Abstract
The transcription factor pancreatic and duodenal homeobox 1 (PDX1) plays an essential role in pancreatic development and in maintaining proper islet function via target gene regulation. Few intestinal PDX1 targets, however, have been described. We sought to define novel PDX1-regulated intestinal genes. Caco-2 human intestinal epithelial cells were engineered to overexpress PDX1 and gene expression profiles relative to control cells were assessed. Expression of 80 genes significantly increased while that of 49 genes significantly decreased more than 4-fold following PDX1 overexpression in differentiated Caco-2 cells. Analysis of the differentially regulated genes with known functional annotations revealed genes encoding transcription factors, growth factors, kinases, digestive glycosidases, nutrient transporters, nutrient binding proteins, and structural components. The gene for fatty acid binding protein 1, liver, FABP1, is repressed by PDX1 in Caco-2 cells. PDX1 overexpression in Caco-2 cells also results in repression of promoter activity driven by the 0.6 kb FABP1 promoter. PDX1 regulation of promoter activity is consistent with the decrease in FABP1 RNA abundance resulting from PDX1 overexpression and identifies FABP1 as a candidate PDX1 target. PDX1 repression of FABP1, LCT, and SI suggests a role for PDX1 in patterning anterior intestinal development.
Keywords: gene regulation, transcription factors, expression profile
INTRODUCTION
Development of the gastrointestinal tract is associated with programmed patterning of epithelial gene expression along the gut A-P axis. Global expression profiling using microarray analysis has revealed distinct gene expression patterns with sharp segmental boundaries for multiple intestine-specific genes along the length of the small intestine 1. The mechanisms regulating spatial restriction of intestine-specific gene expression along the anterior-posterior axis during gastrointestinal development, however, are largely unknown. Transcription factors expressed in specific regions along the A-P gut axis during gastrointestinal organogenesis have been characterized as spatial regulators. An evolutionarily-conserved group of homeodomain-containing genes that regulate endodermal and mesodermal cell fates during gut ontogeny are grouped in chromosomal regions known as the Hox and Parahox clusters, reviewed by Krumlauf2 and Beck3. The spatial expression domains for several Parahox transcription factors expressed in endoderm-derived organs have been mapped along the A-P gut axis4. The Parahox gene, PDX1, pancreatic duodenal homeobox 1, PDX1 (also known as IPF-1, IDX-1 and STF-1) is expressed in the developing pancreas and duodenum. PDX1 is required for pancreas development and the maintenance of functional islet β cells5–9. Specifically, mice null for the PDX1 gene, PDX1−/−, fail to form a pancreas and die in the neonatal period within a week of birth6, 7.
In addition to the pancreas, PDX1 is maximally expressed in the most anterior duodenal region of the intestinal tract with decreased expression in the distal small intestine 10. In neonatal PDX1−/− null mice, the rostral duodenum shows dilated cystic malformations at the stomach/duodenum junction felt to be abnormal Brunner’s glands and areas of local ectopic GLUT2-positive cuboidal epithelium7. Just distal of the abnormal epithelium, the numbers of enteroendocrine cells in the villi are greatly reduced. In further support of a role for PDX1 in patterning intestinal development and cell differentiation, mice with misexpression of PDX1 targeted to the large intestine manifest an altered midgut-hindgut union11 and immature intestinal epithelial rat IEC-6 cells can differentiate into enteroendocrine cells in response to PDX1 overexpression12. PDX1 is a known regulator of a number of genes essential for maintaining pancreatic cell identity and function including insulin13, glucose transporter 214, glucokinase15, islet amyloid polypeptide16–18 and somatostatin8, 9. Both activator8, 9, 13–26 and repressor11, 27–31 functions have been described for PDX1. Few genes expressed in the intestine, however, have been identified as intestinal targets capable of being regulated by PDX1. In order to obtain further insights into the role of PDX1 in regulating intestinal cell fate determination in epithelial cells, we sought to identify downstream target genes regulated by PDX1. cDNA microarray approaches were used to characterize the effects on gene expression profiles following overexpression of PDX1 in Caco-2 cells, a human adenocarcinoma-derived cell line that mimics a small intestinal enterocyte phenotype with respect to expression of several digestive hydrolases.32
MATERIALS AND METHODS
Plasmid constructs
For use in the transfection experiments, mouse PDX1 cDNA, a gift of C. V. Wright (Vanderbilt University), was subcloned into pAlpha+ (Affymax) downstream of a recombinant SV40/HTLV promoter enhancer to generate pmPDX1 as previously described. 29 The 100 bp rat lactase promoter-luciferase reporter plasmid, pgLac100, has been described previously. 29 The 0.6 kb Fabp1 promoter-reporter plasmid, pFABP1luc, was a kind gift of T. Simon, Washington University.
Transfections and luciferase assays
Caco-2 cells were stably transfected with 1.0 μg of pmPDX1 or empty vector pAlpha+ control and selected for G418 resistant pooled or isolated colonies as previously described. 29 The stably transfected cells were maintained in complete DMEM plus G418 and differentiated for 9 days after reaching 100% confluency prior to harvest. For transient transfections, a DNA transfection mixture was prepared consisting of 0.4 pmol of the luciferase reporter reporter construct, 0.05 pmol of pmPDX1 or pAlpha+ empty vector, and 7.0 fmol of pRL-TK (Promega) as an internal control. The individual DNA mixtures were transfected into cells (50–80% confluent) with lipofectamine reagent (INTROGEN) according to the protocol of the manufacturer. Cells were harvested 48 h after transfection and luciferase activity was measured by the Dual-Luciferase Reporter Assay System (Promega) as described by the manufacturer, in a Monolight 3010 luminometer. Experimental lactase promoter-reporter activities were normalized to the activity of the pRL-TK internal control and expressed as relative luciferase activity (means + SD, n = 4), thereby minimizing experimental variability caused by differences in cell viability or transfection efficiency. Statistical significance (P value) was determined by using Student's unpaired t-test.
Western Blot Assays
Caco-2 cell nuclear extracts were prepared as previously described. 29 20 ug of nuclear extract protein was separated by electrophoresis in 12% SDS polyacrylamide gels. After transfer of the protein to 0.45 micron nitrocellulose membrane, blots were processed using the Fast Western Blot Kit (Pierce) according to the manufacturers protocol. Rabbit polyclonal antibody against PDX1 (gift of C.V. Wright, Vanderbilt University) was used at a 1:3000 dilution. Blots were subjected to enhanced chemiluminescence detection using the Supersignal West Pico substrate (Pierce) and imaged with the ImageLab software on a ChemiDoc XRS+ system (BioRad).
Gene Expression Profiling
Total RNA was purified from the stably transfected Caco-2 cell populations and processed for DNA microarray analysis as previously described.33 Briefly, total RNA was labeled with Cy5–deoxyuridine triphosphate and a human reference RNA (Stratagene) was labeled with Cy3–deoxyuridine triphosphate. Hybridizations were performed using human DNA microarrays produced at the Stanford Functional Genomics Facility containing ~41,000 cDNAs of which over 27,000 features represent unique human genes. The microarrays were scanned and the data was deposited in the Stanford Microarray Database (http://genome-www5.stanford.edu). Three preparations of pooled PDX1 transfected cells and two preparations of pooled cells transfected with the pAlpha+ vector alone were analyzed. The SAM, Significant Analysis of Microarrays, software program was used to identify genes that are significantly differentially expressed.34 The software permits the application of a statistical analysis for significance as a means to reduce the number of false positive candidates. Additional analysis of the genes that were significantly elevated > 4-fold or significantly decreased > 4-fold was performed using software for gene annotation and for the identification of biological processes using DAVID, Database for Annotation, Visualization and Integrated Discovery (http://apps1.niaid.nih.gov/david).35
Real-time quantitative RT-PCR
For quantitative real-time RT-PCR, cDNA was synthesized from total RNA using the High Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA) according to the manufacturer's recommendations. Expression levels for each specific gene were determined by TaqMan gene expression assays (Applied Biosystems) with human gene-specific, predesigned TaqMan primers and probe sets. PCR amplification and fluorescence data collection were performed with the ABI Prism 7900 HT sequence detection system (Applied Biosystems). Each gene was assayed in quadruplicate. The housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (Gapdh) was used to normalize for total RNA amount among samples. Normalized Ct values for each gene (ΔCtgene) were calculated as raw Ct values for the gene (Ctgene) minus corresponding raw Ct values for Gapdh (CtGapdh): ΔCtgene = Ctgene − CtGapdh. A relative quantification approach was used in this study to calculate relative mRNA abundance for each gene: first, a reference ΔCt value (ΔCtreference) was calculated by averaging the ΔCtgene values of individual RNA samples from pAlpha+ control population. The differences (ΔΔCtgene) of the ΔCtgene values relative to the ΔCtreference value were calculated as ΔΔCtgene = ΔCtgene − ΔCtreference. Relative mRNA abundance values were then calculated using the formula 2−ΔΔCtgene.
RESULTS
Caco-2 cells engineered to overexpress PDX1
Few gut-specific target genes capable of being regulated by PDX1 have been identified. In order to characterize global gene expression patterns regulated by PDX1 and to identify novel PDX1 intestinal target genes, Caco-2 human intestinal epithelial cells were engineered to overexpress PDX1 for use in characterization of gene expression analyses. Specifically, Caco-2 cells were stably transfected with the PDX1 expression construct, pmPDX1, or the empty expression vector to generate pooled cell populations. PDX1 overexpression was confirmed by Western blot (Figure 1). Anti-PDX1 antibody detects increased abundance of the roughly 45-kilodalton PDX1 protein in Caco-2 cells stably transfected with pmPDX1 compared to cells transfected with the empty vector.
Figure 1.
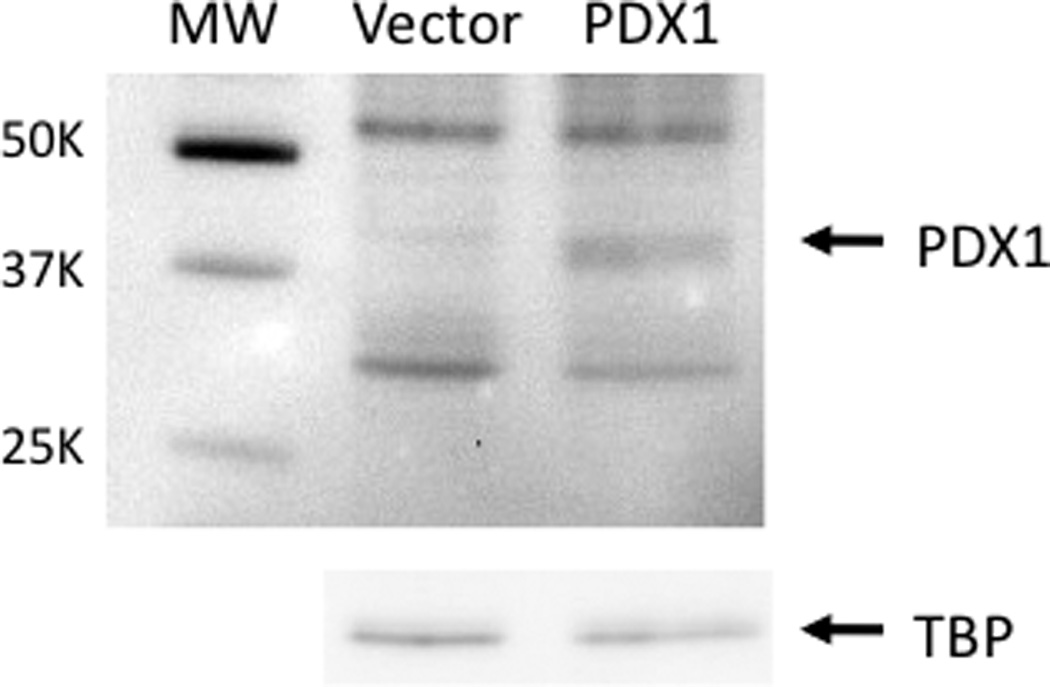
Western blot analysis of PDX1 protein expression in stably transfected Caco-2 cells. A rabbit polyclonal anti-PDX1 antibody detects the roughly 40-kilodalton PDX1 protein in Caco-2 cells stably transfected with pmPDX1 (PDX1) but not in cells transfected with the empty vector pAlpha+ (vector). The membrane was stripped and probed with antibody against TATA binding protein (TBP) to verify loading and transfer.
Gene expression profiles for Caco-2 cells overexpressing PDX1
To identify novel PDX1 intestinal target genes, gene expression profiles for the Caco-2 cells engineered to overexpress PDX1 were assessed using cDNA microarrays. Total RNA was purified from the stably transfected cell populations (PDX1 or empty vector) described above and was processed for DNA microarray analysis. The SAM, Significant Analysis of Microarrays, software program was used to identify genes that are significantly differentially expressed. 806 genes were found to be significantly differentially expressed at least two-fold in the PDX1 expressing cells with a median number of falsely significant genes of 1. A total of 129 genes were found to be significantly elevated or depressed at least 4-fold. The results are graphically displayed using pseudocoloring in Supplementary Data 1.
Additional analysis of the 80 genes that were significantly elevated > 4-fold and the 49 genes that were significantly decreased > 4-fold was performed using software for gene annotation and for the identification of biological processes that are enhanced with PDX1 expression. Annotation of the 129 genes using DAVID, Database for Annotation, Visualization and Integrated Discovery and limited to classifications with 3 or more hits revealed genes that include transcription factors, growth factors, kinases, digestive glycosidases, nutrient transporters, nutrient binding proteins, and structural components.
To validate our initial microarray analysis, we assayed by real-time RT-PCR 13 genes with a >4-fold change in microarray expression levels in pooled PDX1 transfected cells vs. empty vector transfected cells. PDX1 regulation was confirmed by RT-PCR in all 13 genes. In order to investigate variability between different PDX1 overexpression cell populations, two additional distinct clonal PDX1 stably transfected populations were characterized. Additional gene expression analysis was performed for 18 genes (13 genes validated above plus PDX1, ESR1, MAPK8, LCT and SI) in order to compare expression profiles in the three different preparations (pooled and two clonal) of PDX1 expressing cells. Of the 18 genes regulated in the PDX1 pooled population, 11 were similarly regulated in both clonal populations, 6 were similarly regulated in one clone with less than a 2-fold change in the other, and 1 had less than a 2-fold change in both clones (Figure 2). With respect to regulation of the previously reported intestine-specific target genes sucrase-isomaltase (SI) and lactase-phlorizin hydrolase (LCT) quantitative RT-PCR confirmed PDX1-mediated repression of both gene transcripts in the three different populations. While it is presumed that several of the differentially expressed genes identified are candidate targets (direct or indirect) of PDX1, it is possible that differences in gene expression may result from effects independent of PDX1 over-expression. Varying proliferation rates for the cells stably transfected with PDX1 or the control empty vector might affect cell differentiation stage and thus gene expression profiles. In order to determine whether a specific gene could be regulated by PDX1, we therefore proceeded to investigate promoter regulation of a candidate target gene in cells transiently transfected with PDX1.
Figure 2. PDX1 regulation of gene expression in stable transfected Caco-2 cells.
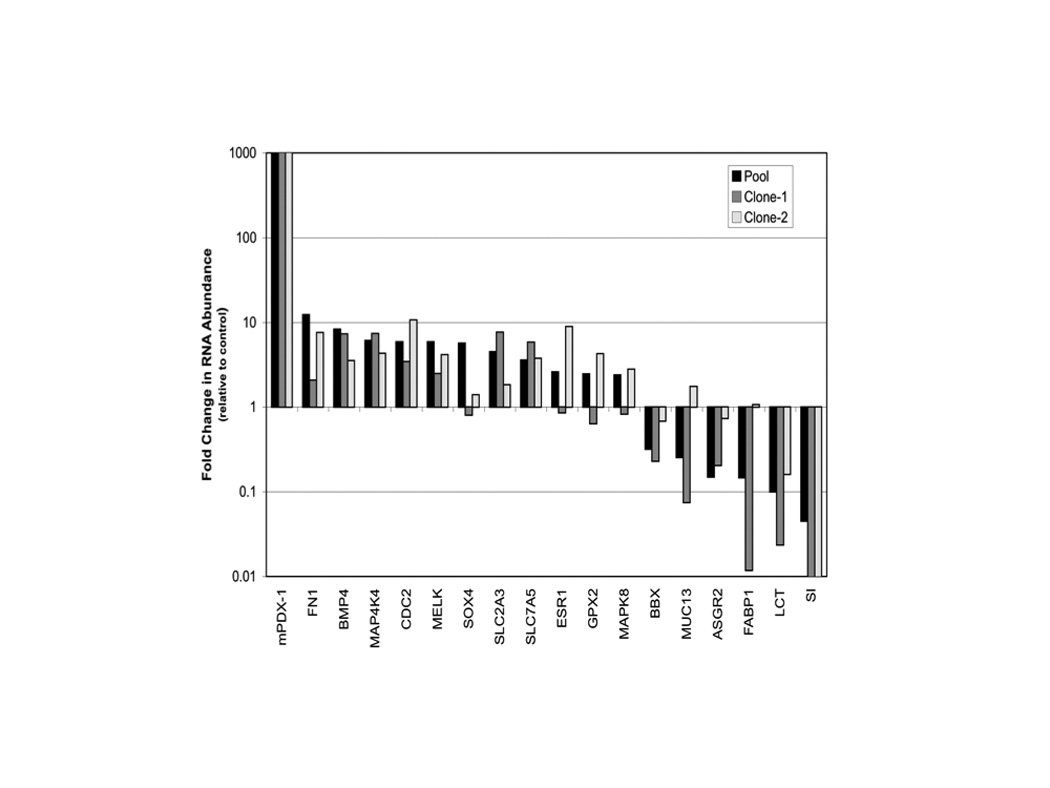
Total RNA was isolated from cell populations stably transfected with PDX1 expression construct or empty vector. Transcript abundance detected by real time RT-PCR for various genes is plotted as fold change relative to the empty vector control population. Means +/− SD. n=4 technical replicates (error bars not detectable at figure resolution).
PDX1 transcriptional regulation of the FABP1 target gene promoter
Fatty acid binding protein 1, liver, FABP1, a candidate gene with known intestinal cell expression, was identified by the microarray screen described above. FABP1 mRNA was decreased ~4-fold in pooled PDX1 expressing Caco-2 cells. In silico analysis of the proximal promoter for the FABP1 gene using the MatInspector (Genomatix, Inc.) transcription factor binding site software, reveals multiple PDX1 consensus binding sites located 5’ to the transcription start-sites (not shown). To determine whether the FABP1 promoter is capable of being regulated by PDX1, Caco-2 cells were co-transfected with a 0.6-kb FABP1 promoter-luciferase construct (pFABP1-luc), or a lactase promoter-luciferase reporter construct (control) and the PDX1 expression construct, pmPDX1, or empty vector. Caco-2 cell extracts were assayed for relative luciferase activity 48 hours after transfection as shown in Figure 3. PDX1 overexpression results in repression of promoter activity driven by the 0.6 kb FABP1 promoter relative to the empty vector control. PDX1 regulation of promoter activity is consistent with the decrease in FABP1 RNA abundance resulting from PDX1 overexpression and identifies FABP1 as a candidate PDX1 target.
Figure 3. PDX1 regulation of FABP1 promoter activity.
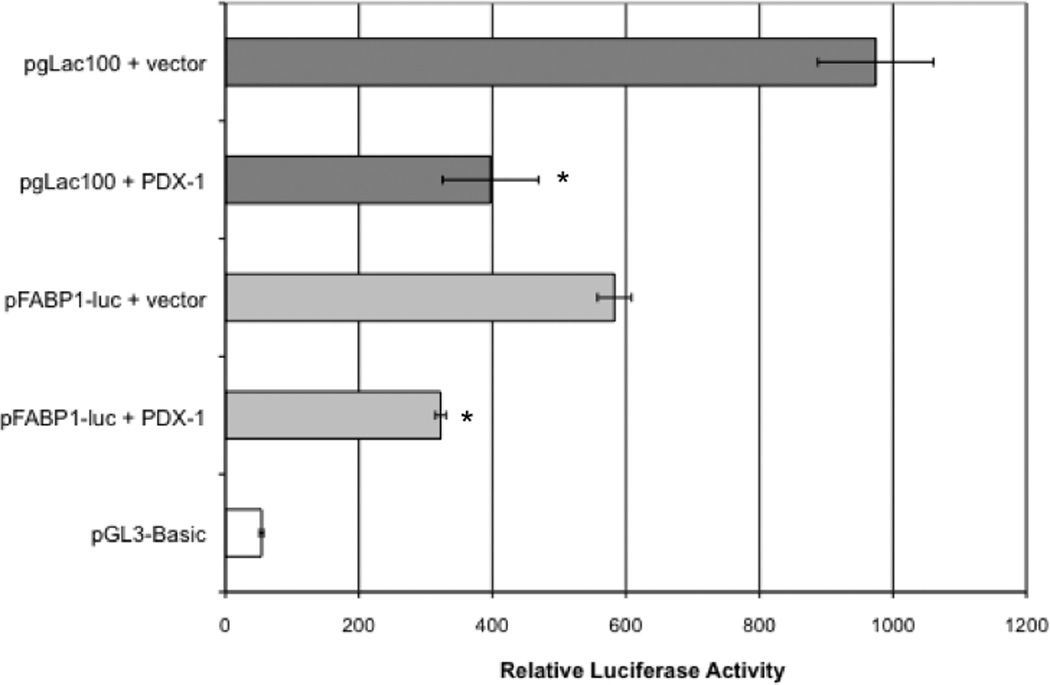
Caco-2 cells were co-transfected with fatty acid binding protein 1 or lactase gene promoter-luciferase constructs (pFABP1-luc, pgLac100, pGL3Basic respectively) along with the pmPDX1 expression construct (PDX1) or pAlpha+ (empty vector). Transfection efficiencies were normalized to renilla luciferase expression of a cotransfected pRL-TK vector and expressed as relative luciferase activity (means +/− SD, n=3). *Significant decrease compared with empty vector (P < 0.05)
DISCUSSION
While PDX1 is a known regulator of a number of genes essential for maintaining pancreatic cell identity and function, few genes expressed in the intestine, including adenosine deaminase (ADA) 19, sucrase-isomaltase (SI) 11 and lactase-phlorizin hydrolase (LCT) 30, have been identified as intestinal target genes capable of being regulated by PDX1. The adenosine deaminase gene is expressed along a defined spatiotemporal pattern in the developing mammalian small intestine. High-level expression of ADA is limited to the villous epithelium of the duodenum similar to the expression profile of PDX1. Dusing et al have shown that PDX1 can interact with a small duodenal enhancer region in the ADA gene 19, 37, 38. Loss of PDX1 binding, via a PDX1 mutated enhancer transgenic construct, resulted in complete loss of highlevel activation in the duodenum. Sucrase-isomaltase gene expression is maximal in the middle segments of the small intestine and decreased in the proximal duodenum. Heller et al have reported that PDX1 is capable of inhibiting transactivation of the SI promoter by the transcription factor CDX2 11. In addition, PDX1 was shown to be capable of physical interaction with CDX2. Similar to sucrase-isomaltase, lactase-phlorizin hydrolase gene expression is maximal in the middle segments of the small intestine and decreased in the proximal duodenum. PDX1 can similarly repress activation of the LCT promoter in intestinal cell culture 30. In support of a role for PDX1 in specifying spatial restriction during intestinal development, Grapin-Botton et al have reported that ectopic expression of PDX1 in chick embryo intestinal epithelial cells extinguishes markers for other non-pancreatoduodenal regions of gut endoderm 39. Specifically, PDX1 expression in the small intestine between the duodenum and yolk stalk turns CdxA off, down-regulates CdxC, and turns off Hex in the bile duct.
In the present study, several candidate PDX1 target genes have been identified by gene expression profiling of intestinal Caco-2 cells engineered to overexpress PDX1. Multiple genes identified were classified as transcription factors, growth factors, kinases, digestive glycosidases, nutrient transporters, nutrient binding proteins, and structural components. The gene encoding fatty acid binding protein 1, liver, FABP1, was among the genes significantly repressed greater than 4-fold by PDX1 overexpression. Fatty acid binding proteins are small cytoplasmic lipid binding proteins involved in intracellular lipid transport. FABP1 binds free fatty acids and their co-enzyme A derivatives. The mouse FABP1 gene homologue, fabpl, is expressed in hepatocytes, enterocytes, and enteroendocrine cells. Along the anterior-posterior axis of the small intestine, FABP1 is expressed in a gradient with jejunal expression greater than that in the duodenum or ileum.40 In that regard, spatial restriction of FABP1 closely mimics that of lactase and sucrase-isomaltase in the intestine. Sweetser et al have mapped distinct cis-acting elements in the 5’ flanking region of the fabpl gene that are necessary for specifying appropriate anteriorposterior spatial patterning of transgene exprssion in mice.40 It is of interest that promoter activity of the FABP1 0.6 kb promoter-reporter construct, pFABP1luc, was repressed by Pdx1 co-transfection in the present study. PDX1 regulated repression of FABP1 promoter activity in intestinal cells is consistent with the reduced abundance of FABP1 protein in the proximal duodenum where PDX1 is most abundant. A similar correlation exists between reports of PDX1 repression of promoter activity of the lactase30 and sucrase-isomaltase genes11, reduced in vivo LCT and SI expression in the proximal duodenum, and decreased LCT and SI transcript levels in Caco-2 cells overexpressing PDX1 (present study). Our findings thus suggest that Pdx1 regulation of candidate target genes including FABP1, LCT, and SI may play a role in specifying the anterior duodenal boundary of spatial expression for enterocytes expressing those genes.
Supplementary Material
Supplementary Data 1. A) Graphical display of genes that are significantly overexpressed (red) or under-expressed (green) compared to an expected distribution of values. 129 genes are identified when the median # of falsely significant genes is set to 0 and a threshold of at least a 4-fold difference in expression is used. B) Pseudocolored image depicting gene expression levels in PDX1 expressing Caco-2 pooled cells (first 3 columns) and vector alone transfected cells (last 2 columns). Only genes that were identified by the SAM analysis are shown.
Highlights.
Fatty acid binding protein 1, liver, FABP1, is identified as a candidate PDX1 target.
FABP1 is repressed in intestinal Caco-2 cells engineered to overexpress PDX1.
PDX1 overexpression results in repression of FABP1 promoter activity.
Repression of FABP1 supports a role for PDX1 in patterning anterior intestinal development.
ACKNOWLEDGEMENTS
This research was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grants DK72416 and DK60715 (to E. Sibley) and DK56339 (to the Stanford Digestive Diseases Center).
We are grateful to Lynne Olds and Ying Hao for technical assistance and advice.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURES
No conflicts of interest are declared by the author(s).
REFERENCES
- 1.Bates MD, Erwin CR, Sanford LP, et al. Novel genes and functional relationships in the adult mouse gastrointestinal tract identified by microarray analysis. Gastroenterology. 2002;122:1467–1482. doi: 10.1053/gast.2002.32975. [DOI] [PubMed] [Google Scholar]
- 2.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 3.Beck F, Tata F, Chawengsaksophak K. Homeobox genes and gut development. Bioessays. 2000;22:431–441. doi: 10.1002/(SICI)1521-1878(200005)22:5<431::AID-BIES5>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 4.Grapin-Botton A, Melton DA. Endoderm development: from patterning to organogenesis. Trends Genet. 2000;16:124–130. doi: 10.1016/s0168-9525(99)01957-5. [DOI] [PubMed] [Google Scholar]
- 5.Ahlgren U, Jonsson J, Jonsson L, Simu K, Edlund H. beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev. 1998;12:1763–1768. doi: 10.1101/gad.12.12.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 7.Offield MF, Jetton TL, Labosky PA, et al. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 8.Leonard J, Peers B, Johnson T, Ferreri K, Lee S, Montminy MR. Characterization of somatostatin transactivating factor-1, a novel homeobox factor that stimulates somatostatin expression in pancreatic islet cells. Mol Endocrinol. 1993;7:1275–1283. doi: 10.1210/mend.7.10.7505393. [DOI] [PubMed] [Google Scholar]
- 9.Miller CP, McGehee RE, Jr, Habener JF. IDX-1: a new homeodomain transcription factor expressed in rat pancreatic islets and duodenum that transactivates the somatostatin gene. Embo J. 1994;13:1145–1156. doi: 10.1002/j.1460-2075.1994.tb06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guz Y, Montminy MR, Stein R, et al. Expression of murine STF-1, a putative insulin gene transcription factor, in beta cells of pancreas, duodenal epithelium and pancreatic exocrine and endocrine progenitors during ontogeny. Development. 1995;121:11–18. doi: 10.1242/dev.121.1.11. [DOI] [PubMed] [Google Scholar]
- 11.Heller RS, Stoffers DA, Hussain MA, Miller CP, Habener JF. Misexpression of the pancreatic homeodomain protein IDX-1 by the Hoxa-4 promoter associated with agenesis of the cecum. Gastroenterology. 1998;115:381–387. doi: 10.1016/s0016-5085(98)70204-5. [DOI] [PubMed] [Google Scholar]
- 12.Yamada S, Kojima H, Fujimiya M, Nakamura T, Kashiwagi A, Kikkawa R. Differentiation of immature enterocytes into enteroendocrine cells by Pdx1 overexpression. Am J Physiol Gastrointest Liver Physiol. 2001;281:G229–G336. doi: 10.1152/ajpgi.2001.281.1.G229. [DOI] [PubMed] [Google Scholar]
- 13.Ohlsson H, Karlsson K, Edlund T. IPF1, a homeodomain-containing transactivator of the insulin gene. Embo J. 1993;12:4251–4259. doi: 10.1002/j.1460-2075.1993.tb06109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waeber G, Thompson N, Nicod P, Bonny C. Transcriptional activation of the GLUT2 gene by the IPF-1/STF-1/IDX-1 homeobox factor. Mol Endocrinol. 1996;10:1327–1334. doi: 10.1210/mend.10.11.8923459. [DOI] [PubMed] [Google Scholar]
- 15.Watada H, Kajimoto Y, Umayahara Y, et al. The human glucokinase gene beta-cell-type promoter: an essential role of insulin promoter factor 1/PDX-1 in its activation in HIT-T15 cells. Diabetes. 1996;45:1478–1488. doi: 10.2337/diab.45.11.1478. [DOI] [PubMed] [Google Scholar]
- 16.Carty MD, Lillquist JS, Peshavaria M, Stein R, Soeller WC. Identification of cis- and trans-active factors regulating human islet amyloid polypeptide gene expression in pancreatic beta-cells. J Biol Chem. 1997;272:11986–11993. doi: 10.1074/jbc.272.18.11986. [DOI] [PubMed] [Google Scholar]
- 17.Bretherton-Watt D, Gore N, Boam DS. Insulin upstream factor 1 and a novel ubiquitous factor bind to the human islet amyloid polypeptide/amylin gene promoter. Biochem J. 1996;313 (Pt 2):495–502. doi: 10.1042/bj3130495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serup P, Jensen J, Andersen FG, et al. Induction of insulin and islet amyloid polypeptide production in pancreatic islet glucagonoma cells by insulin promoter factor 1. Proc Natl Acad Sci U S A. 1996;93:9015–9020. doi: 10.1073/pnas.93.17.9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dusing MR, Florence EA, Wiginton DA. Pdx-1 is required for activation in vivo from a duodenum-specific enhancer. J Biol Chem. 2001;276:14434–14442. doi: 10.1074/jbc.M009249200. [DOI] [PubMed] [Google Scholar]
- 20.Swift GH, Liu Y, Rose SD, et al. An endocrine-exocrine switch in the activity of the pancreatic homeodomain protein PDX1 through formation of a trimeric complex with PBX1b and MRG1 (MEIS2) Mol Cell Biol. 1998;18:5109–5120. doi: 10.1128/mcb.18.9.5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, MacDonald RJ, Swift GH. DNA binding and transcriptional activation by a PDX1.PBX1b.MEIS2b trimer and cooperation with a pancreas-specific basic helix-loop-helix complex. J Biol Chem. 2001;276:17985–17993. doi: 10.1074/jbc.M100678200. [DOI] [PubMed] [Google Scholar]
- 22.Andersen FG, Jensen J, Heller RS, et al. Pax6 and Pdx1 form a functional complex on the rat somatostatin gene upstream enhancer. FEBS Lett. 1999;445:315–320. doi: 10.1016/s0014-5793(99)00144-1. [DOI] [PubMed] [Google Scholar]
- 23.Peshavaria M, Cissell MA, Henderson E, Petersen HV, Stein R. The PDX-1 activation domain provides specific functions necessary for transcriptional stimulation in pancreatic beta-cells. Mol Endocrinol. 2000;14:1907–1917. doi: 10.1210/mend.14.12.0563. [DOI] [PubMed] [Google Scholar]
- 24.Goudet G, Delhalle S, Biemar F, Martial JA, Peers B. Functional and cooperative interactions between the homeodomain PDX1, Pbx, and Prep1 factors on the somatostatin promoter. J Biol Chem. 1999;274:4067–4073. doi: 10.1074/jbc.274.7.4067. [DOI] [PubMed] [Google Scholar]
- 25.Tarling E, Salter A, Bennett A. Transcriptional regulation of human SREBP-1c (sterol-regulatory-element-binding protein-1c): a key regulator of lipogenesis. Biochem Soc Trans. 2004;32:107–109. doi: 10.1042/bst0320107. [DOI] [PubMed] [Google Scholar]
- 26.Peers B, Sharma S, Johnson T, Kamps M, Montminy M. The pancreatic islet factor STF-1 binds cooperatively with Pbx to a regulatory element in the somatostatin promoter: importance of the FPWMK motif and of the homeodomain. Mol Cell Biol. 1995;15:7091–7097. doi: 10.1128/mcb.15.12.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asahara H, Dutta S, Kao HY, Evans RM, Montminy M. Pbx-Hox heterodimers recruit coactivator-corepressor complexes in an isoform-specific manner. Mol Cell Biol. 1999;19:8219–8225. doi: 10.1128/mcb.19.12.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chao SH, Harada JN, Hyndman F, et al. PDX1, a cellular homeoprotein, binds to and regulates the activity of human cytomegalovirus immediate early promoter. J Biol Chem. 2004;279:16111–16120. doi: 10.1074/jbc.M312304200. [DOI] [PubMed] [Google Scholar]
- 29.Seijffers R, Ben-David O, Cohen Y, et al. Increase in PDX-1 levels suppresses insulin gene expression in RIN 1046-38 cells. Endocrinology. 1999;140:3311–3317. doi: 10.1210/endo.140.7.6796. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, Fang R, Olds LC, Sibley E. Transcriptional regulation of the lactase-phlorizin hydrolase promoter by PDX-1. Am J Physiol Gastrointest Liver Physiol. 2004;287:G555–G561. doi: 10.1152/ajpgi.00011.2004. [DOI] [PubMed] [Google Scholar]
- 31.Ritz-Laser B, Gauthier BR, Estreicher A, et al. Ectopic expression of the beta-cell specific transcription factor Pdx1 inhibits glucagon gene transcription. Diabetologia. 2003;46:810–821. doi: 10.1007/s00125-003-1115-7. [DOI] [PubMed] [Google Scholar]
- 32.Hauri HP, Sander B, Naim H. Induction of lactase biosynthesis in the human intestinal epithelial cell line Caco-2. Eur J Biochem. 1994;219:539–546. doi: 10.1111/j.1432-1033.1994.tb19969.x. [DOI] [PubMed] [Google Scholar]
- 33.Hao Y, Triadafilopoulos G, Sahbaie P, Young HS, Omary MB, Lowe AW. Gene expression profiling reveals stromal genes expressed in common between Barrett's esophagus and adenocarcinoma. Gastroenterology. 2006;131:925–933. doi: 10.1053/j.gastro.2006.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Dusing MR, Brickner AG, Lowe SY, Cohen MB, Wiginton DA. A duodenum-specific enhancer regulates expression along three axes in the small intestine. Am J Physiol Gastrointest Liver Physiol. 2000;279:G1080–G1093. doi: 10.1152/ajpgi.2000.279.5.G1080. [DOI] [PubMed] [Google Scholar]
- 38.Dusing MR, Florence EA, Wiginton DA. High-level activation by a duodenum-specific enhancer requires functional GATA binding sites. Am J Physiol Gastrointest Liver Physiol. 2003;284:G1053–G1065. doi: 10.1152/ajpgi.00483.2002. [DOI] [PubMed] [Google Scholar]
- 39.Grapin-Botton A, Majithia AR, Melton DA. Key events of pancreas formation are triggered in gut endoderm by ectopic expression of pancreatic regulatory genes. Genes Dev. 2001;15:444–454. doi: 10.1101/gad.846001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sweetser DA, Birkenmeier EH, Hoppe PC, McKeel DW, Gordon JI. Mechanisms underlying generation of gradients in gene expression within the intestine: an analysis using transgenic mice containing fatty acid binding protein-human growth hormone fusion genes. Genes Dev. 1988;2:1318–1332. doi: 10.1101/gad.2.10.1318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data 1. A) Graphical display of genes that are significantly overexpressed (red) or under-expressed (green) compared to an expected distribution of values. 129 genes are identified when the median # of falsely significant genes is set to 0 and a threshold of at least a 4-fold difference in expression is used. B) Pseudocolored image depicting gene expression levels in PDX1 expressing Caco-2 pooled cells (first 3 columns) and vector alone transfected cells (last 2 columns). Only genes that were identified by the SAM analysis are shown.


