Abstract
Retroelements with long-terminal repeats (LTRs) inhabit nearly all eukaryotic genomes. During the time of their rich evolutionary history they have developed highly diverse forms, ranging from ordinary retrotransposons to complex pathogenic retroviruses such as HIV-I. Errantiviruses are a group of insect endogenous LTR elements that share structural and functional features with vertebrate endogenous retroviruses. The errantiviruses illustrate one of the evolutionary strategies of retrotransposons to become infective, which together with their similarities to vertebrate retroviruses make them an attractive object of research promising to shed more light on the evolution of retroviruses.
Keywords: Drosophila, Gag, endogenous retroviruses, errantivirus, flamenco, gypsy, retroelement
Introduction
The evolutionary relationships of retroelements, their polymorphism, transposition and proliferation strategies as well as the cell control mechanisms of their activity are of great interest due to their major impact on the genome stability in eukaryotes. They participate in insertional mutagenesis, epigenetic processes and are able to provoke chromosomal aberrations.1-5 On the other hand, retroelements have been shown to interact with genome in a mutualistic manner and act as a source of new genes.6,7
LTR-retroelements include several groups: exogenous retroviruses, endogenous retroviruses (ERVs) and LTR-retrotransposons. During evolution three possible strategies of retroelement existence were formed. The first one is based on the “copy and paste” mechanism of genome colonization. It is the most ancient strategy which is used by classic LTR-retrotransposons. They become integrated into the genome and are usually insufficiently genetically equipped to form functional viral particles, enveloped structures, that contain element’s genome and can bud from one cell and enter another, and behave as infectious retroviruses.8 Genetic material of LTR-retrotransposons is transferred vertically via the germ line of the host organism. Exogenous retroviruses demonstrate the opposite approach—they reproduce via infection and their genetic material is transferred horizontally. The reproduction of ERVs can potentially occur by both mechanisms.9 They transfer via host germ line and at the same time they may still be transferred horizontally.10
Exogenous retroviruses are widespread among vertebrates and are quite rare among the invertebrate species. The first invertebrate retrovirus-like infective agent was identified in Drosophila melanogster genome when it was shown that thoroughly studied retrotransposon gypsy has the infective ability.11,12 Subsequently similar properties were reported for other Drosophila retrotransposons such as ZAM and Idefix.13 The latter ones along with gypsy became first members of the group of insect ERVs—errantiviruses.14
Some ERVs evolved from retroviruses by losing infectivity and occupying host germ line. Another group of ERVs gained the retrovirus-like features, such as ability to leave one cell and enter another cell de novo and didn’t have infectious retroviral ancestors. (Fig. 1) Apparently errantiviruses belong to the second category.
Figure 1.
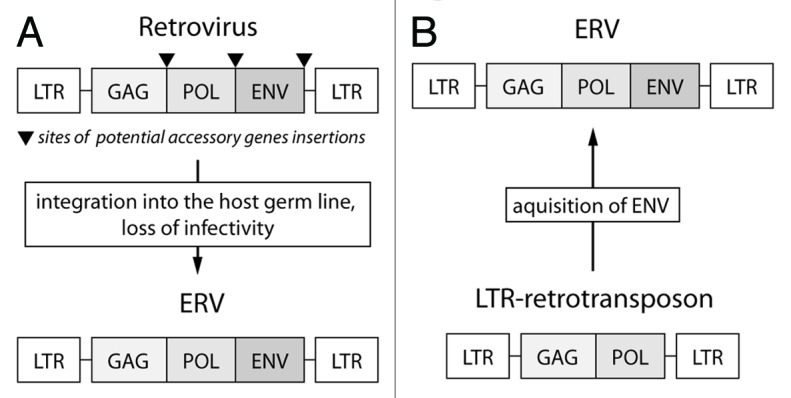
Evolutionary pathways that can lead to the origin of endogenous retroviruses. (A) Strategy that leads to formation of the most vertebrate endogenous retroviruses. In this case infectious retrovirus is loosing its infectivity, enters the germline and transfers vertically. (B) Aquisition of the env gene—a mechanism, by which errantiviruses gained the properties of endogenous retroviruses.
According to the International Committee on Taxonomy of Viruses (ICTV) both exogenous and endogenous retroviruses of vertebrates are part of the Retroviridae family whereas invertebrate retroviruses (as well as some LTR-retrotransposons of Ty3/Gypsy family, plant and fungi ERVs) belong to the Metaviridae.15 Ty1/Copia retrotransposons fall into the Pseudoviridae family.15
Errantiviruses and other representatives of Metaviridae did not evolve to full-fledged viruses, keeping their reproductive strategy more similar to that used by retrotransposons. However, these elements are abundant in insect genomes and their life cycle and interactions with cell control mechanisms draw attention and in a perspective may shed light on the origin, evolution and behavior of all retroviruses.
Structure and Classification
The structure and interactions of errantiviral proteins are not as thoroughly described as for the human retroviral proteins. This can largely be explained by the fact that human HIV is the dominant focus of research in retroviruses. Thus, studies of errnativiruses provide new insight into the evolution of retroviruses beyond studies of HIV.
Genes and proteins
The ability of the LTR-retroelement to infect different cells depends on the functionality of a basic set of the major genes, and on whether the element has additional genes and mechanisms that increase its infectivity.
A typical retrovirus contains two direct long-terminal repeats, some untranslated fragments and three open reading frames (ORFs)—gag, pol and env, which are responsible for the element virus-like particle formation, its retrotranscription ability, and potential infectivity, respectively.16 Gag gene is responsible for the synthesis of structural proteins that fold and protect element’s RNA and form a virus-like particle. Pol is a gene that produce enzymes required for the reverse transcription and integration of the element into the host cell genome. Env encodes surface protein that is integrated into the viral particle membrane and helps virus to enter the host cell.
Gag
The first function of Gag protein is the formation of the complex with retroelement RNA where the reverse transcription takes place. Also, Gag proteins mediate the nuclear transfer of the preintegration complex of the retroelement. This basic set of functions is sufficient for retrotransposition. But in retroviruses Gag proteins also bind to the cell membrane and mediate the virus budding.17
Canonical retroviral Gag is usually expressed as a precursor protein, which is cleaved by virus-encoded protease into the three separate proteins during the virus maturation. The first one is MA (matrix). This domain is commonly myristoylated which makes its binding to the membrane effective. In mature viral particles MA forms the outer shell that lies right beneath the membrane, which is captured from the host cell. CA (capsid) protein forms a hydrophobic cone-shaped shell surrounding the ribonucleoprotein complex that contains the genomic RNA, reverse transcriptase, integrase and tRNA primer. Viral RNA packaging is mediated by the NC (nucleocapsid) protein which is also one of the gag products.18
Gag proteins of most retroviruses carry several conservative structural motifs. The CA domain usually contains so called major homology region, which is represented by QG-X2-E-X5-F-X2-L-X2-H aminoacid consensus.18 MHR is sufficient for the correct capsid formation and virus particle assembly.19 NC domain is characterized by the presence of the zinc-finger motif - Cys-X2-Cys-X4-His-X4-Cys, necessary for the RNA binding.20
Errantiviral Gag is capable of mediating the assembly of extracellular virus-like particles21 but demonstrates some significant differences from the canonical Gag protein. First of all, it lacks the conservative motifs that are commonly found in retroviral Gag protein, and instead incorporates alternative fragments that allow to maintain required functions. In particular, the N-terminal domain of gypsy Gag is not myristoylated, but, according to some data,22 gypsy Gag carries the motif that may interact with proteins that mediate clathrin-dependent protein sorting in the endocytic and secretory pathways.23 It was shown that an equivalent motif is required for the budding of the equine infectious anemia virus (EIAV).24 Second, instead of zinc-finger motif nucleocapsid-like (NC) domain of errantivituses contains the arginine rich region (RRNSSERSTGPRRQR). It is considered that this consensus in NC domain is responsible for RNA binding.25
Finally, the remarkable feature of the errantiviral Gag is that it is apparently not cleaved into the separate МА, СА and NC proteins although some studies show that it may be processed in some other way.26,27 All these peculiarities are also inherent to foamy viruses that are the only members of the retroviral subfamily Spumaretrovirinae. Foamy viruses infect nonhuman primates and are both highly prevalent and highly transmissible. However, these viruses are not known to cause any disease in infected host.28,29 The biology of foamy viruses is more well studied than the biology of errantiviruses (for the review see ref. 30). Spumaretroviral Gag is not cleaved and not myristoylated and also does not carry MHR and zinc fingers. At the same time at the C-terminus of this protein, three boxes rich in glycines and arginines have been identified. It was recently proposed that the first box plays a role in the encapsidation of foamy viral Pol protein.31 The second box was identified to be a nuclear localization signal and also have the ability to interact with histones and chromatin.32-34
There is an additional similarity between foamy viruses and errantiviruses. The assembly of the most retroviral Gag monomers into capsids takes place at a host membrane, while some retroviruses employ different pathway that is characterized by formation of capsids in the cytoplasm of the host cell, independent of membranes. The second variant is utilized by foamy viruses and some orthoretroviruses such as Mason-Pfizer monkey virus (MPMV). Some data indicates that errantiviruses use the same strategy. For example, it was shown that the Gag protein of gypsy retroelement can form multimeric complexes and virus-like structures in vitro.35 Foamy viruses Gag contains a cytoplasmic targeting/retention signal (CTRS) located within the N terminus of the Gag protein.36 It directs Gag assembly to a pericentriolar location, as in MPMV, and is crucial for the production of infective virus particles.37-39 Unfortunately little is known about the existing of such signal sequences in errantiviruses.
Gag protein of errantiviruses contains regions reflecting its possible posttranslational modifications. In particular it carries a number of glycosylation and phosphorylation sites. It also contains the potential site for interacting with SUMO (Small Ubiquitin-like Modifier) protein.22 This protein participates in the intracellular trafficking and mediates the nuclear-cytosolic transport. Interaction with SUMO was demonstrated for some retroviruses.40 The exact role of potential SUMOylation in the errantiviral life cycle remains unknown. Also it is not known whether errantiviruses interact with ESCRT (endosomal sorting complex required for transport) and plasma membrane microdomains such as lipid rafts during their budding and VLP release, as it was shown for retroviruses.41,42 However some data concerning ZAM element indicate that transfer of ZAM particles rely on the use of the endosomal and exosomal pathways that in Drosophila ovaries are normally employed for vitellogenin release and uptake.43
Pol
Pol encodes the enzymatic machinery of the retroviruses. The major component of this reading frame is the reverse transcriptase (revertase) domain. Pol products are responsible for the reverse transcription, integration into the host genome and processing of the structural proteins during virus particle maturation. This ORF is necessarily present in all active retroelements. In view of its great importance pol is the most conservative reading frame in retroviral genomes.
Pol consists of protease, reverse transcriptase, RNase H and integrase. In contrast to Ty1/copia retroelements (Pseudoviridae), retroviruses carry these domains in this order. In Pseudoviridae integrase domain stands after the protease (for the review see refs. 44 and 45). In some cases protease may be encoded separately or within the Gag precursor.16
Protease is necessary for the Gag and Gag-Pol precursors cleavage. As it was previously mentioned for the Gag, the protease of errantiviruses shows high similarity with the protease of foamy viruses. The most conservative motifs of this enzyme in errantiviruses are closer to the foamy viruses than to the pseudoviruses.46
Other domains of pol ORF are represented by the ancient enzymes. It is generally accepted that reverse transcriptase (RT) arose in the period of the transition from the RNA world to the DNA-based genomes. In LTR-retroelements reverse transcriptase usually works in complex with RNase H domain. Phylogenetic trees based on sequences of these domains suggest that all LTR-retroelements are a monophyletic group.47 RT has a core of 180 aminoacids that is highly conservative for all retroelements.48 However the RNase H sequences of Retroviridae members are highly divergent from those of all groups of LTR retrotransposons.49 In retroviruses RNase H domain and reverse transcriptase are separated by a tether domain, which may indicate that the retroviruses have acquired a new RNase H domain downstream of the ancestral domain with the ancestral domain degenerating to become the tether (discussed in ref. 44). Published data suggests that RT and RNAase H sequences of errantiviruses are closer to TY3/Gypsy LTR-retrotransposons than to Retroviridae members.46 RT and RNase H of errantiviruses are well-adapted to the temperature of its host.50
Integrase of errantiviruses is characterized by the absence of the GPY/F motif that is inherent to the most of retroviral families.46 This is another illustration of the errantiviruses and spumaviruses relatedness. It is believed that these two groups of LTR-retroelements lost this motif during evolution. It is noteworthy, that integrase of several errantiviruses demostrates entry-site specificity, that is not the common feature of LTR-retroelements.51,52 Moreover, it was shown that gypsy integrase, unlike the integrase of spumaviruses, can carry out the reverse reaction: precise excision of the element DNA from the genome.53 This fact may indicate the evolutionary relation between retroelement integrases and transposases from DNA-transposons.54,55
Env
The third important ORF in retroviral genome is env. This ORF encodes the transmembrane glycoprotein that is capable of target cell receptor recognition and mediates the viral and host membrane fusion. It is considered that the LTR-retroelement without this gene cannot propagete via infection. Env protein consists of transmembrane and surface domains. In mature viral particles they are not-covalently connected. Env cleavage is mediated by cellular endopeptidases which recognize the conservative RIAR motif in its primary structure.56
Env gene has a complicated evolutionary history. It is considered that this activity could be obtained by retroviruses as a result of fusion proteins capturing from other infectious viruses. Furthermore there is some evidence that this event might took place several times during evolution. This could be the reason why the env genes of distant retroelements may not share the common ancestor.57,58
Some data indicates that env of errantiviruses was captured from the genome of baculoviruses, the large group of insect viruses, via some recombination event. This conclusion is based on the sequence similarity between baculoviral fusion protein Ld130 and errantiviral Env.57 It is shown that Env of errantiviruses contains potential glycosylation sites, however it is apparently cleaved at the non-canonical site.59
It is noteworthy that not all errantiviruses carry functional third ORF. In some elements it is either broken or completely lost. This suggests that this gene is not crucial for the life cycle and propagation strategy of errantiviruses.60
Accessory Genes and Regulatory Elements
Most of the retroviruses carry additional genes and ORFs, which enhance their infectious ability. For example, HIV-1 carries genes, responsible for regulation of viral proteins expression, protection from the cellular anti-viral mechanisms (such as APOBEC 3G protein—a human innate factor, restricting viral propagation),61,62 or may even interact with a cell cycle and differentiation regulation.63 Activity of such genes provides LTR-retroelements with a major advance in effectiveness in comparison with the elements lacking such regulatory genes.
Existence of such “upgrades” is not shown for the errantiviruses, however some of them carry IRES sequences (internal ribosome entry site—a sequence initiating translation without scanning all of the prior 5′ mRNA),64 which are well known for being used by viruses.65 The most well-studied regulating sequence found in errantiviral genomes is the insulator in 5′ untranslated region (UTR) of gypsy element.66 The gypsy insulator is a 340 bp long sequence, containing 12 binding motifs for the 12 zinc finger Supressor of Hairy-wing [Su(Hw)] protein.67,68 Gypsy insulator performs both enhancer blocking and barrier functions,69 that require recruitment of different set of proteins (for the review, see ref. 70), and using them provides gypsy insulator with a versatile capacity of defining regulatory interchanges throughout the Drosophila genome.
One of the most well studied mutations, caused by the gypsy insulator, is the y2 allele of the yellow gene. This gene expression is controlled by a number of tissue-specific enhancers. y2 mutants contain an insertion of gypsy 700 bp upstream from the transcription start, which leads to blockage in the interaction of promoter and body and wing enhancers of the gene. Several works demonstrate, that mutation in su(Hw) gene or insertion of some element into the insulator sequence of gypsy leads to reversion of mutation.71,72 The gypsy insulator also interacts with a Mod(mdg4) and Cp190 proteins.73,74 This interaction is very strong and has been exploited through usage of the gypsy insulator as a powerful instrument for transgene shielding.75
It is possible, that the presence of insulator sequence, within the mobile element, may give this element some advantage in case if the element is inserted in the region of the genome, where the insulator appears to be useful to the host. But insulator is more likely to affect element’s ability to propagate and spread throughout the genome negatively, since increase in copy number of the element, that is able to cause mutations not only by insertion into the coding region but also by effecting the expression of neighboring genes, can be much more deleterious for the genome, then transpositions of the element carrying no regulatory sequences.
Other errantaviruses, besides gypsy, carry additional regulatory sequences, for example ZAM and Idefix.76 The gypsy-like element gtwin also contains a series of short repeats in it’s 5′ UTR, identified as zinc-finger motifs, but there are only 6 of them, which is not enough to compose an insulator. Supposedly, gtwin ancestor had a regulatory element, which was lost throughout the evolution of the element. Some data indicates, that ZAM element carries not an insulator, but an enhancer sequence.76 As opposed to the gypsy retroelement, for which the insulator interacts with Su(Hw), Mod(mdg4), and Cp190 proteins, ZAM's regulatory element interacts with HP1 protein.77 It is also known, that Idefix insulatior has not only enhancer-blocking, but also a barrier activity.78
Classification, Distribution and Individual Features of Errantiviruses
The place of Drosophila errantiviruses in systematics may be discussed in terms of both virus classification15 and transposable element classification.79 Neither system fully reflects the evolution of retroelements, and both tend to use functional criteria in classification. Systematics of mobile elements is still a subject for discussion (see refs. 80 and 81), and a final, well-established classification system reflective of phylogeny has yet to emerge. However, there are several works that allow us to estimate the place of errantiviruses in the evolution of retroelements, which combines processes of phylogenetic divergence with episodes of modular, saltatory, and reticulate evolution.82 As it was mentioned earlier, according to ICTV, errantiviruses are representatives of the Metaviridae family, which also includes semotiviruses (also reffered to as Bel/Pao retrotransposon family) and metaviruses (retroelements, spread among plants and fungi, related to Saccharomyces cerevisiae Ty3 retrotransposon).15 From the transposable elements classification system point of view, errantiviruses are part of the large Ty3/Gypsy superfamily of LTR-retrotransposons.79 The closest relatives of errantiviruses among retrotransposons, according to the phylogeny based on pol ORF, are Drosophila retrotransposon families 412 and Mdg1.82 Representatives of these families do not carry the env gene.83,84
Classification of errantiviruses is not entirely straightforward. Some of the difficulty can be explained by the fact that the errantivirus classification system is based mostly on the Pol phylogeny and that the system largely ignores the fact that some elements do not carry a functional env gene (see Table 1) and thus may not formally be considered Metaviridae members. Moreover, copies of one element found in genomes of different species may exhibit marked differences from one another; their similarity is usually not much greater than that of different elements occupying one genome. According to some phylogenetic studies, one element may be placed within the phylogeny of another, as demonstrated by gypsy and gtwin.87 In fact, we observe the unstructured set of accumulated orthologous and paralogous genetic elements that are subjected to various recombination events and deleterious effects. Sometimes it may be difficult to distinguish a functional copy of the element from a divergent “genomic fossil” that has lost its ability to transpose, which is fixed in the genome and passed from generation to generation like a pseudogene. However, LTR-retroelements demonstrate relatively low frequency of fixed insertions in Drosophila melanogaster.88
Table 1. LTR-retroelements referred to as errantiviruses by different authors.
| Element | Group | tRNA primer | Demonstrated infectivity | Env is lost | Presence in flamenco locus | Described in different Drosophila species | FlyBase ID | References |
|---|---|---|---|---|---|---|---|---|
| Nomad |
Gypsy |
Lys |
|
|
* |
|
FBte0000918 |
85, 86, 140 |
| Burdock |
Gypsy |
Lys |
|
* |
|
* |
FBte0000739 |
86 |
| Springer |
Gypsy |
Lys |
|
|
|
|
FBte0000333 |
86 |
| Gypsy |
Gypsy |
Lys |
* |
|
* |
* |
FBte0000021 |
15, 81, 82, 140 |
| Gypsy 2 |
Gypsy |
Lys |
|
|
* |
|
FBte0001040 |
127 |
| Gypsy 3 |
Gypsy |
Lys |
|
|
* |
|
FBte0001030 |
127 |
| Gypsy 4 |
Gypsy |
Lys |
|
|
* |
|
FBte0000688 |
127 |
| Gypsy 6 |
Gypsy |
Lys |
|
|
* |
|
FBte0001175 |
127 |
| Gtwin |
Gypsy |
Lys |
|
|
* |
* |
FBte0001062 |
127 |
| Beagle |
Gypsy |
Lys |
|
* |
* |
|
FBte0000726 |
86 |
| |
|
|
|
|
|
|
|
|
| ZAM |
17.6 |
Ser |
* |
|
* |
* |
FBte0000217 |
15, 85, 86, 140 |
| Rover |
17.6 |
Ser |
|
|
|
* |
FBte0000692 |
140 |
| Idefix |
17.6 |
Ser |
* |
|
* |
|
FBte0000104 |
15, 85, 86, 140 |
| 17.6 |
17.6 |
Ser |
|
|
* |
|
FBte0000109 |
15, 85, 86, 140 |
| 297 |
17.6 |
Ser |
|
|
* |
|
FBte0000675 |
15, 85, 86, 140 |
| Tv1 |
17.6 |
Ser |
|
|
|
|
FBte0000783 |
15, 86, 86 |
| Tom |
17.6 |
Ser |
|
|
* |
* |
FBte0000773 |
15, 85, 86, 140 |
| Tirant |
17.6 |
Ser |
|
|
* |
* |
FBte0000179 |
15, 85, 140 |
| Quasimodo |
17.6 |
Ser |
|
|
* |
* |
FBte0000640 |
140 |
| Yoyo |
Non-drosophila element |
Lys |
|
|
n/a |
n/a |
n/a |
15, 85, 86 |
| Ted | Non-drosophila element | Ser | n/a | n/a | n/a | 15, 85, 86 |
Errantiviruses are not limited to Drosophila; they are found in the genomes of various arthropods, as well.89-91 These elements are, on average, found in low copy number, often represented by only one or two copies. Even in a case of element amplification, the number of copies usually does not exceed two or three dozen. Some errantiviruses may be absent in certain species or strains of Drosophila.92 For example, elements 297, Tom and rover seem to be restricted to species from the D. melanogaster group.93 Furthermore, no homologous sequences to the env gene of the gypsy, gypsy2, gypsy3, gypsy4, and gypsy6 retroelements were found in D. grimshawi and D. ananassae.87
Errantiviruses, as well as other LTR-retroelements, contain the short sites necessary for reverse transcription priming and the beginning of DNA (+) strand synthesis. These are the tRNA primer binding site (PBS) located downstream to 5′LTR and the polypurine tract (PPT) observed upstream from 3′LTR. PBS is not conservative. Among errantiviruses there are two groups of elements: the Gypsy group and the 17.6 group, which use tRNALys and tRNASer, respectively, as primers for reverse transcription.85 Currently, approximately 15 to 20 (according to different authors) different errantiviruses are described in Drosophila (Table 1).
In light of the ever-increasing number of whole genome sequences, we expect the list of LTR-retroelements and errantiviruses to continue expanding. Currently there are a number of software tools that allow the prediction and annotation of LTR-containing elements in genomes. The search strategy is based on the annotation of LTRs, PBS, and conservative domains of the gag, pol and env genes.94-97
Activity and Regulation
Activity
Despite the fact that errantiviruses carry the env gene which is responsible for the retroviral infectious ability, the evidence for their infectivity are indirect because it is difficult to manipulate errantiviruses with commonly used virological techniques such as titering or neutralizing with antibodies. Also no cytopathogenic effects of errantiviruses have been described. Apparently errantiviruses are significantly less infective than the majority of insect and Drosophila viruses.98,99
Potential errantiviral infectivity was demonstrated as a result of experiments with feeding larvae that carried no active gypsy element in their genomes with the extract from embryos of “gypsy positive” flies or the purified virus-like particles of this element.11,12 In addition, using cell cultures it was shown that gypsy particles from Drosophila melanogaster are able to enter the cells of distinct species such as Drosophila hydei with subsequent amplification of the element and the reinfection of new cells.100 The role that Env protein plays in the errantivirus life cycle and infectivity is not entirely clear because in cell culture experiments the ability to enter the cytoplasm is demonstrated by the particles that carried no surface glycoproteins.101 Furthermore it is generally accepted that Env is not necessary for the retrotransposition.102
Errantiviruses attract attention not only because of their potential infectivity. They can also multiply inside the genomes via “copy and paste” pathway. These commonly low-copied elements in some cases escape from the host control mechanisms and start to transpose with a significant frequency.103,104 Some data indicate that there are so called hot spots for the integration of several elements. For example gypsy, in case of its activation in the genome, enters cut, forked and ovo loci with a much higher frequency than other places in genome.105-107
Retrotransposition activity of errantiviruses may be altered as a response to the structural changes. Thus for the gypsy element the existence of two functionally distinct forms was demonstrated.108 At the nucleotide level, differences between two variants of the element involve single substitutions, deletions and insertions that does not dramatically affect the proteins structure, however the activity of these variants differ fundamentally.109 The similar situation was demonstrated for the two variants of the gtwin element. They carry polymorphic nucleotides in the PBS. Remarkable feature is that the element with functional tRNALys PBS did not transpose in the genome of the Drosophila melanogaster strain G32 but the variant with two substitutions in this site, decreasing the primer annealing efficiency, is amplified.104 Phylogenetic studies of the 17.6, 297, rover and Tom retroelements demonstrate that these errantiviruses also have different subfamilies.93
Extensive data concerning the distribution and polymorphism of errantiviruses in different Drosophila strains and species is accumulated.110,111,46,112 Moreover, there is a growing amount of data indicating the significant rate of horizontal transfer of errantiviruses between evolutionary distant Drosophila species.113,114 The role of the env gene in this process is yet unclear. The analysis of the reported and putative horizontal transfer events does not demonstrate any increase of its rate for the elements with the env ORF. The possible mechanisms of the horizontal transfer may not be associated with retroviral infectivity, although virus-like particles may be considered as a transfer vectors (for the review see ref. 115).
The host genome can be broadly considered as the environmental niche for ERVs and transposable elements.116,117 According to the dominant existing model, the proliferation of transposable elements in genomes is controlled by number of factors. The introduction of a new element to the cell is commonly followed by its transpositions, amplification and possible alterations. The element cannot propagate permanently because TEs are likely to cause deleterious effects and decrease the host fitness.118-121
Species with high amount of novel TE insertions are commonly eliminated from the population by the natural selection (for review, see ref. 122). Furthermore, the host cell has its own pathways allowing to restrict the TEs activity. For this reason elements always reach equlibrium copy number and after that tend to be eliminated from the genome. In some cases, elements can be subjected to a positive natural selection and remains in the genome if it appears to be somehow useful for the host.123
For a long time little was known about the specific pathways of cellular response to the transposable elements activation and it was thought that the equilibrium copy number of mobile elements in the genome is achieved solely by a balance between the forces of transposition, deletion, and natural selection. The discovery of RNA interference and its profound study in different fields and objects shed light on the question of how the cell takes mobile elements’ transpositions under the control. In this context study of errantiviruses biology is of great interest because they demonstrate the peculiar mechanism of propagation in the germ line. Moreover, some Drosophila species have developed the special pathway to control their activity.
Such errantiviruses as gypsy, ZAM and Idefix are not produced in oocytes or nurse cells like many other transposable elements do.124,125,13 Instead, their expression is restricted to somatic follicle cells. Transcripts of these elements are accumulated inside the follicle cells and then targeted to the female germ line cells. This process appears to be independent of the env gene. Chalvet and co-authors demonstrated that env-defective gypsy element, introduced to the naive fly stock by P-element mediated transformation is able to mobilize in this stock, and that production of the gypsy Env in follicle cells does not increase the mobilization of env-defective gypsy element.126 Apparently errantiviruses use the endosomal and exosomal pathways that in Drosophila ovaries are normally employed for the yolk protein precursors transfer to the oocyte. Impairment of the endocytic traffic in the oocyte disturbs ZAM viral particles transit to the germline cells.43 Its noteworthy that the action of the cell control mechanisms are also restricted to the somatic follicle cells that surround the maternal germline.124
Regulation
The exact mechanism that allows a cell to develop the adaptive immunity against active transposons remained unclear for a long time. There was an opinion that there should be special genes to control each element or a group of elements. In case of gypsy, the flamenco locus was found. Flamenco is a large locus on the Drosophila X chromosome. The activity of the gypsy errantivirus monitored and quantified by detecting ovo gene insertional mutants was shown to depend on the flamenco allele.127 Two classes of flamenco alleles, restrictive and permissive, were found in natural populations in about the same proportions.128 Strains containing flam+ alleles were stable with regard to gypsy copy number, and permissive alleles, flam-, allowed gypsy retrotranspositions. Also it was shown that flamenco is capable of controlling ZAM and Idefix activity.129 Nonetheless, early attempts to clone this gene and determine its molecular function failed.
The nature of TE repression and flamenco function in particular became more clear with the accumulation of the data concerning RNA interference pathways. It was shown that there are special small RNAs 23–29 nucleotides long that interact with the proteins of PIWI subfamily and form a small RNA-based system that silences transposable elements by sequence-specific recognition and cleavage of TE transcripts (for the review, see refs. 130 and 131). The Drosophila genome was found to have discreet heterochromatic loci termed piRNA clusters, which contain truncated copies and fragments of transposable elements and serve as a source of piRNAs.132
In Drosophila, piRNAs bind three different Argonaute proteins of PIWI subfamily: Piwi, Aubergine and AGO3.133-135 These proteins interact with different piRNAs: Piwi and Aub bind predominantly antisense-strand (for the element) piRNAs, while AGO3 binds mainly sense-strand piRNAs.133,134 Sense-strand piRNAs are generated on the transposable element template and antisense-strand molecules are produced at the piRNA clusters that extend up to 240 kilobases in length.132 It is considered, that long RNA molecules produced at piRNA clusters undergo processing to become a set of antisense-strand piRNAs that are loaded into Piwi or Aubergine. These RNA-protein complexes cleave sense RNAs, produced from transposable elements. The cleavage of TE transcripts generate the pool of sense-stranded piRNAs that are loaded onto AGO3 protein. AGO3 associated with sense piRNAs cleaves antisense piRNA precursors and the cycle repeats. This model was termed the ping-pong mechanism. The ping-pong amplification loop is mostly restricted to the germline and does not work in the follicle cells.132,134
The flamenco locus appeared to be one of the piRNA clusters.136,129,132 Interesting, that flamenco is active only in follicle cells and its piRNAs are exclusively loaded onto Piwi protein because this is the only member of PIWI subfamily that is expressed in the Drosophila ovarian soma.137 Transcripts encoded by flamenco could be cleaved by the putative nuclease encoded by the zucchini locus, producing RNA fragments that bind to Piwi.138-140
In Drosophila melanogaster flamenco contains not only gypsy, Idefix and ZAM truncated copies but fragments of more than ten errantiviruses and encode antisense piRNAs that can efficiently target these LTR-retroelements in the absence of an active ping-pong mechanism.140 Malone and co-authors identified putative flamenco loci in D. yakuba and D. erecta and showed that the enrichment for sequences derived from errantiviruses is conserved in these species.140 However currently there is no evidence that this particular mechanism with follicle cell expression and transferring into the germ line balanced by flamenco-like piRNA cluster works for other errantiviruses in other insect species.
Conclusions
Errantiviruses are endogenous retroviruses and show some infectivity potential. But in contrast to the vertebrate ERVs that are commonly originated by the degradation of infective retroviruses (for the review, see refs. 9 and 10) they have obtained their incipient infectivity de novo and never had infectious retroviruses as ancestors. Origin of errantiviruses may be an independent case of gaining the infective properties by retroelements.
Many questions concerning errantiviruses remain unclear. It is still hard to estimate the abundance of errantiviruses among insects and arthropods. Functions, adaptive value and reasons for the conservation of the env gene in errantiviral genome is still discussable. Also it is not clear whether env has the ability to enhance the horizontal transfer of elements. The origin and abundance of the specific errantivirus life cycle with the expression exclusively in the somatic follicle cells needs to be studied extensively.
We cannot exclude that the study of errantiviruses may lead to some practical applications. For example foamy viruses are successfuly used as transformation vectors. But the most intriguing point is the evolution and origin of LTR-retroelements and a place of insect endogenous retroviruses in this process. Careful and detailed analysis of errantivirus biology may shed light on the conversion between “copy and paste” strategy of retrotransposons and propagation via infection that is used by retroviruses.
Acknowledgments
The authors would like to thank Drs Anna Voznessenskaya, Anastasia Stolyarenko, Amy Gordon and Yury V. Ilyin for editing the manuscript and useful comments. We are also grateful to Ivan Konstantinov from Visual Science studio for his assistance with figures preparation.
Glossary
Abbreviations:
- LTR
long terminal repeat
- TE
transposable element
- ERV
endogenous retrovirus
- ORF
open reading frame
- RT
reverse transcriptase
- piRNA
PIWI-interacting RNA
- PBS
primer binding site
- PPT
polypurine tract
- MHR
major homology region
- IRES
internal ribosome entry site
- ICTV
International Committee on Taxonomy of Viruses
Glossary
Retroelement: non-systematic group that include retroviruses, retrotransposons and LINE-elements. All these species use reverse transcription in their life cycle
Retrovirus: enveloped RNA virus with the genome that contains gag, pol and env genes and the life cycle characterized by the presence of reverse transcription step and the integration in the host genome with the formation of provirus. The retroviral provirus in the host genome has long-terminal repeats at the both sides of it.
Infectious virus: virus that can be transmitted horizontally via infection
Exogenous retrovirus: infectious retrovirus that is transmitted mostly via infection rather than from ancestors to the offspring inside the germ line.
Endogenous retrovirus: retrovirus that exist mostly in form of a provirus integrated into the host genome.
Viral particle (virion, virus particle): A complete virus particle with its DNA or RNA core and protein and lipid coat as it exists outside the cell.
Virus-like particle: non-infectious virions, with aberrant structure or not containing virus genetic material.
LTR-retrotransposon: mobile genetic element that uses a replicative mechanism of transposition through RNA intermediate and contains reverse transcriptase and integrase domains. Retrotransposons carry direct long-terminal repeats at the both ends of the element’s integrated DNA copy.
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/19234
References
- 1.Corces VG, Geyer PK. Interactions of retrotransposons with the host genome: the case of the gypsy element of Drosophila. Trends Genet. 1991;7:86–90. doi: 10.1016/0168-9525(91)90277-W. [DOI] [PubMed] [Google Scholar]
- 2.Conte C, Dastugue B, Vaury C. Coupling of enhancer and insulator properties identified in two retrotransposons modulates their mutagenic impact on nearby genes. Mol Cell Biol. 2002;22:1767–77. doi: 10.1128/MCB.22.6.1767-1777.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurth R, Bannert N. Beneficial and detrimental effects of human endogenous retroviruses. Int J Cancer. 2010;126:306–14. doi: 10.1002/ijc.24902. [DOI] [PubMed] [Google Scholar]
- 4.Mugnier N, Gueguen L, Vieira C, Bie´mont C. The heterochromatic copies of the LTR retrotransposons as a record of the genomic events that have shaped the Drosophila melanogaster genome. Gene. 2008;411:87–93. doi: 10.1016/j.gene.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 5.Lee J, Han K, Meyer TJ, Kim HS, Batzer MA. Chromosomal inversions between human and chimpanzee lineages caused by retrotransposons. PLoS One. 2008;3:e4047. doi: 10.1371/journal.pone.0004047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jern P, Coffin JM. Effects of retroviruses on host genome function. Annu Rev Genet. 2008;42:709–32. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- 7.Volff JN. Cellular genes derived from Gypsy/Ty3 retrotransposons in mammalian genomes. Ann N Y Acad Sci. 2009;1178:233–43. doi: 10.1111/j.1749-6632.2009.05005.x. [DOI] [PubMed] [Google Scholar]
- 8.Boeke JD, Stoye JP. Retrotransposons, Endogenous Retroviruses, and the Evolution of Retroelements. In: Coffin JM, Hughes SH, Varmus HE, ed. Retroviruses. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 9.Gifford R, Tristem M. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes. 2003;26:291–315. doi: 10.1023/A:1024455415443. [DOI] [PubMed] [Google Scholar]
- 10.Weiss RA. The discovery of endogenous retroviruses. Retrovirology. 2006;3:67. doi: 10.1186/1742-4690-3-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim A, Terzian C, Santamaria P, Pe´lisson A, Purd’homme N, Bucheton A. Retroviruses in invertebrates: the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1994;91:1285–9. doi: 10.1073/pnas.91.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song SU, Gerasimova T, Kurkulos M, Boeke JD, Corces VG. An env-like protein encoded by a Drosophila retroelement: evidence that gypsy is an infectious retrovirus. Genes Dev. 1994;8:2046–57. doi: 10.1101/gad.8.17.2046. [DOI] [PubMed] [Google Scholar]
- 13.Leblanc P, Desset S, Giorgi F, Taddei AR, Fausto AM, Mazzini M, et al. Life cycle of an endogenous retrovirus, ZAM, in Drosophila melanogaster. J Virol. 2000;74:10658–69. doi: 10.1128/JVI.74.22.10658-10669.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boeke JD, Eickbush TH, Sandmeyer SB, Voytas DF. Metaviridae. In: Murphy FA, ed. Virus Taxonomy: ICTV VIIth Report. Springer-Verlag, New York, 1999. [Google Scholar]
- 15.Virus Taxonomy. VIIIth Report of the International Committee on Taxonomy of Viruses”, 2005, Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (Eds), Elsevier Academic Press. [Google Scholar]
- 16.Vogt VM. Retroviral Virions and Genomes. In: Coffin JM, Hughes SH, Varmus HE, ed. Retroviruses. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 17.Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 18.Wills JW, Craven RC. Form, function, and use of retroviral gag proteins. AIDS. 1991;5:639–54. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 19.Mammano F, Ohagen A, Höglund S, Göttlinger HG. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J Virol. 1994;68:4927–36. doi: 10.1128/jvi.68.8.4927-4936.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green LM, Berg JM. A retroviral Cys-Xaa2-Cys-Xaa4-His-Xaa4-Cys peptide binds metal ions: spectroscopic studies and a proposed three-dimensional structure. Proc Natl Acad Sci U S A. 1989;86:4047–51. doi: 10.1073/pnas.86.11.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Syomin BV, Kandror KV, Semakin AB, Tsuprun VL, Stepanov AS. Presence of the gypsy (MDG4) retrotransposon in extracellular virus-like particles. FEBS Lett. 1993;323:285–8. doi: 10.1016/0014-5793(93)81358-7. [DOI] [PubMed] [Google Scholar]
- 22.Syomin BV, Ilyin YV. The functional motifs that are revealed in the gypsy Gag amino acid sequence. Dokl Biochem Biophys. 2004;398:291–3. doi: 10.1023/B:DOBI.0000046640.60059.f9. [DOI] [PubMed] [Google Scholar]
- 23.Ohno H, Stewart J, Fournier MC, Bosshart H, Rhee I, Miyatake S, et al. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science. 1995;269:1872–5. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- 24.Puffer BA, Parent LJ, Wills JW, Montelaro RC. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J Virol. 1997;71:6541–6. doi: 10.1128/jvi.71.9.6541-6546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gabus C, Ivanyi-Nagy R, Depollier J, Bucheton A, Pelisson A, Darlix JL. Characterization of a nucleocapsid-like region and of two distinct primer tRNALys,2 binding sites in the endogenous retrovirus Gypsy. Nucleic Acids Res. 2006;34:5764–77. doi: 10.1093/nar/gkl722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hajek KL, Friesen PD. Proteolytic processing and assembly of gag and gag-pol proteins of TED, a baculovirus-associated retrotransposon of the gypsy family. J Virol. 1998;72:8718–24. doi: 10.1128/jvi.72.11.8718-8724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Semin BV, Turanov OA, Stepanov AS, Il’in IuV. [Binding of nucleic acids by a protein, coding the first open reading frame of the gypsy retrotransposon (MDG4)] Mol Biol (Mosk) 1999;33:423–7. [PubMed] [Google Scholar]
- 28.Switzer WM, Salemi M, Shanmugam V, Gao F, Cong ME, Kuiken C, et al. Ancient co-speciation of simian foamy viruses and primates. Nature. 2005;434:376–80. doi: 10.1038/nature03341. [DOI] [PubMed] [Google Scholar]
- 29.Meiering CD, Linial ML. Historical perspective of foamy virus epidemiology and infection. Clin Microbiol Rev. 2001;14:165–76. doi: 10.1128/CMR.14.1.165-176.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rethwilm A. Molecular biology of foamy viruses. Med Microbiol Immunol. 2010;199:197–207. doi: 10.1007/s00430-010-0158-x. [DOI] [PubMed] [Google Scholar]
- 31.Lee EG, Linial ML. The C terminus of foamy retrovirus Gag contains determinants for encapsidation of Pol protein into virions. J Virol. 2008;82:10803–10. doi: 10.1128/JVI.00812-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee EG, Linial ML. The C terminus of foamy retrovirus Gag contains determinants for encapsidation of Pol protein into virions. J Virol. 2008;82:10803–10. doi: 10.1128/JVI.00812-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schliephake AW, Rethwilm A. Nuclear localization of foamy virus Gag precursor protein. J Virol. 1994;68:4946–54. doi: 10.1128/jvi.68.8.4946-4954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tobaly-Tapiero J, Bittoun P, Lehmann-Che J, Delelis O, Giron ML, de The´ H, et al. Chromatin tethering of incoming foamy virus by the structural Gag protein. Traffic. 2008;9:1717–27. doi: 10.1111/j.1600-0854.2008.00792.x. [DOI] [PubMed] [Google Scholar]
- 35.Semin BV, Popenko VI, Malikova MA, Turapov OA, Stepanov AS, Ilyin Y. The polyprotein gag of retroelement gypsy can form multimeric complexes when expressed in bacterial system. Dokl Biochem Biophys. 2001;380:322–4. doi: 10.1023/A:1012336008371. [DOI] [PubMed] [Google Scholar]
- 36.Eastman SW, Linial ML. Identification of a conserved residue of foamy virus Gag required for intracellular capsid assembly. J Virol. 2001;75:6857–64. doi: 10.1128/JVI.75.15.6857-6864.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sfakianos JN, LaCasse RA, Hunter E. The M-PMV cytoplasmic targeting-retention signal directs nascent Gag polypeptides to a pericentriolar region of the cell. Traffic. 2003;4:660–70. doi: 10.1034/j.1600-0854.2003.00125.x. [DOI] [PubMed] [Google Scholar]
- 38.Yu SF, Eastman SW, Linial ML. Foamy virus capsid assembly occurs at a pericentriolar region through a cytoplasmic targeting/retention signal in Gag. Traffic. 2006;7:966–77. doi: 10.1111/j.1600-0854.2006.00448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Life RB, Lee EG, Eastman SW, Linial ML. Mutations in the amino terminus of foamy virus Gag disrupt morphology and infectivity but do not target assembly. J Virol. 2008;82:6109–19. doi: 10.1128/JVI.00503-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gurer C, Berthoux L, Luban J. Covalent modification of human immunodeficiency virus type 1 p6 by SUMO-1. J Virol. 2005;79:910–7. doi: 10.1128/JVI.79.2.910-917.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat Cell Biol. 2011;13:394–401. doi: 10.1038/ncb2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ono A. Relationships between plasma membrane microdomains and HIV-1 assembly. Biol Cell. 2010;102:335–50. doi: 10.1042/BC20090165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brasset E, Taddei AR, Arnaud F, Faye B, Fausto AM, Mazzini M, et al. Viral particles of the endogenous retrovirus ZAM from Drosophila melanogaster use a pre-existing endosome/exosome pathway for transfer to the oocyte. Retrovirology. 2006;3:25. doi: 10.1186/1742-4690-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eickbush TH, Jamburuthugoda VK. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 2008;134:221–34. doi: 10.1016/j.virusres.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peterson-Burch BD, Voytas DF. Genes of the Pseudoviridae (Ty1/copia retrotransposons) Mol Biol Evol. 2002;19:1832–45. doi: 10.1093/oxfordjournals.molbev.a004008. [DOI] [PubMed] [Google Scholar]
- 46.Llorens C, Fares MA, Moya A. Relationships of gag-pol diversity between Ty3/Gypsy and Retroviridae LTR retroelements and the three kings hypothesis. BMC Evol Biol. 2008;8:276. doi: 10.1186/1471-2148-8-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malik HS. Ribonuclease H evolution in retrotransposable elements. Cytogenet Genome Res. 2005;110:392–401. doi: 10.1159/000084971. [DOI] [PubMed] [Google Scholar]
- 48.Xiong Y, Eickbush TH. Similarity of reverse transcriptase-like sequences of viruses, transposable elements, and mitochondrial introns. Mol Biol Evol. 1988;5:675–90. doi: 10.1093/oxfordjournals.molbev.a040521. [DOI] [PubMed] [Google Scholar]
- 49.Malik HS, Eickbush TH. Phylogenetic analysis of ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res. 2001;11:1187–97. doi: 10.1101/gr.185101. [DOI] [PubMed] [Google Scholar]
- 50.Arnaud F, Peyretaillade E, Dastugue B, Vaury C. Functional characteristics of a reverse transcriptase encoded by an endogenous retrovirus from Drosophila melanogaster. Insect Biochem Mol Biol. 2005;35:323–31. doi: 10.1016/j.ibmb.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 51.Leblanc P, Dastugue B, Vaury C. The integration machinery of ZAM, a retroelement from Drosophila melanogaster, acts as a sequence-specific endonuclease. J Virol. 1999;73:7061–4. doi: 10.1128/jvi.73.8.7061-7064.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nefedova LN, Mannanova MM, Kim AI. Integration specificity of LTR-retrotransposons and retroviruses in the Drosophila melanogaster genome. Virus Genes. 2011;42:297–306. doi: 10.1007/s11262-010-0566-4. [DOI] [PubMed] [Google Scholar]
- 53.Kuzin AB, Khudaĭbergenova BM, Liubomirskaia NV, Kim AI, Il’in IuV. [Precise excision of the MDG4 (gypsy) transposon from the Drosophila melanogaster cut and forked locus] Dokl Akad Nauk. 1995;340:135–7. [PubMed] [Google Scholar]
- 54.Nowotny M. Retroviral integrase superfamily: the structural perspective. EMBO Rep. 2009;10:144–51. doi: 10.1038/embor.2008.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bao W, Kapitonov VV, Jurka J. Ginger DNA transposons in eukaryotes and their evolutionary relationships with long terminal repeat retrotransposons. Mob DNA. 2010;1:3. doi: 10.1186/1759-8753-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swanstrom R, Wills JW. Synthesis, Assembly, and Processing of Viral Proteins. In: Coffin JM, Hughes SH, Varmus HE, ed. Retroviruses. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 57.Malik HS, Henikoff S, Eickbush TH. Poised for contagion: evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res. 2000;10:1307–18. doi: 10.1101/gr.145000. [DOI] [PubMed] [Google Scholar]
- 58.Kim FJ, Battini JL, Manel N, Sitbon M. Emergence of vertebrate retroviruses and envelope capture. Virology. 2004;318:183–91. doi: 10.1016/j.virol.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 59.Misseri Y, Cerutti M, Devauchelle G, Bucheton A, Terzian C. Analysis of the Drosophila gypsy endogenous retrovirus envelope glycoprotein. J Gen Virol. 2004;85:3325–31. doi: 10.1099/vir.0.79911-0. [DOI] [PubMed] [Google Scholar]
- 60.Pearson MN, Rohrmann GF. Envelope gene capture and insect retrovirus evolution: the relationship between errantivirus and baculovirus envelope proteins. Virus Res. 2006;118:7–15. doi: 10.1016/j.virusres.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 61.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–50. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 62.Henriet S, Mercenne G, Bernacchi S, Paillart JC, Marquet R. Tumultuous relationship between the human immunodeficiency virus type 1 viral infectivity factor (Vif) and the human APOBEC-3G and APOBEC-3F restriction factors. Microbiol Mol Biol Rev. 2009;73:211–32. doi: 10.1128/MMBR.00040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laguette N, Bre´gnard C, Benichou S, Basmaciogullari S. Human immunodeficiency virus (HIV) type-1, HIV-2 and simian immunodeficiency virus Nef proteins. Mol Aspects Med. 2010;31:418–33. doi: 10.1016/j.mam.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 64.Kozak M. A second look at cellular mRNA sequences said to function as internal ribosome entry sites. Nucleic Acids Res. 2005;33:6593–602. doi: 10.1093/nar/gki958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meignin C, Bailly JL, Arnaud F, Dastugue B, Vaury C. The 5′ untranslated region and Gag product of Idefix, a long terminal repeat-retrotransposon from Drosophila melanogaster, act together to initiate a switch between translated and untranslated states of the genomic mRNA. Mol Cell Biol. 2003;23:8246–54. doi: 10.1128/MCB.23.22.8246-8254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ramos E, Ghosh D, Baxter E, Corces VG. Genomic organization of gypsy chromatin insulators in Drosophila melanogaster. Genetics. 2006;172:2337–49. doi: 10.1534/genetics.105.054742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Savitskaya E, Melnikova L, Kostuchenko M, Kravchenko E, Pomerantseva E, Boikova T, et al. Study of long-distance functional interactions between Su(Hw) insulators that can regulate enhancer-promoter communication in Drosophila melanogaster. Mol Cell Biol. 2006;26:754–61. doi: 10.1128/MCB.26.3.754-761.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parkhurst SM, Harrison DA, Remington MP, Spana C, Kelley RL, Coyne RS, et al. The Drosophila su(Hw) gene, which controls the phenotypic effect of the gypsy transposable element, encodes a putative DNA-binding protein. Genes Dev. 1988;2:1205–15. doi: 10.1101/gad.2.10.1205. [DOI] [PubMed] [Google Scholar]
- 69.Gdula DA, Gerasimova TI, Corces VG. Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila. Proc Natl Acad Sci U S A. 1996;93:9378–83. doi: 10.1073/pnas.93.18.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–13. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 71.Geyer PK, Green MM, Corces VG. Reversion of a gypsy-induced mutation at the yellow (y) locus of Drosophila melanogaster is associated with the insertion of a newly defined transposable element. Proc Natl Acad Sci U S A. 1988;85:3938–42. doi: 10.1073/pnas.85.11.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Geyer PK, Corces VG. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 1992;6:1865–73. doi: 10.1101/gad.6.10.1865. [DOI] [PubMed] [Google Scholar]
- 73.Golovnin A, Mazur A, Kopantseva M, Kurshakova M, Gulak PV, Gilmore B, et al. Integrity of the Mod(mdg4)-67.2 BTB domain is critical to insulator function in Drosophila melanogaster. Mol Cell Biol. 2007;27:963–74. doi: 10.1128/MCB.00795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pai CY, Lei EP, Ghosh D, Corces VG. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell. 2004;16:737–48. doi: 10.1016/j.molcel.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 75.Silicheva M, Golovnin A, Pomerantseva E, Parshikov A, Georgiev P, Maksimenko O. Drosophila mini-white model system: new insights into positive position effects and the role of transcriptional terminators and gypsy insulator in transgene shielding. Nucleic Acids Res. 2010;38:39–47. doi: 10.1093/nar/gkp877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Conte C, Dastugue B, Vaury C. Coupling of enhancer and insulator properties identified in two retrotransposons modulates their mutagenic impact on nearby genes. Mol Cell Biol. 2002;22:1767–77. doi: 10.1128/MCB.22.6.1767-1777.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Minervini CF, Marsano RM, Casieri P, Fanti L, Caizzi R, Pimpinelli S, et al. Heterochromatin protein 1 interacts with 5’UTR of transposable element ZAM in a sequence-specific fashion. Gene. 2007;393:1–10. doi: 10.1016/j.gene.2006.12.028. [DOI] [PubMed] [Google Scholar]
- 78.Brasset E, Hermant C, Jensen S, Vaury C. The Idefix enhancer-blocking insulator also harbors barrier activity. Gene. 2010;450:25–31. doi: 10.1016/j.gene.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 79.Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–82. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 80.Kapitonov VV, Jurka J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat Rev Genet. 2008;9:411–2, author reply 414. doi: 10.1038/nrg2165-c1. [DOI] [PubMed] [Google Scholar]
- 81.Seberg O, Petersen G. A unified classification system for eukaryotic transposable elements should reflect their phylogeny. Nat Rev Genet. 2009;10:276. doi: 10.1038/nrg2165-c3. [DOI] [PubMed] [Google Scholar]
- 82.Llorens C, Muñoz-Pomer A, Bernad L, Botella H, Moya A. Network dynamics of eukaryotic LTR retroelements beyond phylogenetic trees. Biol Direct. 2009;4:41. doi: 10.1186/1745-6150-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cizeron G, Bie´mont C. Polymorphism in structure of the retrotransposable element 412 in Drosophila simulans and D. melanogaster populations. Gene. 1999;232:183–90. doi: 10.1016/S0378-1119(99)00126-2. [DOI] [PubMed] [Google Scholar]
- 84.Costas J, Valade´ E, Naveira H. Structural features of the mdg1 lineage of the Ty3/gypsy group of LTR retrotransposons inferred from the phylogenetic analyses of its open reading frames. J Mol Evol. 2001;53:165–71. doi: 10.1007/s002390010206. [DOI] [PubMed] [Google Scholar]
- 85.Terzian C, Pe´lisson A, Bucheton A. Evolution and phylogeny of insect endogenous retroviruses. BMC Evol Biol. 2001;1:3. doi: 10.1186/1471-2148-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Llorens C, Futami R, Covelli L, Domi´nguez-Escribá L, Viu JM, Tamarit D, et al. The Gypsy Database (GyDB) of mobile genetic elements: release 2.0. Nucleic Acids Res. 2011;39(Database issue):D70–4. doi: 10.1093/nar/gkq1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ludwig A, Valente VL, Loreto EL. Multiple invasions of Errantivirus in the genus Drosophila. Insect Mol Biol. 2008;17:113–24. doi: 10.1111/j.1365-2583.2007.00787.x. [DOI] [PubMed] [Google Scholar]
- 88.Petrov DA, Fiston-Lavier AS, Lipatov M, Lenkov K, González J. Population genomics of transposable elements in Drosophila melanogaster. Mol Biol Evol. 2011;28:1633–44. doi: 10.1093/molbev/msq337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jin-Shan X, Qing-You X, Jun L, Guo-Qing P, Ze-Yang Z. Survey of long terminal repeat retrotransposons of domesticated silkworm (Bombyx mori) Insect Biochem Mol Biol. 2005;35:921–9. doi: 10.1016/j.ibmb.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 90.Gladyshev EA, Meselson M, Arkhipova IR. A deep-branching clade of retrovirus-like retrotransposons in bdelloid rotifers. Gene. 2007;390:136–45. doi: 10.1016/j.gene.2006.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tubio JM, Tojo M, Bassaganyas L, Escaramis G, Sharakhov IV, Sharakhova MV, et al. Evolutionary dynamics of the Ty3/gypsy LTR retrotransposons in the genome of Anopheles gambiae. PLoS One. 2011;6:e16328. doi: 10.1371/journal.pone.0016328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kaminker JS, Bergman CM, Kronmiller B, Carlson J, Svirskas R, Patel S, et al. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 2002;3:H0084. doi: 10.1186/gb-2002-3-12-research0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vidal NM, Ludwig A, Loreto EL. Evolution of Tom, 297, 17.6 and rover retrotransposons in Drosophilidae species. Mol Genet Genomics. 2009;282:351–62. doi: 10.1007/s00438-009-0468-0. [DOI] [PubMed] [Google Scholar]
- 94.McCarthy EM, McDonald JF. LTR_STRUC: a novel search and identification program for LTR retrotransposons. Bioinformatics. 2003;19:362–7. doi: 10.1093/bioinformatics/btf878. [DOI] [PubMed] [Google Scholar]
- 95.Rho M, Choi JH, Kim S, Lynch M, Tang H. De novo identification of LTR retrotransposons in eukaryotic genomes. BMC Genomics. 2007;8:90. doi: 10.1186/1471-2164-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Minervini CF, Viggiano L, Caizzi R, Marsano RM. Identification of novel LTR retrotransposons in the genome of Aedes aegypti. Gene. 2009 Jul 1;440(1-2):42-9; PMID: 19362135; DOI; 10.1016/j.gene.2009.03.021. [DOI] [PubMed]
- 97.Steinbiss S, Willhoeft U, Gremme G, Kurtz S. Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res. 2009;37:7002–13. doi: 10.1093/nar/gkp759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Passarelli AL. Barriers to success: how baculoviruses establish efficient systemic infections. Virology. 2011;411:383–92. doi: 10.1016/j.virol.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huszar T, Imler JL. Drosophila viruses and the study of antiviral host-defense. Adv Virus Res. 2008;72:227–65. doi: 10.1016/S0065-3527(08)00406-5. [DOI] [PubMed] [Google Scholar]
- 100.Syomin BV, Fedorova LI, Surkov SA, Ilyin YV. The endogenous Drosophila melanogaster retrovirus gypsy can propagate in Drosophila hydei cells. Mol Gen Genet. 2001;264:588–94. doi: 10.1007/s004380000344. [DOI] [PubMed] [Google Scholar]
- 101.Syomin BV, Leonova TY, Ilyin YV. Evidence for horizontal transfer of the LTR retrotransposon mdg3, which lacks an env gene. Mol Genet Genomics. 2002;267:418–23. doi: 10.1007/s00438-002-0678-1. [DOI] [PubMed] [Google Scholar]
- 102.Pelisson A, Mejlumian L, Robert V, Terzian C, Bucheton A. Drosophila germline invasion by the endogenous retrovirus gypsy: involvement of the viral env gene. Insect Biochem Mol Biol. 2002;32:1249–56. doi: 10.1016/S0965-1748(02)00088-7. [DOI] [PubMed] [Google Scholar]
- 103.Ilyin YV, Lyubomirskaya NV, Kim AI. Retrotransposon Gypsy and genetic instability in Drosophila (review) Genetica. 1991;85:13–22. doi: 10.1007/BF00056102. [review] [DOI] [PubMed] [Google Scholar]
- 104.Stefanov YE, Glukhov IA, Kotnova AP, Salenko VB, Pasyukova EG, Lyubomirskaya NV, et al. Amplification of “defective” retrotransposon gtwin in D. melanogaster strain carrying large complex chromosomal aberration. Mol Genet Genomics. 2010;284:373–81. doi: 10.1007/s00438-010-0573-0. [DOI] [PubMed] [Google Scholar]
- 105.Hoover KK, Gerasimova TI, Chien AJ, Corces VG. Dominant effects of suppressor of Hairy-wing mutations on gypsy-induced alleles of forked and cut in Drosophila melanogaster. Genetics. 1992;132:691–7. doi: 10.1093/genetics/132.3.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kuzin AB, Liubomirskaia NV, Khudaĭbergenova BM, Kim AI, Il’in IuV. [Retrotransposon MDG4 (gypsy) integration hot spot and its precise excision from the D. melanogaster forked locus] Dokl Akad Nauk. 1994;335:656–8. [PubMed] [Google Scholar]
- 107.Me´vel-Ninio M, Mariol MC, Gans M. Mobilization of the gypsy and copia retrotransposons in Drosophila melanogaster induces reversion of the ovo dominant female-sterile mutations: molecular analysis of revertant alleles. EMBO J. 1989;8:1549–58. doi: 10.1002/j.1460-2075.1989.tb03539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lyubomirskaya NV, Arkhipova IR, Ilyin YV, Kim AI. Molecular analysis of the gypsy (mdg4) retrotransposon in two Drosophila melanogaster strains differing by genetic instability. Mol Gen Genet. 1990;223:305–9. doi: 10.1007/BF00265067. [DOI] [PubMed] [Google Scholar]
- 109.Lyubomirskaya NV, Avedisov SN, Surkov SA, Ilyin YV. Two Drosophila retrotransposon gypsy subfamilies differ in ability to produce new DNA copies via reverse transcription in Drosophila cultured cells. Nucleic Acids Res. 1993;21:3265–8. doi: 10.1093/nar/21.14.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Here´dia F, Loreto EL, Valente VL. Distribution and conservation of the transposable element gypsy in drosophilid species. Genet Mol Biol. 2007;30:133–8. doi: 10.1590/S1415-47572007000100023. [DOI] [Google Scholar]
- 111.Salenko VB, Kotnova AP, Karpova NN, Lyubomirskaya NV, Ilyin YV. Polymorphism of canonical and noncanonical gypsy sequences in different species of Drosophila melanogaster subgroup: possible evolutionary relations. Mol Genet Genomics. 2008;279:463–72. doi: 10.1007/s00438-008-0325-6. [DOI] [PubMed] [Google Scholar]
- 112.Vieira C, Lepetit D, Dumont S, Bie´mont C. Wake up of transposable elements following Drosophila simulans worldwide colonization. Mol Biol Evol. 1999;16:1251–5. doi: 10.1093/oxfordjournals.molbev.a026215. [DOI] [PubMed] [Google Scholar]
- 113.Ludwig A, Loreto EL. Evolutionary pattern of the gtwin retrotransposon in the Drosophila melanogaster subgroup. Genetica. 2007;130:161–8. doi: 10.1007/s10709-006-9003-y. [DOI] [PubMed] [Google Scholar]
- 114.Kotnova AP, Glukhov IA, Karpova NN, Salenko VB, Lyubomirskaya NV, Ilyin YV. Evidence for recent horizontal transfer of gypsy-homologous LTR-retrotransposon gtwin into Drosophila erecta followed by its amplification with multiple aberrations. Gene. 2007;396:39–45. doi: 10.1016/j.gene.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 115.Loreto EL, Carareto CM, Capy P. Revisiting horizontal transfer of transposable elements in Drosophila. Heredity (Edinb) 2008;100:545–54. doi: 10.1038/sj.hdy.6801094. [DOI] [PubMed] [Google Scholar]
- 116.Kidwell MG, Lisch D. Transposable elements as sources of variation in animals and plants. Proc Natl Acad Sci U S A. 1997;94:7704–11. doi: 10.1073/pnas.94.15.7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet. 2011;12:615–27. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Montgomery E, Charlesworth B, Langley CH. A test for the role of natural selection in the stabilization of transposable element copy number in a population of Drosophila melanogaster. Genet Res. 1987;49:31–41. doi: 10.1017/S0016672300026707. [DOI] [PubMed] [Google Scholar]
- 119.Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–20. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- 120.Mackay TF. What prevents transposable elements from taking over the genome? A commentary on ‘A test for the role of natural selection in the stabilization of transposable element copy number in a population of Drosophila melanogaster’ by Elizabeth Montgomery, Brian Charlesworth and Charles H. Langley. Genet Res. 2007;89:433–4. doi: 10.1017/S0016672308009646. [DOI] [PubMed] [Google Scholar]
- 121.Lee YC, Langley CH. Transposable elements in natural populations of Drosophila melanogaster. Philos Trans R Soc Lond B Biol Sci. 2010;365:1219–28. doi: 10.1098/rstb.2009.0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Brookfield JF. The ecology of the genome - mobile DNA elements and their hosts. Nat Rev Genet. 2005;6:128–36. doi: 10.1038/nrg1524. [DOI] [PubMed] [Google Scholar]
- 123.Pardue ML, Rashkova S, Casacuberta E, DeBaryshe PG, George JA, Traverse KL. Two retrotransposons maintain telomeres in Drosophila. Chromosome Res. 2005;13:443–53. doi: 10.1007/s10577-005-0993-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pe´lisson A, Song SU, Prud’homme N, Smith PA, Bucheton A, Corces VG. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J. 1994;13:4401–11. doi: 10.1002/j.1460-2075.1994.tb06760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tcheressiz S, Calco V, Arnaud F, Arthaud L, Dastugue B, Vaury C. Expression of the Idefix retrotransposon in early follicle cells in the germarium of Drosophila melanogaster is determined by its LTR sequences and a specific genomic context. Mol Genet Genomics. 2002;267:133–41. doi: 10.1007/s00438-002-0641-1. [DOI] [PubMed] [Google Scholar]
- 126.Chalvet F, Teysset L, Terzian C, Prud’homme N, Santamaria P, Bucheton A, et al. Proviral amplification of the Gypsy endogenous retrovirus of Drosophila melanogaster involves env-independent invasion of the female germline. EMBO J. 1999;18:2659–69. doi: 10.1093/emboj/18.9.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Prud’homme N, Gans M, Masson M, Terzian C, Bucheton A. Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics. 1995;139:697–711. doi: 10.1093/genetics/139.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pe´lisson A, Teysset L, Chalvet F, Kim A, Prud’homme N, Terzian C, et al. About the origin of retroviruses and the co-evolution of the gypsy retrovirus with the Drosophila flamenco host gene. Genetica. 1997;100:29–37. doi: 10.1023/A:1018336303298. [DOI] [PubMed] [Google Scholar]
- 129.Me´vel-Ninio M, Pelisson A, Kinder J, Campos AR, Bucheton A. The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics. 2007;175:1615–24. doi: 10.1534/genetics.106.068106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–58. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 131.Khurana JS, Theurkauf W. piRNAs, transposon silencing, and Drosophila germline development. J Cell Biol. 2010;191:905–13. doi: 10.1083/jcb.201006034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 133.Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–22. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–90. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 135.Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–42. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- 136.Sarot E, Payen-Groschêne G, Bucheton A, Pe´lisson A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166:1313–21. doi: 10.1534/genetics.166.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–27. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, et al. A broadly conserved pathway generates 3’UTR-directed primary piRNAs. Curr Biol. 2009;19:2066–76. doi: 10.1016/j.cub.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, et al. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–9. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- 140.Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–35. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]


