Abstract
Regulation of the actin cytoskeleton is crucial for cell morphology and migration. One of the key molecules that regulates actin remodeling is the small GTPase Rho. Rho shuttles between the inactive GDP-bound form and the active GTP-bound form, and works as a molecular switch in actin remodeling in response to both extra- and intra-cellular stimuli. Mammalian homolog of Diaphanous (mDia) is one of the Rho effectors and produces unbranched actin filaments. While Rho GTPases activate mDia, the mechanisms of how the activity of mDia is downregulated in cells remains largely unknown. In our recent paper, we identified Liprin-α as an mDia interacting protein and found that Liprin-α negatively regulates the activity of mDia in the cell by displacing it from the plasma membrane through binding to the DID-DD region of mDia. Here, we review these findings and discuss how Liprin-α regulates the Rho-mDia pathway and how the mDia-Liprin-α complex functions in vivo.
Keywords: Liprin, Rho, actin cytoskeleton, formin, mDia
Introduction
Actin nucleators
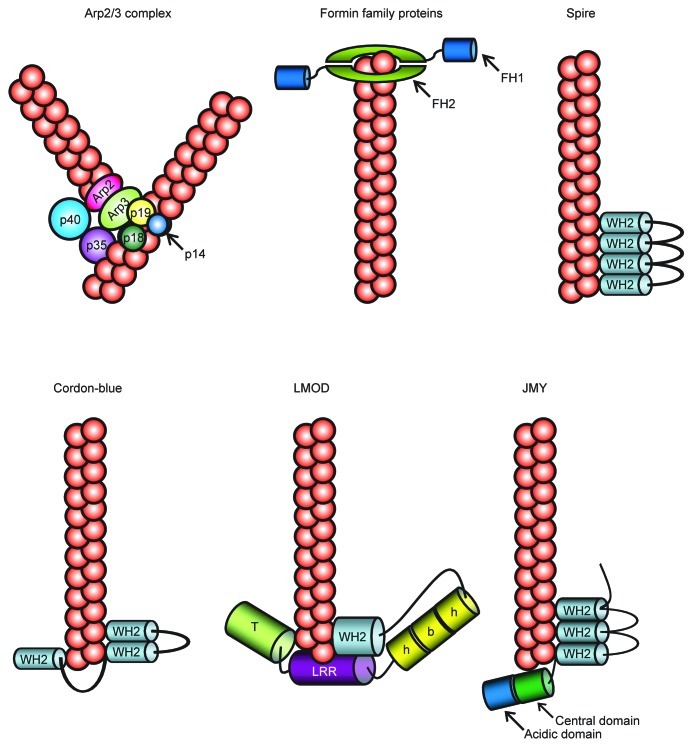
Temporal and spatial remodeling of the actin cytoskeleton plays a central role in cell morphology, polarity, migration and cytokinesis. However, since there are many factors that inhibit de novo actin nucleation in cells, spontaneous nucleation of G-actin (globular-actin) to form actin filaments is very slow. Thus, distinct nucleators are required to promote actin filament formation in cells. Recent works have shown that there are various proteins that are capable of stimulating actin nucleation and/or polymerization, including formins, Arp2/3 complex, spire, cordon-blue (COBL), leiomodin (LMOD), junction-mediating and -regulatory protein (JMY) (Fig. 1).1,2 These distinct actin nucleators employ different actin nucleating mechanisms. Arp2/3 binds to the side of pre-existing actin filaments and generates branched actin filament networks. Spire, COBL and LMOD are new classes of proteins that contain between one and four G-actin-binding motifs, including WASP-homology 2 (WH2) domain. Their nucleation mechanisms of unbranched actin filaments are related, but each may generate an actin nucleus with distinct properties. Spire stably associates with four actin monomers in a prenucleation complex, and remains associated at the side of the pointed end of an actin filament after nucleation, allowing for free barbed end elongation. COBL uses three WH2 domains and stabilizes both short-pitch and long-pitch associations to generate actin polymerization nuclei while, LMOD uses a single WH2 domain along with two unrelated actin binding domains to assemble trimeric actin nuclei and might remain associated at their pointed end. Furthermore JMY nucleates actin by both activating Arp2/3 and assembling filaments directly using a spire-like mechanism. Additionally, formin family proteins nucleate and accelerate unbranched actin filaments through binding to the barbed-end of actin filaments.
Figure 1. A scheme of actin polymerization by Arp2/3, formin, and new classes of actin nucleators. Arp2/3 binds to the side of pre-existing actin filaments and generates branched actin filament networks. FH2 domain of Formin family proteins nucleate and accelerate unbranched actin filaments through binding to the barbed-end of actin filaments. Spire associates with four actin monomers in a prenucleation complex, and remains associated at the side of the pointed end of an actin filament after nucleation. COBL stabilizes both short-pitch and long-pitch associations to generate actin polymerization nuclei through three WH2 domains. LMOD assembles trimeric actin nuclei and might remain associated at their pointed end. JMY nucleates actin by both activating Arp2/3 and assembling filaments directly using a Spire-like mechanism. FH, formin homology; WH2, WASP homology 2; LRR, Leucin-rich repeats; T, tropomyosin- and actin-binding helices; h-b-h, helix-basic-helix.
Formin family
Formins are defined by the presence of a highly conserved C-terminal formin homology (FH) 1 and 2 domains and comprise a large family that is conserved from yeast to mammals.3 The FH2 domain catalyzes de novo actin nucleation and polymerization, being persistently associated with the barbed end of nascent filaments and protecting them from capping proteins.3-5 The adjacent FH1 domain recruits profilin-actin complexes and accelerates filament elongation. More than 30 formins have been described to date, with more than 15 members in vertebrates (Fig. 2).5 The phylogenetic analysis of FH2 domains reveals that the 15 formins can be classified into seven groups: the Dia, formin (FMN), Dishevelled-associated activator of morphogenesis (DAAM), FMN-like (FMNL, also known as FRL), inverted formin (INF), Delphilin and FH1/FH2 domain-containing protein (FHOD) groups.
Figure 2. The seven subfamilies of mammalian formins.
Regulation of mDia
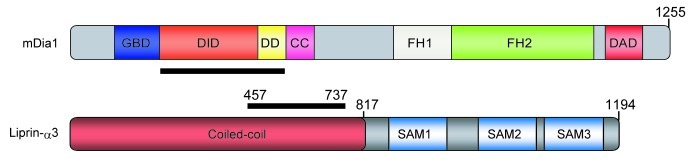
mDia is an effector of the small GTPase Rho, and is involved in the Rho-mediated formation of actin stress fibers, formation of the contractile ring, and formation of filopodia.6-9 Three mDia isoforms, mDia1, mDia2 and mDia3, are expressed in mammalian cells. mDia is composed of the GTPase-binding domain (GBD), Dia-inhibitory domain (DID), dimerization domain (DD), and coiled-coil domain (CC) in the N-terminus, and FH1-FH2 domain and Dia auto-regulatory domain (DAD) in the C-terminus (Fig. 3). In the resting state, mDia is auto-inhibited via intra-molecular interaction between DID and DAD, which inhibits the ability of FH1-FH2 to nucleate and elongate actin filaments in vitro and in vivo. In response to intra- or extra-cellular stimuli the inactive GDP-bound form of Rho is converted to active GTP-bound form and binding of activated Rho to GBD disrupts the DID-DAD interaction and leads to activation of mDia.6 Activated mDia then induces actin polymerization through the FH1-FH2 domain.
Figure 3. Domain structure of mDia1 and Liprin-α3. The bold lines represent the mDia-Liprin-α interacting regions. Amino acid number for the N- and C- terminal residues of each mutant protein are shown.
Although activation mechanisms of formins are thus well characterized, inactivation mechanisms of formins remain poorly understood. Intriguingly, whereas purified formins remain bound to and elongate the barbed end for many minutes to produce long filaments, actin cables and the cytokinetic ring assembled by yeast formins in vivo are comprised of relatively short filaments of 0.3–2.3 μm in length.10-14 These findings indicate that the function of mDia is crucially regulated in the cell. Recently, three formin-binding proteins with inhibitory actions have been identified, they are, Dia-interacting protein (DIP), Saccharomyces cerevisiae Bud14 and Drosophila melanogaster Spire.15-17 DIP specifically inhibits mDia2-mediated actin nucleaction and bundling activity in vitro by direct binding to FH2 domain. Bud14 also binds to the FH2 domain of yeast formin Bnr1 and displaces Bnr1 from growing barbed ends in vitro, and regulates actin cable length and architecture in vitro. The kinase noncatalytic C-lobe domain of Spire binds to the FH2 domain of D. melanogaster formin Cappuccino and strongly inhibits actin nucleation by the FH2 domain in vitro. Thus, all of these formin binding proteins inhibit formin-mediated actin polymerization by blocking the FH2 domain.
Notably, release of auto-inhibition by activated Rho is suggested to induce not only actin polymerizing activity but also membrane localization of mDia in the cell.3 Because endogenous mDia1 is mainly distributed diffusely in the cytoplasm, membrane targeting of mDia molecules has been studied using truncation fragments.18,19 Such studies have revealed that membrane localization of mDia is mediated through its DID-DD-CC regions in addition to its binding to Rho.19-22 These results suggest that, in addition to regulation by Rho, the activity and localization of mDia are regulated by its interaction with DID-DD-CC-binding proteins.
In our recent paper, we searched for mDia-binding proteins by pull-down assay with GBD-DID-DD regions of mDia1 as a bait in mouse brain lysates, and identified Liprin-α as an mDia-binding protein.23 Moreover, we showed that Liprin-α controls mDia-mediated actin stress fiber formation through regulating its membrane localization.
Liprin Family Proteins
Liprin family
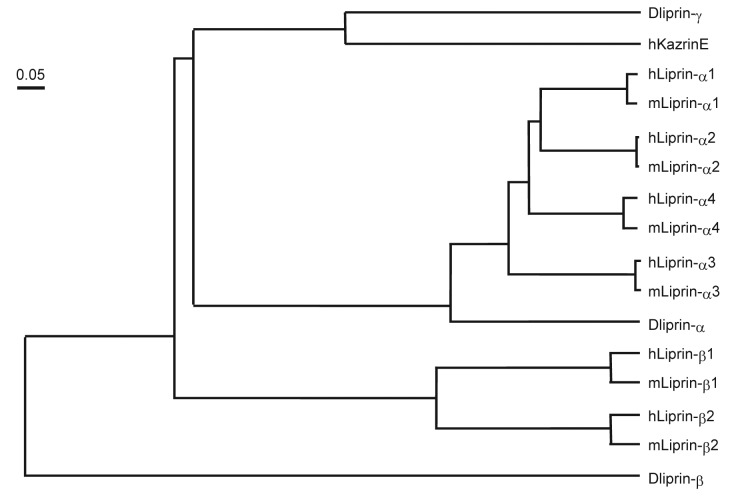
Liprin was first identified as an interacting protein of the leukocyte common antigen-related (LAR) family of receptor protein tyrosine phosphatase in 1995.24 Liprin is composed of the coiled-coil domain in the N-terminus and three consecutive sterile-α-motif (SAM) domains (often referred to as Liprin homology domain) in the C-terminus (Fig. 3). The second SAM domain of liprin-α interacts with the intracellular domain of LAR. Liprin is a large family of proteins that are classified into three types: Liprin-α, Liprin-β and KazrinE (Fig. 4). Vertebrate contain four Liprin-αs (α1–4), two Liprin-βs (β1, 2) and KazrinE. Drosophila and Xenopus also have three types of Liprins: Liprin-α, β, and γ. KazrinE and Liprin-γ have been recently identified as a third type of protein related to the Liprin family.25-28 Among four isoforms of Kazrin (KazrinA, B, C and E) only KazrinE contains the Liprin-α homology domain.27 In mammals, Liprin-α1, Liprin-β1 and β2 are expressed ubiquitously, whereas, Liprin-α2 and α3 are mainly expressed in the brain, and Liprin-α4 is in the brain, heart and skeletal muscle.25 In contrast, C. elegans contains only one Liprin-α, called syd-2 (synapse defective-2), which shares 50% identity to human Liprin-α1.
Figure 4. Liprin phylogenetic tree. Phylogram was generated by Genetyx of the evolutionary distances between the three Drosophila Liprins and their closest human and mouse homologs.
Function of Liprin-α
In cultured cells, Liprin-α was suggested to be involved in LAR distribution and clustering.25 Recent studies have shown that depletion of endogenous Liprin-α1 decreases migration and spreading of cultured cells, whereas the overexpression of the protein increased cell migration and spreading.29,30
Functions of Liprin-α have been well studied in neuronal cells. Liprin-α works as a scaffold and is important for presynaptic as well as postsynaptic development. Presynaptically, Liprin-α interacts with a diverse array of proteins such as RIM1α and ERC family that regulate the formation of the active zone.31,32 Loss of function mutants of the C. elegans syd-2 result in a diffuse distribution of presynaptic markers, lengthening of the presynaptic active zone, and impairment of synaptic transmission in the presynapse.33 Similarly, Dliprin-α, a Drosophila homolog of Liprin-α, mutant shows doubling of active zone length and defects in neurotransmitter release.34 Liprin-α also binds the MALS/Veli-Cask-Mint1 scaffolding complex that regulates cycling of reverse synaptic vesicles into the readily releasable pool of synaptic vesicles that are primed to fuse with the presynaptic membrane.35 These results demonstrate that Liprin-α is important for formation of the presynaptic active zone and regulation of neurotransmitter release. In addition, Dliprin-α is shown to be important for the formation of new synaptic boutons at the neuromuscular junction (NMJ) and R1-R6 and R7 photoreceptor axon targeting.36,37 Postsynaptically, Liprin-α is necessary for proper postsynaptic targeting of AMPA receptor and dendritic spine morphology.38 Liprin-α is also known to interact with KIF1A, a neuron-specific kinesin-3, and promotes the delivery of synaptic material by a direct increase in kinesin processivity and an indirect suppression of dynein activation.39,40
About Our Paper
In our recent work, we identified Liprin-α as a novel mDia-binding protein.23 The binding is mediated through the central region of Liprin-α and through the DID-DD regions of mDia (Fig. 3). Liprin-α competes with DAD for binding to DID, and binds preferably to the open form of mDia. Pull down analysis suggests that Liprin-α does not form a ternary complex with active RhoA and mDia. Therefore, we suggest a model for the mDia-Liprin-α interaction, that Liprin-α binds to mDia after active RhoA opens mDia and is dissociated from it. Given the popular opinion that mDia isoforms are maintained in an inactive state in the cytosol and then recruited to the plasma membrane upon activation where they catalyze actin polymerization, we examined the effect of Liprin-α on the actin cytoskeleton and the membrane localization of mDia.3 Overexpression of a Liprin-α fragment containing the mDia1-binding region attenuated the Rho-mDia mediated formation of stress fibers, and decreased localization of mDia at the plasma membrane. Conversely, depletion of Liprin-α by RNA interference enhanced formation of actin stress fibers and increased the amount of mDia in the membrane fraction. Taken together, we conclude that Liprin-α negatively regulates the activity of mDia in the cell by displacing it from the plasma membrane through binding to the DID-DD region and regulates the amounts of Rho-mDia-mediated actin stress fibers in the cell.
Remaining Questions
Molecular mechanisms
As we have now shown, mDia1 is recruited to the membrane in response to serum stimulation. We also found recruitment of Liprin-α to the membrane fraction at a slower late than mDia1 (data not shown). These results imply that Liprin-α activity may also be regulated downstream of serum stimuli. Overexpression of Liprin-α3(1–817) suppresses formation of Rho-mDia-induced stress fibers. By contrast, overexpression of full length Liprin-α3 had a slight effect and Liprin-α3(1–417) lacking the mDia-binding region or Liprin-α3(457–737) only containing mDia-binding regions, had no effect on the actin cytoskeleton. These results suggest that suppression of stress fiber formation by Liprin-α3(1–817) requires not only its binding site for mDia but also functional domain(s) or another binding site for other protein(s). Accordingly, we postulated that Liprin-α would be activated by serum or other yet to be identified factors following which the functional region is exposed and available to bind mDia or other proteins. As we described above, although Liprin-α preferably binds to an open, active form of mDia, it cannot bind to mDia in complex with the GTP-bound active form of RhoA. This means that Liprin-α binds to an open form of mDia after GTP-RhoA dissociates from it. This working model raises new questions: when does active-Rho dissociate from mDia, and how is dissociation of Rho from GBD regulated? Recently, Cho and coworkers reported that Xenopus Kazrin (xKazrinA) binds to p190B RhoGAP directly.41 They also showed that xKazrinA depletion significantly increases RhoA activity, and induces actin cytoskeleton reorganization. As KazrinA and KazrinE share the N-terminal coiled-coil domain, Liprin-α would also likely share this sequence. Hence, RhoGAP will be one of the candidates for interaction with Liprin-α, and subsequently regulate Rho activity and Rho-mDia-interaction, together with Liprin-α. If the N-terminal region of Liprin-α potentially associates with RhoGAP, we would propose the following hypothesis which would answer the questions described above. (1) Liprin-α binds to RhoGAP, and then RhoGAP accelerates the intrinsic GTPase activity of Rho and inactivates active Rho in complex with mDia. (2) GDP-bound inactive Rho dissociates from mDia and, after that, Liprin-α binds to mDia and dissociates it from the plasma membrane. To further analyze how the Rho-mDia signaling is regulated in cells, examination of these hypotheses will be necessary.
Physiological significance
In immature neurons, Liprin-α is necessary for the development of synapses. In Drosophila, both Liprin-α homolog Dliprin-α and LAR homolog Dlar localize presynaptically at the NMJ. Dliprin-α mutants display significant reduction in the number of synaptic boutons, an increase in the length of active zones, and a reduction in synaptic vesicle release at the NMJ.34 Dlar mutants have a nearly identical phenotype, which is consistent with a model in which Liprin-α and LAR work together to control synapse formation.34 LAR and Liprin-α appear to control synaptic growth through cytoskeletal rearrangement however, the precise mechanism remains unknown. Recently, Drosophila Diaphanous was also reported to localize presynaptically at the NMJ.42 Diaphanous mutants showed a decrease in the number of synaptic boutons as well as the number of actin puncta per bouton, and an increase in the rate of microtubule (MT) polymerization. Reduction of bouton number in Diaphanous mutants were similar to that of each Dlar/Diaphanous, Dlar/Trio (Rac1/RhoG and RhoA guanine-nucleotide exchange factor: GEF) or Diaphanous/Trio double heterozygous mutants, and neuronal expression of an active Diaphanous transgene which lacks both the N-terminal GBD and the C-terminal DAD domains, rescued bouton numbers in the Dlar mutant. In addition, Astigarraga and coworkers also reported that Trio expression in neurons is sufficient to rescue the NMJ phenotype of Liprin-α mutants.28 These results together suggest that Trio functions downstream of Liprin-α and Diaphanous functions downstream of Trio, and regulates synaptic growth through the regulation of synaptic actin polymerization and MT stabilization. However, how these proteins interact with each other remains unknown. In our work, we showed the direct and novel interaction between Liprin-α and mDia. Thus, Liprin-α may control synaptic growth at the NMJ downstream of LAR, by accurately regulating the activity of mDia.
Taken together, Liprin-α works not only as a scaffold protein but also as a regulatory protein of the actin cytoskeleton through the control of mDia activity. Thus, Liprin-α may now be included among the key molecules which link signaling molecules with the cytoskeleton.
Acknowledgments
We thank Aliza Ehrlich for critical reading of the manuscript. This work was supported in part by Grants-in-Aid for Scientific Research (grant numbers 18002015, 22501011, 23121514 and 23229003) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and the Global COE program “The Center of Frontier Medicine.”
Footnotes
Previously published online: www.landesbioscience.com/journals/BioArchitecture/article/20442
Reference
- 1.Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–51. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zuchero JB, Belin B, Mullins RD. Actin binding to WH2 domains regulates nuclear import of the multifunctional actin regulator JMY. Mol Biol Cell. 2012;23:853–63. doi: 10.1091/mbc.E11-12-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- 4.Moseley JB, Sagot I, Manning AL, Xu Y, Eck MJ, Pellman D, et al. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol Biol Cell. 2004;15:896–907. doi: 10.1091/mbc.E03-08-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Higgs HN. Formin proteins: a domain-based approach. Trends Biochem Sci. 2005;30:342–53. doi: 10.1016/j.tibs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 6.Watanabe N, Kato T, Fujita A, Ishizaki T, Narumiya S. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat Cell Biol. 1999;1:136–43. doi: 10.1038/11056. [DOI] [PubMed] [Google Scholar]
- 7.Hotulainen P, Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J Cell Biol. 2006;173:383–94. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watanabe S, Ando Y, Yasuda S, Hosoya H, Watanabe N, Ishizaki T, et al. mDia2 induces the actin scaffold for the contractile ring and stabilizes its position during cytokinesis in NIH 3T3 cells. Mol Biol Cell. 2008;19:2328–38. doi: 10.1091/mbc.E07-10-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schirenbeck A, Bretschneider T, Arasada R, Schleicher M, Faix J. The Diaphanous-related formin dDia2 is required for the formation and maintenance of filopodia. Nat Cell Biol. 2005;7:619–25. doi: 10.1038/ncb1266. [DOI] [PubMed] [Google Scholar]
- 10.Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, et al. Role of formins in actin assembly: nucleation and barbed-end association. Science. 2002;297:612–5. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- 11.Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol. 2002;4:626–31. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- 12.Higashida C, Miyoshi T, Fujita A, Oceguera-Yanez F, Monypenny J, Andou Y, et al. Actin polymerization-driven molecular movement of mDia1 in living cells. Science. 2004;303:2007–10. doi: 10.1126/science.1093923. [DOI] [PubMed] [Google Scholar]
- 13.Kamasaki T, Arai R, Osumi M, Mabuchi I. Directionality of F-actin cables changes during the fission yeast cell cycle. Nat Cell Biol. 2005;7:916–7. doi: 10.1038/ncb1295. [DOI] [PubMed] [Google Scholar]
- 14.Kamasaki T, Osumi M, Mabuchi I. Three-dimensional arrangement of F-actin in the contractile ring of fission yeast. J Cell Biol. 2007;178:765–71. doi: 10.1083/jcb.200612018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chesarone M, Gould CJ, Moseley JB, Goode BL. Displacement of formins from growing barbed ends by bud14 is critical for actin cable architecture and function. Dev Cell. 2009;16:292–302. doi: 10.1016/j.devcel.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisenmann KM, Harris ES, Kitchen SM, Holman HA, Higgs HN, Alberts AS. Dia-interacting protein modulates formin-mediated actin assembly at the cell cortex. Curr Biol. 2007;17:579–91. doi: 10.1016/j.cub.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 17.Quinlan ME, Hilgert S, Bedrossian A, Mullins RD, Kerkhoff E. Regulatory interactions between two actin nucleators, Spire and Cappuccino. J Cell Biol. 2007;179:117–28. doi: 10.1083/jcb.200706196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, et al. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 1997;16:3044–56. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brandt DT, Marion S, Griffiths G, Watanabe T, Kaibuchi K, Grosse R. Dia1 and IQGAP1 interact in cell migration and phagocytic cup formation. J Cell Biol. 2007;178:193–200. doi: 10.1083/jcb.200612071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seth A, Otomo C, Rosen MK. Autoinhibition regulates cellular localization and actin assembly activity of the diaphanous-related formins FRLalpha and mDia1. J Cell Biol. 2006;174:701–13. doi: 10.1083/jcb.200605006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe S, Okawa K, Miki T, Sakamoto S, Morinaga T, Segawa K, et al. Rho and anillin-dependent control of mDia2 localization and function in cytokinesis. Mol Biol Cell. 2010;21:3193–204. doi: 10.1091/mbc.E10-04-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorelik R, Yang C, Kameswaran V, Dominguez R, Svitkina T. Mechanisms of plasma membrane targeting of formin mDia2 through its amino terminal domains. Mol Biol Cell. 2011;22:189–201. doi: 10.1091/mbc.E10-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakamoto S, Ishizaki T, Okawa K, Watanabe S, Arakawa T, Watanabe N, et al. Liprin-α controls stress fiber formation by binding to mDia and regulating its membrane localization. J Cell Sci. 2012;125:108–20. doi: 10.1242/jcs.087411. [DOI] [PubMed] [Google Scholar]
- 24.Serra-Pagès C, Kedersha NL, Fazikas L, Medley Q, Debant A, Streuli M. The LAR transmembrane protein tyrosine phosphatase and a coiled-coil LAR-interacting protein co-localize at focal adhesions. EMBO J. 1995;14:2827–38. doi: 10.1002/j.1460-2075.1995.tb07282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serra-Pagès C, Medley QG, Tang M, Hart A, Streuli M. Liprins, a family of LAR transmembrane protein-tyrosine phosphatase-interacting proteins. J Biol Chem. 1998;273:15611–20. doi: 10.1074/jbc.273.25.15611. [DOI] [PubMed] [Google Scholar]
- 26.Spangler SA, Hoogenraad CC. Liprin-alpha proteins: scaffold molecules for synapse maturation. Biochem Soc Trans. 2007;35:1278–82. doi: 10.1042/BST0351278. [DOI] [PubMed] [Google Scholar]
- 27.Nachat R, Cipolat S, Sevilla LM, Chhatriwala M, Groot KR, Watt FM. KazrinE is a desmosome-associated liprin that colocalises with acetylated microtubules. J Cell Sci. 2009;122:4035–41. doi: 10.1242/jcs.047266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Astigarraga S, Hofmeyer K, Farajian R, Treisman JE. Three Drosophila liprins interact to control synapse formation. J Neurosci. 2010;30:15358–68. doi: 10.1523/JNEUROSCI.1862-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen JC, Unoki M, Ythier D, Duperray A, Varticovski L, Kumamoto K, et al. Inhibitor of growth 4 suppresses cell spreading and cell migration by interacting with a novel binding partner, liprin alpha1. Cancer Res. 2007;67:2552–8. doi: 10.1158/0008-5472.CAN-06-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asperti C, Astro V, Totaro A, Paris S, de Curtis I. Liprin-alpha1 promotes cell spreading on the extracellular matrix by affecting the distribution of activated integrins. J Cell Sci. 2009;122:3225–32. doi: 10.1242/jcs.054155. [DOI] [PubMed] [Google Scholar]
- 31.Schoch S, Castillo PE, Jo T, Mukherjee K, Geppert M, Wang Y, et al. RIM1alpha forms a protein scaffold for regulating neurotransmitter release at the active zone. Nature. 2002;415:321–6. doi: 10.1038/415321a. [DOI] [PubMed] [Google Scholar]
- 32.Ko J, Na M, Kim S, Lee JR, Kim E. Interaction of the ERC family of RIM-binding proteins with the liprin-alpha family of multidomain proteins. J Biol Chem. 2003;278:42377–85. doi: 10.1074/jbc.M307561200. [DOI] [PubMed] [Google Scholar]
- 33.Zhen M, Jin Y. The liprin protein SYD-2 regulates the differentiation of presynaptic termini in C. elegans. Nature. 1999;401:371–5. doi: 10.1038/43886. [DOI] [PubMed] [Google Scholar]
- 34.Kaufmann N, DeProto J, Ranjan R, Wan H, Van Vactor D. Drosophila liprin-alpha and the receptor phosphatase Dlar control synapse morphogenesis. Neuron. 2002;34:27–38. doi: 10.1016/S0896-6273(02)00643-8. [DOI] [PubMed] [Google Scholar]
- 35.Olsen O, Moore KA, Fukata M, Kazuta T, Trinidad JC, Kauer FW, et al. Neurotransmitter release regulated by a MALS-liprin-alpha presynaptic complex. J Cell Biol. 2005;170:1127–34. doi: 10.1083/jcb.200503011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choe KM, Prakash S, Bright A, Clandinin TR. Liprin-alpha is required for photoreceptor target selection in Drosophila. Proc Natl Acad Sci U S A. 2006;103:11601–6. doi: 10.1073/pnas.0601185103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hofmeyer K, Maurel-Zaffran C, Sink H, Treisman JE. Liprin-alpha has LAR-independent functions in R7 photoreceptor axon targeting. Proc Natl Acad Sci U S A. 2006;103:11595–600. doi: 10.1073/pnas.0604766103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dunah AW, Hueske E, Wyszynski M, Hoogenraad CC, Jaworski J, Pak DT, et al. LAR receptor protein tyrosine phosphatases in the development and maintenance of excitatory synapses. Nat Neurosci. 2005;8:458–67. doi: 10.1038/nn1416. [DOI] [PubMed] [Google Scholar]
- 39.Shin H, Wyszynski M, Huh KH, Valtschanoff JG, Lee JR, Ko J, et al. Association of the kinesin motor KIF1A with the multimodular protein liprin-alpha. J Biol Chem. 2003;278:11393–401. doi: 10.1074/jbc.M211874200. [DOI] [PubMed] [Google Scholar]
- 40.Miller KE, DeProto J, Kaufmann N, Patel BN, Duckworth A, Van Vactor D. Direct observation demonstrates that Liprin-alpha is required for trafficking of synaptic vesicles. Curr Biol. 2005;15:684–9. doi: 10.1016/j.cub.2005.02.061. [DOI] [PubMed] [Google Scholar]
- 41.Cho K, Vaught TG, Ji H, Gu D, Papasakelariou-Yared C, Horstmann N, et al. Xenopus Kazrin interacts with ARVCF-catenin, spectrin and p190B RhoGAP, and modulates RhoA activity and epithelial integrity. J Cell Sci. 2010;123:4128–44. doi: 10.1242/jcs.072041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pawson C, Eaton BA, Davis GW. Formin-dependent synaptic growth: evidence that Dlar signals via Diaphanous to modulate synaptic actin and dynamic pioneer microtubules. J Neurosci. 2008;28:11111–23. doi: 10.1523/JNEUROSCI.0833-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]