The secondary structure of encapsidated MS2 genomic RNA poses an interesting RNA folding challenge. Cryoelectron microscopy has demonstrated that encapsidated MS2 RNA is well-ordered. Models of MS2 assembly suggest that the RNA hairpin–protein interactions and the appropriate placement of hairpins in the MS2 RNA secondary structure can guide the formation of the correct icosahedral particle. The RNA hairpin motif that is recognized by the MS2 capsid protein dimers, however, is energetically unfavorable, and thus free energy predictions are biased against this motif. Computer programs called Crumple, Sliding Windows, and Assembly provide useful tools for prediction of viral RNA secondary structures when the traditional assumptions of RNA structure prediction by free energy minimization may not apply. This approach thoroughly explores conformational space and generates an ensemble of secondary structures. The predictions from this new approach can test hypotheses and models of viral assembly and guide construction of complete three-dimensional models of virus particles.
Keywords: viral RNA, RNA secondary structure predictions, RNA ensemble
Abstract
The secondary structure of encapsidated MS2 genomic RNA poses an interesting RNA folding challenge. Cryoelectron microscopy has demonstrated that encapsidated MS2 RNA is well-ordered. Models of MS2 assembly suggest that the RNA hairpin–protein interactions and the appropriate placement of hairpins in the MS2 RNA secondary structure can guide the formation of the correct icosahedral particle. The RNA hairpin motif that is recognized by the MS2 capsid protein dimers, however, is energetically unfavorable, and thus free energy predictions are biased against this motif. Computer programs called Crumple, Sliding Windows, and Assembly provide useful tools for prediction of viral RNA secondary structures when the traditional assumptions of RNA structure prediction by free energy minimization may not apply. These methods allow incorporation of global features of the RNA fold and motifs that are difficult to include directly in minimum free energy predictions. For example, with MS2 RNA the experimental data from SELEX experiments, crystallography, and theoretical calculations of the path for the series of hairpins can be incorporated in the RNA structure prediction, and thus the influence of free energy considerations can be modulated. This approach thoroughly explores conformational space and generates an ensemble of secondary structures. The predictions from this new approach can test hypotheses and models of viral assembly and guide construction of complete three-dimensional models of virus particles.
INTRODUCTION
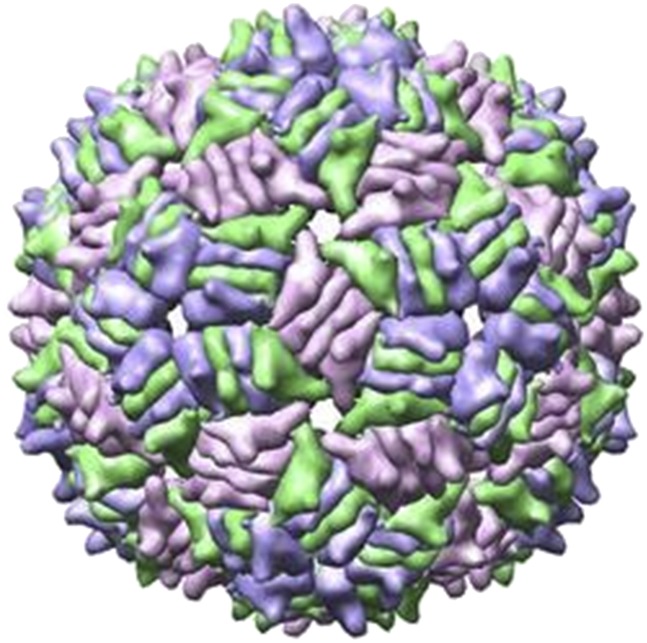
The MS2 RNA genome has a significant, functional structure when encapsidated inside the virus particle. Cryoelectron microscopy of MS2 particles attached to the Escherichia coli pilus at the fivefold vertex and analyzed with only fivefold averaging reveals that 90% of the encapsidated RNA is well-ordered and forms two layers inside the icosahedral viral particle (Toropova et al. 2008, 2011). The MS2 bacteriophage is a T = 3 icosahedral viral particle composed of 180 copies of the same coat protein (Valegard et al. 1990; Golmohammadi et al. 1993). These coat proteins form dimers, and dimers that bind an RNA hairpin have an allosteric conformational change in the FG loop region of one protein, making an asymmetric A/B dimer rather than a symmetric C/C dimer (Stockley et al. 2007; Dykeman and Twarock 2010; Dykeman et al. 2010; Rolfsson et al. 2010). The FG loops of the B conformation of the coat protein surround the fivefold vertices of the icosahedrons, and the FG loops of the A and C conformations of the coat protein interdigitate at the threefold vertices of the icosahedrons (Fig. 1) in order to make an icosahedron of 60 A/B dimers and 30 C/C dimers (Valegard et al. 1990; Golmohammadi et al. 1993). The symmetry of the icosahedron and the locations of the RNA hairpins in the A/B dimers suggest possible paths for the series of hairpins in the MS2 genome (Dykeman et al. 2011). Thus, binding RNA hairpins may play an important role in assembling a virus particle with the correct size and symmetry (Koning et al. 2003; Stockley et al. 2007; Basnak et al. 2010; Morton et al. 2010). Predictions of the MS2 bacteriophage RNA secondary structures can facilitate modeling of the assembly pathways and the complete three-dimensional structure of the MS2 virus particle.
FIGURE 1.
Surface representation of MS2 bacteriophage from Protein Data Bank file 1ZDH (Valegard et al. 1997), a crystal structure of in vitro assembled MS2 coat protein with synthetic RNA hairpins. A/B dimers are shown in blue and green. C/C dimers are shown in purple. The fivefold vertices appear as blue stars. The sixfold vertices appear as three purple and three green points.
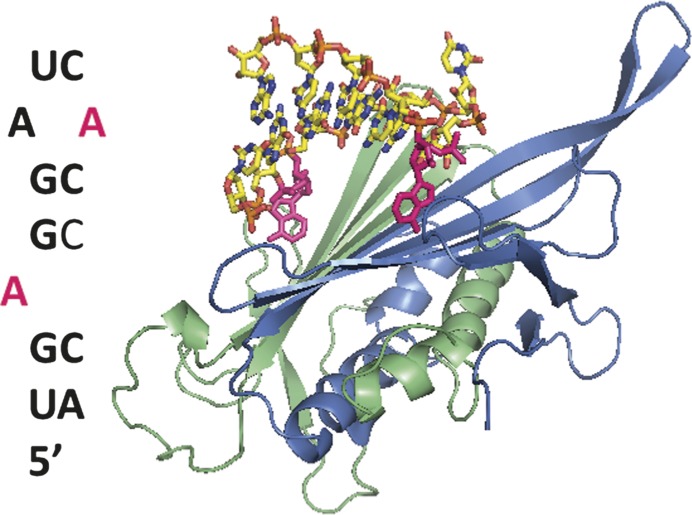
The MS2 bacteriophage RNA genome has a greater propensity for hairpin formation than other viral nucleic acid sequences (Nussinov and Sussman 1980). The trigger hairpin in MS2 initiates assembly of the viral particle and regulates translation (Beckett and Uhlenbeck 1988; Beckett et al. 1988; Pickett and Peabody 1993; Peabody 1997). Sequence comparisons to similar bacteriophage and SELEX experiments have identified a consensus motif for the trigger hairpin–coat protein dimer (TR-CP2) interaction (Schneider et al. 1992; Witherell et al. 1991; Hirao et al. 1999; Shtatland et al. 2000). The crystal structure of the virus particle formed in vitro with RNA trigger hairpins and coat protein reveals clear electron density for the RNA (Valegard et al. 1994, 1997). Crystallographic studies of several hairpins identified by SELEX experiments and hairpins with site-specific chemical modifications reveal the hydrogen bonding and stacking interactions that stabilize this RNA–protein interaction (Fig. 2; Valegard et al. 1994; Convery et al. 1998; van den Worm et al. 1998; Rowsell et al. 1998; Grahn et al. 1999, 2000, 2001; Helgstrand et al. 2002; Horn et al. 2004, 2006). The binding pocket creates space for hairpins of only 3–4 nucleotides (nt). The stacking interactions and hydrogen bonding to the amino group of the A in the 3′ position of the hairpin (which is often referred to as the −4 position relative to the AUG start sequence in MS2 RNA) favor an A nucleotide in that position in the SELEX experiments. A cytosine can also be selected in that position. The bulged nucleotide in the stem helix (the −10 position relative to the AUG start sequence) reaches across the protein dimer interface to make stabilizing hydrogen bonding and stacking interactions. The free energy of the trigger hairpin–coat protein dimer interaction is −11.9 kcal/mol (Beckett and Uhlenbeck 1988), and mutations and deviations from the consensus motif alter binding affinity (Carey et al. 1983; Lowary and Uhlenbeck 1987; Wu and Uhlenbeck 1987; Witherell et al. 1991; LeCuyer et al. 1996; Romaniuk et al. 1997; Johansson et al. 1998).
FIGURE 2.
Trigger RNA hairpin-coat protein dimer interaction. The asymmetric A/B dimer of two coat proteins is shown in blue and green ribbons. The RNA helix stretches across the β-sheet dimer interface. Two adenines that make important hydrogen bonds with each coat protein are shown in bright pink. The adenine in the 3′ position of the hairpin hydrogen bonds with lysine, threonine, and serine amino acids on the blue dimer. The single bulge adenine interacts with lysine, threonine, and serine amino acids on the green dimer. Figure created from Protein Data Bank file 1ZDH (Valegard et al. 1997), ViPER (Shepherd et al. 2006), and Pymol (Schrodinger 2008).
Although the TR-CP2 interaction is very energetically and structurally stabilizing, the consensus RNA secondary structure motif contains energetically unfavorable motifs. Each hairpin initiation and each bulge loop initiation adds a free energy penalty to secondary structure predictions (Mathews et al. 2004). RNA secondary structure predictions based on traditional free energy minimization do not include RNA–protein interactions and assume that the lowest free energy structure is the functional structure. In the case of encapsidated MS2 RNA, RNA–protein interactions provide significant energetic stability (60 × −11.9 kcal/mol = −732.0 kcal/mol maximum amount of additional free energy from protein binding) that creates an enormous energy window over which to search for possible suboptimal RNA structures. The free energy of the RNA–protein interaction (−11.9 kcal/mol) is larger than the free energy of RNA hairpin formation (−9.3 kcal/mol) (Witherell et al. 1991). Thus, the free energy from TR-CP2 interactions could overcome hairpin initiation and suboptimal RNA hairpin folding. Furthermore, viral assembly may be a kinetically driven rather than a thermodynamically determined process. Thus, traditional RNA secondary structure predictions based on free energy minimization may not be the best tool to predict possible structures for encapsidated MS2 RNA.
The Sliding Windows and Assembly approach offers an alternative method to predict RNA secondary structures when the assumptions of traditional free energy minimization may not apply (Schroeder et al. 2011). The basic idea is to predict all possible hairpins within small windows of the nucleotide sequence and then filter and score the possible hairpins based on experimental data. The Crumple program rapidly calculates all possible secondary structures for each window using only base-pairing rules and does not consider thermodynamics. The scoring function is flexible and can accommodate a wide variety of experimental data, including crystallography, cryoelectron microscopy, SELEX, chemical modification, and phylogenic comparisons. Thermodynamic parameters can also be included in the scoring function, but the advantage of Crumple and Sliding Windows is the ability to modulate the influence of thermodynamics on predictions. Crumple and Sliding Windows generate the elements of an ensemble of secondary structures. The Assembly step then finds combination of hairpins with the best possible overall score and can include constraints based on predicted paths of the genome on the icosahedral lattice. Assembly generates only one subset of the ensemble of possible solutions. This study presents predictions for encapsidated MS2 bacteriophage RNA based on the minimum number of helices from crystallography and cryoelectron microscopy (Valegard et al. 1990, 1994, 1997; Toropova et al. 2008, 2011), the sequence motifs identified by SELEX (Hirao et al. 1999; Shtatland et al. 2000), and predicted Hamiltonian paths of the RNA genome on the icosahedral lattice (Dykeman et al. 2011).
RESULTS
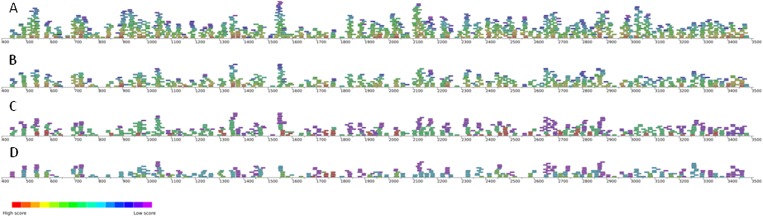
Scoring functions based on experimental data can reduce the possible conformational space of possible folds for an RNA sequence. In order to demonstrate how SELEX data can reduce the possible folds for MS2 RNA, each criterion, or characteristic of the SELEX consensus motif, is added one at a time, and a plot of the possible hairpins satisfying these criteria are shown in Figure 3. The x-axis is the nucleotide number, and the bars represent the nucleotides forming a helix. The color of the bar represents the score for that hairpin, or how well that hairpin satisfies the criteria. The scoring functions used for Figure 3, A through D, consolidate the results of several SELEX experiments done under several conditions with different selection stringency and competition (Witherell et al. 1991; Schneider et al. 1992; Hirao et al. 1999; Shtatland et al. 2000). The scoring functions describe motifs or global features of RNA that are difficult to directly incorporate into traditional RNA structure prediction programs.
FIGURE 3.
Predictions of MS2 RNA secondary structure ensembles using Crumple and Sliding Windows. Increasingly strict scoring functions reduce the conformational space of ensembles of RNA hairpins in encapsidated MS2 RNA. MS2 RNA nucleotide numbers between 415 and 3509 are along the x-axis. The bars represent nucleotides forming a hairpin that satisfies the experimental constraints as defined by a scoring function. The color of the bar shows how well the hairpin satisfies the scoring criteria. The scoring function for each ensemble A–D has progressively more detailed criteria. (A) Hairpins of 3–4 nt with at least five pairs in the stem. (B) Hairpins with an A or C in the 3′ position. (C) Hairpins with an A or U in the 5′ position of 4- or 3-nt loops, respectively. (D) Hairpin stems with bulged nucleotides preferred 2 or 3 nt below 4- or 3-nt loops, respectively.
For MS2 RNA, the least restrictive scoring function identifies all possible hairpins of 3 or 4 nt with at least 5 bp in the stem. Watson-Crick or GU pairs have the best score. Helices with fewer mismatches have better scores, and terminal mismatches are scored better than internal mismatches. Helices containing bulges and internal loops are also allowed. The scoring for helices thus also favors helices that would likely be thermodynamically stable, but does not directly calculate predicted free energies. This collection of possible hairpins in the MS2 sequence is shown in Figure 3A.
The scoring can be further refined by including the preferences for hairpin binding sequences determined by SELEX (Witherell et al. 1991; Schneider et al. 1992; Hirao et al. 1999; Shtatland et al. 2000). An A nucleotide is strongly selected in the 3′ position of the hairpin for both 3- and 4-nt hairpins. A C nucleotide is also allowed with a lower score. The set of hairpins scored with this additional criterion is shown in Figure 3B. The preferences for an A in the 5′ position of 4-nt hairpins and a U in the 5′ position of 3-nt hairpins can also be added to the scoring function (Fig. 3C).
In the case of MS2 RNA, helices with a single nucleotide bulge 2 bp below a 4-nt hairpin or 3 bp below a 3-nt hairpin bind the coat protein more favorably (Fig. 2). The bulged nucleotide reaches across the dimer interface and stacks and hydrogen bonds with several amino acids. One monomer of the protein dimer interacts with the bulged nucleotide, and the other interacts with the hairpin nucleotides. An adenine is the most selected nucleotide for the bulge, followed by a guanine. A pyrimidine is ranked lower. A mismatch (1 × 1 nucleotide loop) or a 1 × 2 nucleotide asymmetric loop that would have a similar bulged nucleotide is ranked next. Helices with no bulge are ranked lowest. The results from the scoring function that includes these preferences for bulged nucleotides are shown in Figure 3D.
All the scoring functions restrict the conformational space of possible RNA foldings. The amount of restriction depends on how many criteria are applied and on whether the criteria is defined as an absolute requirement or a preference with higher rank. The scoring function is, therefore, extremely important in determining which helices and which assemblies will emerge from the assembly process.
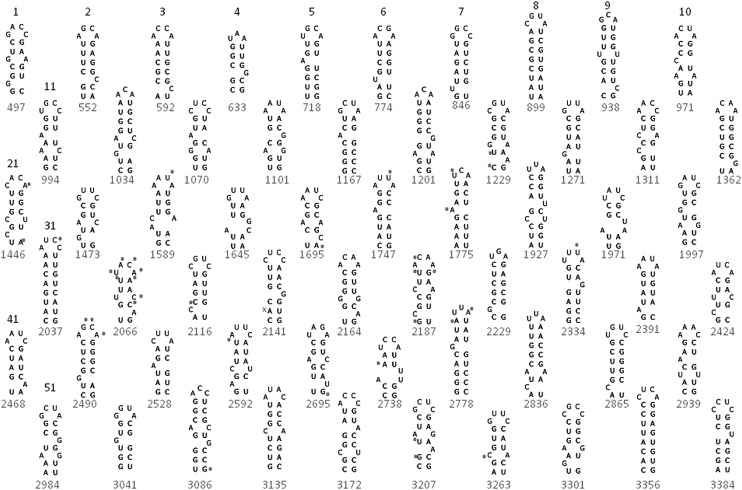
The Assembly step identifies 60 hairpins that best satisfy the criteria and give the best score. Sixty hairpins are proposed to bind the 60 A/B coat protein dimers. This constraint is derived from crystallography and cryoelectron microscopy data on MS2 viral particles (Valegard et al. 1990; Golmohammadi et al. 1993; Stockley et al. 2007; Toropova et al. 2008, 2011; Dykeman and Twarock 2010; Dykeman et al. 2010; Rolfsson et al. 2010). The predicted Hamiltonian path for the series of hairpins is based on the geometry of the icosahedral lattice (Dykeman et al. 2011). This provides an additional constraint for the spacing between hairpins that is included in the Assembly step. The hairpins identified in the Assembly step from the ensemble in Figure 3D are shown in Figure 4. The ensembles in Figure 3 best represent the set of predicted structures for encapsidated MS2. The hairpins resulting from the Assembly step (Fig. 4) are the best representatives of the ensemble for a given scoring function and follow the best proposed Hamiltonian path. The set of hairpins in Figure 4 requires only 10 short steps and can fit more than one possible Hamiltonian path. Any nonoverlapping set of 60 hairpins that follows any of the proposed Hamiltonian paths in either direction would satisfy all the experimental constraints.
FIGURE 4.
Set of 60 hairpins that best satisfy the scoring function in Figure 3D and the best-predicted Hamiltonian path for MS2 RNA (Dykeman et al. 2011). The helix numbers are listed only for the top row (hairpins 1–10) and then the first hairpin on the left in each row. The number below the hairpin represents the first 5′ nucleotide in the helix. Hairpins are listed in 5′-3′ sequence order. Small circles indicate strong chemical modification hits on in vitro–transcribed free MS2 RNA (Fiers et al. 1976; Skripkin et al. 1990). An x marks the single chemical modification hit at A2142 that is inconsistent with the predicted hairpins. Chemical modification is allowed at helix ends, in GU pairs, adjacent to GU pairs, and adjacent to bulges and loops (Mathews et al. 2004).
The representative single secondary structure (Fig. 4) has 60 hairpins of 3–4 nt. All the hairpins have either an A or C in the 3′ position. Thirty-four of the hairpins have a bulged nucleotide as a single bulge, a single mismatch, or a 1 × 2 loop in the appropriate position to stabilize protein dimer binding. Bulges further down the helix may also potentially stabilize the RNA-protein interaction, and all but four of the loops have at least one bulged nucleotide. Thus, these hairpins match the SELEX data well. In contrast, the ensemble centroids predicted by SFOLD (Ding et al. 2004a,b) or Vienna software (Gruber et al. 2008) have many hairpins but few that would form stabilizing RNA-coat protein interactions (Table 1). GNRA tetraloops are a common motif found in secondary structures predicted with free energy minimization and would satisfy many of the criteria, but very few of the hairpins predicted with standard software include bulged nucleotides. Thus, the flexibility of the scoring functions in the Sliding Windows and Assembly approach enables the generation of secondary structures that best match the experimental data.
TABLE 1.
Predicted hairpins in MS2 RNA sequences

The unrestrained Vienna predicted centroid structure comes very close to generating 60 hairpins that would satisfy the dimer protein binding criteria. Sixteen of the 60 hairpins in Figure 4 are also present in the Vienna predicted structure. This may result from the stability of GNRA tetraloops, stable 1 × 1 loop thermodynamic parameters, or the inclusion of thermodynamic stability as the last criterion (as a tie-breaker when all other scores are equal) in the scoring functions used to generate Figures 3 and 4. The Vienna predicted structure, however, contains many multibranch loops and does not satisfy constraints from possible Hamiltonian predicted paths. Although the structures generated by SFOLD and Vienna software have substantially lower free energies, the energetic stabilization when forming virus particles would be less.
In addition, the secondary structure resulting from Assembly is intentionally underpredicted; there are many more possible pairing interactions that can form within the 2542 nt between the 60 hairpins. These additional pairs may be better identified in future modeling studies of the RNA and protein in the three-dimensional virus particle. The assembly program produces only the minimum hairpins necessary to satisfy the crystallography and cryoelectron microscopy data. Future studies and more asymmetric constraints will be necessary to continue modeling the remaining encapsidated MS2 RNA.
DISCUSSION
The approach of using Crumple, Sliding Windows, and Assembly to generate an ensemble of RNA structures is a useful tool when the assumptions of traditional free energy minimization may not hold. In the case of encapsidated MS2 RNA structure, the motif that binds the coat protein dimers has a very energetically favorable RNA–protein interaction but a thermodynamically unfavorable RNA secondary structure. The ensemble in Figure 3D and the set of hairpins in Figure 4 provide a useful starting point for modeling the remaining RNA structures and the full virus structure. The hairpins presented here provide a better starting point than traditional secondary structure methods for the following reasons: The motif for RNA–protein binding interaction is a better match, the set of hairpins satisfies a Hamiltonian path and thus guides the relative placement of hairpins in the particle, and the RNA structure is inherently underpredicted in order to allow flexible fitting with and around the remaining protein.
The hairpins identified in the Assembly step (Fig. 4) are consistent with all but one of the 110 strongest chemical modification hits in previous studies of MS2 RNA free in solution (Fiers et al. 1976; Skripkin et al. 1990). These data were not included in the prediction, but the scoring function could easily include this kind of data. Chemical modification can occur in unpaired nucleotides or flexibly paired nucleotides adjacent to GU pairs, loops, or the end of a helix (Mathews et al. 2004). The one exception in the MS2 prediction is A2142, which is hit strongly by dimethyl sulfate but is predicted to occur between two Watson-Crick base pairs in hairpin 34. Hairpin 32 has many chemical modification hits that are computationally acceptable but may make hairpin formation unlikely in the absence of capsid proteins. Weak hits were not considered because these hits are not usually used in RNA structure predictions and, in the case of an ensemble of RNA structures, do not distinguish between strong hits in a minority conformation versus weak hits in a majority of conformations. The MS2 RNA in solution may also exist in an ensemble of RNA secondary structures. Predictions can be made about changes in chemical modification patterns for the ensemble in solution and the ensemble of structures inside the virus particle. For example, the nucleotides in the hairpin 42 loop are hit strongly, and the prediction is that these nucleotides would be protected upon capsid protein dimer binding.
The predictions from Sliding Windows and Assembly also generate testable hypotheses for virus assembly. One model of MS2 assembly poses that the TR-CP2 interaction directs the formation of pentamers and guides assembly of the correct T = 3 virus particle (Koning et al. 2003; Stockley et al. 2007; Basnak et al. 2010; Morton et al. 2010). If the TR-CP2 interaction is an essential interaction throughout the assembly process of the entire genome, then the predicted ensemble with the most constraints would best represent the encapsidated RNA conformations. This ensemble predicts that the adenines in RNA hairpins (233 adenines in the 60 hairpins in Fig. 4) would most often be protected. There are clear gaps between possible hairpins in Figure 3D. This ensemble of conformations would generate a chemical probing protection pattern with distinct groups of hits with very different intensities. On the other hand, if the TR-CP2 interaction is necessary only for initiation of assembly and then the cooperativity of viral assembly is strong enough that the specific TR-CP2 interactions become less important as assembly continues, then very loose restraints for hairpins, as shown in Figure 3A, may better represent the ensemble of encapsidated RNA conformations. This model predicts that very few adenines would not be involved in hairpins at least some of the time and thus have reduced DMS modification. One would expect weak to moderate DMS hits to occur with similar intensity across the entire genome without clusters of strong and weak hits in this model.
Previous models of MS2 bacteriophage secondary structure have been generated from chemical and enzymatic probing of the in vitro–transcribed MS2 RNA free in solution (Fiers et al. 1976; Skripkin et al. 1990). The MS2 RNA secondary structures may also change in the presence of the capsid proteins. Comparisons of chemical probing on in vitro and in capsa viral RNA genomes in cucumber mosaic virus, HIV, and xenotropic murine leukemia virus–related virus (XMRV) show some regions of structure conservation and some regions where chemical accessibility changes (Rodrigues-Alvarado and Roossinck 1997; Paillart et al. 2004; Wilkinson et al. 2008; Watts et al. 2009; Grohman et al. 2011). The previous MS2 RNA secondary structure predictions also predict a single minimum free energy structure. Long RNAs of 3569 nt are unlikely to adopt a single conformation in the absence of proteins and may better be described by a Boltzmann centroid (Ding and Lawrence 2001) or an ensemble of RNA structures (Quarrier et al. 2010; Marek et al. 2011; Schroeder et al. 2011). When folding the MS2 RNA sequence from nucleotides 415–3509 in a Vienna package (Gruber et al. 2008), the probability of the minimum free energy structure occurring is less than 0.005. In contrast, the predictions generated by Crumple and Sliding Windows represent the many possible hairpins that satisfy the experimental data as an ensemble of structures.
The ensemble model of encapsidated viral RNA implies structural heterogeneity inside the virus particle, which is consistent with crystallographic observations. The electron density for hairpins bound to the coat protein dimers is not observed in crystal structures of native virus particles (Valegard et al. 1990) but is observed in crystal structures of particles reconstituted with 19-mer RNA hairpins and coat proteins (Valegard et al. 1994). The crystal structures are icosahedrally averaged, which complicates the estimation of the true structural heterogeneity. The sequence variation in populations of native virus particles contributes to internal structural heterogeneity. Both structural heterogeneity within the virus particle and multiple orientations of the virus particle within the crystal lattice contribute to the lack of electron density for the RNA genome in the native virus crystal structure (Fisher and Johnson 1993; Larson et al. 1993, 1998; Larson and McPherson 2001; Schneemann 2006). The cryoelectron microscopy of native virus particles bound to the E. coli pilus with only fivefold averaging at 9 Å, however, shows RNA density at all dimer interfaces (Toropova et al. 2008, 2011). The most loosely constrained ensemble (Fig. 3A) contains the most variation in the RNA hairpins and contains many suboptimal binding hairpins with more structural heterogeneity. This loosely constrained ensemble may partly explain the lack of RNA density in the native virus crystal structure.
An additional possible explanation is less than full occupancy for an RNA hairpin at each A/B dimer interface. The trigger hairpin is important to initiate assembly (Beckett and Uhlenbeck 1988; Beckett et al. 1988; Pickett and Peabody 1993; Peabody 1997), but as assembly proceeds, the cooperativity of the coat protein assembly may provide sufficient energy to induce the A/B dimer transition and thus relax the requirement for RNA hairpin binding. The coat proteins alone form virus particles, although the efficiency of assembling the correct T = 3 capsid is lower. The fivefold averaging and 9 Å resolution in the cryoelectron microscopy may not reveal inhomogeneous occupancy. If less than 60 hairpins are necessary and the virus particle randomly adopts different orientations in the crystal lattice, then the RNA electron density may be averaged away. If less than 60 hairpins are necessary, the predictions for possible hairpins in Figure 3 does not change. In the Assembly step, if less than 60 hairpins are necessary, then the effect on the prediction is that the hairpins with the lowest scores drop out of the set. Future asymmetric reconstructions and extensive modeling are required to test different hypotheses of virus assembly and reconcile the crystallographic and current electron microscopy data. Crumple and Sliding Windows provide tools to generate possible structures to test hypotheses of viral RNA structure and assembly.
The Crumple, Sliding Windows, and Assembly approach facilitates incorporating a wide variety of experimental data into predictions of RNA secondary structures. The data about the minimum number and minimum length of helices in RNA conformations provided by cryoelectron microscopy and crystallography of viruses are a global constraint on RNA folding that is difficult to incorporate as a constraint in traditional RNA folding programs. SELEX data provide information of sequence preferences in motifs that may be contrary to free energy minimization rules. The approach is not limited by sampling methods or the assumptions of free energy minimization. Crumple is computationally fast because thermodynamic calculations are not included in every step of the prediction. The conformations of encapsidated viral genomic RNA involve significant protein–RNA interactions, and the viral assembly process may be kinetically rather than thermodynamically determined. Thus, Crumple, Sliding Windows, and Assembly provide a useful tool for prediction of viral RNA secondary structures when the traditional assumptions of RNA structure prediction by free energy minimization may not apply.
Many RNA do not fold into a unique single structure and are better described as an ensemble of structures (Uhlenbeck 1995; Solomatin et al. 2010; Marek et al. 2011). In the case of encapsidated viral RNA, the hypothesis is that the RNA is neither completely disordered nor a single unique structure but rather an ensemble of secondary structures with similar characteristics that facilitate viral assembly. The Crumple, Sliding Windows, and Assembly computations can generate an ensemble model of encapsidated viral RNA. The resulting predictions can test hypotheses and models of viral assembly, suggest new experiments, and guide construction of complete three-dimensional models of virus particles.
MATERIALS AND METHODS
Sliding windows
A collection of hairpin structures are generated using the Crumple program (Schroeder et al. 2011). All software for Crumple, Sliding Windows, and Assembly is freely available at http://adenosine.chem.ou.edu. The calculation is done with each window of 30 nt in the MS2 RNA genome sequence (GenBank file NC001417) between nucleotides 415–3509. The 5′ and 3′ ends of MS2 RNA interact with the maturase protein and thus are not included in the Crumple computation. The possible hairpin secondary structures are organized within the computation by their final 3′ base and length.
A scoring function specific for the MS2 folding problem is used to rank each helix, and a “best” helix is chosen for each possible position and length. A “better” helix has a lower score. The scoring function can include a variety of different types of experimental data and can be readily modified for each folding problem and the types of experimental data available. Several different scoring functions were developed to incorporate the characteristics of motifs identified by SELEX experiments. For example, one scoring function ranked hairpins according to the binding constant or the natural log of the binding constant for hairpins in one SELEX experiment. In order to accommodate the results of several SELEX experiments in different selection conditions, highly selected motifs and nucleotides preferred at specific positions are scored according to a combined selection order. Penalties several orders of magnitude larger are applied to disallow structures that are selected against in the SELEX experiments. The hairpin's thermodynamic stability is the last criteria considered as a tie-breaker when all other scores for other characteristics are equivalent. The series of scoring functions with increasingly specific criteria in Figure 3 demonstrate how the possible conformational space can be narrowed or fine-tuned. The definition of the possible conformational space is determined by the user and is limited only by the ability to define a scoring function that represents the experimental data. The scoring functions and files used to generate the results in Figures 3 and 4 are available at http://adenosine.chem.ou.edu/software.html#sliding.
Assembly
A dynamic algorithm for assembly is used to assemble 60 hairpins from a set of possible hairpins. A partial assembly is here defined as a set of mutually acceptable hairpins, i.e., hairpins that do not overlap or pseudoknot. A partial assembly has a score, equal to the sum of the scores of the hairpins it contains. It also has a count, equal to the number of hairpins it contains. A two-dimensional table of empty partial assemblies is created, with a width equal to the length of the sequence and a depth equal to the desired number of helices in the final assembly.
The calculation in the assembly algorithm begins at the first column of the first row. This is the “one-helix” row. At each entry in the row, the “best” helix that starts at the corresponding sequence location is chosen and compared in score to the helix in the previous entry in the row. Entries with no helices are considered to have infinite scores. The helix with the lower score is then added to the partial assembly of the current entry. In this way, each entry in the row represents the best-scoring single helix that occurs at or before that position.
To fill out the next row (the “two-helix” row), for each entry in order, the best helix of each length is examined for the corresponding sequence position. This time, however, the score of each helix is summed with a score from the previous row, which is the entry occurring just before the beginning of the current helix (the current column minus the width of the helix). This ensures that the helices are mutually compatible and do not overlap each other. Again, the best of these scores is compared to the score of the previous assembly on the two-helix row, and the lower scoring, i.e., the better of the two, is kept as the assembly for the current entry in the table. In this way, each entry in the second row represents the best-scoring two-helix assembly that can occur at or before that sequence position. This means that the final entry in the row represents the best scoring assembly of two helices in the whole sequence. Calculation continues, each row adding a single helix and creating the best-scoring assembly of n helices. When the entire table is filled out, the final entry of the final row will contain the best-scoring assembly of the desired number of helices.
The spacing between helices is a variable that can be used to include predictions of the path that the hairpins may follow to place a hairpin on each twofold axis with an A/B asymmetric dimer in the virus particle. These predictions are based on an analysis of Hamiltonian paths and estimates of the most efficient folding pathway (Dykeman et al. 2011). The step from an adjacent dimer, a short step, or a dimer that is part of another pentagon or hexagon, a long step, would have a defined minimum distance based on the geometry of the icosahedral particle. This distance can be used to estimate the minimum number of nucleotides required between helices. Different possible Hamiltonian paths have a different pattern of short and long steps. This pattern can be included in the assembly process as a further constraint on the best-scoring predicted structure. In Figure 4, the best proposed Hamiltonian path proposed by Dykeman et al. (2011) was used as a constraint.
For the MS2 analysis on an Athalon +6400 computer at 3.2 GHz with 4 GB RAM, crumpling 3094 windows of 30 nt required approximately 1 min and 4 MB of memory. Scoring and filtering the Crumple output requires 10–20 min depending on the scoring function. The assembly algorithm runs as N × M, where N is the length of the sequence and M is the number of helices. The limiting factor is the research time to define an appropriate scoring function. Thus, this approach to predicting an ensemble of secondary structures is fast and efficient.
ACKNOWLEDGMENTS
This work was supported by a National Science Foundation CAREER grant 0844913. All software used in this work is freely available at http://adenosine.chem.ou.edu/. We thank Adam Heck, Jonathan Stone, Jui-Wen Lui, Xiaobo Gu, Erik Dykemann, Reidun Twarock, and Peter Stockley for discussions about this work.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.032326.112.
REFERENCES
- Basnak G, Morton VL, Rolfsson O, Stonehouse N, Ashcroft AE, Stockley PG 2010. Viral genomic single-stranded RNA directs the pathway toward a T=3 capsid. J Mol Biol 395: 924–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett D, Uhlenbeck OC 1988. Ribonucleoprotein complexes of R17 coat protein and translational operator analog. J Mol Biol 204: 927–938 [DOI] [PubMed] [Google Scholar]
- Beckett D, Wu H-N, Uhlenbeck OC 1988. Roles of operatory and non-operator RNA sequences in bacteriophage R17 capsid assembly. J Mol Biol 204: 939–947 [DOI] [PubMed] [Google Scholar]
- Carey J, Lowary PT, Uhlenbeck OC 1983. Interaction of R17 coat protein with synthetic variants of its ribonucleic acid binding site. Biochemistry 22: 4723–4730 [DOI] [PubMed] [Google Scholar]
- Convery MA, Rowsell S, Stonehouse NJ, Ellington AD, Hirao I, Murray JB, Peabody DS, Phillips SE, Stockley PG 1998. Crystal structure of an RNA aptamer-protein complex at 2.8 A resolution. Nat Struct Biol 5: 133–139 [DOI] [PubMed] [Google Scholar]
- Ding Y, Lawrence CE 2001. Statistical prediction of single-stranded regions in RNA secondary structure and application to predicting effective antisense target sites and beyond. Nucleic Acids Res 29: 1034–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Chan C, Lawrence C 2004a. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res 31: 7280–7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Chan CY, Lawrence CE 2004b. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res 32: W135–W141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykeman EC, Twarock R 2010. All-atom normal-mode analysis reveals an RNA-induced allostery in a bacteriophage coat protein. Phys Rev E Stat Nonlin Soft Matter Phys 81: 031908 doi: 10.1103/PhysRevE.81.031908 [DOI] [PubMed] [Google Scholar]
- Dykeman EC, Stockley PG, Twarock R 2010. Dynamic allostery controls coat protein conformer switching during MS2 phage assembly. J Mol Biol 395: 916–923 [DOI] [PubMed] [Google Scholar]
- Dykeman EC, Grayson NE, Toropova K, Ranson NA, Stockley PG, Twarock R 2011. Simple rules for efficient assembly predict the layout of a packaged viral RNA. J Mol Biol 408: 399–407 [DOI] [PubMed] [Google Scholar]
- Fiers W, Contreras R, Duerinck F, Haegeman G, Iserentant D, Merregaert J, Min Jou W, Molemans F, Raeymaekers A, Van den Berghe A, et al. 1976. Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene. Nature 260: 500–507 [DOI] [PubMed] [Google Scholar]
- Fisher AJ, Johnson JE 1993. Ordered duplex RNA controls capsid architecture in an icosahedral animal virus. Nature 361: 176–179 [DOI] [PubMed] [Google Scholar]
- Golmohammadi R, Valegard K, Fridborg K, Liljas L 1993. The refined structure of bacteriophage MS2 at 2.8 A resolution. J Mol Biol 234: 620–639 [DOI] [PubMed] [Google Scholar]
- Grahn ES, Stonehouse NJ, Murray JB, van den Worm S, Valegard K, Fridborg K, Stockley PG, Liljas L 1999. Crystallographic studies of RNA hairpin in complexes with recombinant MS2 capsids: implications for binding requirements. RNA 5: 131–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grahn ES, Stonehouse NJ, Adams CJ, Fridborg K, Beigleman L, Matulic-Adamic J, Warriner SL, Stockley PG, Liljas L 2000. Deletion of a single hydrogen bonding atom from the MS2 RNA operator leads to dramatic rearrangements at the RNA-coat protein interface. Nucleic Acids Res 28: 4611–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grahn E, Moss T, Helgstrand C, Fridborg K, Sundaram M, Tars K, Lago H, Stonehouse NJ, Davis DR, Stockley PG, et al. 2001. Structural basis of pyrimidine specificity in the MS2 RNA hairpin-coat-protein complex. RNA 7: 1616–1627 [PMC free article] [PubMed] [Google Scholar]
- Grohman JK, Kottegoda S, Gorelick RJ, Allbritton NL, Weeks KM 2011. Femtomole SHAPE reveals regulatory structures in the authentic XMRV RNA genome. J Am Chem Soc 133: 20326–20334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A, Lorenz R, Bernhart S, Neubock R, Hofacker I 2008. The Vienna RNA Websuite. Nucleic Acids Res 36: W70–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgstrand C, Grahn E, Moss T, Stonehouse NJ, Tars K, Stockley PG, Liljas L 2002. Investigating the structural basis of purine specificity in the structures of MS2 coat protein RNA translational operator hairpins. Nucleic Acids Res 30: 2678–2685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao I, Spingola M, Peabody D, Ellington AD 1999. The limits of specificity: An experimental analysis with RNA aptamers to MS2 coat protein variants. Mol Divers 4: 75–89 [DOI] [PubMed] [Google Scholar]
- Horn WT, Convery MA, Stonehouse NJ, Adams CJ, Liljas L, Phillips SE, Stockley PG 2004. The crystal structure of a high affinity RNA stem-loop complexed with the bacteriophage MS2 capsid: further challenges in the modeling of ligand–RNA interactions. RNA 10: 1776–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn WT, Tars K, Grahn E, Helgstrand C, Baron AJ, Lago H, Adams CJ, Peabody DS, Phillips SE, Stonehouse NJ, et al. 2006. Structural basis of RNA binding discrimination between bacteriophages Qβ and MS2. Structure 14: 487–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson HE, Dertinger D, LeCuyer K, Behlen LS, Greef CH, Uhlenbeck OC 1998. A thermodynamic analysis of the sequence-specific binding of RNA by bacteriophage MS2 coat protein. Proc Natl Acad Sci 95: 9244–9249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning R, van den Worm S, Plaiser JR, van Duin J, Abrahams JP, Koerten H 2003. Visualization by cryo-electron microscopy of genomic RNA that binds to the protein capsid inside bacteriophage MS2. J Mol Biol 332: 415–422 [DOI] [PubMed] [Google Scholar]
- Larson SB, McPherson A 2001. Satellite tobacco mosaic virus RNA: structure and implications for assembly. Curr Opin Struct Biol 11: 59–65 [DOI] [PubMed] [Google Scholar]
- Larson SB, Koszelak S, Day J, Greenwood A, Dodds JA, McPherson A 1993. Double helical RNA in satellite tobacco mosaic virus. Nature 361: 179–182 [DOI] [PubMed] [Google Scholar]
- Larson NB, Day J, Greenwood A, McPherson A 1998. Refined structure of satellite tobacco mosaic virus at 1.8A resolution. J Mol Biol 277: 37–59 [DOI] [PubMed] [Google Scholar]
- LeCuyer KA, Behlen LS, Uhlenbeck OC 1996. Mutagenesis of a stacking contact in the MS2 coat protein–RNA complex. EMBO J 15: 6847–6853 [PMC free article] [PubMed] [Google Scholar]
- Lowary PT, Uhlenbeck OC 1987. An RNA mutation that increases the affinity of an RNA-protein interaction. Nucleic Acids Res 15: 10483–10493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek MS, Johnson-Buck A, Walter NG 2011. The shape-shifting quasispecies of RNA: one sequence, many functional folds. Phys Chem Chem Phys 13: 11524–11537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH 2004. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci 101: 7287–7292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton VL, Dykemna EC, Stonehouse NJ, Ashcroft AE, Twarock R, Stockley PG 2010. The impact of viral RNA on assembly pathway selection. J Mol Biol 401: 298–208 [DOI] [PubMed] [Google Scholar]
- Nussinov R, Sussman JL 1980. MS2 RNA has a potential to form an unusually large number of stable hairpins. J Theor Biol 85: 481–486 [DOI] [PubMed] [Google Scholar]
- Paillart J-C, Dettenhofer M, Yu X, Ehresmann C, Ehresmann B, Marquet R 2004. First snapshots of the HIV-1 RNA structure in infected cells and in virions. J Biol Chem 279: 48397–48403 [DOI] [PubMed] [Google Scholar]
- Peabody DS 1997. Role of the coat protein–RNA interaction in the life cycle of the bacteriophage MS2. Mol Gen Genet 254: 358–364 [DOI] [PubMed] [Google Scholar]
- Pickett GG, Peabody DS 1993. Encapsidation of heterologous RNAs by bacteriophage MS2 coat protein. Nucleic Acids Res 21: 4621–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarrier S, Martin JS, Davis-Neulander L, Beauregard A, Laederach A 2010. Evaluation of the information content of RNA structure mapping data for secondary structure prediction. RNA 16: 1108–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Alvarado G, Roossinck MJ 1997. Structural analysis of a necrogenic strain of cucumber mosaic cucumovirus satellite RNA in planta. Virology 236: 155–166 [DOI] [PubMed] [Google Scholar]
- Rolfsson O, Torpova K, Ranson NA, Stockley PG 2010. Mutually-induced conformational switching of RNA and coat protein underpins efficient assembly of a viral capsid. J Mol Biol 401: 309–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniuk PJ, Lowary P, Wu H-N, Stormo G, Uhlenbeck OC 1997. RNA binding site of R17 coat protein. Biochemistry 26: 1563–1568 [DOI] [PubMed] [Google Scholar]
- Rowsell S, Stonehouse NJ, Convery MA, Adams CJ, Ellington AD, Hirao I, Peabody DS, Stockley PG, Phillips SE 1998. Crystal structures of a series of RNA aptamers complexed to the same protein target. Nat Struct Biol 5: 970–975 [DOI] [PubMed] [Google Scholar]
- Schneemann A 2006. The structural and functional role of RNA in icosahedral virus assembly. Annu Rev Microbiol 60: 51–67 [DOI] [PubMed] [Google Scholar]
- Schneider D, Tuerk C, Gold L 1992. Selection of high affinity RNA ligands to the bacteriophage R17 coat protein. J Mol Biol 228: 682–689 [DOI] [PubMed] [Google Scholar]
- Schrodinger L. 2008. The PyMOL Molecular Graphics System Version 1.2r3pre. http://www.pymol.org.
- Schroeder SJ, Stone JW, Bleckley S, Gibbons T, Mathews DM 2011. Ensemble of secondary structures for encapsidated satellite tobacco mosaic virus RNA consistent with chemical probing and crystallography constraints. Biophys J 101: 167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd CM, Borelli IA, Lander G, Natarajan P, Siddavanahalli V, Bajaj C, Johnson JE, Brooks CL, Reddy VS 2006. VIPERdb: a relational database for structural virology. Nucleic Acids Res 34: D386–D389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtatland T, Gill SC, Javornik BE, Johansson HE, Singer BS, Uhlenbeck OC, Zichi DA, Gold L 2000. Interactions of Escherichia coli RNA with bacteriophage MS2 coat protein: genomic SELEX. Nucleic Acids Res 28: E93 doi: 10.1093/nar/28.21.e93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skripkin EA, Adhin MR, de Smit MH, van Duin J 1990. Secondary structure of the central region of the bacteriophage MS2 RNA: conservation and biological significance. J Mol Biol 211: 447–463 [DOI] [PubMed] [Google Scholar]
- Solomatin SV, Greenfield M, Chu S, Herschlag D 2010. Multiple native states reveal persistent ruggedness of an RNA folding landscape. Nature 463: 681–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockley PG, Rolfsson O, Thompson GS, Basnak G, Francese S, Stonehouse NJ, Homans SW, Ashcroft AE 2007. A simple, RNA-mediated allosteric switch controls the pathway to formation of a T=3 viral capsid. J Mol Biol 369: 541–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toropova K, Basnak G, Twarock R, Stockley PG, Ranson NA 2008. The three-dimensional structure of genomic RNA in bacteriophage MS2: implications for assembly. J Mol Biol 375: 824–836 [DOI] [PubMed] [Google Scholar]
- Toropova K, Stockley PG, Ranson NA 2011. Visualising a viral RNA genome poised for release from its receptor complex. J Mol Biol 408: 408–419 [DOI] [PubMed] [Google Scholar]
- Uhlenbeck OC 1995. Keeping RNA happy. RNA 1: 4–6 [PMC free article] [PubMed] [Google Scholar]
- Valegard K, Liljas L, Fridborg K, Unge T 1990. The three-dimensional structure of the bacterial virus MS2. Nature 345: 36–41 [DOI] [PubMed] [Google Scholar]
- Valegard K, Murray JB, Stockley PG, Stonehouse NJ, Liljas L 1994. Crystal structure of an RNA bacteriophage coat protein-operator complex. Nature 371: 623–626 [DOI] [PubMed] [Google Scholar]
- Valegard K, Murray JB, Stonehouse NJ, van den Worm S, Stockley PG, Liljas L 1997. The three-dimensional structures of two complexes between recombinant MS2 capsids and RNA operator fragments reveals sequence-specific protein-RNA interactions. J Mol Biol 270: 724–738 [DOI] [PubMed] [Google Scholar]
- van den Worm SH, Stonehouse NJ, Valegard K, Murray JB, Walton C, Fridborg K, Stockley PG, Liljas L 1998. Crystal structures of MS2 coat protein mutants in complex with wild-type RNA operator fragments. Nucleic Acids Res 26: 1345–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Swanstrom R, Burch CL, Weeks KM 2009. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460: 711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM 2008. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol 6: e96 doi: 10.1371/journal.pbio.0060096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witherell GW, Gott JM, Uhlenbeck OC 1991. Specific interaction between RNA phage coat proteins and RNA. Prog Nucleic Acid Res Mol Biol 40: 185–220 [DOI] [PubMed] [Google Scholar]
- Wu H-N, Uhlenbeck OC 1987. Role of a bulged a residue in a specific RNA-Protein interaction. Biochemistry 26: 8221–8227 [DOI] [PubMed] [Google Scholar]