Abstract
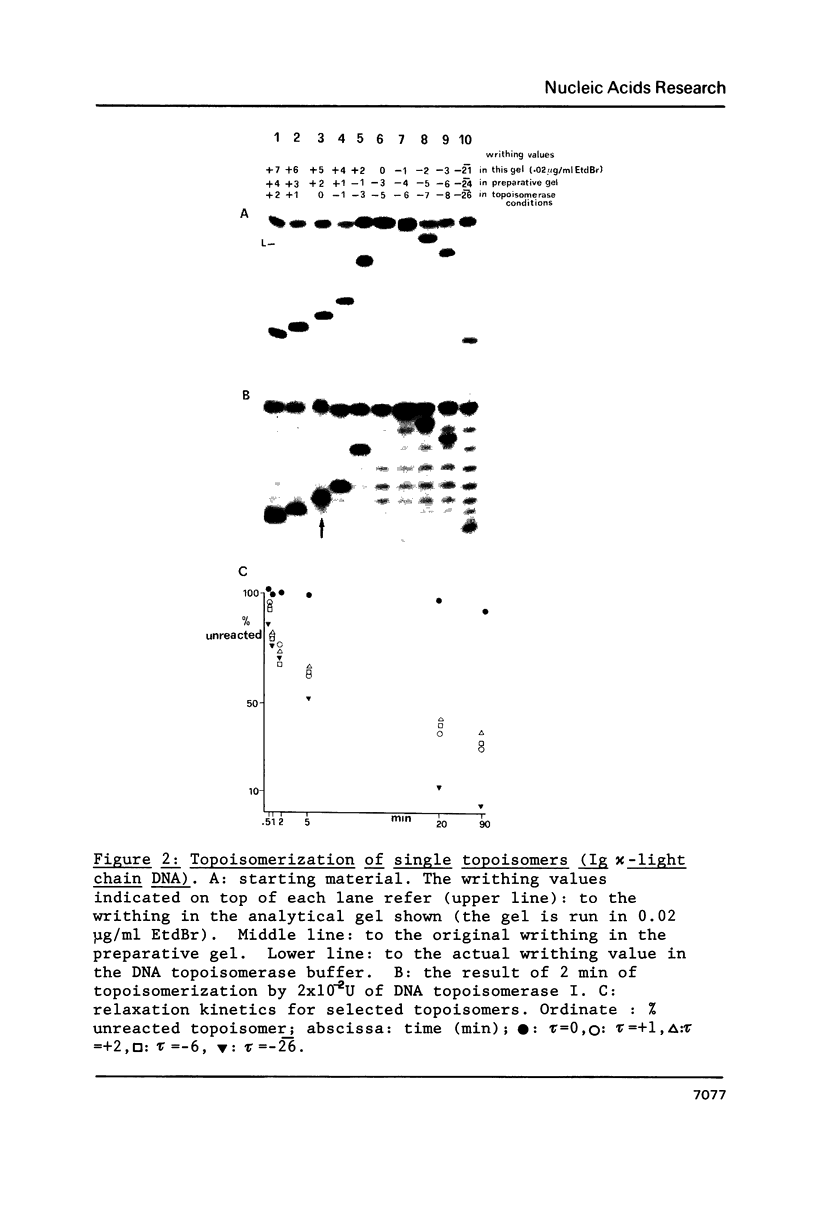
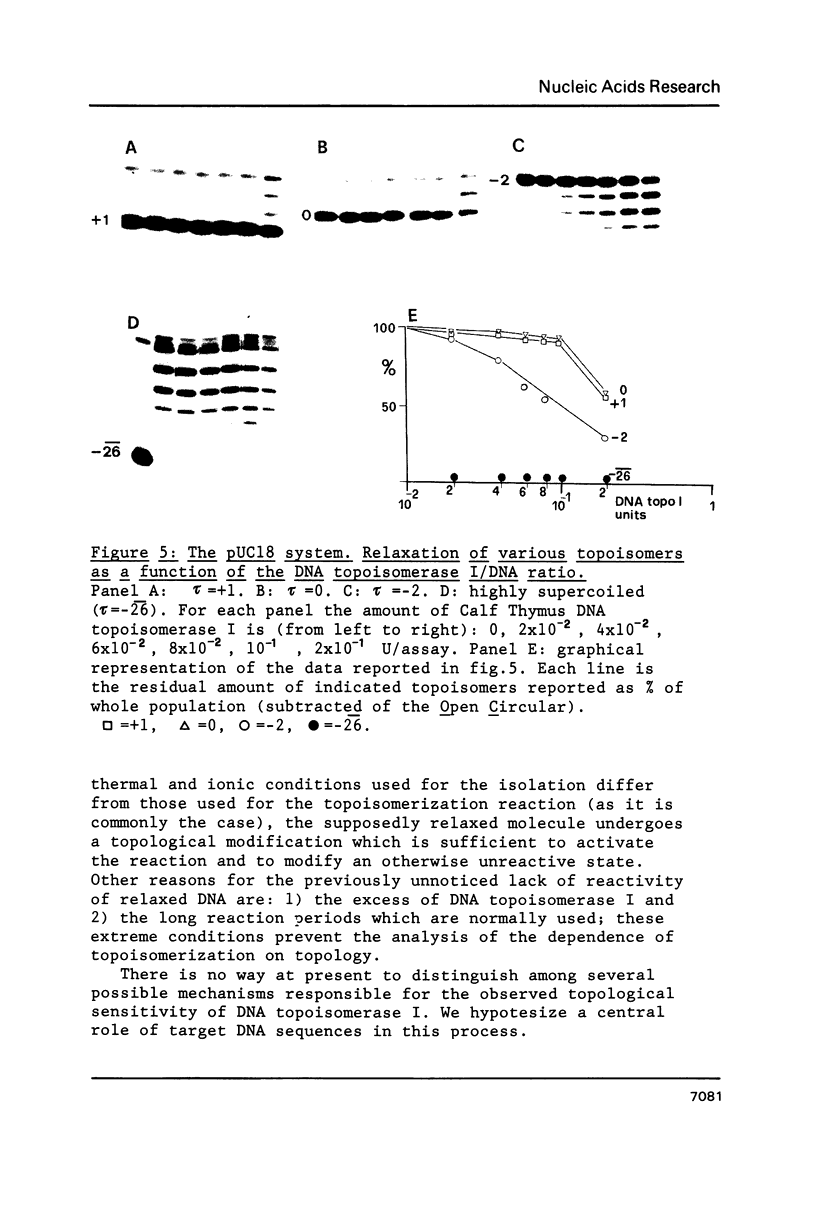
The effects of supercoiling on the topoisomerization reaction by eukaryotic DNA topoisomerases I have been analyzed. The systems used were: DNA topoisomerase I from wheat germ, chicken erythrocyte and calf thymus on a 2.3 kb DNA fragment which encompasses the immunoglobulin kappa-light chain (L kappa) promoter of the mouse plasmacytoma MPC11; S. cerevisiae DNA topoisomerase I on a 2.2 kb DNA fragment from the same organism which encompasses the regulatory and the coding region of the ADH II gene; wheat germ DNA topoisomerase I on the plasmid pUC18. It was found in every system that lack of torsional stress prevents topoisomerization of the substrate. A simple regulatory model of DNA topoisomerase I function, based on topological considerations, is presented.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Been M. D., Burgess R. R., Champoux J. J. DNA strand breakage by wheat germ type 1 topoisomerase. Biochim Biophys Acta. 1984 Jul 18;782(3):304–312. doi: 10.1016/0167-4781(84)90066-6. [DOI] [PubMed] [Google Scholar]
- Been M. D., Burgess R. R., Champoux J. J. Nucleotide sequence preference at rat liver and wheat germ type 1 DNA topoisomerase breakage sites in duplex SV40 DNA. Nucleic Acids Res. 1984 Apr 11;12(7):3097–3114. doi: 10.1093/nar/12.7.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonven B. J., Gocke E., Westergaard O. A high affinity topoisomerase I binding sequence is clustered at DNAase I hypersensitive sites in Tetrahymena R-chromatin. Cell. 1985 Jun;41(2):541–551. doi: 10.1016/s0092-8674(85)80027-1. [DOI] [PubMed] [Google Scholar]
- Busk H., Thomsen B., Bonven B. J., Kjeldsen E., Nielsen O. F., Westergaard O. Preferential relaxation of supercoiled DNA containing a hexadecameric recognition sequence for topoisomerase I. Nature. 1987 Jun 18;327(6123):638–640. doi: 10.1038/327638a0. [DOI] [PubMed] [Google Scholar]
- Camilloni G., Della Seta F., Grazia Ficca A., Di Mauro E. Purified Saccharomyces cerevisiae RNA polymerase II interacts homologously with two different promoters as revealed by P1 endonuclease analysis. Mol Gen Genet. 1986 Aug;204(2):249–257. doi: 10.1007/BF00425506. [DOI] [PubMed] [Google Scholar]
- Champoux J. J., Dulbecco R. An activity from mammalian cells that untwists superhelical DNA--a possible swivel for DNA replication (polyoma-ethidium bromide-mouse-embryo cells-dye binding assay). Proc Natl Acad Sci U S A. 1972 Jan;69(1):143–146. doi: 10.1073/pnas.69.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E., Kuehl M., Wall R. RNA splicing generates a variant light chain from an aberrantly rearranged kappa gene. Nature. 1980 Aug 21;286(5775):776–779. doi: 10.1038/286776a0. [DOI] [PubMed] [Google Scholar]
- Dean F. B., Cozzarelli N. R. Mechanism of strand passage by Escherichia coli topoisomerase I. The role of the required nick in catenation and knotting of duplex DNA. J Biol Chem. 1985 Apr 25;260(8):4984–4994. [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew R. E., Liu L. F., Wang J. C. Interaction between DNA and Escherichia coli protein omega. Formation of a complex between single-stranded DNA and omega protein. J Biol Chem. 1978 Jan 25;253(2):511–518. [PubMed] [Google Scholar]
- Edwards K. A., Halligan B. D., Davis J. L., Nivera N. L., Liu L. F. Recognition sites of eukaryotic DNA topoisomerase I: DNA nucleotide sequencing analysis of topo I cleavage sites on SV40 DNA. Nucleic Acids Res. 1982 Apr 24;10(8):2565–2576. doi: 10.1093/nar/10.8.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour D. S., Elgin S. C. Localization of specific topoisomerase I interactions within the transcribed region of active heat shock genes by using the inhibitor camptothecin. Mol Cell Biol. 1987 Jan;7(1):141–148. doi: 10.1128/mcb.7.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto T., Laipis P., Wang J. C. The purification and characterization of DNA topoisomerases I and II of the yeast Saccharomyces cerevisiae. J Biol Chem. 1984 Aug 25;259(16):10422–10429. [PubMed] [Google Scholar]
- Javaherian K., Liu L. F. Association of eukaryotic DNA topoisomerase I with nucleosomes and chromosomal proteins. Nucleic Acids Res. 1983 Jan 25;11(2):461–472. doi: 10.1093/nar/11.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D. E., Coleclough C., Perry R. P. Functional significance and evolutionary development of the 5'-terminal regions of immunoglobulin variable-region genes. Cell. 1982 Jun;29(2):681–689. doi: 10.1016/0092-8674(82)90184-2. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K., Pflugfelder G., Wang J. C. The cleavage of DNA by type-I DNA topoisomerases. Cold Spring Harb Symp Quant Biol. 1984;49:411–419. doi: 10.1101/sqb.1984.049.01.047. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K., Wang J. C. Bacterial DNA topoisomerase I can relax positively supercoiled DNA containing a single-stranded loop. J Mol Biol. 1985 Oct 5;185(3):625–637. doi: 10.1016/0022-2836(85)90075-0. [DOI] [PubMed] [Google Scholar]
- Martin S. R., McCoubrey W. K., Jr, McConaughy B. L., Young L. S., Been M. D., Brewer B. J., Champoux J. J. Multiple forms of rat liver type I topoisomerase. Methods Enzymol. 1983;100:137–144. doi: 10.1016/0076-6879(83)00050-6. [DOI] [PubMed] [Google Scholar]
- Max E. E., Maizel J. V., Jr, Leder P. The nucleotide sequence of a 5.5-kilobase DNA segment containing the mouse kappa immunoglobulin J and C region genes. J Biol Chem. 1981 May 25;256(10):5116–5120. [PubMed] [Google Scholar]
- Maxwell A., Gellert M. Mechanistic aspects of DNA topoisomerases. Adv Protein Chem. 1986;38:69–107. doi: 10.1016/s0065-3233(08)60526-4. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Shure M., Tang D., Vinograd J., Vosberg H. P. Action of nicking-closing enzyme on supercoiled and nonsupercoiled closed circular DNA: formation of a Boltzmann distribution of topological isomers. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4280–4284. doi: 10.1073/pnas.72.11.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell D. W., Smith M., Williamson V. M., Young E. T. Nucleotide sequence of the yeast alcohol dehydrogenase II gene. J Biol Chem. 1983 Feb 25;258(4):2674–2682. [PubMed] [Google Scholar]
- Schon E., Evans T., Welsh J., Efstratiadis A. Conformation of promoter DNA: fine mapping of S1-hypersensitive sites. Cell. 1983 Dec;35(3 Pt 2):837–848. doi: 10.1016/0092-8674(83)90116-2. [DOI] [PubMed] [Google Scholar]
- Shishido K., Noguchi N., Ando T. Correlation of enzyme-induced cleavage sites on negatively superhelical DNA between prokaryotic topoisomerase I and S1 nuclease. Biochim Biophys Acta. 1983 May 20;740(1):108–117. doi: 10.1016/0167-4781(83)90127-6. [DOI] [PubMed] [Google Scholar]