Abstract
The vertebrate neural retina contains seven major cell types, which arise from a common multipotent progenitor pool. During neurogenesis, these cells stop cycling, commit to a single fate, and differentiate. The mechanism and order of these steps remain unclear. The first-born type of retinal neurons, ganglion cells (RGCs), develop through the actions of Math5 (Atoh7), Brn3b (Pou4f2) and Islet1 (Isl1) factors, whereas inhibitory amacrine and horizontal precursors require Ptf1a for differentiation. We have examined the link between these markers, and the timing of their expression during the terminal cell cycle, by nucleoside pulse-chase analysis in the mouse retina. We show that G2 phase lasts 1–2 hours at embryonic (E) 13.5 and E15.5 stages. Surprisingly, we found that cells expressing Brn3b and/or Isl1 were frequently co-labeled with EdU after a short chase (<1 hr) in early embryos (<E14), indicating that these factors, which mark committed RGCs, can be expressed during S or G2 phases. However, during late development (>E15), Brn3b and Isl1 were exclusively expressed in post-mitotic cells, even as new RGCs are still generated. In contrast, Ptf1a and amacrine marker AP2α were detected only after terminal mitosis, at all developmental stages. Using a retroviral tracer in embryonic retinal explants (E12–E13), we identified two-cell clones containing paired ganglion cells, consistent with RGC fate commitment prior to terminal mitosis. Thus, although cell cycle exit and fate determination are temporally correlated during retinal neurogenesis, the order of these events varies according to developmental stage and final cell type.
Keywords: retinal ganglion cell (RGC), clonal analysis, optic nerve, retinal cell cycle, cell fate specification, Brn3b, Pou4f2, Isl1, cell cycle exit, Ptf1a
INTRODUCTION
The vertebrate neural retina is populated by seven major cell types, which are generated in an invariant histotypic order from a common progenitor pool (Livesey and Cepko, 2001; Wong and Rapaport, 2009). Individual cell fates are specified by intrinsic transcriptional programs and the retinal microenvironment. At the onset of retinal neurogenesis, on embryonic day 11 (E11) in the mouse, the first neurons exit the cell cycle and differentiate as retinal ganglion cells (RGCs), whose axons form the optic nerves. Mouse RGC birthdates extend from E11 to P0 with a peak at E14 (Drager, 1985; Young, 1985a). This temporal profile significantly overlaps the distribution of birthdates for cone photoreceptor, horizontal and amacrine cells. Rod photoreceptors, bipolar cells and Müller glia generally have later birthdates.
RGC development is controlled by a complex transcriptional network. Math5 (Atoh7) is expressed transiently in multipotent precursors (Brzezinski et al., 2012; Yang et al., 2003; Skowronska-Krawczyk et al., 2009) and is necessary for RGC fate specification (Brown et al., 2001; Wang et al., 2001). The Brn3b (Pou4f2) and Islet1 (Isl1) homeodomain genes form two regulatory nodes that are downstream of Math5 in the RGC differentiation hierarchy (Mu et al., 2008; Pan et al., 2008). Both factors are thought to be expressed in newly post-mitotic cells, and are abundant in differentiated ganglion cells (Pan et al., 2008; Pan et al., 2005; Xiang, 1998). Brn3b appears to mark committed RGC precursors, as it is expressed exclusively in RGCs and required for terminal differentiation (Erkman et al., 1996; Gan et al., 1996; Qiu et al., 2008; Xiang, 1998). Brn3b is made in most, but not all, differentiated RGCs. Paralog Brn3a, and likely Brn3c, are functionally interchangeable with Brn3b at the protein level (Pan et al., 2005). These three genes have highly overlapping but distinct spatiotemporal expression patterns (Xiang et al., 1995; Xiang et al., 1993), which confer unique roles in RGC differentiation (Badea et al., 2009; Badea and Nathans, 2011; Wang et al., 2002a). Isl1 is also required for RGC development (Mu et al., 2008; Pan et al., 2008), but is expressed in a wider population of cells (Elshatory et al., 2007a). The Brn3b+ population is essentially contained within the Isl1 lineage (Pan et al., 2008). Amacrine and horizontal neurons are specified in part by Ptf1a, a transiently expressed factor that acts downstream of FoxN4 (Fujitani et al., 2006; Li et al., 2004). Brn3b and Ptf1a are likely to assemble in opposing transcriptional complexes that regulate RGC or amacrine/horizontal cell differentiation, respectively (Fujitani et al., 2006; Qiu et al., 2008).
The sequential birth order of different retinal cell types reflects a shift in the fate bias of progenitors during development (Livesey and Cepko, 2001). In addition, the progenitor cell cycle length increases progressively (Alexiades and Cepko, 1996; Sinitsina, 1971; Young, 1985b). Because neurons do not divide, and differentiation occurs after the final division, it is generally thought that cell cycle exit is strictly coupled to fate specification. However, it remains unclear how cycle length, terminal division, and neurogenic fate are linked, and precisely when fate commitment occurs (Dyer and Cepko, 2001; Ohnuma and Harris, 2003). To explore these questions, we have determined the onset of expression of three key regulators of RGC, horizontal and amacrine cell fate specification (Brn3b, Isl1 and Ptf1a), relative to the terminal cell cycle. Unexpectedly, during early development (E11.5–E13.5), we found that Brn3b and Isl1, unlike Ptf1a, can be coexpressed prior to cell cycle exit, during late S or G2 phases. Retroviral lineage analysis revealed multiple two-cell clones containing paired RGCs, suggesting that these terminal progenitors represent committed ganglion cell precursors that undergo a final mitosis. Surprisingly, the timing changes significantly during late embryonic stages (after E15), such that Brn3b and Isl1 are expressed exclusively in post-mitotic cells. Our findings suggest that cell fate commitment can occur before or after cell cycle exit. The order of these events is not rigidly fixed.
MATERIALS AND METHODS
Pulsed EdU/BrdU and cell cycle labeling
All mouse studies were approved by the University of Michigan Committee on the Use and Care of Animals (UCUCA). Pregnant dams were injected with 5-bromo-2-deoxyuridine (BrdU, 100 μg/g body mass) or 5-ethynyl-2-deoxyuridine (EdU, 6.7 μg/g body mass) on embryonic (E) day 11.5, 13.5, 15.5 or 16.0. BrdU is rapidly absorbed within minutes after an intraperitoneal injection (Kriss and Revesz, 1962). EdU and BrdU have comparable incorporation kinetics and give similar results (Zeng et al., 2010), but EdU affords better detection sensitivity for multi-labeling experiments and was thus used for the majority of our studies. Embyros were harvested after a variable chase period, between 0.5 to 12 hours, in order to follow cells after they complete DNA synthesis (S phase). The time of onset of a particular transcription factor expression was defined by the first chase period in we detected cells positive for the particular marker and EdU/BrdU.
Histology
Embryonic eyes were fixed for 0.5–1 hour in 2–4% paraformaldehyde (PFA) at 22°C, cryoprotected with 30% sucrose in phosphate-buffered saline (PBS), and frozen in OCT compound (Tissue-Tek, Torrance, CA). Short fixation was critical for immunodetection of Brn3b and cyclinD1. Cryosections (10 μm) were blocked with 10% normal donkey serum (NDS) and 1% bovine serum albumin (BSA) in PBTx (0.1 M NaPO4 pH 7.3, 0.5% Triton X-100) for 4 hours at 22°C. Sections were incubated overnight at 4°C with primary antisera diluted in PBTx with 3% NDS, 1% BSA. Sections were then washed in PBS 0.5% BSA, incubated for 2 hours at 22°C with Dylight 488- (1:1000), Dylight 549- (1:1000), or Dylight 649- (1:500) conjugated secondary antibodies (Jackson Immunoresearch, West Grove, PA), mounted in Prolong Gold Antifade media (Invitrogen, Carlsbad, CA), and imaged using the Zeiss LSM510 Meta confocal microscope at the University of Michigan Microscopy and Image Analysis Core Facility (MIL).
The primary antibodies were: mouse anti-AP2α (1:1000, 3B5, DSHB, Iowa City, Iowa); rat anti-BrdU (1:100, BU1/75, Harlan Seralab, Indianapolis, IN); rabbit anti-βgal (1:5000, ICN Cappel, Aurora, OH); rat anti-βgal (1:500) (Saul et al., 2008); mouse anti-Brn3a (1:50, 14A6, Santa Cruz Biotechnology, Santa Cruz, CA); goat anti-Brn3b (1:200, sc31987, Santa Cruz); mouse anti-cyclin D1 (1:100, A-12, Santa Cruz); chicken anti-GFP (1:2000, Abcam, Cambridge, MA); mouse anti-Isl1 (1:200, 39.4D5, DSHB); rabbit anti-phosphohistone H3 (1:200, Upstate, Lake Placid, NY); rabbit anti-Ptf1a (1:800) (Fujitani et al., 2006).
Following immunostaining, EdU was detected using an azide-alkyne cycloaddition reaction (Buck et al., 2008) (Click-iT-647, Invitrogen). For detection of BrdU and other markers, cryosections were fully stained with primary and secondary antibodies for the marker protein. Sections were then treated with 2.4 N HCl in PBTx for 1 hour at 22°C, washed, and immunostained for BrdU.
Quantitative analysis of Brn3b onset
The percentage of EdU+Brn3b+ cells among the Brn3b+ cohort was determined for eight different EdU chase times, at E13.5 (0.5, 1, 2, 4, 12 hrs) and E15.5 (1, 4, 12 hrs). Double-positive cells were definitively identified by visual examination of 3-dimensional confocal Z-stacks (225 μm × 225 μm × 8–12 μm). Two to six sections were analyzed for each time point, from at least 2 embryos, representing 440 to 930 Brn3b+ cells. Error is reported as the standard deviation among sections. The total Brn3b+ population represents both newly neurogenic cells and existing ganglion cells. To selectively analyze neurogenic cells, we also normalized percentages to the 12-hr chase value at E13.5. At this stage, 12 hrs is sufficient time for any EdU-labeled neurogenic cell to progress from S to G0 phase. This single EdU pulse thus captures all newly Brn3b+ cells born within a 5–6 hr period (length of S phase). The normalized value at 1–2 hrs reflects new Brn3b+ cells that initiate Brn3b expression in terminal G2 phase.
Retroviral clone analysis in retinal explants
Retinal explant cultures and retroviral infections were performed as described (Brzezinski et al., 2012), using established methods (Hatakeyama and Kageyama, 2002; Wang et al., 2002b). Briefly, Math5-lacZ/+ retinas were dissected from E12.5 or E13.5 embryos, flattened onto Nucleopore polycarbonate membranes (0.4 μm pore size, GE Healthcare, Piscataway, NJ), and placed into Transwell culture dishes containing neurobasal media (Invitrogen) with B27 and N2 supplements, glutamine (0.4 mM), brain-derived neurotrophic factor (BDNF, 50 ng/mL, Peprotech, Rocky Hill, NJ), ciliary neurotrophic factor (CNTF, 10 ng/mL, Peprotech), penicillin (50 U/mL), streptomycin (50 μg/mL), and gentamicin (0.5 μg/mL).
MIG (bicistronic murine stem cell virus driving expression of GFP) retroviral stocks were prepared in advance by calcium phosphate transfection of the Phoenix ecotropic packaging cell line (Pear, 2001; Swift et al., 2001) with pMSCV-IRES-GFP plasmid DNA (Van Parijs et al., 1999). Filtered viral preparations containing polybrene (hexadimethrine bromide, 0.8 μg/mL, Sigma Aldrich, St. Louis, MO) were titered on NIH3T3 fibroblasts and diluted to ~8×105 colony-forming units per mL. One drop of retrovirus (25 μL) was added on top of each fresh explant to sparsely mark dividing cells (Roe et al., 1993) and their descendants.
Retinal explants were cultured for 3 days at the gas-media interface at 37°C under 5% CO2. After 2 days, half of the media was replaced with fresh media. On day 3, explants were fixed for 30 minutes in 4% PFA, and processed for cryosectioning or stained directly as wholemounts. Thick (30 μm) sections, or wholemount retinas, were immunostained for GFP and Brn3a/3b, and imaged by confocal microscopy. The size and composition of clonal clusters were determined by 3-dimensional analysis of Z-stacks. A clone was defined as a group of directly apposed GFP+ cells that were separated by at least 4 cell bodies from other GFP+ cells. For assessing symmetry of fate, only clones with at least one Brn3+ RGC were scored. To assess the nonrandom distribution of Brn3+ cells among clones, expected values were calculated based on the Poisson distribution, using the number of Brn3+ cells among the total population of clones to estimate μ (mean) for the distribution. Observed and expected distributions were compared using the χ2 test.
RESULTS
G2 phase is 1–2 hours long in most retinal cells
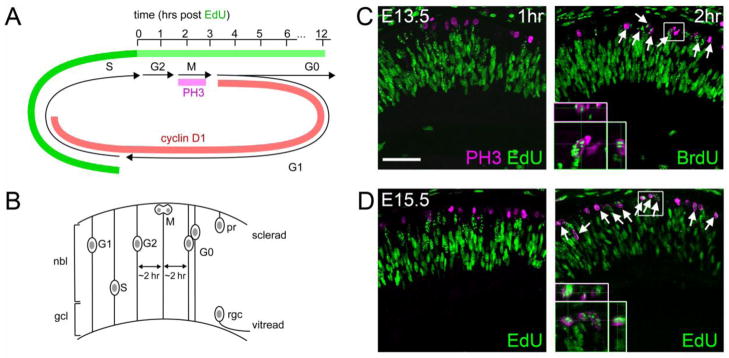
The eukaryotic cell cycle is anchored by M phase, when cytokinesis occurs, and S phase when DNA synthesis occurs. These are separated by G1 and G2 ‘gap’ phases, respectively (Fig. 1A) (Nurse, 2000). As a first step to evaluate the timing of the transcription factor expression relative to cell cycle exit, we systematically determined the onset of M phase in cycling progenitors at different developmental stages using EdU or BrdU pulse-chase experiments. In this experimental paradigm, cycling progenitors incorporate nucleoside analogs EdU or BrdU during DNA replication (S phase), and these labeled progenitors are then followed as they progress through G2 and M phases (Fig. 1A). During the cell cycle, retinal progenitors undergo stereotyped interkinetic nuclear migration, such that M phase occurs at the sclerad surface (Fig. 1B) (Baye and Link, 2008). To assess the onset of M phase, we co-stained embryonic sections for phosphohistone H3 (PH3) and EdU or BrdU to identify the shortest chase that yielded double-labeled cells. Histone H3 is transiently phosphorylated for the entire duration of M phase (Bradbury, 1992). No EdU+ PH3+ cells were detected after a 1 hr EdU chase, while most PH3+ had punctate EdU labeling after a 2 hr chase (Fig. 1C). These results suggest that M phase initiates at 1–2 hrs after the end of S phase, in the majority of progenitors. Similar results were observed at E15.5 (Fig. 1D), and are consistent with measurements of G2 length at other embryonic times (Sinitsina, 1971; Young, 1985b).
Fig. 1.
Timing of cell cycle progression in the mouse retinal neuroepithelium at E13.5 and E15.5. (A) EdU pulse-chase experiments. Phosphohistone H3 (PH3) is expressed during M phase, while cyclin D1 is primarily expressed during G1 and early S phases. After an EdU pulse, labeled S phase cells progress into G2, M and G1/G0 phases. (B) Diagram of interkinetic nuclear migration, showing the positions of progenitor nuclei at each stage of the cell cycle. Following terminal M phase, G0 daughters segregate vertically according to their cell fate. Ganglion cells (rgc) migrate toward the base (vitread), while photoreceptors (pr) migrate toward the apex (sclerad). (C–D) E13.5 and E15.5 embryos stained for PH3 and EdU or BrdU following a 1–2 hr chase. No PH3+ cells are EdU+ after a 1 hr chase, but most are EdU+ or BrdU+ after a 2 hr chase. Therefore, M phase initiates at 1–2 hrs after S phase in most cells, at both of developmental stages. nbl, neuroblastic layer; gcl, ganglion cell layer. Scale bar, 50 μm.
Expression of Brn3b and Isl1, but not Ptf1a or AP2α, prior to cell cycle exit
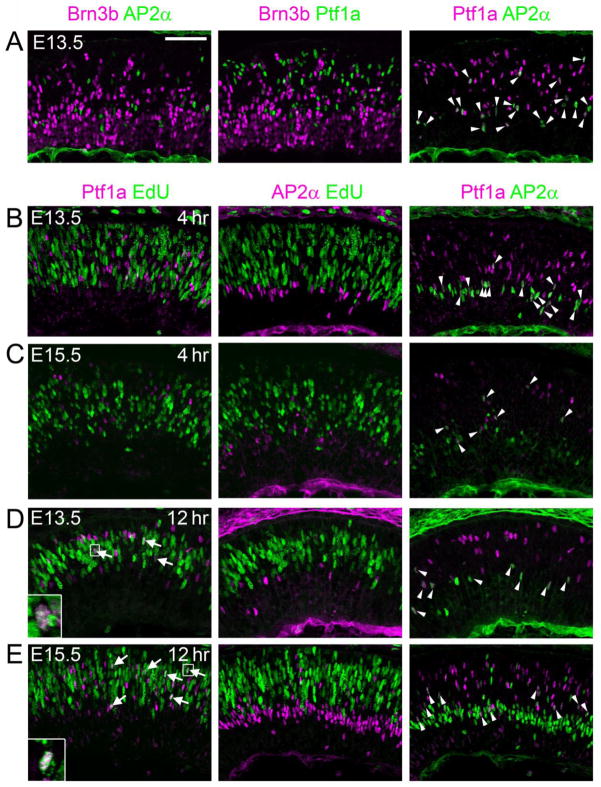
Using the pulse-chase paradigm, it is possible to unambiguously identify the onset of expression relative to the end of S phase. Previous lineage analysis and marker co-staining have shown that Brn3b is restricted to RGCs (Badea et al., 2009; Qiu et al., 2008; Xiang et al., 1995), and that Ptf1a is confined to inhibitory horizontal and amacrine cells (Fujitani et al., 2006). As a first step in this analysis, we tested whether Brn3b and Ptf1a truly mark mutually exclusive populations of committed precursors during early retinal development, by co-staining sections from E13.5 retinas for Brn3b, Ptf1a, and the amacrine marker AP2α (Bassett et al., 2007). We observed no overlap of Ptf1a or AP2α with Brn3b (Fig 2A) consistent with previous results observed during late gestation (Bassett et al., 2007; Fujitani et al., 2006; Qiu et al., 2008). As expected, many AP2α-expressing cells were contained within the Ptf1a population, and are presumed to be precursors of inhibitory amacrine neurons.
Fig. 2.
Co-expression and onset analysis of amacrine and horizontal markers Ptf1a and AP2α. (A) Developing horizontal or amacrine neurons do not express Brn3b. Sections from E13.5 retinas coimmunostained for Brn3b, AP2α and Ptf1a. Although many Brn3b+ cells are present, none co-label with the horizontal and inhibitory amacrine marker Ptf1a, or the pan-amacrine marker AP2α, suggesting that Brn3b specifically marks RGCs during early development. (B–E) Sections from E13.5 (B,D) or E15.5 (C,E) embryos co-stained with Ptf1a, AP2α and EdU following a 4 hr (B,C) or 12 hr (D,E) chase. No Ptf1a+ or AP2α+ cells are EdU+ after a 4 hr chase, suggesting that these factors are expressed exclusively in post-mitotic (G0) cells. Many Ptf1a+ EdU+ cells are apparent after a 12 hr chase at both E13.5 and E15.5 (arrows, insets), indicating that this factor initiates expression in migratory cells. Many AP2α+ cells are Ptf1a+ (arrowheads), marking the inhibitory amacrine population. Scale bar, 50 μm.
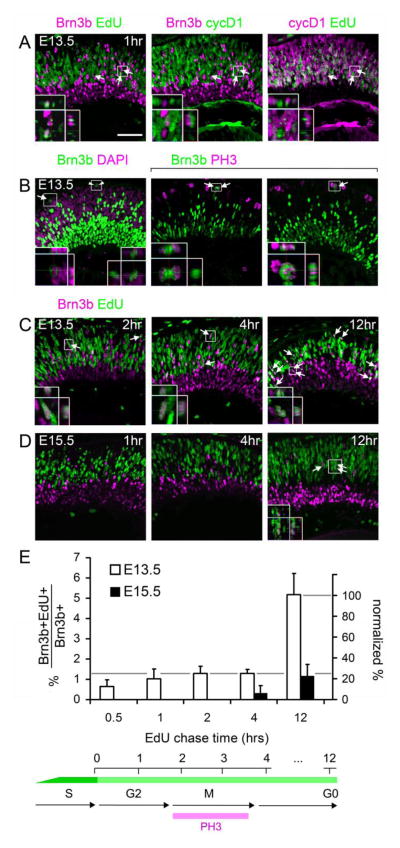
We next determined the onset of Brn3b, Ptf1a and AP2α with respect to the terminal cell cycle as outlined (Fig. 1A). At both E13.5 and E15.5, we observed no overlap between Ptf1a or AP2α and EdU after a 4 hr chase (Fig. 2B,C), but Ptf1a+EdU+ cells were apparent after a 12 hr chase (Fig. 2D,E). Although Ptf1a is transiently expressed and vital for development of horizontal and amacrine cells, its actions are apparently delayed until after cell cycle exit, most likely during cell migration. In contrast, at E13.5, we found that Brn3b+ EdU+ cells were present after a 1 hr chase (2–5 per high-power field, Fig. 3A, Supplemental File). All Brn3b+ cells were cyclinD1− (cycD1), including the EdU+ cohort, whereas the vast majority of EdU+ cells (>95%) were cycD1+ (Fig. 3A). In most cycling cells, cycD1 is made during G1 and down-regulated during S phase, and it controls the G1/S transition. CycD1 is reactivated in G2 (Stacey, 2003), except in terminally mitotic cells (Yang et al., 2006). In the developing retina, cycD1 can be detected in all cells, except for post-mitotic neurons (G0) and a small subpopulation of G2 cells that are likely represent the neurogenic cohort (Barton and Levine, 2008). Accordingly, we interpret the Brn3b+ EdU+ cycD1− precursors at E13.5 as neurogenic cells in late S or G2 phase of the terminal division. Consistent with interpretation, we observed rare Brn3b+ cells at the scleral margin (apex), in M phase (PH3+, arrows) or in adjacent pairs (arrowheads) suggestive of a recent division (Fig. 3B). Furthermore, Brn3b+ EdU+ cells can be followed through the cell cycle, with symmetric Brn3b segregation (Fig. 3B), and migration into the GCL with a 12 hr chase at E13.5 (Fig. 3C). Many of the resulting Brn3b+ EdU+ cells are in close proximity, as presumptive daughter pairs. At E15.5, essentially no Brn3b+ EdU+ cells were observed after 1- or 4-hr chase periods, but were readily detected throughout the retina after a 12 hr chase (1–4 per high-power field, Fig. 3D). Although new Brn3b+ cells continue to be generated throughout the retina, Brn3b is expressed exclusively in post-mitotic cells at this stage. Quantitative analysis revealed that only 4.8 ± 0.9% of Brn3b+ cells at E13.5, and 1.2 ± 0.6% at E15.5, were EdU+ after a 12 hr chase (Fig. 3E). These cells represented the sum of neurogenic cells initiating Brn3b expression during S/G2 and G0 phase. To estimate the fraction of Brn3b+ cells at E13.5 that initiate expression during S/G2, we normalized the Brn3b+EdU+ fractions observed after a 0.5–4 hr chase to the 12 hr chase value (Fig. 3E). This analysis revealed that 50–60% of new Brn3b+ cells (accounting for mitotic doubling) initiated expression prior to cell cycle exit (<4 hr chase).
Fig. 3.
The onset of Brn3b and Isl1 expression within individual cells is progressively delayed during retinal development. (A) Sections from E13.5 embryos co-stained for Brn3b, cyclinD1 (cycD1) and EdU following a 1 hr chase. In some cells, Brn3b is co-localized with EdU (arrows), but not with the G1/early-S phase marker cycD1, indicating that Brn3b expression initiates in late S or G2 phase. Insets show two cells that are Brn3b+ EdU+ cyclinD1−. The orthogonal (Y–Z) views demonstrate the importance of the confocal Z-stack for distinguishing adjacent cells. One cell has a punctate EdU staining pattern, indicating incorporation in late-replicating DNA regions at the end of S phase. (B) Sections from E13.5 embryos stained for Brn3b and the mitotic marker PH3 or the nuclear counterstain DAPI. Brn3b+ cells in M phase (arrows) and presumptive daughter pairs (arrowheads) are indicated. (C–D) Sections from E13.5 or E15.5 embryos co-stained for Brn3b and EdU following the indicated chase period. At E13.5 (C), many newly generated Brn3b+ cells are co-labeled with EdU after a 2, 4 or 12 hr chase. By 12 hrs, some EdU-labeled cells migrate to the nascent GCL, and many Brn3b+ EdU+ cells are in close proximity as presumptive daughter pairs. At E15.5 (D), no Brn3b+ EdU+ cells are observed after 1 hr chase, and very few are detected after a 4 hr chase, but Brn3b+ EdU+ are readily detected after a 12 hr chase (arrows). (E) Quantitative analysis of Brn3b and EdU co-labeling. The abundance of co-labeled cells is plotted as a percentage of all Brn3b+ cells (left scale) or neurogenic Brn3b+ cells (right scale, normalized to the 12 hr chase value at E13.5). Approximately 30% of newly Brn3b+ cells initiated expression prior to cell cycle exit at E13.5 (gray line). In contrast, few Brn3b+ cells are co-labeled with EdU prior to 12 hrs at E15.5, indicating that RGCs are specified later at this developmental stage. Scale bar, 50 μm.
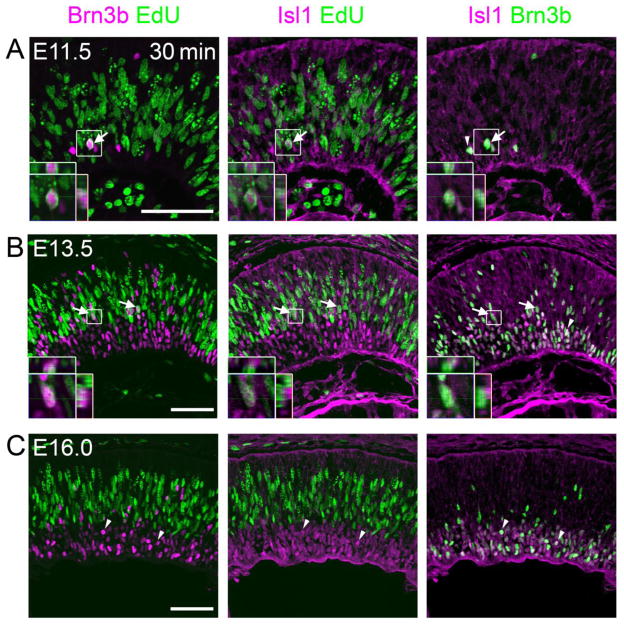
To confirm these observations regarding the timing of RGC marker expression relative to terminal M phase, we evaluated a second regulator of RGC differentiation, Islet-1 (Isl1). We co-stained sections for Brn3b, Isl1 and EdU following a 30 min chase. We observed numerous Brn3b+ Isl1+ cells that were EdU+ at E11.5 or E13.5 (Fig. 4A,B), whereas no Brn3b+ Isl1+ cells were EdU+ at E16.0 (Fig. 4C). These results indicate that at least two key regulators of RGC fate are expressed prior to cell cycle exit during early (<E14), but not during late (>E15), developmental stages.
Fig. 4.
Co-expression of Brn3b and Isl1 during or shortly after the terminal cell cycle. (A–C) Sections from E11.5, E13.5, and E16.0 embryos co-stained for Brn3b, Isl1 and EdU following a 30 min chase. At E11.5 (A) and E13.5 (B), multiple Brn3b+ Isl1+ EdU+ cells are observed (arrows), indicating that RGC markers can initiate expression prior to cell cycle exit, in late S or G2 phase. In contrast, at E16.0 (C), all Brn3b+ Isl1+ cells are EdU− (examples marked by arrowheads). The insets show orthogonal Z-stack views of magnified Brn3b+ Isl1+ EdU+ cells. Scale bar, 50 μm.
RGC fate symmetry in marked clones
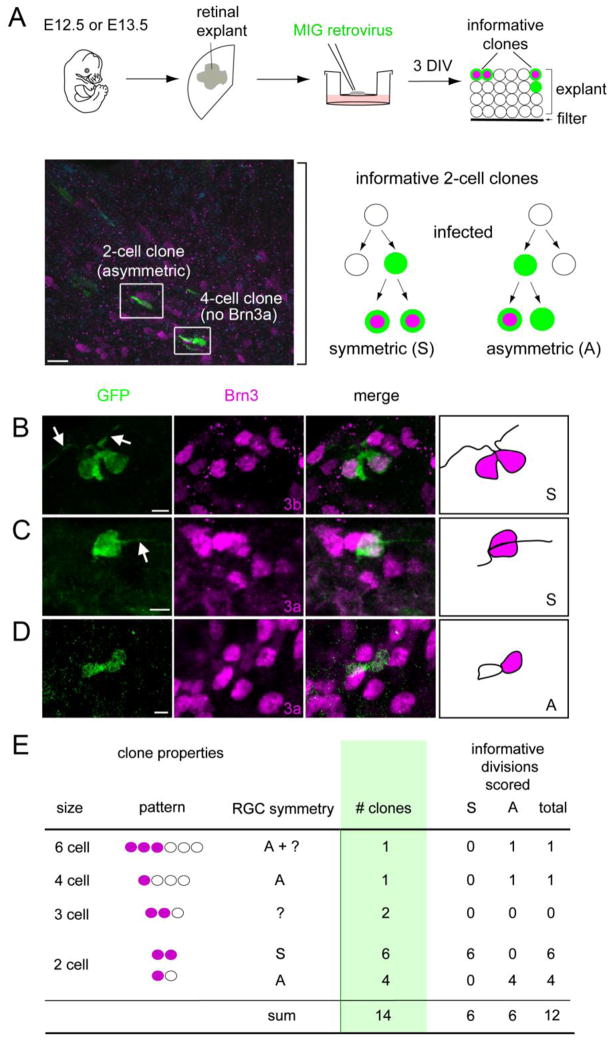
If progenitors express Brn3b and Isl1 before terminal M phase, they should give rise to paired RGCs. To test this hypothesis, we performed a clonal analysis. Retinal explants from E12.5 or E13.5 embryos were infected with MSCV-IRES-GFP (MIG) retrovirus at low density to generate independent clones. After culturing 3 days in vitro (DIV), we scored the resulting GFP+ clones for their size and number of RGCs, using Brn3a or 3b immunoreactivity and cellular morphology to identify ganglion cells (Fig. 5A). Given the divergent roles of Brn3a and Brn3b in RGC development, despite functional redundancy at the protein level (Badea et al., 2009; Badea and Nathans, 2011; Pan et al., 2005), we compared the overlap and onset of these two markers during the cell cycle at different embryonic ages. At E15.5, most Brn3b+ cells (~80%) also expressed Brn3a (Suppl Fig. 1). However, Brn3a was only detected in postmigratory RGCs within the GCL, consistent with previous results (Pan et al., 2005), and did not overlap with EdU following a 12 hr chase. We therefore used Brn3a primarily in the clonal analysis to identify mature RGCs.
Fig. 5.
Paired ganglion cells can be generated from retinal progenitors by symmetric terminal division. (A) Retinas were explanted from E12.5 or E13.5 embryos, infected at clonal density with MSCV-IRES-GFP (MIG) retrovirus (green), and cultured for 3 days in vitro (DIV). The micrograph shows a cross-section from a representative explant (bracket) coimmunostained for Brn3a (magenta) and GFP (green). The schema shows informative two-cell GFP+ clones: a symmetric [S] clone containing two Brn3+ (3a or 3b) RGCs, and an asymmetric [A] clone with one Brn3+ RGC. (B–D) Confocal Z-stack projections and drawings from representative symmetric (B, C) or asymmetric (D) clones containing RGCs. Most Brn3+ cells have long processes (arrows), confirming that they are differentiated RGCs. (E) Summary of observed clones containing at least one Brn3+ cell. In these experiments, RGCs were identified using Brn3a (12 clones) or Brn3b (2 clones) antisera interchangeably. Among 12 informative divisions, an equal number with symmetric and asymmetric fates was observed. Scale bars, 10 μm in A; 5 μm in B–D.
In twelve explanted retinas, we observed approximately 250 GFP+ clones, which were 1–16 cells in size. Fourteen (~5%) included at least one Brn3+ RGC. Among these informative clones, we observed two-cell clones with paired (Fig. 5B,C) or single (Fig. 5D) RGCs. Of the 12 neurogenic divisions that could unambiguously be scored among 14 clones, six were symmetric, resulting in paired RGCs (Fig. 5E). In principle, paired RGCs could arise if both daughter cells independently adopt the RGC fate, stochastically or in response to the local retinal environment. Given the very small number of GFP+ ganglion cells observed in these explants (n = 24), and the high frequency of symmetric fating (6 of 12 divisions), this pattern is unlikely to reflect independent events paired by chance (χ2 > 40, df = 2, P < 0.001 for a Poisson distribution). Instead, this statistical clustering, and the expression of definitive RGC markers in G2 and through cytokinesis (Fig. 3), strongly argue that many RGCs arise in pairs and descend from progenitors that commit to the ganglion fate during their final cell cycle, prior to terminal mitosis.
DISCUSSION
The kinetics of the retinal progenitor cell cycle have been extensively characterized in rodents and other vertebrate species by window and cumulative nucleoside labeling, cell counting, and percent labeled mitosis (PLM) methods (Alexiades and Cepko, 1996; Fujita, 1962; Li et al., 2000; Sinitsina, 1971; Young, 1985b). In addition, a variety of nuclear factors responsible for generating histotypic diversity in the retina have been identified through loss- and gain-of-function genetic analysis, and expression studies (Ohsawa and Kageyama, 2008; Swaroop et al., 2010). However, the basic question of when the cell fate decision is made relative to the terminal cell cycle has remained largely unanswered. In this paper, we have integrated expression and cell cycle analysis to determine the precise time that transcription factors controlling RGC, amacrine and horizontal fates appear during the last (neurogenic) cell cycle.
RGC fate commitment can occur before cell cycle exit
At early developmental times (<E14), we detected Isl1 and Brn3b in many neurogenic cells during late S or G2 phases of the terminal cell cycle (Figs. 3,4), similar to Math5-lacZ (Brzezinski et al., 2012). These results contrast with previous reports (Pan et al., 2008; Xiang et al., 1993), most likely due to differences in sampling and improved detection sensitivity. Our interpretation is based on three convergent results: the precise timing of PH3 (Fig. 1), Brn3b and Isl1 (Fig. 4) onset relative to the end of S phase, the mutually exclusive pattern of cycD1 and Brn3b expression (Fig. 3A), and the presence of Brn3b+ M phase cells (Fig. 3B).
There are three possible explanations for the observation of RGC markers in dividing cells. First, the immunopositive cells may reflect low-level leaky transcription of Brn3b or Isl1 that is not biologically meaningful. This seems unlikely given that the expression in S/G2 phase progenitors is often as strong as that observed in post-mitotic cells (e.g. arrows in Fig 4A,B). Second, Isl1 and Brn3b may not accurately mark committed RGC precursors. Indeed, Isl1 is also expressed in differentiating amacrine and bipolar precursors, and has a role in the development of both of these interneuron classes (Elshatory et al., 2007a; Elshatory et al., 2007b). In contrast, Brn3b is not co-expressed with Ptf1a or AP2α, which mark nascent horizontal and amacrine interneurons, and it is made in most, but not all RGCs (Fig. 2A). Complementary co-expression and lineage analyses, using Cre recombinase or stable histogenic reporters, have also demonstrated that Brn3b is made only in RGCs (Badea et al., 2009; Bassett et al., 2007; Fujitani et al., 2006; Qiu et al., 2008). In the absence of Brn3b, progenitors adopt non-RGC fates (Badea et al., 2009; Qiu et al., 2008). Overexpression of Brn3b in progenitors promotes RGC fate and suppresses amacrine differentiation (Feng et al., 2011; Liu et al., 2000; Qiu et al., 2008). Taken together, these results strongly suggest that Brn3b identifies committed RGCs, and does not label multipotent progenitors. Third, the RGC fate decision may precede terminal mitosis, and occur during G2. According to this interpretation, Brn3b+ Isl1+ EdU+ cells represent progenitors committed to develop as RGCs. In general, progenitors may undergo symmetric or asymmetric divisions to generate one or two post-mitotic daughters, respectively (Huttner and Kosodo, 2005). Our pulse-chase (Fig. 3) and clonal analyses (Fig. 5) suggest that many divisions giving rise to RGCs are symmetric and terminal. This observation is somewhat surprising considering that the retina is rapidly expanding during the period of early development. However, G2 fate commitment may be necessary for prompt deployment of RGCs, which establish a pioneering scaffold for axon pathfinding (Pittman et al., 2008; Raper and Mason, 2010). Alternatively, the relatively early onset of Brn3b expression may reflect the position of RGCs as the first sampled fate in the histogenetic sequence (Wong and Rapaport, 2009).
Our clonal data are generally consistent with previous observations of terminal divisions in the early retina. In rodent and frog lineage analysis, many two-cell clones have been observed during early development, but typically these cells have discordant fates (Holt et al., 1988; Turner et al., 1990; Wong and Rapaport, 2009). Notably, among 114 two-cell clones analyzed in these reports, only one contained paired RGCs (Holt et al., 1988). During early zebrafish development, most ath5-expressing progenitors, monitored by time-lapse imaging of mosaic ath5-GFP embryos, undergo a terminal division and give rise to at least one ganglion cell (Poggi et al., 2005). The daughters typically have discordant fates, but paired RGCs may arise from wild-type cells in an ath5 mutant environment.
Similar to our finding of mitotic Brn3b+ cells (Fig. 3B), RA4+ cells were observed in terminal mitotic anaphase in the chick retina (Waid and McLoon, 1995), and migrating in pairs shortly after mitosis (McLoon and Barnes, 1989). Although originally interpreted as RGC precursors, it is no longer clear that the RA4 neurofilament antigen marks ganglion cells exclusively (Gutierrez et al., 2011). Beyond these observations, there is precedent for G2 commitment during neurogenesis of other cell types. In the chick retina, committed horizontal cell precursors typically arrest in G2 and undergo a final non-apical mitosis (Boije et al., 2009), which may explain the presence of paired horizontal cell clones of a single subtype (Rompani and Cepko, 2008). In the ferret cerebral cortex, progenitors lose their competence to respond to environmental signals following terminal G2 phase (McConnell and Kaznowski, 1991). Similarly, heterochronic analysis of dissociated rat retinal cells suggests that progenitors commit to the amacrine fate (VC1.1+) during G2/M of the last cell cycle (Belliveau and Cepko, 1999).
Post-mitotic fate plasticity in the retina
In this study, the amacrine marker Ptf1a was only detected in post-mitotic cells, at both E13.5 and E15.5 (Fig. 2B–E). In Ptf1a mutant mice carrying Cre knock-in alleles, many lineage-marked cells expressed Brn3b and adopted RGC fates (Fujitani et al., 2006). Taken together, these results suggest a degree of post-mitotic plasticity, such that some amacrine cells solidify their identity after the terminal division. Likewise, photoreceptor and bipolar precursors retain the ability to switch fates after cell cycle exit (Adler and Hatlee, 1989; Brzezinski et al., 2010; Ng et al., 2011; Oh et al., 2007). Indeed, the bZIP factor NRL, which instructs rod photoreceptor fate, initiates expression soon after terminal M phase, throughout development (Akimoto et al., 2006; data not shown). In comparison to Ptf1a, AP2α expression was further delayed, suggesting that it is involved in downstream differentiation processes and not fate specification.
We observed a gradual restriction in the expression of Brn3b and Isl1, as well as Math5-lacZ (Brzezinski et al., 2012), to post-mitotic cells during late gestation (Fig. 3D). At these ages (>E15), many RGCs are still generated, including migrating EdU+ Brn3b+ cells (Drager, 1985; Young, 1985a). In the early (<E14) retina, we observed both symmetric and asymmetric patterns of RGC marker expression among daughter cells in clones, suggesting that some cells commit to the RGC fate after terminal mitosis in the early retina (Fig. 5E) (Brzezinski et al., 2012). The difference in the timing of RGC marker onset between the early and late retina cannot be explained by a change in the length of G2, which is relatively static (Fig. 1), or by the progressive lengthening of the cell cycle during development (Alexiades and Cepko, 1996; Sinitsina, 1971; Young, 1985b). Indeed, if transcription initiates after G1 or S phase with a fixed time delay, then the onset of expression following terminal S phase should be advanced (closer to S), rather than delayed as we observed during the course of development. It is more likely that progenitor fate decisions occur before or after cell cycle exit, depending on the lineage or developmental stage. Alternatively, the expression of differentiation markers may be delayed at late developmental stages while the timing of RGC fate commitment is relatively fixed.
What could explain these shifts in expression kinetics? Environmental signals elaborated by differentiating neurons, such as Delta ligand or Shh, increase as RGCs accumulate (Ahmad et al., 1997; Wang et al., 2005; Yang, 2004; Yu et al., 2006) and could delay the onset of RGC transcription factor expression. Alternatively, the onset may be intrinsically programmed to shift over time, and is potentially regulated by the same upstream cascades that control cell cycle exit. Further studies are needed to distinguish these mechanisms. Nonetheless, it is clear that cell fate determination and cell cycle exit, though correlated, are not strictly causally related events.
Supplementary Material
Brn3a and Brn3b mark highly overlapping populations of differentiated ganglion cells during early development. E15.5 retinal sections were stained for Brn3b, Brn3a, and EdU following a 12 hour chase. Each single (top) and double (bottom) labeling combination is shown. Approximately 80% of Brn3b+ cells also express Brn3a (example marked by arrows). Migrating Brn3b+ EdU+ are present (arrowheads, inset), while Brn3a is only made in post-migratory cells in the GCL. Scale bar, 50 μm
Acknowledgments
The authors are grateful to Dellaney Rudolph and Melinda Nagy for technical support; to Helena Edlund for Ptf1a antisera; to Chris Edwards, and the UM microscopy and image analysis laboratory staff, for technical advice; to Sean Morrison for the MIG retroviral construct; to Joe Brzezinski, Nadean Brown, Chris Chou, Sally Camper and David Turner for valuable discussions and critical reading of the manuscript. This research was funded by National Institutes of Health (NIH) R01 grant EY14259 (TG). LP was supported by NIH T32 grants EY13934 and GM07863.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adler R, Hatlee M. Plasticity and differentiation of embryonic retinal cells after terminal mitosis. Science. 1989;243:391–393. doi: 10.1126/science.2911751. [DOI] [PubMed] [Google Scholar]
- Ahmad I, Dooley CM, Polk DL. Delta-1 is a regulator of neurogenesis in the vertebrate retina. Dev Biol. 1997;185:92–103. doi: 10.1006/dbio.1997.8546. [DOI] [PubMed] [Google Scholar]
- Akimoto M, Cheng H, Zhu D, Brzezinski JA, Khanna R, Filippova E, Oh EC, Jing Y, Linares JL, Brooks M, Zareparsi S, Mears AJ, Hero A, Glaser T, Swaroop A. Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors. Proc Natl Acad Sci U S A. 2006;103:3890–3895. doi: 10.1073/pnas.0508214103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexiades MR, Cepko C. Quantitative analysis of proliferation and cell cycle length during development of the rat retina. Dev Dyn. 1996;205:293–307. doi: 10.1002/(SICI)1097-0177(199603)205:3<293::AID-AJA9>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Badea TC, Cahill H, Ecker J, Hattar S, Nathans J. Distinct roles of transcription factors brn3a and brn3b in controlling the development, morphology, and function of retinal ganglion cells. Neuron. 2009;61:852–864. doi: 10.1016/j.neuron.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea TC, Nathans J. Morphologies of mouse retinal ganglion cells expressing transcription factors Brn3a, Brn3b, and Brn3c: analysis of wild type and mutant cells using genetically-directed sparse labeling. Vision Res. 2011;51:269–279. doi: 10.1016/j.visres.2010.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton KM, Levine EM. Expression patterns and cell cycle profiles of PCNA, MCM6, cyclin D1, cyclin A2, cyclin B1, and phosphorylated histone H3 in the developing mouse retina. Dev Dyn. 2008;237:672–682. doi: 10.1002/dvdy.21449. [DOI] [PubMed] [Google Scholar]
- Bassett EA, Pontoriero GF, Feng W, Marquardt T, Fini ME, Williams T, West-Mays JA. Conditional deletion of activating protein 2alpha (AP-2alpha) in the developing retina demonstrates non-cell-autonomous roles for AP-2alpha in optic cup development. Mol Cell Biol. 2007;27:7497–7510. doi: 10.1128/MCB.00687-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baye LM, Link BA. Nuclear migration during retinal development. Brain Res. 2008;1192:29–36. doi: 10.1016/j.brainres.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belliveau MJ, Cepko CL. Extrinsic and intrinsic factors control the genesis of amacrine and cone cells in the rat retina. Development. 1999;126:555–566. doi: 10.1242/dev.126.3.555. [DOI] [PubMed] [Google Scholar]
- Boije H, Edqvist PH, Hallbook F. Horizontal cell progenitors arrest in G2-phase and undergo terminal mitosis on the vitreal side of the chick retina. Dev Biol. 2009;330:105–113. doi: 10.1016/j.ydbio.2009.03.013. [DOI] [PubMed] [Google Scholar]
- Bradbury EM. Reversible histone modifications and the chromosome cell cycle. Bioessays. 1992;14:9–16. doi: 10.1002/bies.950140103. [DOI] [PubMed] [Google Scholar]
- Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128:2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski JA, Lamba DA, Reh TA. Blimp1 controls photoreceptor versus bipolar cell fate choice during retinal development. Development. 2010;137:619–629. doi: 10.1242/dev.043968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski JA, Prasov L, Glaser T. Math5 defines the ganglion cell competence state in a subpopulation of retinal progenitor cells exiting the cell cycle. Dev Biol. 2012;365:395–413. doi: 10.1016/j.ydbio.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck SB, Bradford J, Gee KR, Agnew BJ, Clarke ST, Salic A. Detection of S-phase cell cycle progression using 5-ethynyl-2′-deoxyuridine incorporation with click chemistry, an alternative to using 5-bromo-2′-deoxyuridine antibodies. Biotechniques. 2008;44:927–929. doi: 10.2144/000112812. [DOI] [PubMed] [Google Scholar]
- Drager UC. Birth dates of retinal ganglion cells giving rise to the crossed and uncrossed optic projections in the mouse. Proc R Soc Lond B Biol Sci. 1985;224:57–77. doi: 10.1098/rspb.1985.0021. [DOI] [PubMed] [Google Scholar]
- Dyer MA, Cepko CL. Regulating proliferation during retinal development. Nat Rev Neurosci. 2001;2:333–342. doi: 10.1038/35072555. [DOI] [PubMed] [Google Scholar]
- Elshatory Y, Deng M, Xie X, Gan L. Expression of the LIM-homeodomain protein Isl1 in the developing and mature mouse retina. J Comp Neurol. 2007a;503:182–197. doi: 10.1002/cne.21390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshatory Y, Everhart D, Deng M, Xie X, Barlow RB, Gan L. Islet-1 controls the differentiation of retinal bipolar and cholinergic amacrine cells. J Neurosci. 2007b;27:12707–12720. doi: 10.1523/JNEUROSCI.3951-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkman L, McEvilly RJ, Luo L, Ryan AK, Hooshmand F, O’Connell SM, Keithley EM, Rapaport DH, Ryan AF, Rosenfeld MG. Role of transcription factors Brn-3.1 and Brn-3.2 in auditory and visual system development. Nature. 1996;381:603–606. doi: 10.1038/381603a0. [DOI] [PubMed] [Google Scholar]
- Feng L, Eisenstat DD, Chiba S, Ishizaki Y, Gan L, Shibasaki K. Brn-3b inhibits generation of amacrine cells by binding to and negatively regulating DLX1/2 in developing retina. Neuroscience. 2011;195:9–20. doi: 10.1016/j.neuroscience.2011.08.015. [DOI] [PubMed] [Google Scholar]
- Fujita S. Kinetics of cellular proliferation. Exp Cell Res. 1962;28:52–60. doi: 10.1016/0014-4827(62)90311-7. [DOI] [PubMed] [Google Scholar]
- Fujitani Y, Fujitani S, Luo H, Qiu F, Burlison J, Long Q, Kawaguchi Y, Edlund H, MacDonald RJ, Furukawa T, Fujikado T, Magnuson MA, Xiang M, Wright CV. Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development. 2006;133:4439–4450. doi: 10.1242/dev.02598. [DOI] [PubMed] [Google Scholar]
- Gan L, Xiang M, Zhou L, Wagner DS, Klein WH, Nathans J. POU domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc Natl Acad Sci U S A. 1996;93:3920–3925. doi: 10.1073/pnas.93.9.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C, McNally M, Canto-Soler MV. Cytoskeleton proteins previously considered exclusive to ganglion cells are transiently expressed by all retinal neuronal precursors. BMC Dev Biol. 2011;11:46. doi: 10.1186/1471-213X-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama J, Kageyama R. Retrovirus-mediated gene transfer to retinal explants. Methods. 2002;28:387–395. doi: 10.1016/s1046-2023(02)00257-8. [DOI] [PubMed] [Google Scholar]
- Holt CE, Bertsch TW, Ellis HM, Harris WA. Cellular determination in the Xenopus retina is independent of lineage and birth date. Neuron. 1988;1:15–26. doi: 10.1016/0896-6273(88)90205-x. [DOI] [PubMed] [Google Scholar]
- Huttner WB, Kosodo Y. Symmetric versus asymmetric cell division during neurogenesis in the developing vertebrate central nervous system. Curr Opin Cell Biol. 2005;17:648–657. doi: 10.1016/j.ceb.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Kriss JP, Revesz L. The distribution and fate of bromodeoxyuridine and bromodeoxycytidine in the mouse and rat. Cancer Res. 1962;22:254–265. [PubMed] [Google Scholar]
- Li S, Mo Z, Yang X, Price SM, Shen MM, Xiang M. Foxn4 controls the genesis of amacrine and horizontal cells by retinal progenitors. Neuron. 2004;43:795–807. doi: 10.1016/j.neuron.2004.08.041. [DOI] [PubMed] [Google Scholar]
- Li Z, Hu M, Ochocinska MJ, Joseph NM, Easter SS., Jr Modulation of cell proliferation in the embryonic retina of zebrafish (Danio rerio) Dev Dyn. 2000;219:391–401. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1063>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Liu W, Khare SL, Liang X, Peters MA, Liu X, Cepko CL, Xiang M. All Brn3 genes can promote retinal ganglion cell differentiation in the chick. Development. 2000;127:3237–3247. doi: 10.1242/dev.127.15.3237. [DOI] [PubMed] [Google Scholar]
- Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- McConnell SK, Kaznowski CE. Cell cycle dependence of laminar determination in developing neocortex. Science. 1991;254:282–285. doi: 10.1126/science.254.5029.282. [DOI] [PubMed] [Google Scholar]
- McLoon SC, Barnes RB. Early differentiation of retinal ganglion cells: an axonal protein expressed by premigratory and migrating retinal ganglion cells. J Neurosci. 1989;9:1424–1432. doi: 10.1523/JNEUROSCI.09-04-01424.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X, Fu X, Beremand PD, Thomas TL, Klein WH. Gene regulation logic in retinal ganglion cell development: Isl1 defines a critical branch distinct from but overlapping with Pou4f2. Proc Natl Acad Sci U S A. 2008;105:6942–6947. doi: 10.1073/pnas.0802627105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L, Lu A, Swaroop A, Sharlin DS, Forrest D. Two transcription factors can direct three photoreceptor outcomes from rod precursor cells in mouse retinal development. J Neurosci. 2011;31:11118–11125. doi: 10.1523/JNEUROSCI.1709-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P. A long twentieth century of the cell cycle and beyond. Cell. 2000;100:71–78. doi: 10.1016/s0092-8674(00)81684-0. [DOI] [PubMed] [Google Scholar]
- Oh EC, Khan N, Novelli E, Khanna H, Strettoi E, Swaroop A. Transformation of cone precursors to functional rod photoreceptors by bZIP transcription factor NRL. Proc Natl Acad Sci U S A. 2007;104:1679–1684. doi: 10.1073/pnas.0605934104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuma S, Harris WA. Neurogenesis and the cell cycle. Neuron. 2003;40:199–208. doi: 10.1016/s0896-6273(03)00632-9. [DOI] [PubMed] [Google Scholar]
- Ohsawa R, Kageyama R. Regulation of retinal cell fate specification by multiple transcription factors. Brain Res. 2008;1192:90–98. doi: 10.1016/j.brainres.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Pan L, Deng M, Xie X, Gan L. ISL1 and BRN3B co-regulate the differentiation of murine retinal ganglion cells. Development. 2008;135:1981–1990. doi: 10.1242/dev.010751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Yang Z, Feng L, Gan L. Functional equivalence of Brn3 POU-domain transcription factors in mouse retinal neurogenesis. Development. 2005;132:703–712. doi: 10.1242/dev.01646. [DOI] [PubMed] [Google Scholar]
- Pear W. Transient transfection methods for preparation of high-titer retroviral supernatants. Curr Protoc Mol Biol. 2001;Chapter 9(Unit 9):11. doi: 10.1002/0471142727.mb0911s36. [DOI] [PubMed] [Google Scholar]
- Pittman AJ, Law MY, Chien CB. Pathfinding in a large vertebrate axon tract: isotypic interactions guide retinotectal axons at multiple choice points. Development. 2008;135:2865–2871. doi: 10.1242/dev.025049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poggi L, Vitorino M, Masai I, Harris WA. Influences on neural lineage and mode of division in the zebrafish retina in vivo. J Cell Biol. 2005;171:991–999. doi: 10.1083/jcb.200509098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu F, Jiang H, Xiang M. A comprehensive negative regulatory program controlled by Brn3b to ensure ganglion cell specification from multipotential retinal precursors. J Neurosci. 2008;28:3392–3403. doi: 10.1523/JNEUROSCI.0043-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raper J, Mason C. Cellular strategies of axonal pathfinding. Cold Spring Harb Perspect Biol. 2010;2:a001933. doi: 10.1101/cshperspect.a001933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe T, Reynolds TC, Yu G, Brown PO. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompani SB, Cepko CL. Retinal progenitor cells can produce restricted subsets of horizontal cells. Proc Natl Acad Sci U S A. 2008;105:192–197. doi: 10.1073/pnas.0709979104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul SM, Brzezinski JA, Altschuler RA, Shore SE, Rudolph DD, Kabara LL, Halsey KE, Hufnagel RB, Zhou J, Dolan DF, Glaser T. Math5 expression and function in the central auditory system. Mol Cell Neurosci. 2008;37:153–169. doi: 10.1016/j.mcn.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinitsina VF. DNA synthesis and cell population kinetics in embryonal histogenesis of the retina in mice. Arkh Anat Gistol Embriol. 1971;61:58–67. [PubMed] [Google Scholar]
- Skowronska-Krawczyk D, Chiodini F, Ebeling M, Alliod C, Kundzewicz A, Castro D, Ballivet M, Guillemot F, Matter-Sadzinski L, Matter JM. Conserved regulatory sequences in Atoh7 mediate non-conserved regulatory responses in retina ontogenesis. Development. 2009;136:3767–3777. doi: 10.1242/dev.033449. [DOI] [PubMed] [Google Scholar]
- Stacey DW. Cyclin D1 serves as a cell cycle regulatory switch in actively proliferating cells. Curr Opin Cell Biol. 2003;15:158–163. doi: 10.1016/s0955-0674(03)00008-5. [DOI] [PubMed] [Google Scholar]
- Swaroop A, Kim D, Forrest D. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci. 2010;11:563–576. doi: 10.1038/nrn2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift S, Lorens J, Achacoso P, Nolan GP. Rapid production of retroviruses for efficient gene delivery to mammalian cells using 293T cell-based systems. Curr Protoc Immunol. 2001;Chapter 10(Unit 10):17C. doi: 10.1002/0471142735.im1017cs31. [DOI] [PubMed] [Google Scholar]
- Turner DL, Snyder EY, Cepko CL. Lineage-independent determination of cell type in the embryonic mouse retina. Neuron. 1990;4:833–845. doi: 10.1016/0896-6273(90)90136-4. [DOI] [PubMed] [Google Scholar]
- Van Parijs L, Refaeli Y, Lord JD, Nelson BH, Abbas AK, Baltimore D. Uncoupling IL-2 signals that regulate T cell proliferation, survival, and Fas-mediated activation-induced cell death. Immunity. 1999;11:281–288. doi: 10.1016/s1074-7613(00)80103-x. [DOI] [PubMed] [Google Scholar]
- Waid DK, McLoon SC. Immediate differentiation of ganglion cells following mitosis in the developing retina. Neuron. 1995;14:117–124. doi: 10.1016/0896-6273(95)90245-7. [DOI] [PubMed] [Google Scholar]
- Wang SW, Kim BS, Ding K, Wang H, Sun D, Johnson RL, Klein WH, Gan L. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SW, Mu X, Bowers WJ, Kim DS, Plas DJ, Crair MC, Federoff HJ, Gan L, Klein WH. Brn3b/Brn3c double knockout mice reveal an unsuspected role for Brn3c in retinal ganglion cell axon outgrowth. Development. 2002a;129:467–477. doi: 10.1242/dev.129.2.467. [DOI] [PubMed] [Google Scholar]
- Wang SW, Mu X, Bowers WJ, Klein WH. Retinal ganglion cell differentiation in cultured mouse retinal explants. Methods. 2002b;28:448–456. doi: 10.1016/s1046-2023(02)00264-5. [DOI] [PubMed] [Google Scholar]
- Wang Y, Dakubo GD, Thurig S, Mazerolle CJ, Wallace VA. Retinal ganglion cell-derived sonic hedgehog locally controls proliferation and the timing of RGC development in the embryonic mouse retina. Development. 2005;132:5103–5113. doi: 10.1242/dev.02096. [DOI] [PubMed] [Google Scholar]
- Wong LL, Rapaport DH. Defining retinal progenitor cell competence in Xenopus laevis by clonal analysis. Development. 2009;136:1707–1715. doi: 10.1242/dev.027607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M. Requirement for Brn-3b in early differentiation of postmitotic retinal ganglion cell precursors. Dev Biol. 1998;197:155–169. doi: 10.1006/dbio.1998.8868. [DOI] [PubMed] [Google Scholar]
- Xiang M, Zhou L, Macke JP, Yoshioka T, Hendry SH, Eddy RL, Shows TB, Nathans J. The Brn-3 family of POU-domain factors: primary structure, binding specificity, and expression in subsets of retinal ganglion cells and somatosensory neurons. J Neurosci. 1995;15:4762–4785. doi: 10.1523/JNEUROSCI.15-07-04762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M, Zhou L, Peng YW, Eddy RL, Shows TB, Nathans J. Brn-3b: a POU domain gene expressed in a subset of retinal ganglion cells. Neuron. 1993;11:689–701. doi: 10.1016/0896-6273(93)90079-7. [DOI] [PubMed] [Google Scholar]
- Yang K, Hitomi M, Stacey DW. Variations in cyclin D1 levels through the cell cycle determine the proliferative fate of a cell. Cell Div. 2006;1:32. doi: 10.1186/1747-1028-1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ. Roles of cell-extrinsic growth factors in vertebrate eye pattern formation and retinogenesis. Semin Cell Dev Biol. 2004;15:91–103. doi: 10.1016/j.semcdb.2003.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Ding K, Pan L, Deng M, Gan L. Math5 determines the competence state of retinal ganglion cell progenitors. Dev Biol. 2003;264:240–254. doi: 10.1016/j.ydbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Young RW. Cell differentiation in the retina of the mouse. Anat Rec. 1985a;212:199–205. doi: 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]
- Young RW. Cell proliferation during postnatal development of the retina in the mouse. Brain Res. 1985b;353:229–239. doi: 10.1016/0165-3806(85)90211-1. [DOI] [PubMed] [Google Scholar]
- Yu C, Mazerolle CJ, Thurig S, Wang Y, Pacal M, Bremner R, Wallace VA. Direct and indirect effects of hedgehog pathway activation in the mammalian retina. Mol Cell Neurosci. 2006;32:274–282. doi: 10.1016/j.mcn.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Zeng C, Pan F, Jones LA, Lim MM, Griffin EA, Sheline YI, Mintun MA, Holtzman DM, Mach RH. Evaluation of 5-ethynyl-2′-deoxyuridine staining as a sensitive and reliable method for studying cell proliferation in the adult nervous system. Brain Res. 2010;1319:21–32. doi: 10.1016/j.brainres.2009.12.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Brn3a and Brn3b mark highly overlapping populations of differentiated ganglion cells during early development. E15.5 retinal sections were stained for Brn3b, Brn3a, and EdU following a 12 hour chase. Each single (top) and double (bottom) labeling combination is shown. Approximately 80% of Brn3b+ cells also express Brn3a (example marked by arrows). Migrating Brn3b+ EdU+ are present (arrowheads, inset), while Brn3a is only made in post-migratory cells in the GCL. Scale bar, 50 μm