently similar in the central ribosome-rich region, but quantitative estimates are more difficult for that less well defined volume. In E. coli, the best quantitative estimates of mean, total biopolymer volume fraction (including protein, ribosomes, and nucleic acid) were obtained for K-12 cells in minimal growth conditions that do not match the current study (Konopka et al., 2007). The result at normal growth osmolality was 0.16, about half of which was due to ribosomes. If we add the protein volume fraction of ~0.08 from minimal growth conditions to the ribosome volume fraction of ~0.22 from the present work, the total biopolymer volume fraction in the ribosome-rich regions could easily be ~0.30. This is large enough to exert a substantial excluded-volume effect on protein and ribosome diffusion (Bakshi et al., 2011).
These estimates enable rough comparison of our results to a recent model diffusion calculation treating cytoplasmic elements as appropriately sized hard spheres and including hydrodynamic interactions (Ando & Skolnick, 2010). The biopolymer mass density in the model cytoplasm varied from 250–350 mg/ml, with the mole fraction of each component (proteins and ribosomes) derived from content measurements and kept constant. This model obtained good agreement with experimental GFP diffusion coefficients without recourse to strong inter-particle attractions or unusually high viscosity of the cytoplasmic fluid. The estimate for single 70S ribosomes at 350 mg/ml (somewhat larger than our estimated volume fraction of ~0.30) is Dribo = 0.2 μm2-s−1, which is five times larger than the experimental Dribo ~ 0.04 μm2-s−1 for our majority component. However, this is probably not an appropriate comparison. The model assumed a well-mixed cytoplasm of proteins and 70S ribosome monomers. That is, the model did not account for the strong segregation of ribosomes documented here or for the likely presence of polysomes. A more realistic model of the ribosome-rich regions would have a larger fraction of the biopolymer mass carried by ribosomes and would explore the effects of monosomes vs polysomes. Both factors would slow diffusion in the model and bring the calculations into closer agreement with our experimental results. As a final cautionary note, the diffusing entity in our study may not even be appropriately modeled as bare polysomes. Real polysomes are trailing long, nascent polypeptide chains. It is not known if these nascent proteins are unfolded, partially folded, enveloped by chaperones, or fully folded. Long polypeptide chains and large chaperones might further hinder diffusion in the real experimental system.
The mean copy number of RNAP is estimated to be about 4600 per cell. An estimate from early work on B/r cells is about 4000, interpolated between results for 40-min and 60-min doubling times (Dennis & Bremer, 1974). The spatial distribution of RNA polymerase closely mimics that of the DNA itself (Figs. 2 and 4), consistent with strong non-specific and specific binding to DNA. Significantly, the widefield RNAP radial distribution in the vicinity of the nucleoids is slightly wider than the DRAQ5-DNA signal and extends all the way to the cytoplasmic membrane (Fig. S4C and S4D). This suggests somewhat stronger association of RNAP to the outer regions of DNA lobes, in rough agreement with earlier images of fixed and immuno-stained cells (Durrenberger et al., 1988). Transcription may occur preferentially near the interface between nucleoid and ribosome-rich regions. Finally, there is very little evidence of either RNAP (Fig. 4D) or DNA (Fig. 1B) proximal to the endcap membranes.
Co-transcriptional translation in E. coli
By co-transcriptional translation, we mean the translation of mRNA by ribosomes while the message is being transcribed, thus forming a large, branched biopolymer complex. The same phenomenon is often referred to as coupled transcription and translation. Early EM studies of E. coli cell lysate by electron microscopy observed DNA decorated with multiple RNAP molecules, each attached to a strand of mRNA which was in turn coated with a chain of ribosomes (Miller et al., 1970).
The high degree of DNA-ribosome and RNAP-ribosome segregation exhibited by our images does not prohibit co-transcriptional translation, but it strongly suggests that most protein is translated from free mRNA messages that have diffused into the ribosome-rich regions. That is, most translation is uncoupled from transcription (Llopis et al., 2010). The reason is that we expect mRNA messages to be compact in space and thus unable to span typical distances of ~0.5 μm between DNA and the ribosome-rich regions. Polymers tend to adopt compact configurations because they lose configurational entropy when stretched. There is direct evidence for compact polysome configurations from a recent cryoelectron tomography study; indeed, polysomes appear to adopt definite compact three-dimensional structures (Brandt et al., 2009). It remains plausible that many of the ~10–15% of ribosomes located within the dense part of the nucleoid are carrying out co-transcriptional translation. These copies may also be undergoing ribosome assembly. However, the ~85–90% majority in the ribosome-rich regions are quite distant from the DNA and presumably unable to couple to DNA-tethered, partially synthesized transcripts.
Consistent with this argument, we found no clear evidence of tethered diffusion of ribosomes. The mild curvature in the MSD(τ) plots could be reasonably well explained by confinement within the ribosome-rich regions on the limited timescale of the tracking experiments. Furthermore, in these moderate growth conditions, the two endcaps contain very little of the DNA (as best we can discern from widefield images, Fig. 1B), some 17,000 of the 55,000 ribosomes, and at most 180 of the 4,600 RNAP copies (only 4%). Even in the unlikely event that all these RNAP copies were transcribing DNA, there would be at most 180 tethered partial messages in the endcaps at a given moment. That is far too little mRNA to engage the 17,000 ribosomes there. We suspect that the few “stray” RNAP copies in the endcaps are probably not transcribing. Instead, they are “lost” copies in search of a transcription initiation site. A future detailed study of RNAP diffusion will test this suggestion more critically.
The estimate of ~7000 ribosomes lying within the dense nucleoid regions is consistent with the suggestion that most of these are 70S ribosomes engaged in coupled transcription and translation. If ~1200 RNAP copies (25% of the total) are actively transcribing at a given moment, then some 1200 nascent mRNA copies are coupled to DNA. If the mean length of a complete transcript is ~1000 bp (contour length of 350 nm), then the mean contour lengths of these partially completed transcripts is ~175 nm. If 70S ribosomes of 20-nm diameter were close-packed along the nascent mRNA, there would be sufficient total contour length to accommodate some 10,000 ribosomes. It is thus plausible that many of the copies within the dense nucleoid are 70S ribosomes actively translating.
Based on these arguments, we suggest that complete mRNA copies typically find their way to the ribosome-rich regions by diffusion. They are likely decorated by ribosomes and perhaps cold-shock proteins that protect them from degradation en route (El-Sharoud & Graumann, 2007, Brandt et al., 2009). Based on Dribo = 0.04 μm2-s−1, a ribosome-decorated mRNA copy diffuses a root-mean-square distance of 0.5 μm (about half the size of a nucleoid lobe) in only 1 s. The time to transcribe an average-length protein message of 1000 nt is τtrx ~ 20 s (Dennis & Bremer, 1974, Llopis et al., 2010). The mean lifetime of an mRNA copy relative to degradation is τmRNA ~ 5 min (Bernstein et al., 2002). After transcription and release a complete message will find the ribosome-rich region very quickly. The fraction of protein production coupled to transcription would be roughly τtrx /τmRNA ~ 0.07. This is roughly comparable to our estimate of the fraction of ribosomes found within the nucleoids, ~0.10–0.15.
Our evidence that the majority of translation is uncoupled with transcription agrees with earlier conclusions based on the ratio of τtrx /τmRNA. It stands in apparent conflict with the early EM study of E. coli cell extracts (Miller et al., 1970), in which no free polysomes were found. Essentially all the ribosome-decorated mRNAs were attached to chromosomal DNA.
The spatiobiology of transcription/translation in C. crescentus may be very different from that in E. coli and B. subtilis. Using both FISH (fluorescence in situ hybridization, which requires fixation and permeabilization) and live-cell imaging methods, Jacobs-Wagner and coworkers recently found that mRNA remains localized near the site of transcription for long periods (Llopis et al., 2010). Images of mRNA were punctal and co-localized with markers of the corresponding gene. Most ribosomes did not diffuse in photobleaching/recovery experiments, as if they were translating mRNA that is somehow tethered to the DNA. In addition, the ribosomes and chromosomal DNA are distributed homogeneously in the C. crescentus cytoplasm. Even if messages do not diffuse long distances, they are already in close proximity to ribosomes (Llopis et al., 2010). We do not presently understand the underlying causes of the different DNA-ribosome morphologies in C. crescentus vs E. coli and B. subtilis.
Transertion in E. coli
A substantial body of evidence indicates that most membrane proteins are inserted co-translationally by the signal recognition particle SRP in conjunction with the Sec translocase (Driessen & Nouwen, 2008). The transertion hypothesis refers to co-transcriptional translation and simultaneous insertion of membrane proteins (Woldringh, 2002). Because transertion would tether the chromosomal DNA to the cytoplasmic membrane, Woldringh and others have suggested that transertion acts as an “expanding force” on the DNA itself (Woldringh & Nanninga, 2006, Woldringh, 2002, Cabrera et al., 2009). In this view, the spatial extent of the chromosomal DNA is a compromise between expanding forces (primarily transertion) and compacting forces (supercoiling, inter-strand coupling by architectural proteins, and the tendency of branched polymers to avoid walls and maximize configurational entropy) (Woldringh & Nanninga, 2006). Woldringh further suggested that serial transertion of proteins into the crowded membrane might assist DNA segregation (Woldringh, 2002).
The present work adds circumstantial, quantitative support for the transertion mechanism. First, we have directly observed time-dependent radial shrinkage of the nucleoids at t ~ 8 min after treatment with either rifampicin or chloramphenicol (data not shown). Both treatments eventually halt translation, which will break the link between the chromosomal DNA and the cytoplasmic membrane. Second, in our moderate growth conditions we find clear evidence that the radial distributions of both ribosomes and RNAP extend all the way to the cytoplasmic membrane in close proximity to the nucleoid lobes (Figs. S4B and S4D). While these membrane-proximal copies are plausible candidates for the transertion mechanism, we have no evidence as yet that these RNAP copies are transcribing DNA or that these ribosomes are translating protein.
To the extent that our results support the feasibility of the transertion mechanism, they do so only at the cylindrical part of the cytoplasmic membrane near the flanks of the dense nucleoid lobes. All the evidence argues against transertion at the endcaps. There are very few if any RNAP copies near the endcap membrane regions (Fig. 4D), and the (lower resolution) widefield DNA axial distribution dies off well before reaching the endcap membranes (Fig. 1B). Extensive coupling of the endcap membranes to DNA would evidently require stretching an mRNA message some 300–500 nm. In our view, transertion might well supply a direct radially expanding force on the nucleoids, but probably cannot supply a direct axially expanding force. Thus halting of transertion can readily explain radial compaction of the nucleoids (which occurs for both Rif and Chl treatment at t = 8 min), but not the observed axial contraction under Chl treatment. It remains possible, however, that direct insertion of proteins into a crowded cytoplasmic membrane in effect pulls the nucleoid outward as the cell grows (Woldringh, 2002). The correlation between the outward movement of segregated DNA lobes and the cell growth as observed in Fig. 1D provides some support for such a mechanism.
EXPERIMENTAL PROCEDURES
Bacterial strains and plasmids
The construct used for imaging ribosomes, AFS55, which contains a translational fusion of yfp to the C-terminus of rpsB (the gene encoding S2), was constructed by lambda-Red mediated recombination. The YFP variant here is GFPmut3.1, having the substitutions V68L Q69K Q80R T203Y. A DNA segment containing yfp and a kanamycin resistance gene was amplified by PCR from the plasmid pEB45 (Batchelor & Goulian, 2006) with the primers 5′–CCCAGGCGGAAGAAAGCTTCGTAGAAGCTGAGVNNVNNVNNVNNVNNVNNCGTAA AGGAGAAGAAC-3′ and 5′-ACTCGAACTATTTTGGGGGAGTTATCAAGCGTGTAGGCTGGAGCTGCTTC-3′.The underlined portions in the first and second primers anneal to the C-terminus of rpsB just before the stop codon, and the region beginning five bases after the stop codon, respectively. The first primer encodes an 18 base randomized linker connecting S2 and YFP. The PCR product was electroporated into MG1655/pKD46 as described earlier (Datsenko & Wanner, 2000). A kanamycin resistant colony was selected and verified to express YFP. The rpsB-yfp fusion was then moved to a clean MG1655 background by P1 transduction, resulting in AFS55. The linker between rpsB and yfp was sequenced and determined to be: 5′-GAGCAGGAAAGGCGACAGGAGCGT-3′. The underlined portion is the linker and the first and last codons are the last codon of rpsB and the first codon of yfp, respectively.
For the RNAP studies, the rpoc::ygfp strain (RLG7470) was constructed in the same way as the rpoc::gfp strain as described elsewhere (Bratton et al., 2011). The construct was transferred to a VH1000 background using P1 transduction. The protein yGFP is a variant of GFP developed by Nielsen et al (Nielsen et al., 2006); it has similar absorption and emission spectra to those of YFP, in spite of its name. The labeled entity is then called β′-yGFP. For the construct expressing bare YFP, we used colony PCR to extract the YFP gene from the S2-YFP construct. This fragment was ligated to a similarly digested fragment of pASK-IBA3plus (Invitrogen, Carlsbad, CA). DH5α cells were transformed with the resulting plasmid (pJW2), and transformants were selected on ampicillin-containing plates. MG1655 cells were subsequently transformed with the same plasmid. For measurement of the radius of MG1655 and VH1000 cells, we transformed both cell lines with a pASK-IBA3plus a plasmid expressing Kaede.
Cell growth and preparation
In bulk EZ rich defined medium (EZRDM, (Neidhardt. F. C. et al., 1974)) at 30°C with 50 μg/mL Kanamycin, the S2-YFP-expressing strain AFS55 grow in bulk medium with doubling times of 54 min (Fig. S14). For wild-type MG1655 under the same growth conditions, the doubling time in bulk medium is 51 min. The β′-yGFP-expressing strain RLG7470 has a doubling time of 43 min (Fig. S14). This is comparable to the 45 min doubling time of the background strain VH1000.
Cells were grown overnight with shaking in 30°C water bath. We subsequently made subcultures by diluting the stationary phase culture at least 1:250 into 3 mL of fresh EZRDM. When cells had grown to midlog phase (OD600 = 0.4–0.6), 7 μL of culture was placed on poly-L-lysine-coated coverslips to immobilize cells for observation. The assembly was sealed with nail polish. Imaging by widefield epifluorescence or by single-molecule photoactivation was carried out at 30°C for no longer than 40 min after plating. During that time, cells continue to grow. As a control, we have also imaged the ribosome distribution in cells plated on agar pads. The distribution for cells in agar is indistinguishable from that for cells plated on polylysine (Fig. S17). For imaging the time course of cell growth and the action of antibiotics, cells were plated in a temperature-controlled sample chamber (RC-20H; Warner Instruments, Hamden, CT). The whole assembly was brought into contact with the microscope objective and warmed to 30°C.
Doubling times for cell growth in sealed coverslip-slide chambers and in the flow chamber were estimated by measuring cell length vs time using phase contrast microscopy (Fig. S10). The constructs AFS55 and RLG7470 grow with a doubling time of 75–80 min for coverslip-slide chambers and of 60–65 min in the flow chamber with aerated EZRDM growth medium flowing. A few experiments were carried out in the sealed chamber for cells grown in a chemically defined MOPS-buffered minimal medium (0.28 Osm MBM, (Konopka et al., 2007, Neidhardt. F. C. et al., 1974)) and in the undefined rich medium (LB).
To examine the distribution of the label by itself, the bare protein YFP was imaged using the strain JCW112, which contains the YFP expressing plasmid in the same MG1655 strain used for the ribosome studies. Cells were grown overnight with shaking at 30°C in EZRDM with 100 μg/mL ampicillin. We subsequently made subcultures of these cells by diluting the stationary phase in fresh EZRDM. When cells had grown to midlog phase, anhydrotetracycline was added to a final concentration of 45 nM to induce YFP expression. After 6 min of induction the cells were centrifuged and resuspended in fresh EZRDM twice to remove the inducer. The cells were then incubated again in growth medium for at least 30 min at 30°C to enable maturation of the fluorescent protein and then plated on poly-L-lysine coated coverslips for imaging.
Rifampicin and chloramphenicol stock solutions were prepared by dissolving 1 mg and 10 mg of the drugs in ethanol, respectively. Stock solutions of the drugs were added to the midlog cell culture to attain a final concentration of 200 μg/mL. For imaging of ribosome and DNA static spatial distributions, the cultures were shaken for 30 min before plating and imaging. For widefield imaging of the time course of drug action, we injected drug solution in EZRDM into the flow chamber already containing plated cells in growth medium. Superresolution imaging of ribosomes in the presence of drugs was carried out in sealed coverslip-slide chambers.
DNA staining with DRAQ5
To image the DNA distribution of the cells, we used the red fluorescent DNA stain DRAQ5 (Biostatus Limited). The MIC of DRAQ5 was determined to be 5 μM for MG1655 cells growing at 30°C in EZRDM. For lower concentrations growth was maintained. A DRAQ5 concentration of 200 nM provided adequate fluorescence intensity. For these studies 2 μL of 100 μM stock solution of DRAQ5 was added to a 1 mL culture of cells in midlog phase and the culture was shaken for 10 minutes, after which 7 μL of culture was plated on a cover slip for imaging.
Widefield epifluorescence imaging
The fluorophore in the S2-YFP constructs both absorb green light (λmax = 514 nm in absorption) and fluoresce green-yellow (λmax = 533 nm in emission). The β′-yGFP construct has similar spectra. A 514 nm Ar+ laser (Melles Griot, Carlsbad, CA) was used for imaging. The yellow emission was collected using a 560/50 emission filter (Chroma). The DNA stain DRAQ5 absorbs red light (λmax = 647 nm in absorption). When intercalated into double-strand DNA it fluoresces with emission maximum at 691 nm. A 633 nm laser (HeNe, Coherent, Santa Clara, CA) was used to image the DRAQ5-DNA distribution inside cells. Emission from the DRAQ5 excitation was collected using a 700/75 emission filter (Chroma). Widefield fluorescence images were acquired with a Nikon Eclipse Ti inverted microscope with oil immersion objectives (either Nikon Plan Fluor 100X, NA 1.30 or Nikon APO TIRF 100X, NA 1.49) and a 1.5X tube lens. For time-lapse imaging, fast shutters (Uniblitz LS2, Vincent Associates, New York) were used to synchronize illumination and image acquisition. Images were recorded by a back-illuminated EMCCD camera with 16 μm × 16 μm pixels (iXon DV-887, Andor Technology, Connecticut). Each pixel corresponds to 105×105 nm2 at the sample with 150X overall magnification.
Superresolution imaging of ribosome-YFP and RNAP-yGFP
Technical details of the imaging method are given in Supplemental Information. In cells as small as E. coli, accurate single-molecule localization can begin only when the rate of return of molecules to the fluorescent state becomes small enough that at most 2–3 copies per camera frame are fluorescent in each cell, enabling single molecules to be distinguished from each other. This limits the ability to vary the time between camera frames, which in turn limits our ability to obtain MSD plots for long lag times.
As a cautionary note, we have directly observed laser-induced changes in DNA morphology. In both wild-type and S2-YFP expressing cells, strong laser illumination at 514 nm or 561 nm for minutes expands the nucleoid spatial distribution, diminishes ribosome-DNA segregation, increases the ribosome diffusion coefficient (much like Rif treatment), and slows growth (Fig. S12). Laser-driven nucleoid expansion occurs even in wild-type (unlabeled) cells. Such effects were readily observed following continuous illumination of cells at 6–7 kW/cm2 for 10–15 min. The mechanism is unclear; perhaps local heating or photodamage affects important cellular constituents which determine DNA morphology. Such high laser dosages were avoided in the study of single molecule imaging of both ribosome and RNAP molecules.
Single-molecule image analysis
Images were analyzed using a MATLAB GUI developed in our lab (Bakshi et al., 2011). Images were smoothed and filtered to obtain a zero-based image. Bright spots were located with pixel level accuracy by a peakfinding algorithm which finds local maxima in an image. A user defined intensity threshold was used as the minimum brightness of a pixel arising from a single molecule. Centroids of the bright spots were calculated from an 8 × 8 pixel square centered on the local maxima determined by the peak finding algorithm.
We prefer localization by centroid rather than fitting to model point-spread functions because the images are asymmetrically blurred by diffusion during each camera frame. Centroid is much faster than Gaussian fitting and is easily implemented for analysis of Monte Carlo results as well. Importantly, the smoothing and filtering places single-molecule images on a near-zero background, which is essential for accurate tracking by centroid. Earlier tests (Cheezum et al., 2001) found large bias and standard deviations using a centroid algorithm to locate stationary images. However, that study calculated the centroid using a large (80 px × 80 px) grid without high-pass filtering, so that the numerous distant pixels had inordinate weight in the centroid result. Our own numerical tests on an 8 × 8 grid for smoothed and filtered images mimicking ribosome and RNAP single molecule diffusion are described in Supporting Information. The results in Fig. S15 show that the centroid gives dynamic localization precision σx,y varying from 15–30 nm in each dimension as the peak signal to noise ratio (PSNR) varies from 7 to 14, the relevant range in our experiments. This is only 10–15% larger localization standard deviation than that of Gaussian fitting under the same conditions.
For single-molecule tracking we store the (x,y) positions of the centroid. Using a modified MATLAB version of the tracking program written by Crocker and Grier (Crocker & Grier, 1996) we connect the centroids of a bright feature from consecutive frames into a trajectory. Analysis of trajectories was carried as described before (Bakshi et al., 2011). To determine the distribution of molecule positions along the long and short axis of the cell, we have used a mask generated from the white light image (Supporting information). Analyzed positions were rotated to project along the two axes using the orientation of the mask (Fig. S9).
Monte Carlo simulations
Monte Carlo simulations of free diffusion were performed in truncated spherocylindrical volumes that mimic the shape of the space occupied by ribosomes within the E. coli cytoplasm. Because there are few ribosomes within the nucleoid, the dense region of nucleoid is represented by two impenetrable cylinders placed within the larger spherocylinder representing the entire cytoplasm (Fig. 5). The distribution of initial positions is chosen randomly within the available space. This model problem is characterized by four parameters, the length and radius of the outer spherocylinder (L1 and R1 = 380 nm, with L1 the length of the straight, cylindrical part) and the length and radius of the inner spherocylinders (L2 = 800 nm and R2 = 380 nm). The end caps of each spherocylinder have the same radius as the cylindrical part. These parameters were varied to mimic the experimental ribosome spatial distribution. Random walk simulations were carried out in MATLAB as described before (Bakshi et al., 2011). The diffusion coefficient was varied until experimental and simulated MSD plots were similar.
Supplementary Material
Acknowledgments
SB and JCW thank the National Institute of General Medical Sciences for support under grant R01-GM086468 (an American Recovery and Reinvestment Act grant). AS and MG were supported by NSF grant MCB0615957. We acknowledge Dr. Benjamin Bratton and Dr. Colin Ingram for help with data analysis and Mr. Heejun Choi for construction of the YFP-expressing plasmid. We are thankful to Rachel Anne Mooney and Robert Landick for the RLG7470 strain. Scientific discussions with Dr. Tamas Gaal, Prof. Richard Gourse, and Prof. Christine Jacobs-Wagner have been invaluable.
References
- Ando T, Skolnick J. Crowding and hydrodynamic interactions likely dominate in vivo macromolecular motion. Proc Natl Acad Sci U S A. 2010;107:18457–18462. doi: 10.1073/pnas.1011354107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam TA, Hiraga S, Ishihama A. Two types of localization of the DNA-binding proteins within the Escherichia coli nucleoid. Genes Cells. 2000;5:613–626. doi: 10.1046/j.1365-2443.2000.00350.x. [DOI] [PubMed] [Google Scholar]
- Bakshi S, Bratton BP, Weisshaar JC. Subdiffraction-Limit Study of Kaede Diffusion and Spatial Distribution in Live Escherichia coli. Biophys J. 2011;101:2535–2544. doi: 10.1016/j.bpj.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor E, Goulian M. Imaging OmpR localization in Escherichia coli. Molecular Microbiology. 2006;59:1767–1778. doi: 10.1111/j.1365-2958.2006.05048.x. [DOI] [PubMed] [Google Scholar]
- Bernstein JA, Khodursky AB, Lin PH, Lin-Chao S, Cohen SN. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc Natl Acad Sci U S A. 2002;99:9697–9702. doi: 10.1073/pnas.112318199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Brandt F, Etchells SA, Ortiz JO, Elcock AH, Hartl FU, Baumeister W. The Native 3D Organization of Bacterial Polysomes. Cell. 2009;136:261–271. doi: 10.1016/j.cell.2008.11.016. [DOI] [PubMed] [Google Scholar]
- Bratton BP, Mooney RA, Weisshaar JC. Spatial Distribution and Diffusive Motion of RNA Polymerase in Live Escherichia coli. Journal of Bacteriology. 2011;193:5138–5146. doi: 10.1128/JB.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera JE, Cagliero C, Quan S, Squires CL, Jin DJ. Active Transcription of rRNA Operons Condenses the Nucleoid in Escherichia coli: Examining the Effect of Transcription on Nucleoid Structure in the Absence of Transertion. J Bacteriol. 2009;191:4180–4185. doi: 10.1128/JB.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera JE, Jin DJ. The distribution of RNA polymerase in Escherichia coli is dynamic and sensitive to environmental cues. Molecular Microbiology. 2003;50:1493–1505. doi: 10.1046/j.1365-2958.2003.03805.x. [DOI] [PubMed] [Google Scholar]
- Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 2001;104:901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- Cheezum MK, Walker WF, Guilford WH. Quantitative comparison of algorithms for tracking single fluorescent particles. Biophysical Journal. 2001;81:2378–2388. doi: 10.1016/S0006-3495(01)75884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker JC, Grier DG. Methods of digital video microscopy for colloidal studies. J Colloid Interface Sci. 1996;179:298–310. [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis PP, Bremer H. Macromolecular-composition during steady-state growth of Escherichia coli B/r. Journal of Bacteriology. 1974;119:270–281. doi: 10.1128/jb.119.1.270-281.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessen AJM, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annual Review of Biochemistry. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- Durrenberger M, Bjornsti MA, Uetz T, Hobot JA, Kellenberger E. Intracellular location of the histone-like protein HU in Escherichia coli. Journal of Bacteriology. 1988;170:4757–4768. doi: 10.1128/jb.170.10.4757-4768.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sharoud WM, Graumann PL. Cold shock proteins aid coupling of transcription and translation in bacteria. Sci Prog. 2007;90:15–27. doi: 10.3184/003685007780440549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English BP, Hauryliuk V, Sanamrad A, Tankov S, Dekker NH, Elf J. Single-molecule investigations of the stringent response machinery in living bacterial cells. Proc Natl Acad Sci U S A. 2011;108:E365–E373. doi: 10.1073/pnas.1102255108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forschhamer J, Lindahl L. Growth rate of polypeptide chains as a function of the cell growth rate in a mutant of Escherichia coli 15. Journal of Molecular Biology. 1971;55:563–568. doi: 10.1016/0022-2836(71)90337-8. [DOI] [PubMed] [Google Scholar]
- Golding I, Cox EC. RNA dynamics in live Escherichia coli cells. Proc Natl Acad Sci U S A. 2004;101:11310–11315. doi: 10.1073/pnas.0404443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D. Analysis and interpretation of two-dimensional single-particle tracking microscopy measurements: Effect of local surface roughness. Anal Biochem. 2008;377:24–32. doi: 10.1016/j.ab.2008.02.019. [DOI] [PubMed] [Google Scholar]
- Hess ST, Girirajan TPK, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka MC, Weisshaar JC, Record MT., Jr Methods of changing biopolymer volume fraction and cytoplasmic solute concentrations for in vivo biophysical studies. Osmosensing and Osmosignaling. 2007;428:487–504. doi: 10.1016/S0076-6879(07)28027-9. [DOI] [PubMed] [Google Scholar]
- Lee SF, Thompson MA, Schwartz MA, Shapiro L, Moerner WE. Super-Resolution Imaging of the Nucleoid-Associated Protein HU in Caulobacter crescentus. Biophys J. 2011;100:L31–L33. doi: 10.1016/j.bpj.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PJ. Bacterial subcellular architecture: recent advances and future prospects. Mol Microbiol. 2004;54:1135–1150. doi: 10.1111/j.1365-2958.2004.04343.x. [DOI] [PubMed] [Google Scholar]
- Lewis PJ, Thaker SD, Errington J. Compartmentalization of transcription and translation in Bacillus subtilis. EMBO J. 2000;19:710–718. doi: 10.1093/emboj/19.4.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl L. Intermediates and time kinetics of in vivo assembly of Escherichia coli ribosomes. Journal of Molecular Biology. 1975;92:15–37. doi: 10.1016/0022-2836(75)90089-3. [DOI] [PubMed] [Google Scholar]
- Llopis PM, Jackson AF, Sliusarenko O, Surovtsev I, Heinritz J, Emonet T, Jacobs-Wagner C. Spatial organization of the flow of genetic information in bacteria. Nature. 2010;466:77–U90. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas J, Weber MHW, Graumann PL. Specific polar localization of ribosomes in Bacillus subtilis depends on active transcription. EMBO Rep. 2001;2:685–689. doi: 10.1093/embo-reports/kve160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller OL, Hamkalo BA, Thomas CA. Visualization of bacterial genes in action. Science. 1970;169:392. doi: 10.1126/science.169.3943.392. [DOI] [PubMed] [Google Scholar]
- Mondal J, Bratton BP, Li Y, Yethiraj A, Weisshaar JC. Entropy-Based Mechanism of Ribosome-Nucleoid Segregation in E. coli Cells. Biophysical Journal. 2011;100:2605–2613. doi: 10.1016/j.bpj.2011.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Neidhardt FC, Bloch PL, Smith DF. Culture Medium for Enterobacteria. Journal of Bacteriology. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo-Dinur K, Nussbaum-Shochat A, Ben-Yehuda S, Amster-Choder O. Translation-Independent Localization of mRNA in E. coli. Science. 2011;331:1081–1084. doi: 10.1126/science.1195691. [DOI] [PubMed] [Google Scholar]
- Nielsen HJ, Ottesen JR, Youngren B, Austin SJ, Hansen FG. The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol Microbiol. 2006;62:331–338. doi: 10.1111/j.1365-2958.2006.05346.x. [DOI] [PubMed] [Google Scholar]
- Norris V, Madsen MS. Autocatalytic Gene Expression OccursviaTransertion and Membrane Domain Formation and Underlies Differentiation in Bacteria: A Model. Journal of Molecular Biology. 1995;253:739–748. doi: 10.1006/jmbi.1995.0587. [DOI] [PubMed] [Google Scholar]
- Robinow C, Kellenberger E. The Bacterial Nucleoid Revisited. Microbiol Rev. 1994;58:211–232. doi: 10.1128/mr.58.2.211-232.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang XW. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Meth. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L, McAdams HH, Losick R. Why and How Bacteria Localize Proteins. Science. 2009;326:1225–1228. doi: 10.1126/science.1175685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski JA, Lappe JW, Patel HN, Huffman HA, Wachter RM. Base catalysis of chromophore formation in Arg(96) and Glu(222) variants of green fluorescent protein. Journal of Biological Chemistry. 2005;280:26248–26255. doi: 10.1074/jbc.M412327200. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Choi PJ, Li GW, Chen HY, Babu M, Hearn J, Emili A, Xie XS. Quantifying E-coli Proteome and Transcriptome with Single-Molecule Sensitivity in Single Cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WQ, Li GW, Chen CY, Xie XS, Zhuang XW. Chromosome Organization by a Nucleoid-Associated Protein in Live Bacteria. Science. 2011;333:1445–1449. doi: 10.1126/science.1204697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldringh CL. The role of co-transcriptional translation and protein translocation (transertion) in bacterial chromosome segregation. Mol Microbiol. 2002;45:17–29. doi: 10.1046/j.1365-2958.2002.02993.x. [DOI] [PubMed] [Google Scholar]
- Woldringh CL, Nanninga N. Structural and physical aspects of bacterial chromosome segregation. J Struct Biol. 2006;156:273–283. doi: 10.1016/j.jsb.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Young R, Bremer H. Polypeptide-chain-elongation rate in Escherichia coli B/r as a function of growth rate. Biochem J. 1976;160:185–194. doi: 10.1042/bj1600185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.