Abstract
Plants are the basis of life on earth. We cannot overemphasize their importance. The value of plant genome initiatives is self-evident. The need is to identify priorities for action. The angiosperm genome is highly variable, but the extent of this variability is unknown. Uncertainties remain about the number of genes and the number of species living. Many plants will become extinct before they are discovered. We risk losing both genes and vital information about plant uses. There are also major gaps in our karyotypic knowledge. No chromosome count exists for >70% of angiosperm species. DNA C values are known for only ≈1% of angiosperms, a sample unrepresentative of the global flora. Researchers reported new relationships between genome size and characters of major interest for plant breeding and the environment and the need for more data. In 1997, a Royal Botanic Gardens Kew workshop identified gaps and planned international collaboration to fill them. An electronic version of the Angiosperm DNA C value database also was published. Another initiative, which will make a very significant contribution to the conservation of plant genetic diversity on a global scale is Kew’s Millennium Seed Bank, partly funded by the U.K. Millennium Commission, celebrating the year 2000. Costing up to £80 million (£1 = $1.62), its main aims are to collect and conserve the seed of almost all of the U.K. spermatophyte flora by the year 2000, to collect and conserve a further 10% of the world spermatophyte flora principally from the drylands by 2009, and to provide a world class building as the focus of this activity by 2000.
Plants are the basis of life on earth. We cannot overemphasize their importance. They affect the global environment, acting to prevent soil erosion, as a major carbon sink and as a reservoir of moisture, and they provide the oxygen we breathe. They are environmentally friendly factories making our fibers, fuels, pharmaceuticals, forages, and food. To meet the challenges of the next millennium—managing the global environment sustainably and of feeding ourselves—we will need increased knowledge of plant genes and genomes and how to manipulate them. The value of plant genome initiatives is self-evident. The need is to identify priorities for action.
Basic Data on Plant Genomes.
Scientists often speak and write of “the plant genome,” but this entity is an ideal concept. In practice, it is highly variable, but the extent of its variation is often still unknown. Thus, it is important to highlight the need for more work on many basic aspects of angiosperm genomes.
The Number of Different Genes.
First, there remains uncertainty about the number of genes. An article in Science recently described a disagreement about the number of genes in the human genome (1). Estimates differ more than twofold from a conservative 60,000 to a liberal 150,000. The situation for angiosperm plants is similar; estimates for the gene number range from 25,000 to 50,000 (2, 3). These issues will be resolved with precision in a few years as genome sequencing projects in both kingdoms are completed.
The Number of Different Angiosperm Species.
Second, there is similar uncertainty about the number of species living today. The number of validly named angiosperm species is ≈250,000, but this number differs from the number alive today, for two important reasons. First, many angiosperm species (perhaps 50,000) are undiscovered and unnamed. It is not difficult to find such species nor are they restricted to obscure families of little economic potential. The number of new names at species level on Index Kewensis (4) since 1986 is 28,378, including 1,416 for grasses. The Palms of Madagascar (5) lists all of the 171 species known, including 70 (i.e., 41%) unknown before 1986. The discovery of new species is not limited to distant jungles, as the finding of Zea diploperennis in Mexico, a close relative of corn, showed (6). Of >700 grass species known in Bolivia, 40 (i.e., 5%) were named since 1986.
The second factor making the number of angiosperm species uncertain is species extinction. Many species known to science have been lost, whereas others will become extinct before they are discovered. Species extinction rates are hard to predict. In 1992, the World Conservation Monitoring Centre predicted the average loss of plant species as 5.4% per decade or 27% in the next 50 years (7). Other predictions are more conservative, yet they predict even higher losses in particular groups, e.g., 50% of palm species lost in the next 50–100 years (8). When a species becomes extinct, its loss is irreversible and forever. However, we do not know how present or projected species losses translate into a loss of coding genes unique to a species or a loss of economic value. Work to quantify these losses is needed urgently.
The Number of Plant Uses.
We risk losing not only genes but also information about plant uses. Plant genome initiatives should link to strategies to conserve and make available not only genotypes but also knowledge of useful characters built up over centuries of experience. With >200 years of world-wide involvement in economic botany, the Royal Botanic Gardens (RBG) Kew has a unique role as a global plant information center. One example is the Survey of Economic Plants for Arid and Semi-Arid Lands (SEPASAL). This database (9), begun at RBG Kew in 1981, contains information on >6,000 useful dryland species, excluding major crops. Table 1 shows the breakdown of information for 766 grass species in the SEPASAL database. Ability to search SEPASAL on several search criteria allows detailed inquiries to be answered, e.g., “useful salt tolerant grasses of Namibia.”
Table 1.
Breakdown of information for 766 grass species in the SEPASAL database (9)
| Level 1 use | Number of species | Level 2 use | Number of species |
|---|---|---|---|
| Food | 137 | leaves | 14 |
| seeds | 113 | ||
| Food additives | 3 | ||
| Animal food | 636 | leaves and stems | 604 |
| seeds and flowers | 38 | ||
| Invertebrate food | 2 | ||
| Materials | 106 | fibers | 88 |
| essential oils | 9 | ||
| Fuels | 14 | ||
| Vertebrate poisons | 43 | ||
| Nonvertebrate poisons | 3 | ||
| Medicines | 19 | ||
| Environmental uses | 258 | erosion control | 184 |
| revegetators | 81 | ||
| soil improvers | 28 |
Descriptive work on plant nuclear genomes is >100 years old. This work has produced a great mass of quantitative data that together reveals plant genomes as strikingly variable in many different respects. However, it is assumed often wrongly that all of the basic work on plant genomes has been done so that in this area our knowledge base is complete and secure. In fact, there are still major gaps in our knowledge of gross karyotypic characters, such as chromosome number, ploidy level, and genome size.
Chromosome Number (2n).
A chromosome count exists for only ≈25% of angiosperm species. Even this information is incomplete or incorrect for many species because many counts were made for only one individual or population, and many species were misnamed. Arabidopsis thaliana (2n = 2x = 10) has five pairs of chromosomes, but counts for a related British crucifer Cardamine pratensis include 61 numbers from 16 to 96 (10). Clearly, one count may give a very incomplete picture for a species.
Low and Minimum Chromosome Numbers.
Cytology still can yield big surprises as the history of chromosome numbers shows. In 1956, the lowest chromosome number known for an angiosperm was 2n = 6. Since then, five taxa in three different families were found with 2n = 4, including two grasses (11).
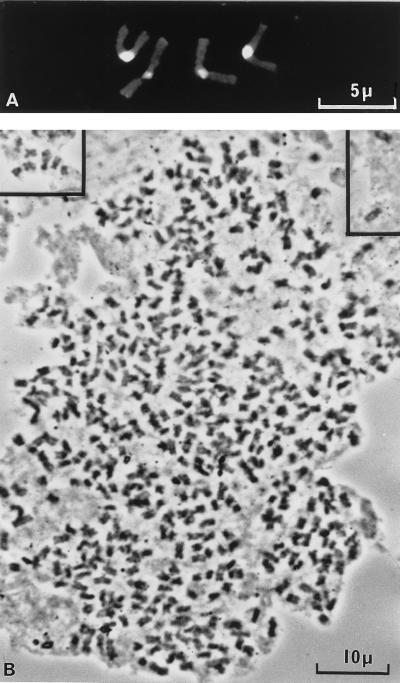
As recently as 1986, Crossland and Crozier (12) reported that sibling species of the primitive Australian ant Myrmecia pilosa has the lowest chromosome number possible, namely 2n = 2. Only two animal species with this lowest possible chromosome number are known, but no angiosperm with 2n = 2 is known. Because 200,000 species are uncounted, it seems possible that at least one may have 2n = 2. If so, it may have much more than curiosity value. A. thaliana became a model genetic organism partly because of its very small genome size (13). Any angiosperm with only one chromosome almost certainly would become another interesting model system for genome studies. The physical gene map in such a taxon would be of considerable interest for studies of synteny and may provide useful indications of what the ancestral angiosperm gene map was like. Meanwhile, it will be interesting to compare genome architectures in related species with 2n = 4 from the same dicot family, such as Haplopappus gracilis and Brachychome dichromosomatica, both from the Asteraceae family, with that in an unrelated monocot species with 2n = 4, such as the grasses Zingeria biebersteiniana (Fig. 1A) and Colpodium versicola (11).
Figure 1.
Root-tip mitotic metaphases in (A) the grass Z. biebersteiniana (2n ≈ 4) and (B) the palm V. gerardii (2n ≈ 600), new chromosome counts that increased the range of chromosome numbers known for monocots over fourfold since 1960 (11, 16). Chromosomes are C-banded by using a fluorochrome (A) or stained with Feulgen and aceto-orcein (B).
High and Maximum Chromosome Numbers.
Cytology also can yield surprises regarding high chromosome numbers. Until recently, the highest count for a monocot was 2n ≈ 266 for the grass Poa literosa (14), whereas the highest count for a palm was 2n ≈ 200 (15). However, cytological work at RBG Kew on Voanioala gerardii (Fig. 1B), a rare rainforest palm from Madagascar described as a new monotypic genus by John Dransfield in 1989, showed a count of 2n ≈ 600 (16), three times the previous maximum for a palm and double that previously known for a monocot. It is important to recognize that the full range of chromosome numbers in angiosperms (and large families) is uncertain and yet may be several times that already reported. So there is a real need for more basic work.
Genome Size.
A similar picture emerges for genome size variation. Several large reference lists of angiosperm nuclear DNA amounts have been published since 1976 (17, 18). It has been possible to estimate genome size since the 1950s, yet DNA C values are known only for <3,000 species or ≈1% of angiosperms. Many values are first approximations; most are unverifiable, and some are for misidentified taxa. Recent analysis of this sample (19) showed that it is very unrepresentative of the world’s angiosperm flora for taxonomic groups, geographic regions, and plant life forms. Thus, genome size data are rare for palms and noxious weeds. Of the 269 known sources, none was from China and only two were from Africa. DNA estimates are known for <30% of families (≈118 of 450). A first estimate is yet to be reported for most families, including many large tropical groups like the Begoniaceae, with >800 species. Not only is genome size unknown for many economically important grasses (e.g., bamboos), but the full range of genome sizes in angiosperms is uncertain. Here, too, there is still much to do.
The Importance of Genome Size.
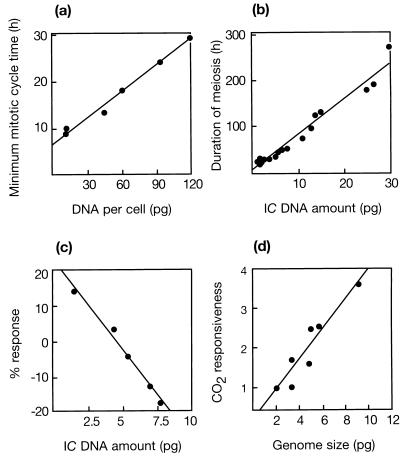
Comparative studies of angiosperms have played a leading role in showing that DNA C value is correlated with a wide range of phenotypic characters at the cellular level. Thus, genome size shows strikingly precise positive correlations with many cell size and mass characters and with rates of cell development. For example, in 1963, Van’t Hof and Sparrow (20) showed that the duration of the minimum mitotic cell cycle in six diploid species grown at 23°C is related closely with genome size (Fig. 2a). Fig. 2b shows a similar plot for the duration of meiosis on genome size in 18 diploid species grown at 20°C (21). Clearly, there is a close relationship between the two characters. Genome size is a major variable with far-reaching consequences. Each relationship seen individually is strikingly close, but when several plots for widely different characters are all viewed together, this impression is strongly reinforced (Fig. 2). DNA amount correlates closely with many important phenotypic characters, and many of the relationships between DNA amount and various cellular characters are strikingly precise for biological phenomena and more reminiscent of physical relationships.
Figure 2.
Relationships between genome size and four developmental or environmental characters in angiosperms: (a) minimum root-tip mitotic cycle time in six diploids grown at 23°C (redrawn from ref. 20); (b) the duration of meiosis in 18 diploid species (including three grass species) grown at 20°C (replotted from ref. 21); (c) response in the absolute growth rate of the shoot in a mixture of five herbaceous species (including three grass species) to elevated temperature—winter warming of +5°C above ambient (replotted from ref. 27); and (d) enhanced growth (responsiveness in final biomass to elevated CO2) in eight annual grass species (replotted from ref. 25).
The importance of nucleotypic correlations based on biophysical absolutes is that they apply to all of the species, irrespective of chromosome number or genome size, and set limits on the range of phenotypes that can be expressed by genic control. As such, they can help considerably to unify our understanding of genomes (22). For example, the duration of meiosis in A. thaliana has not been estimated. However, the regression in Fig. 2b predicts that it would take only ≈10 hr at 20°C. Note also that meiosis in species with very large genomes is of similar duration to the entire life cycle (≈4 weeks) in species with very small genomes, such as A. thaliana. From this evidence alone, it was predicted in 1972 that all of the species that can complete a life cycle in <6 weeks (known as ephemerals) must have very small genomes (23). Still, 25 years later, no exception to this prediction has been found. So knowledge of genome size and understanding of nucleotypic effects have considerable predictive values, which can help to unify our understanding of genomes.
Researchers continue to report new relationships between genome size and characters of interest for plant breeders and ecologists including frost resistance (24), biomass production (25), and the effects of a nuclear winter (26) or of global warming (27). For example, Fig. 2c shows the response of a mixture of five herbaceous species (including three grasses) with different genome sizes to winter warming of +5°C above ambient. Such results led Grime (27) to suggest that species with low DNA amounts will derive an advantage from warmer winter conditions. Another relationship (Fig. 2d) between responsiveness to elevated CO2 and genome size in seven annual grasses was noted by Jasienski and Bazzaz (25). They concluded that genome size potentially both can influence the responsiveness of a plant species to CO2 and be affected by CO2. Such relationships suggest that there may be significant practical benefits from an ability to manipulate genome size in crops. Surprising results reported recently by Price and Johnston (28) show that the 2C DNA amount in sunflowers (Helianthus annuus) varied 2.8-fold from 2.8 to 7.9 pg, depending on light quantity and quality. They concluded that the major factor responsible for inducing a change was the ratio of red to far-red light, and they suggested that phytochromes might be involved in the instability of DNA content in sunflowers, which may have adaptive significance. Such results, if confirmed, suggest a need to understand both the mechanisms responsible for modulating genome size and how to use them to advantage in crop plants.
Angiosperm DNA C Value Database.
Because genome size data impact on many fields, including systematics, molecular biology, and plant breeding, the available data must be easily accessible for analysis in a user-friendly form. Information on DNA amounts in different angiosperm taxa is often hard to locate because published data are widely scattered in diverse journals. Consequently, since 1976, several collected lists of DNA amounts in angiosperms have been published for reference purposes (17–19, 29–31).
Finding whether an estimate for a particular species is listed took longer as the number of such lists rose. To overcome this problem, it was decided to pool all of the data from the five separate lists into one combined list and to make the list available in both paper and electronic forms (31). A first version of the pooled list for ≈3,000 species was published electronically in 1997†. This list gives the 4C DNA amount for taxa in alphabetical order within genera, but it lacks many details given in the five separate lists, although users are directed to where these can be found. The Convention on Biological Diversity (32) noted the need to make biodiversity data available, despite imperfections, a view that merits support. Consequently, this database has been published in a limited form while more work is done.
Angiosperm Genome Size Workshop.
C value data have been used in a wide range of analyses, and several authors noted the need for more data. For example, Jasienski and Bazzaz (25) stated, “The analysis is limited to grasses due to paucity of published … measurements of DNA amounts in plants,” whereas Bennett and Leitch (19) noted that no DNA C value yet is known for >70% of angiosperm families.
There is clearly a need for more data on genome size in angiosperms, which must be accurate, of verifiable origin, and more representative of the global angiosperm flora. Meeting this challenge was one aim of a workshop on angiosperm genome size held at RBG Kew in September 1997. This workshop addressed aspects of best practice including: (i) the choice and supply of optimum calibration standards, (ii) standard protocols for estimating genome size, and (iii) ensuring that species identities are correct and verifiable. It also sought to identify and prioritize the major gaps (systematic, regional, and plant type, etc.) in our knowledge and to plan to fill them while measuring DNA C values for the next 1% of species.
The Angiosperm DNA C values database is one of several new plant genome initiatives at RBG Kew that involves international networks of partners. The most exciting, which will make a very significant contribution to the conservation of plant genetic diversity on a global scale, is the Millennium Seed Bank.
The Millennium Seed Bank Project.
We live in a golden age for understanding genes but a darkening age for biodiversity. It would be tragic if, just as we learn to transfer useful genes between genomes, we lose the species that contain them! Thus, plant genome initiatives must not be seen in isolation from plant conservation.
In 1972, at the first United Nations conference on the human environment, the conservation of crop genetic resources was identified as a necessary target to underwrite food security. Early experience of seed banking had focused mainly on a few crop species (33). It was then unknown whether the same technology would apply more widely to seed of wild species. RBG Kew’s Seed Bank was established at Wakehurst Place in Sussex to discover whether the seed banking of the ≈250,000 wild species, not addressed elsewhere, was feasible and appropriate (34). This initiative was a far-sighted action in 1974 when conservation was a voice crying in the wilderness. By 1992, when Agenda 21 at the Rio Earth Summit called for improved methods for the ex situ conservation of all the plant kingdom (35), RBG Kew had some answers, and the world’s largest and most diverse seed bank devoted to wild species conservation operating at internationally approved standards (36–38). Thus, the Bank contained 10,000 accessions of seeds from ≈4,000 species (39), representing 10 and 53% of the world’s plant genera and families, respectively.
Despite such progress, action about an order of magnitude greater clearly was needed to do justice to the problem. RBG Kew should not act alone, given heightened sensitivities on the ownership of genetic resources. An international approach was needed, in the spirit of the Convention on Biological Biodiversity that would build on RBG Kew’s experience and expertise in the field. After the U.K.’s ratification of the Convention on Biological Biodiversity (32) and the publication of a U.K. national action plan for biodiversity conservation (40), Kew concluded that a significant increase in efforts to collect, conserve, and research seeds of wild species in the U.K. and the world’s drylands was vital and decided to explore the feasibility of developing a global repository for seeds. The project would cost up to £80 million, but where might such large sums of money be raised? Then, in 1994, it was announced that money from the Millennium Commission, one of the distributors of proceeds from the new U.K. national lottery, would be available to fund large projects and visionary buildings to celebrate the year 2000. The timing and the sums available were highly opportune. RBG Kew bid to the Millennium Commission in April 1995. News of success—a grant now worth up to £30 million came in December 1995, subject to matching funds being raised from elsewhere.
RBG Kew’s Foundation and Friends organization was tasked with fund-raising for the project. In May 1996, H.R.H. The Prince of Wales launched the Millennium Seed Bank Appeal (Patron Sir David Attenborough) with a target of raising £7.3 million. Commitments of support from companies and trusts soon totaled >£5 million, including £2.5 million from the telecommunications company Orange. Over £300,000 also has been raised from Friends and individuals. In addition, the Wellcome Trust announced a grant of £9.2 million toward the Millenium Seed Bank project in March 1997. Thus, ≈£15 million of matching funding has been raised within one year.
This encouraging experience suggests an important lesson. When considering new initiatives, scientists should not limit their thinking to government funding. Companies, charitable trusts, and philanthropists can all have key roles to play, and a wide public involvement is desirable. Moreover, the Millennium Seed Bank project shows that a plant project can excite public interest and support. Plant science needs to relate more directly to the people, and a plant genome initiative can provide both a challenge and the opportunity for this relationship.
The Millennium Seed Bank project is one of a very few landmark projects, partly funded by the U.K. Millennium Commission, to celebrate the year 2000. The project is planned to cost up to £80 million over 14 years from 1997 to 2010. Its main aims, in brief, are:
- 1.
to collect and conserve living seed of almost all of the U.K. spermatophyte flora by the year 2000;
- 2.
to collect and conserve seed of an additional 10% of the world’s spermatophyte flora principally from the drylands by 2010;
- 3.
to carry out research to improve all of the aspects of seed conservation;
- 4.
to make seeds available for species reintroduction into the wild, for academic research, and for screening for potential new uses of plants;
- 5.
to encourage plant conservation throughout the world by facilitating access to, and transfer of, technology;
- 6.
to educate and involve the public in plant conservation; and,
- 7.
to provide a world class building, as the focus of this activity, by 2000.
The Wellcome Trust Millennium Building.
The project will construct the Wellcome Trust Millennium Building at RBG, Wakehurst Place, Sussex as the focus of the whole project. It will represent the foremost center in the world for research into seed conservation. Construction should begin in 1998, and the building should be completed and open to the public by 2000.
The Millennium Seed Bank building includes the following main elements:
- 1.
an underground vault for storing and protecting seeds of thousands of endangered species. Within this vault, cold stores will offer space for collections of ≈25,000 species with space for further cold stores allowing considerable expansion for perhaps an additional 10–20% of the world’s flora;
- 2.
laboratory facilities for receiving, cleaning, and processing seeds before their long term storage in the Millennium Seed Bank vault;
- 3.
laboratories for research aiming to improve seed collecting and storage techniques and for germinating plants from seed stored in the Bank; and
- 4.
facilities for visiting researchers, including accommodation for 30 visiting scientists from overseas.
Collecting the U.K. Spermatophyte Flora.
The project will first collect and conserve representative samples of living seed of all of the U.K.’s native plants, which set viable seed, by the year 2000. Excluding some microspecies that are not morphologically distinct in the genera Rubus, Hieraceum, and Taraxacum, the U.K. native spermatophyte flora numbers ≈1,442 species (43) of which Kew already holds 552, leaving a key target of 890 (of which a few may prove very difficult or impossible to collect or conserve). This collection will involve collaboration with many conservation organizations including English Nature, Scottish Natural Heritage, the Countryside Council for Wales, and the local Wildlife Trusts. Field work will span three field seasons (1997–1999).
No other country has conserved its entire flora in this way, and so this effort will not only allow the U.K. to fulfill its obligations to conserve its flora agreed under the Biodiversity Convention and to make it available for research but also will provide a valuable experience and a model from which others can benefit. It is hoped that this project will stimulate other countries to follow suit and act to conserve their own flora.
Collecting 10% of the World Spermatophyte Flora.
It would be impossible to collect seed of every plant species in the near future, so the project will concentrate on collecting 10% of the world’s spermatophyte flora (≈250,000 species; ref. 43) by the year 2010. The main planning phase will be 1997–1999; the main collecting phase will be 1999–2009. The project will increase the number of collectors from 2 (currently) to 27 (in the year 2000) including 5 (and a coordinator) based at the Millennium Seed Bank and 22 collectors overseas in partner countries. Collecting will be carried out under bilateral agreements that cover profit-sharing as a result of intellectual property rights, in accordance with the Convention on Biological Diversity (32).
Targeting.
The seed collecting will be highly targeted. Countries have been targeted for partnership on the basis of existing collaboration, ease of access, extent of arid, semi-arid, and dry subhumid land, number of endemic plant species, and the floristic region in which the collecting occurs. On this basis, 18 countries have been identified as priorities. One key target will be species of known value to human welfare adapted to dryland conditions, including those listed by RBG Kew’s SEPASAL database (9). Another will be species of medicinal value, whereas a third key target will be endemic species.
Seed Research.
The seed research program of the Millennium Seed Bank will support the aims of the global seed collecting program and both elements of the conservation program: processing and storage. Seed banking is often still an act of faith. For such taxonomically and geographically wide seed banking activity, strong research support is essential because seed of most species that RBG Kew collects and banks has not been examined before by science. A compendium of known storage behavior of the world’s seeds published recently (38) listed only 6,000 species (just >2% of 250,000). Of those, ≈40% are known only through research at RBG, Wakehurst Place.
Most species are desiccation tolerant (or orthodox) and will be bankable. However, two other categories (recalcitrant and intermediate) are recognized, which may account for 20% of the world’s species, for which banking is difficult or impossible because drying kills their seed. Finding diagnostics for them and developing new techniques to overcome these problems are two highly important research objectives. Whereas research will focus strongly on supporting practical and applied project targets and collaborations, the Millennium Seed Bank also will provide unique opportunities for other exciting developments in seed science and should become an important resource center for new research leading to greatly increased knowledge of seed biology and understanding of the molecular mechanisms that determine different patterns of seed development, germination, and storage behavior.
Distribution of Seeds to Users.
Apart from the conservation role, the Millennium Seed Bank will act as a global resource for research and trials. Seed material held in the Millennium Seed Bank will be made available to all bona fide researchers under the terms and conditions of the Convention on Biological Diversity. Fair and equitable sharing of any benefits arising out of the use of seed will be guaranteed.
Technology Transfer.
While acting as the central facility, the Millennium Seed Bank also will promote the ex situ conservation of plant genetic resources in their country of origin by encouraging access to and transfer of technology for collaborating countries that currently lack adequate facilities or expertise. This technology transfer will involve two main elements: first, advisory visits by the Millennium Seed Bank staff to collaborating countries and second, opportunities for scientists from such countries to study and use the facilities in the Millennium Seed Bank.
Public Education.
Educating the public as to the importance of preserving the Earth’s plant life is essential, and this aim forms a crucial aspect of the project. Over 250,000 people currently visit RBG, Wakehurst Place each year. Visitors to the Millennium Seed Bank will be able to use public walkways to observe seed scientists at work, use interactive exhibits that emphasize the vital role of seed banking in conservation, and view a glasshouse displaying some of the U.K. and dryland plants stored in the Bank.
Values and Vision.
What is the value of each banked plant genome? Time will show. The answer certainly will transcend purely economic terms. However, the cost per seed banked will be ≈5 pence (≈8 cents). The cost of maintenance will be ≈£15 (≈$25) per species per decade, whereas the cost of collecting, processing, and banking the seed of a species will be ≈£950 (≈$1,500). So for the same cost, it is possible to insure one or two automobiles for one year or a species for centuries and, in some cases, for the entire next millennium.
The Millennium Seed Bank will make a very significant contribution to the conservation of plant genetic diversity on a global scale and will provide an immensely valuable resource for future generations. The project is timely and truly of the millennium. Thus, the Millennium Seed Bank is a plant genome initiative that marks our concern and hope today and may become a visionary gift of life to the peoples of tomorrow.
ABBREVIATIONS
- RBG
Royal Botanic Gardens
- SEPASAL
Survey of Economic Plants for Arid and Semi-Arid Lands
Footnotes
Bennett, M.D., Cox, A.V. & Leitch, I.J. (1997) http://www.rbgkew.org.uk/cval/database1.html.
References
- 1.Cohen J. Science. 1997;275:769. [Google Scholar]
- 2.Heslop-Harrison J S. J Cell Sci. 1991;100:15–21. [Google Scholar]
- 3.Goodman H M, Ekers J R, Dean C. Proc Natl Acad Sci USA. 1995;92:10831–10835. doi: 10.1073/pnas.92.24.10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Index Kewensis (1997) Oxford Univ. Press, Oxford.
- 5.Dransfield J, Beentje H. The Palms of Madagascar. Kew and the International Palm Society, Kew, U.K.: Royal Botanic Gardens; 1995. [Google Scholar]
- 6.Iltis H H, Doebley J F, Guzman M, Pazy B. Science. 1979;203:186–188. doi: 10.1126/science.203.4376.186. [DOI] [PubMed] [Google Scholar]
- 7.World Conservation Monitoring Centre. Global Biodiversity: Status of the Earth’s Living Resources. London: Chapman & Hall; 1992. [Google Scholar]
- 8.Smith, D. M., May, R. M., Pellew, R., Johnson, T. H. & Walter, K. S. Nature (London) 364, 494–496.
- 9.Davis S D, Sinclair N J, Cook F E M. Proceedings of the Fifth International Rangeland Congress. UT: Salt Lake City; 1995. 1, 111–112. [Google Scholar]
- 10.Clapham A R, Tutin T G, Warburg E F. Flora of the British Isles. Cambridge, U.K.: Cambridge Univ. Press; 1952. [Google Scholar]
- 11.Bennett S T, Leitch I J, Bennett M D. Chromosome Res. 1995;3:101–108. doi: 10.1007/BF00710670. [DOI] [PubMed] [Google Scholar]
- 12.Crossland M W J, Crozier R H. Science. 1986;231:1278. doi: 10.1126/science.231.4743.1278. [DOI] [PubMed] [Google Scholar]
- 13.Leutwiler L S, Hough-Evans B R, Meyerowitz E M. Mol Gen Genet. 1984;194:15–23. [Google Scholar]
- 14.Hair J B, Beuzenberg E L. Nature (London) 1961;189:160. [Google Scholar]
- 15.Robertson B L. J South Afr Bot. 1976;42:97–108. [Google Scholar]
- 16.Johnson M A T, Kenton A Y, Bennett M D, Brandham P E. Genome. 1989;32:328–333. [Google Scholar]
- 17.Bennett M D, Smith J B. Philos Trans R Soc London B. 1976;274:227–274. doi: 10.1098/rstb.1976.0044. [DOI] [PubMed] [Google Scholar]
- 18.Bennett M D, Smith J B. Philos Trans R Soc London B. 1991;334:309–345. [Google Scholar]
- 19.Bennett M D, Leitch I J. Ann Bot (London) 1995;76:113–176. [Google Scholar]
- 20.Van’t Hof J, Sparrow A H. Proc Natl Acad Sci USA. 1963;49:897–902. doi: 10.1073/pnas.49.6.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bennett M D. Philos Trans R Soc London B. 1977;277:201–277. doi: 10.1098/rstb.1977.0012. [DOI] [PubMed] [Google Scholar]
- 22.Bennett M D. In: Unifying Plant Genomes: Symposia of the Society for Experimental Biology. Heslop-Harrison J S, editor. Cambridge, U.K.: Society for Experimental Biology; 1996. , No. L, pp. 45–52. [Google Scholar]
- 23.Bennett M D. Proc R Soc London B. 1972;181:109–135. doi: 10.1098/rspb.1972.0042. [DOI] [PubMed] [Google Scholar]
- 24.MacGillivray C W, Grime J P. Funct Ecol. 1995;9:320–325. [Google Scholar]
- 25.Jasienski M, Bazzaz F A. Nature (London) 1995;376:559–560. [Google Scholar]
- 26.Grime J P. Int J Environ Studies. 1986;28:11–19. [Google Scholar]
- 27.Grime J P. Aspects Appl Biol. 1996;45:3–13. [Google Scholar]
- 28.Price H J, Johnston J S. Proc Natl Acad Sci USA. 1996;93:11264–11267. doi: 10.1073/pnas.93.20.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennett M D, Smith J P, Heslop-Harrison J. Proc R Soc London B. 1982;216:179–199. [Google Scholar]
- 30.Arumuganathan K, Earl E D. Plant Molec Biol Rep. 1991;9:208–219. [Google Scholar]
- 31.Bennett M D, Leitch I J. Ann Bot (London) 1997;80:169–196. [Google Scholar]
- 32.United Nations Environment Programme. Convention on Biological Diversity. Nairobi, Kenya: United Nations Environment Programme; 1992. [Google Scholar]
- 33.Frankel O H. The Significance, Utilisation and Conservation of Crop Genetic Resources. Rome: Food Agric. Org. U.N.; 1971. [Google Scholar]
- 34.Thompson P A. Biol Conserv. 1974;6:15–19. [Google Scholar]
- 35.Quarrie J, editor. Earth Summit ‘92. The United Nations Conference on Environment and Development. London: Regency Press; 1992. [Google Scholar]
- 36.Prendergast H D V, Linington S, Smith R D. In: Desertified Grasslands—Their Biology and Management. Chapman G P, editor. London: Academic; 1992. pp. 235–250. [Google Scholar]
- 37.Smith R D. In: Collecting Plant Genetic Diversity: Technical Guidelines. Guarino L, Ramanatha Rao V, Reed R, editors. Wallingford: CAB International; 1995. pp. 419–456. [Google Scholar]
- 38.Hong T D, Linington S, Ellis R H. Seed Storage Behavior: A Compendium Handbooks for Genebanks. No. 4. Rome: International Plant Genetic Resources Institute; 1996. [Google Scholar]
- 39.Linington S H. List of Seeds 1994. Kew, U.K.: The Royal Botanic Gardens; 1994. [Google Scholar]
- 40.Anonymous. Biodiversity, The UK Action Plan. London: Her Majesty’s Stationery Office; 1994. p. 188. [Google Scholar]
- 41.Kent D A. List of Vascular Plants of the British Isles. London: Botanical Society of the British Isles; 1992. [Google Scholar]
- 42.Mabberly D J. The Plant Book. Cambridge, U.K.: Cambridge Univ. Press; 1990. [Google Scholar]