Abstract
The chemoreceptors of Escherichia coli localize to the cell poles and form a highly ordered array in concert with the CheA kinase and the CheW coupling factor. However, a high-resolution structure of the array has been lacking, and the molecular basis of array assembly has thus remained elusive. Here, we use cryoelectron tomography of flagellated E. coli minicells to derive a 3D map of the intact array. Docking of high-resolution structures into the 3D map provides a model of the core signaling complex, in which a CheA/CheW dimer bridges two adjacent receptor trimers via multiple hydrophobic interactions. A further, hitherto unknown, hydrophobic interaction between CheW and the homologous P5 domain of CheA in an adjacent core complex connects the complexes into an extended array. This architecture provides a structural basis for array formation and could explain the high sensitivity and cooperativity of chemotaxis signaling in E. coli.
Keywords: bacterial chemotaxis, protein–protein interactions, receptor clustering, signal transduction
Bacterial chemotaxis is a prototypical model system for understanding basic principles of signal transduction. The core chemotaxis signaling complex, composed of chemoreceptors [also known as methyl-accepting chemotaxis proteins (MCPs)], the histidine kinase CheA, and the coupling protein CheW, senses chemical signals and transduces them to the flagellar motor to control its direction of rotation (1, 2). The signaling complex assembles into interconnected arrays at cell poles and cooperatively regulates kinase activity (3, 4). This clustering has been proposed to be important for signal amplification and the remarkable sensitivity of the system (5, 6). However, there are no high-resolution structures for the core signaling complexes or for the array in the well-characterized Escherichia coli model system.
Two of the four MCPs of E. coli are present in high abundance (7), and they sense serine (Tsr) and aspartate (Tar), respectively. The remaining two are present in lower amounts, and they sense ribose and galactose/glucose (Trg) or dipeptides (Tap) by interacting with periplasmic binding proteins for those compounds. Each MCP consists of a periplasmic ligand binding domain and a cytoplasmic signaling domain, connected by transmembrane helices. The conserved cytoplasmic signaling domain is organized as a trimer of dimers (for simplicity, trimers of dimers will be referred to simply as trimers.) (8). Transmembrane signaling occurs in the context of multicomponent arrays, in which the receptor signaling domains are interconnected by CheW and CheA (9).
The homodimeric histidine kinase CheA and its response regulator CheY transduce signals generated by the receptors to the flagellar switch (10). Each CheA monomer is composed of five structural domains with distinct functions. The N-terminal substrate domain, P1, is a five-helix bundle that contains His48, the site of phosphorylation. The P2 domain contains high-affinity docking surfaces for binding the response regulators CheY and CheB. The P2 domain is highly mobile and plays little structural role in core complex formation (11). Domains P3, P4, and P5 perform dimerization, catalytical, and regulatory functions, respectively. P5 is homologous to CheW and is essential for interaction of CheA with both CheW and the MCPs (12, 13). The CheA gene of E. coli encodes two proteins, CheAL (654 aa) and CheAS (557 aa) (14), made by initiating translation from different start codons. CheAS, which lacks all but helix 5 and part of helix 4 of P1, is sufficient to mediate the clustering of MCPs but cannot support chemotaxis on its own (15).
Significant progress has been made in determining the atomic structures of the individual components of the core signaling complex, including the cytoplasmic domains from E. coli Tsr (8) and Thermotoga maritima MCP1143C (16) and the various domains of CheA (17–19) and CheW (12, 20). The interactions of some of the key components have been characterized by intensive studies using X-ray crystallography, NMR, ESR spectroscopy, chemical interaction mapping, and disulfide cross-linking (11, 16, 21–24). Cryoelectron tomography (cryo-ET) has revealed the most detailed view of intact bacterial receptor arrays currently available (3, 25–27). The 12-nm hexagonal lattice demonstrated by these studies suggests that it is probably a universally conserved pattern (3). The averaged density of the arrays in Caulobacter cells suggests that six MCP trimers are arranged 7.5 nm apart in a hexagonally packed lattice (26). Cryo-ET was also used to visualize the receptor arrays in E. coli (27). However, the amount of extractable information is inadequate to determine the structures of the core signaling complex and the receptor array.
Results and Discussion
Minicell Generation.
A mutation in the shape-determining actin homolog mreB causes many cells to maintain their rod shape but to assume significantly smaller diameters (∼0.5 μm) than the parent (∼1 μm) (28). Introduction of a deletion of minCDE, inactivation of the ClpX gene, and overexpression of the plasmid-borne FlhC/FlhD genes in this strain result in a unique strain (WM4011) that produces flagellated minicells. These round minicells, which lack chromosomal DNA, are frequently generated by cytokinesis near the poles of the small-diameter cells, and they are often less than 0.5 μm in diameter. Their flagellation and large chemoreceptor arrays result from the overexpression of flhDC, which up-regulates the expression of most flagellar and chemotaxis genes (29).
Chemotaxis of Minicell-Producing Strain WM4011.
To determine whether strain WM4011 is normal for chemotaxis, we performed capillary assays (30) with the canonical attractants l-aspartate and l-serine. WM4011 and RP437 (31) cells, the latter serving as a chemotaxis-positive control, were grown in the absence or presence of flhDC overexpression induced with arabinose. In the absence of arabinose, both strains responded robustly to aspartate and serine (SI Appendix, Fig. S1), although more cells accumulated with strain RP437. After induction of flhDC overexpression, which increases the levels of all proteins involved in motility and chemotaxis, strain WM4011 was still able to carry out serine chemotaxis, although its response at high levels of attractant in the capillary (10 mM) was lower than that of strain WM4011 in the absence of arabinose. In contrast, chemotaxis with strain RP437 was just as good, or even better, after the addition of arabinose. We conclude that the skinny-cell phenotype does not impinge on normal chemotactic behavior to any great extent. However, overproduction of flhDC may decrease chemotactic performance, either because of an excess of flagella or overproduction of MCPs and other Che proteins. When expression of the flhDC plasmid in strain RP437 was induced with arabinose, the accumulation of viable bacteria in capillaries containing 10 mM serine also fell sixfold, from 120,000 to 20,000 cfu.
The capillary assay relies on colony counts. Therefore, it cannot directly measure the chemotaxis responses of minicells. However, because receptor arrays are located at the cell poles in E. coli (4), we conclude that the receptor arrays in minicells are probably structurally and functionally intact and normal.
Cryo-ET Reconstructions of Flagellated Minicells.
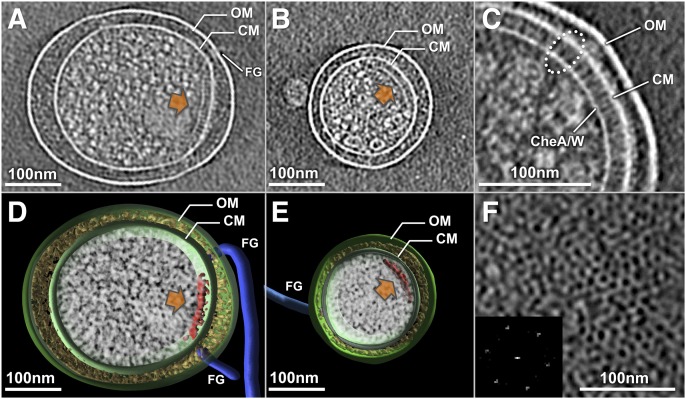
To achieve the best possible cryo-ET reconstructions, we particularly focused on minicells with diameters of 0.2–0.4 μm and with one to four flagellar motors (Fig. 1 A and B). Of 1,000 minicells examined, less than 20% contained readily visible receptor arrays. Tomographic reconstructions revealed the prominent features of an E. coli minicell: the outer membrane, the cytoplasmic membrane, the flagella, and the receptor array (Fig. 1 D and E). At higher magnification, the receptor arrays appear as clusters of pillar-like densities that extend from the cytoplasmic membrane and connect with a layer of high electron density at the their membrane-distal ends, as can be seen in a side view (Fig. 1C). The periplasmic and cytoplasmic domains of MCPs, as well as the distal layer that presumably contains CheA and CheW, can be readily recognized (Fig. 1C). From the top view, the receptor array appears as a hexagonal lattice (Fig. 1F). This pattern is consistent with the receptor arrays observed in many different species of bacteria (3).
Fig. 1.
Cryo-ET of E. coli minicells reveals receptor arrays. (A and B) Central slices of tomographic reconstructions show that the receptor arrays (orange arrows) in typical minicells varied from 200–400 nm in size. (D and E) Corresponding 3D models were generated by manually segmenting the outer membrane (OM; green), cytoplasmic membrane (CM; green), flagella (FG; blue), and receptor arrays (red). Electron densities in the cytoplasm and periplasmic space are shown in white and yellow, respectively. (C) In a zoom-in side view, the periplasmic domain, cytoplasmic domain of the MCPs, and CheA-CheW basal layer are readily discernible. (F) Top view of the receptor array and a power spectrum (Inset) reveal the hexagonal lattice.
Three-Dimensional Map of Receptor Arrays in Situ.
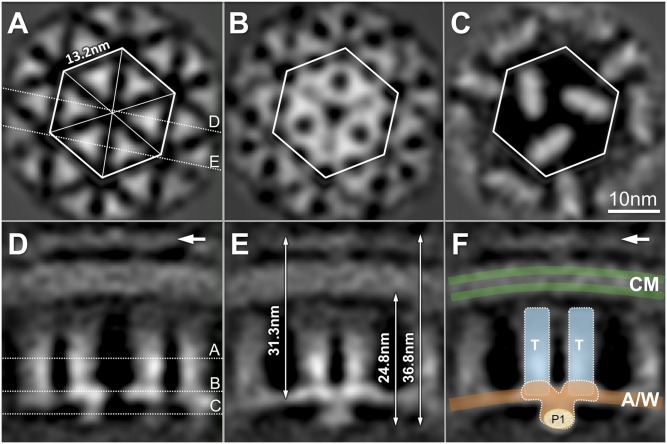
A 3.2-nm resolution map (Fig. 2) of receptor arrays was determined by subvolume analysis. The structure is similar in overall appearance to the structures reported for two known receptor arrays (25, 26), yet it reveals crucial previously undescribed details. In a transverse section parallel to the cytoplasmic membrane, our image shows a hexagonal lattice with 13.2-nm spacing (Fig. 2A). Six triangular densities form a hexagonal unit, and each of these is postulated to represent an MCP trimer. The distance between two adjacent trimeric densities is 7.5 nm. Notably, the flat surfaces of the trimeric densities face each other. This orientation is different from that predicted by other maps and models, which juxtapose the apices of the trimeric densities (23, 25, 26, 32). The densities increase in complexity at the distal end of the receptor array (Fig. 2 B and C). The locations of those slices are shown in a cross-section perpendicular to the cytoplasmic membrane (Fig. 2D).
Fig. 2.
The 3D map of receptor arrays derived from subvolume analysis. (A–C) Three transverse slices parallel to the membrane were taken at the positions shown in a cross-section perpendicular to the membrane. (A) Hexagonal lattice with 13.2-nm spacing contains six triangular densities, corresponding to the expected location of the MCP trimers. The distance between the centers of two adjacent triangular densities is 7.5 nm, and their flat surfaces face each other. (B) Pattern at the transverse slice across the CheA-CheW basal layer reveals complex densities that bridge between neighboring trimers. (C) At the membrane distal surface, three elongated densities predominate and presumably correspond to P1 and P4 domains of CheA dimers. (D and E) In slices perpendicular to the membrane, which are denoted in A, the pillar-like MCP densities are evident. White arrows indicate a density layer consistent with the periplasmic domains of the receptors. (E) Two densities merge together at the distal end of neighboring trimers. The total distance from the apex of the periplasmic domain to the distal end of the receptor array is 36.8 nm, whereas the distal end is 24.8 nm from the surface of the inner leaflet of the cytoplasmic membrane. A continuous-density layer at the distal end of the trimers is visible 31.3 nm away from the apex of the periplasmic domain. (F) Cartoon model of the MCP trimers (T; cyan), CheA/CheW layer (A/W; orange), and cytoplasmic membrane (CM; green) is overlaid onto the electron density map. (Scale bar: 10 nm.)
The pillar-like densities are perpendicular to the membrane (Fig. 2D), and the trimeric densities merge together at their distal ends (Fig. 2E). The distance between the distal end of the complex and the membrane-distal apex of the periplasmic domain is 36.8 nm, and the distal end is about 24.8 nm from the surface of the inner leaflet of the cytoplasmic membrane. The midpoint of the continuous-density layer mentioned above is about 31.3 nm from the apex of the periplasmic domain. We propose that the ends of two neighboring MCP trimers (colored blue in Fig. 2F) are connected by a continuous density layer at the base, which is likely formed by CheA and CheW, as illustrated in a cartoon model (Fig. 2F). Below this layer, an elongated density can be observed (Fig. 2 C and F). It is significantly smaller in the arrays derived from the CheAs strain BC212 (33) (SI Appendix, Fig. S8 D and H), suggesting that the P1 domains of the CheA dimer are located at the bottom of the complex (Fig. 2F).
Molecular Architecture of the Core Signaling Complex.
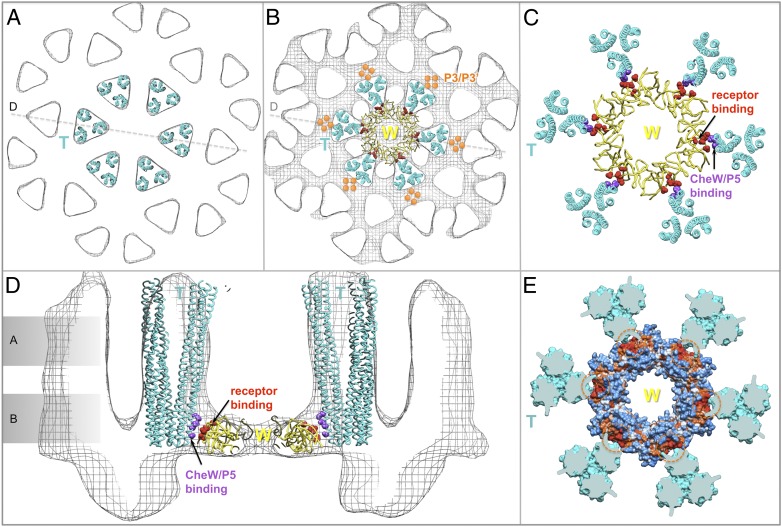
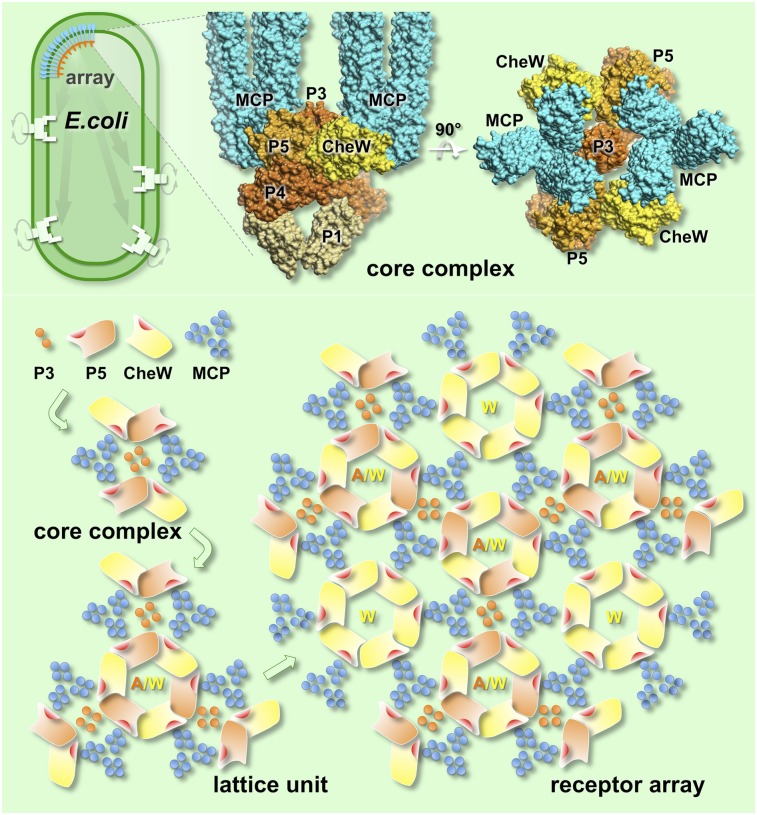
Our in situ density map of the receptor array provides a framework for understanding the architecture of the core complex and its arrangement inside the array at the molecular level. We computationally fit the atomic structures of individual components into the corresponding features of the electron density maps. The most prominent features of the density maps are the MCP trimers. The atomic model for the Tsr trimer (8), comprising residues A300–M480, fits very well into our density map as a rigid body (Fig. 3 A and D). Four dimers from two adjacent trimers face each other and pack closely with similar mutual distances (Fig. 3A). The highly conserved cytoplasmic tips (residues G370–S410) of two trimers are embedded in the continuous layer of densities corresponding to the P3 and P5 domains of CheA and CheW. Both CheW and the P5 domain of CheA contain docking surfaces that interact with the cytoplasmic tips of the Tsr trimers (Fig. 3 B and D). The well-defined trimeric densities disappear at residues A300 and M480, the region at which the Tsr dimers start to separate from each other and have higher atomic temperature B-factors (8). Thus, at this point, there must be a region of intrinsic flexibility (SI Appendix, Fig. S8), possibly corresponding to the glycine hinge (34).
Fig. 3.
Molecular architecture of the MCP/CheW/CheA core complex and the entire receptor array, formed by docking the atomic structures onto the Cryo-ET density map. A core complex with a stoichiometry of 6:2:1 (6 MCP dimers and 2 CheW monomers for each CheA dimer) was constructed by computationally fitting the atomic structures of individual components into the density map from Fig. 2. (A and D) Six structures of the Tsr trimer (residues 300–480; labeled as a cyan T) fit well into the map. (A–C) Progressive slices through the core complex (indicated in D), moving away from the membrane into the cytoplasm, are shown. (A) Four Tsr dimers face each other and form the interface between two adjacent trimers. At level c, only the P1 and P4 domains of CheA are well-resolved, with P2 apparently being too unstructured to provide a coherent density. (D) Cytoplasmic tips of the Tsr trimers are shown embedded into the density layer corresponding to CheA/CheW. Two Tsr trimers, outlined in A, belong to one complex (rendered as ribbons); they join together with one CheA dimer and two CheW (W) monomers in B and C. (B and E) Composite model of the CheA/CheW complex is placed between two cytoplasmic tips of two Tsr trimers, and the P3 domain of CheA is aligned roughly parallel to them. Residues I33, E38, I39, and V87 of CheW, which are critical for receptor binding, are shown in red. The red residues in the P5 domain of CheA are presumptive receptor-binding determinants, based on homology to CheW. The hydrophobic core of the Tsr trimer and hydrophobic residues connecting a Tsr dimer to P5 or CheW are highlighted in purple and/or indicated by a purple dashed circle. (E) These hydrophobic pockets are also shown as orange residues encircled by orange dashes in the hydrophobicity surface model at the top, with hydrophilic residues shown in blue and hydrophobic residues in orange. Both P5 and CheW can interact simultaneously with two different Tsr dimers to form a core complex. (E and F) Two adjacent core complexes are connected by a previously undescribed interaction between P5 and CheW. Three P5/CheW complexes form a ring. Subdomain-2 of CheW and subdomain-1 of P5 are critical for the CheW/P5 interaction (orange arrows), as is observed in the crystal structure of P4/P5/CheW (16). Residues D521, G629, V607, K616, A622, L633, and I634 from P5 and residues V45, T46, T51, K56, I65, M156, and L158 from CheW are colored in orange and yellow, respectively. Subdomain-2 of P5 is adjacent to subdomain-1 from the adjacent CheW. In F, a cyan arrow points to a second interface between P5 and CheW. Some residue substitutions at R555 (green) impair the in vivo chemotactic function of CheA (22). Several hydrophobic residues (L542, L545 and L552; cyan) are also located at this interface.
To correlate our structural model directly with the wealth of information available from mutational analysis and protein-protein interaction studies in E. coli, homology models of corresponding components of the E. coli receptor array were constructed based on atomic structures of P3/P4/P5 (18) and P4/P5/CheW (16) from T. maritima and P1 from Salmonella (19). We then built a large complex of P3/P4/P5 together with CheW, by aligning the three crystal structures (16, 18, 20) together (Materials and Methods).
We docked the P1 domains onto the bottom part of the receptor array, because the truncated P1 structure derived from a CheAS mutant does not contain this extra density (Fig. 3C and SI Appendix, Fig. S8). We then docked the whole P3/P4/P5/CheW complex as a rigid body into the map. The P3 dimer, which is a four-helix bundle, sits in the middle of the density surrounded by four Tsr dimers and is aligned roughly parallel to the distal ends of the trimers (Fig. 3 B and E), as was recently proposed by ESR spectroscopy (23) and chemical mapping (11). This arrangement implies that the P3 dimer and four Tsr dimers are tightly packed. The P4 domain is on top of the P1 domain. Several essential residues for P1 binding, derived from the chemical mapping study (11), are located precisely at the interface of P1/P4. Therefore, our initial model of the core complex is based primarily on available atomic structures of individual components and well-defined protein-protein interactions. Our 3.2-nm resolution map provides the critical interdomain geometries needed for building the core signaling complex model in the context of the in situ receptor array.
Despite a reasonable fit of the P1, P3, and P4 domains, P5 and CheW in our initial model did not fit in the electron density map. In addition, there was no obvious interaction between CheW and the Tsr trimer, although based on mutational analysis (35) and the crystal structures of Tsr (8) and CheW (12), the two proteins should have a distinct hydrophobic interaction. We therefore chose a residue (L509) in the flexible linker region between P4 and P5 (16) as a hinge for further model building. To maintain the extensive binding interface between them seen in the crystal structure (16), P5 and CheW were rotated around the hinge as a rigid body. The optimal fit was achieved when the P5 domain and CheW were placed at the same height relative to the Tsr trimers (Fig. 3D and Movie S1). A hydrophobic receptor binding surface of CheW [residues I33, E38, I39, V87 in E. coli; colored red (21)] closely approaches the hydrophobic residues of an E. coli Tsr dimer (F373, I377, L380, and V384; colored purple in Fig. 3E). The homologous region of the E. coli P5 domain (residues L528, E533, S534, and I581) is also close to the same hydrophobic residues from another Tsr dimer (Fig. 3 B and E). Both P5 and CheW can interact simultaneously with two different Tsr dimers through the hydrophobic interaction that holds CheW and Tsr together. Therefore, a CheA/CheW complex is able to bridge two adjacent Tsr trimers to generate a core complex consisting of two Tsr trimers, two CheW monomers, and one CheA dimer (Fig. 3 D and E).
This configuration is consistent with the observation that a complex having a 6:2:1 stoichiometry (6 MCP dimers and 2 CheW monomers for each CheA dimer) composes the minimal functional unit for receptor-coupled CheA activity (36). The resulting model preserves several key interactions: CheW/Tsr (21, 35), CheW/P5 (16), P1/P4 (11), and P3/MCPs (11, 23). It also provides unique insight into the interaction between Tsr and P5.
Modeling the Extended Receptor Array.
A key element of bacterial chemotaxis is the clustering of chemoreceptors that facilitates cooperativity and enhances sensitivity (2). Our model for the core complex can be extended as a receptor array (Fig. 3). The core complexes appear to be interconnected by a previously unknown interaction between subdomain-1 of CheW and subdomain-2 of P5 (Fig. 3F, cyan arrows). This interaction is distinct from that between subdomain-2 of CheW and subdomain-1 of P5 (Fig. 3F, orange arrows), as previously characterized by pulsed dipolar ESR spectroscopy (16) and cysteine-scanning analysis (13). Despite this distinction, these interfaces are formed by a similar hydrophobic interaction between two adjacent domains (Fig. 3 E and F) because of the structural homology between the P5 domain and CheW. Because several mutations affecting residues in subdomain-2 of P5 impair the in vivo chemotactic function of CheA (22), we favor the idea that suboptimal chemotaxis may be caused by decreased cooperativity within the receptor cluster. Therefore, we propose that the interactions between CheW and the homologous P5 domain are essential, not only for assembling the core complex but for clustering core complexes into a receptor array. Such interactions could allow different signaling complexes, including those containing the two low-abundance MCPs and the Aer redox receptor (37, 38), to be easily incorporated into mixed arrays.
Arrays Contain Hexameric Rings Consisting Only of CheW.
Three core complexes form one unit in the center of the density map, but they are also involved in forming six units surrounding the center (Fig. 2 and SI Appendix, Fig. S7). Three of them are identical to the one in the center, whereas another three are clearly distinct at the level of the CheA/CheW layer (Fig. 2 and SI Appendix, Fig. S8). A different 3D density map was derived from these units (Fig. 4 and SI Appendix, Fig. S8D). Six Tsr trimers fit well into the map (Fig. 4A), but there is insufficient density to form an intact CheA/CheW complex in the middle of the six Tsr trimers (Fig. 4 B and D). Therefore, we propose that six CheW monomers interact with six Tsr trimers to form a CheW-only ring in the middle (Fig. 4C). Notably, the CheW ring is able to maintain molecular interactions similar to those made by P5/CheW in that each monomer interacts with its adjacent Tsr trimer via strong hydrophobic interactions (Fig. 4E). This finding suggests that the CheW ring could reinforce the array as well as provide an explanation for receptor clustering in the absence of CheA (15).
Fig. 4.
CheW-only ring in the receptor array. (A and D) Six Tsr trimers (residues 300–480; labeled as a cyan T) were fitted into the map as a rigid body. (A and B) Progressive slices through the core complex in D, moving into the cytoplasm and away from the membrane, are shown. (B and C) Six CheW (W) molecules form a ring that is structurally similar to the CheW/P5 ring (Fig. 3) but lacks additional density beneath the CheW-only ring (d). Each CheW interacts with one dimer subunit of a Tsr trimer, which is free to form a core complex with an adjacent MCP trimer, and a P3/P3′ dimer serves as the central core. (C and D) At the interface between one Tsr trimer and one CheW monomer, residues I33, E38, I39, and V87 of CheW (colored in red) form a hydrophobic pocket (orange dashed circles) adjacent to hydrophobic residues (F373, I377, L380, and V384; purple) from Tsr. (E) Same residues from different Tsr monomers that form the hydrophobic core of the Tsr trimer (purple dashed circles in Fig. 3E) are shown in orange and highlighted with orange dashed circles in the hydrophobicity map of the CheW ring.
Molecular Basis of Array Formation.
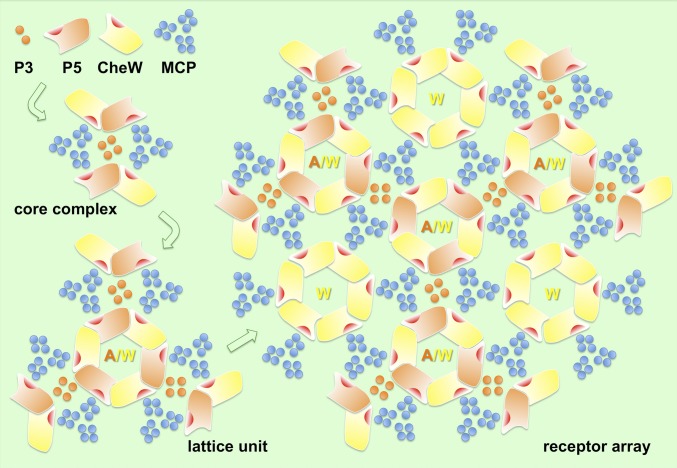
The functional unit of the receptor array is the core complex, which is formed by the interactions among the cytoplasmic tips of MCP trimers and the P3 and P5 domains of CheA and CheW. The P5/CheW interaction connects the core complexes into an array with 6:2:1 stoichiometry. Two of three MCP dimers interact with P5 or CheW, and the P5/CheW ring connects three core complexes (including 6 MCP trimers). This arrangement leaves one MCP dimer in each trimer without a binding partner. Therefore, six of these dimers group together and form a ring with a hole in the middle. Our data suggest that the CheW ring could fill this hole by interacting with these “free” MCP dimers to help stabilize the array (Fig. 5 and Movie S1).
Fig. 5.
Cartoon illustrates the assembly of an extended receptor array. The initial components, consisting of the P3 and P5 domains of CheA (orange), MCP trimers (blue), and CheW (yellow) form a core complex. Three core complexes are interconnected by P5/CheW interactions to form a lattice unit, which can assemble further to form an indefinitely large array. Rings containing only CheW may play a role in reinforcing the network to achieve optimal cooperativity and sensitivity.
The overall stoichiometry of the array shown in Fig. 5 is 6:4:1 (6 MCP dimers, 4 CheW monomers, and 1 CheA dimer). The occupancy of the CheW-only ring in vivo is likely to be variable. Therefore, the stoichiometry of the components in the arrays of living cells may vary from 6:2:1 to 6:4:1. A WT E. coli cell has been reported to contain 2.9 MCP dimers and 2.4 CheW monomers per CheA dimer (7). Thus, both CheA and CheW are in excess relative to the amounts needed to form extended arrays with or without CheW-only rings. The simplest explanation is that virtually all MCPs are captured in arrays, whereas a substantial portion of CheW and CheA is free in solution and able to exchange with their counterparts within the arrays. In fact, fluorescence recovery after photobleaching (FRAP) experiments have indicated an average lifetime at 20 °C of only about 12 min for both CheA and CheW within an E. coli receptor patch in vivo (39). In contrast, the receptors themselves persist within a patch for more than a cell generation (39).
Other studies have observed significantly more CheW monomers than CheA monomers (40, 41). Extra CheW may be helpful in generating the ultrastable networks seen in vitro (41). On the other hand, our model also suggests that hydrophobic residues within the Tsr dimer (F373, I377, L380, and V384) are involved in both Tsr trimer formation (8) and CheW binding (Fig. 4E). Thus, high levels of CheW could interfere with MCP trimer formation, as has been observed experimentally (9, 42). Therefore, formation of a functional array in vivo likely requires a fine balance between the three components.
Implications for the Mechanism of Chemotactic Signaling.
The high connectivity among MCP trimers in our model provides a basis for understanding the emergent properties of the receptor array, such as its high sensitivity, extensive dynamic range, and impressive signal amplification (43–45). In particular, the proposed connection between subdomain-2 of the P5 domain in one core complex and subdomain-1 of a CheW subunit in an adjacent core complex suggests a mechanism for the lateral spread throughout the array of an attractant signal originating at one MCP dimer (46). Our model also suggests experimental approaches to study functionally characterized mutant MCPs, such as those with “locked on” or “locked off” signal output (47–51) to determine the structural basis of those emergent properties. The mechanism of signal transduction through a highly cooperative network of chemoreceptors and other chemotaxis proteins (2) continues to be a fascinating topic to explore.
Our approaches and models complement the recent studies of Briegel et al. (52) and Vu et al. (53). Our P5/CheW ring and two specific protein-protein interactions (CheW/P5 and P5/MCP) are consistent with the 4.5-Å X-ray structure of a receptor/CheA/CheW complex (52) and the NMR results of a CheW/receptor interaction (53). Briegel et al. (52) also provided a high-resolution tomographic reconstruction of the MCPs, in which the dimers within an MCP trimer were resolved. However, their tomographic images did not clearly depict the CheA/CheW layer. Because we collected side-view images (Fig. 1C), our data provide structural evidence that two MCP trimers are connected by a strong density corresponding to the CheW/CheA complex. In addition, our images identify the location of the CheA P1 phosphoryl-transfer domain, which is located on the cytoplasmic side of the continuous CheW/CheA layer. Significantly, our array model presents a stoichiometry of 6:2:1 (6 MCP dimers, 2 CheW monomers, and 1 CheA dimer), which is consistent with the recent observation of a 6:2:1 complex within the minimal functional unit for receptor-coupled CheA activity (36), The existence of CheW-only rings, in addition to the alternating CheA/CheW rings, could contribute to the ultrastable array (41) as well as provide an explanation for the variable MCP/CheW/CheA stoichiometries (from 6:2:1 to 6:4:1) observed in living cells.
Materials and Methods
Preparation of the Flagellated Minicell-Producing E. coli Strain.
An E. coli strain WM3433, described recently (28), produced tiny minicells at a high frequency. We then modified this strain to overexpress flagellar and chemotaxis genes so that minicells would be more likely to contain receptor arrays and flagella. Overexpression was achieved by introducing plasmid pBAD30-flhDC into WM3433 to create strain WM4011. This plasmid contains a DNA segment from 48 bp upstream of flhD to 54 bp downstream of flhC inserted into pBAD30 between the BamHI and XbaI sites to fuse flhDC to the araBAD promoter (54). l-arabinose (0.2%) was added to induce flhDC expression. Bacterial culture grows overnight to stationary phase at 37 °C in tryptone broth. Fresh cultures were prepared from a 1:100 dilution of the overnight growth in tryptone broth and then cultured at 37 °C to late log phase. To enrich the minicells, the bacterial culture was centrifuged at 10,000 × g to remove the large cells, and the supernatant fraction was further centrifuged at 41,000 × g to collect the minicells. By light microscopy, we verified that most minicells were motile.
Cryo-ET Data Collection and 3D Reconstructions.
Bacterial cultures were mixed with 15 nm of colloidal gold (used as fiducial markers in image alignment) and then deposited onto freshly glow-discharged, holey carbon grids for 1 min. The grids were blotted with filter paper and rapidly frozen in liquid ethane, using a gravity-driven plunger apparatus. The resulting frozen-hydrated specimens were imaged at −170 °C using a Polara G2 electron microscope (FEI Company) equipped with a field emission gun and a 4,096 × 4,096 CCD camera (TVIPS). The microscope was operated at 300 kV with a magnification of 31,000×, resulting in an effective pixel size of 5.7 Å after 2 × 2 binning. Using the “batch tomography” program (FEI Company), low-dose, single-axis tilt series were collected from each minicell at −4 to −6 μm defocus with a cumulative dose of ∼100 e−/Å2 distributed over 87 images and covering an angular range of −64° to +64°, with an angular increment of 1.5°. Under these conditions, the first zero of the contrast transfer function (CTF) is in the range of 2.8–3.4 nm. No CTF correction was applied because a “Thon ring” was not detectable under this low-dose condition (1.1 e−/Å2). Tilted images were automatically aligned and reconstructed using a combination of the IMOD (55) and RAPTOR (56) packages. In total, 1,024 reconstructions were generated to provide a sufficient selection of reconstructions for further processing.
Subvolume Average and Correspondence Analysis.
We used the package developed by Winkler and collaborators (57, 58) for the following process. The original tomograms (2,048 × 2,048 × 1,200 voxels) were generally too noisy for direct visualization. Conventional imaging analysis, including 4 × 4 × 4 binning, contrast inversion, and low-pass filtering, was therefore used to enhance the contrast of the binned tomograms. Cryotomograms of 197 minicells containing chemoreceptor arrays were selected after carefully screening all the reconstructions. Good contrast was also critical for identifying the lattice units inside the arrays by visual inspection, using “tomopick” (58) (SI Appendix, Fig. S2). A total of 12,483 subvolumes (128 × 128 × 128 voxels), having one lattice unit and with the surrounding membrane in the center, were computationally extracted from the tomograms. The approximate local orientation of each small patch was estimated based on its location relative to the center of the minicell, therefore providing two of the three Euler angles. The subvolume analysis of 2D arrays was carried out as described previously (57, 59–61).
To accelerate the image analysis, 4 × 4 × 4 binned subvolumes (32 × 32 × 32 voxels) and 2 × 2 × 2 binned subvolumes (64 × 64 × 64 voxels) of the small patch were generated, respectively. Binning increases the signal-to-noise ratio, and thus results in more reliable initial alignment and classification. This process is particularly critical for subvolume analysis because of the large amounts of low-contrast data with missing wedge effects.
As a first step, a global average of all the extracted 4 × 4 × 4 binned subvolumes was formed after application of two Euler angles that were coarsely determined previously. This image revealed the membrane and a layer of high electron density at the membrane-distal end but not the pillar-like densities. After a translational alignment was carried out based on the global average (SI Appendix, Fig. S3), multivariate statistical analysis and hierarchical ascendant classification were performed to analyze the arrays in variable orientations (57, 58). Relevant voxels of the aligned subvolumes were selected by specifying a binary mask. The mask was generated in such a way that it contained mostly voxels of the lattice unit and excluded the cytoplasmic membrane. Eight class averages (SI Appendix, Fig. S4) were computed by averaging Fourier coefficients, such that the missing regions were taken into account explicitly. All class averages were then band pass-filtered (the low-pass limit was chosen to remove high-frequency noise, and the high-pass limit was chosen to remove low-frequency density variations) and further aligned with each other to minimize the difference in their lattice orientation. The aligned class averages were used as alignment references in the subsequent processing cycles.
The in-plane rotational alignment of subvolumes was performed with an orientation search of the third Euler angle by maximizing the constrained correlation (62), in which the missing wedge effects are compensated. Multivariate data analysis and hierarchical ascendant classification were applied to analyze the structural heterogeneity. This process is iterative: Class averages obtained in one cycle are used as alignment references in the subsequent cycle.
Two distinct class averages were generated based on slightly less than 50% of the total data, respectively. Averaging was carried out in Fourier space, such that Fourier coefficients falling in the region of the missing wedge could be excluded. There was no missing wedge visible in reciprocal space (SI Appendix, Fig. S5); therefore, the class averages received complete coverage and their structures were isotropic. The trimeric shape densities in those class averages become apparent in 2 × 2 × 2 binned data (SI Appendix, Fig. S6), and even more so in the original data (Figs. 2–4). Threefold symmetry instead of sixfold symmetry was imposed for the final maps, because the densities at the bottom of the arrays only showed threefold symmetry. Fourier shell correlation coefficients were estimated by comparing the correlation between two randomly divided halves of the aligned images used to generate the final maps. Note that the resolution of the density map in SI Appendix, Fig. S7A, estimated based on Fourier shell correlation, is 3.2 nm at the 0.5 cutoff (SI Appendix, Fig. S7C). It is roughly the same as the resolution corresponding to the first zero of the CTF under the conditions used for data collection.
Homology Modeling.
The atomic structure of intact E. coli CheA has not been determined. Homology modeling of E. coli CheA is based on the corresponding structures from T. maritima P3-P4-P5 and Salmonella P1, using the SWISS-MODEL protein modeling server (63). The sequence identity of CheA P3–P5 domains from E. coli and T. maritima is 43%, and the similarity is 66% (SI Appendix, Fig. S9). The sequence identity of CheA P1 domain from E. coli and Salmonella is 93%. The modeled E. coli CheA is structurally similar to the T. maritima CheA. The minimal rmsd for the backbone is 0.2 Å.
Molecular Modeling.
The atomic structural model (PDB ID code 1QU7) of the Tsr trimer (8), comprising residues A300–M480, was fitted into our density map as a rigid body by using the “fit in map” function of Chimera software (64). The cytoplasmic tips of two trimers merge in the continuous layer of densities corresponding to CheA/CheW. Our model of the CheA/CheW complex was based on four atomic structures (PDB ID codes 1B3Q, 2CH4, 1I5N, and 2HO9). The P5 domain shared by the two structures (PDB ID codes 1B3Q and 2CH4) was aligned to generate a P3/P4/P5/CheW complex (SI Appendix, Fig. S10 and Movie S1). This initial CheA/CheW complex structure (1 CheA dimer and 2 CheW monomers) was placed on top of the P1 domain (PDB ID code 1I5N), which was located at the bottom of the density map (Fig. 3C). The P3 domain sits in the middle of the density and is aligned roughly parallel to the distal ends of the Tsr trimers (Fig. 3 B and E), as was recently proposed (23). The P4 domain is adjacent to the P1 domain, a result consistent with the data from chemical mapping (11). At this stage of modeling, the P2, P3, and P4 domains fit into the density map very well. However, both the P5 domain and CheW did not fit into this map (SI Appendix, Fig. S10 B and C). In addition, CheW was far away from the cytoplasmic tips of two Tsr trimers. Because of the intrinsic flexibility among multiple domains of CheA, a linker residue (L509) between P4 and P5 was selected as a hinge (16). A rotation of 60° was needed to create an optimal fit of P5 and CheW together as a rigid body (SI Appendix, Fig. S10D and Movie S1). In particular, residues I33, E38, I39, and V87 of CheW, which are critical for receptor binding, are able to interact with the cytoplasmic tips of the Tsr trimer (21). The resulting model was initially manually fitted into the EM density map and further refined in the context of a lattice unit using Chimera. This approach is necessary because protein-protein interactions have to be considered in the context of the complex together with adjacent molecules.
Three-Dimensional Visualization.
Tomographic reconstructions were visualized using IMOD (55). The 3D surface renderings of E. coli minicells were segmented manually using Amira (Visage Imaging). Chimera (64) was used for model building, segmentation, visualization, and animation.
Accession Code.
One density map has been deposited in the Electron Microscopy Database (accession no. EMD-5404).
Supplementary Material
Acknowledgments
We thank Drs. Steven Norris, Jeffrey Actor, Ken Taylor, Rasika Harshey, Brian Crane, Jerry Hazelbauer, Mingshan Li, Joe Falke, Annette Erbse, and Sandy Parkinson for their comments. The flhD/C plasmid was kindly provided by Rasika Harshey. Lily Bartoszek proofread the manuscript before submission. M.D.M. thanks the Bartoszek Fund for Basic Biological Science for support and encouragement. J.L., B.H., and D.M. were supported, in part, by Grant R01AI087946 from the National Institute of Allergy and Infectious Diseases and Grant AU-1714 from the Welch Foundation. W.M. was supported by Grant R01GM61074 from the National Institute of General Medical Sciences and a grant from the Human Frontier Science Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Electron Microscopy Database, http://www.emdatabank.org/ [accession no. EMD-5404 (density map)].
See Author Summary on page 8806 (volume 109, number 23).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200781109/-/DCSupplemental.
References
- 1.Sourjik V, Armitage JP. Spatial organization in bacterial chemotaxis. EMBO J. 2010;29:2724–2733. doi: 10.1038/emboj.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: High-performance signaling in networked arrays. Trends Biochem Sci. 2008;33(1):9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briegel A, et al. Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci USA. 2009;106:17181–17186. doi: 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 5.Bray D, Levin MD, Morton-Firth CJ. Receptor clustering as a cellular mechanism to control sensitivity. Nature. 1998;393:85–88. doi: 10.1038/30018. [DOI] [PubMed] [Google Scholar]
- 6.Sourjik V. Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol. 2004;12:569–576. doi: 10.1016/j.tim.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Li M, Hazelbauer GL. Cellular stoichiometry of the components of the chemotaxis signaling complex. J Bacteriol. 2004;186:3687–3694. doi: 10.1128/JB.186.12.3687-3694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim KK, Yokota H, Kim SH. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 9.Studdert CA, Parkinson JS. Insights into the organization and dynamics of bacterial chemoreceptor clusters through in vivo crosslinking studies. Proc Natl Acad Sci USA. 2005;102:15623–15628. doi: 10.1073/pnas.0506040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borkovich KA, Kaplan N, Hess JF, Simon MI. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc Natl Acad Sci USA. 1989;86:1208–1212. doi: 10.1073/pnas.86.4.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller AS, Kohout SC, Gilman KA, Falke JJ. CheA Kinase of bacterial chemotaxis: Chemical mapping of four essential docking sites. Biochemistry. 2006;45(29):8699–8711. doi: 10.1021/bi060580y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griswold IJ, et al. The solution structure and interactions of CheW from Thermotoga maritima. Nat Struct Biol. 2002;9:121–125. doi: 10.1038/nsb753. [DOI] [PubMed] [Google Scholar]
- 13.Zhao J, Parkinson JS. Cysteine-scanning analysis of the chemoreceptor-coupling domain of the Escherichia coli chemotaxis signaling kinase CheA. J Bacteriol. 2006;188:4321–4330. doi: 10.1128/JB.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith RA, Parkinson JS. Overlapping genes at the cheA locus of Escherichia coli. Proc Natl Acad Sci USA. 1980;77:5370–5374. doi: 10.1073/pnas.77.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skidmore JM, et al. Polar clustering of the chemoreceptor complex in Escherichia coli occurs in the absence of complete CheA function. J Bacteriol. 2000;182:967–973. doi: 10.1128/jb.182.4.967-973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park SY, et al. Reconstruction of the chemotaxis receptor-kinase assembly. Nat Struct Mol Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- 17.McEvoy MM, Hausrath AC, Randolph GB, Remington SJ, Dahlquist FW. Two binding modes reveal flexibility in kinase/response regulator interactions in the bacterial chemotaxis pathway. Proc Natl Acad Sci USA. 1998;95:7333–7338. doi: 10.1073/pnas.95.13.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bilwes AM, Alex LA, Crane BR, Simon MI. Structure of CheA, a signal-transducing histidine kinase. Cell. 1999;96:131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- 19.Mourey L, et al. Crystal structure of the CheA histidine phosphotransfer domain that mediates response regulator phosphorylation in bacterial chemotaxis. J Biol Chem. 2001;276:31074–31082. doi: 10.1074/jbc.M101943200. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Hu Y, Fu W, Xia B, Jin C. Solution structure of the bacterial chemotaxis adaptor protein CheW from Escherichia coli. Biochem Biophys Res Commun. 2007;360:863–867. doi: 10.1016/j.bbrc.2007.06.146. [DOI] [PubMed] [Google Scholar]
- 21.Boukhvalova M, VanBruggen R, Stewart RC. CheA kinase and chemoreceptor interaction surfaces on CheW. J Biol Chem. 2002;277:23596–23603. doi: 10.1074/jbc.M202288200. [DOI] [PubMed] [Google Scholar]
- 22.Zhao J, Parkinson JS. Mutational analysis of the chemoreceptor-coupling domain of the Escherichia coli chemotaxis signaling kinase CheA. J Bacteriol. 2006;188:3299–3307. doi: 10.1128/JB.188.9.3299-3307.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhatnagar J, et al. Structure of the ternary complex formed by a chemotaxis receptor signaling domain, the CheA histidine kinase, and the coupling protein CheW as determined by pulsed dipolar ESR spectroscopy. Biochemistry. 2010;49:3824–3841. doi: 10.1021/bi100055m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim SH, Wang W, Kim KK. Dynamic and clustering model of bacterial chemotaxis receptors: Structural basis for signaling and high sensitivity. Proc Natl Acad Sci USA. 2002;99:11611–11615. doi: 10.1073/pnas.132376499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Briegel A, et al. Location and architecture of the Caulobacter crescentus chemoreceptor array. Mol Microbiol. 2008;69:30–41. doi: 10.1111/j.1365-2958.2008.06219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khursigara CM, Wu X, Subramaniam S. Chemoreceptors in Caulobacter crescentus: Trimers of receptor dimers in a partially ordered hexagonally packed array. J Bacteriol. 2008;190:6805–6810. doi: 10.1128/JB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang P, Khursigara CM, Hartnell LM, Subramaniam S. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc Natl Acad Sci USA. 2007;104:3777–3781. doi: 10.1073/pnas.0610106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, Chen CY, Shiomi D, Niki H, Margolin W. Visualization of bacteriophage P1 infection by cryo-electron tomography of tiny Escherichia coli. Virology. 2011;417:304–311. doi: 10.1016/j.virol.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chilcott GS, Hughes KT. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol Mol Biol Rev. 2000;64:694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adler J. A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J Gen Microbiol. 1973;74(1):77–91. doi: 10.1099/00221287-74-1-77. [DOI] [PubMed] [Google Scholar]
- 31.Parkinson JS, Houts SE. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimizu TS, et al. Molecular model of a lattice of signalling proteins involved in bacterial chemotaxis. Nat Cell Biol. 2000;2:792–796. doi: 10.1038/35041030. [DOI] [PubMed] [Google Scholar]
- 33.Cantwell BJ, et al. CheZ phosphatase localizes to chemoreceptor patches via CheA-short. J Bacteriol. 2003;185:2354–2361. doi: 10.1128/JB.185.7.2354-2361.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coleman MD, Bass RB, Mehan RS, Falke JJ. Conserved glycine residues in the cytoplasmic domain of the aspartate receptor play essential roles in kinase coupling and on-off switching. Biochemistry. 2005;44:7687–7695. doi: 10.1021/bi0501479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu JD, Parkinson JS. Genetic evidence for interaction between the CheW and Tsr proteins during chemoreceptor signaling by Escherichia coli. J Bacteriol. 1991;173:4941–4951. doi: 10.1128/jb.173.16.4941-4951.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li M, Hazelbauer GL. Core unit of chemotaxis signaling complexes. Proc Natl Acad Sci USA. 2011;108:9390–9395. doi: 10.1073/pnas.1104824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bibikov SI, Biran R, Rudd KE, Parkinson JS. A signal transducer for aerotaxis in Escherichia coli. J Bacteriol. 1997;179:4075–4079. doi: 10.1128/jb.179.12.4075-4079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rebbapragada A, et al. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc Natl Acad Sci USA. 1997;94:10541–10546. doi: 10.1073/pnas.94.20.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schulmeister S, et al. Protein exchange dynamics at chemoreceptor clusters in Escherichia coli. Proc Natl Acad Sci USA. 2008;105:6403–6408. doi: 10.1073/pnas.0710611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levit MN, Grebe TW, Stock JB. Organization of the receptor-kinase signaling array that regulates Escherichia coli chemotaxis. J Biol Chem. 2002;277:36748–36754. doi: 10.1074/jbc.M204317200. [DOI] [PubMed] [Google Scholar]
- 41.Erbse AH, Falke JJ. The core signaling proteins of bacterial chemotaxis assemble to form an ultrastable complex. Biochemistry. 2009;48:6975–6987. doi: 10.1021/bi900641c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cardozo MJ, Massazza DA, Parkinson JS, Studdert CA. Disruption of chemoreceptor signalling arrays by high levels of CheW, the receptor-kinase coupling protein. Mol Microbiol. 2010;75:1171–1181. doi: 10.1111/j.1365-2958.2009.07032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sourjik V, Berg HC. Functional interactions between receptors in bacterial chemotaxis. Nature. 2004;428:437–441. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- 44.Vaknin A, Berg HC. Direct evidence for coupling between bacterial chemoreceptors. J Mol Biol. 2008;382:573–577. doi: 10.1016/j.jmb.2008.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lai RZ, et al. Cooperative signaling among bacterial chemoreceptors. Biochemistry. 2005;44:14298–14307. doi: 10.1021/bi050567y. [DOI] [PubMed] [Google Scholar]
- 46.Goldman JP, Levin MD, Bray D. Signal amplification in a lattice of coupled protein kinases. Mol Biosyst. 2009;5:1853–1859. doi: 10.1039/B903397a. [DOI] [PubMed] [Google Scholar]
- 47.Swain KE, Gonzalez MA, Falke JJ. Engineered socket study of signaling through a four-helix bundle: Evidence for a yin-yang mechanism in the kinase control module of the aspartate receptor. Biochemistry. 2009;48:9266–9277. doi: 10.1021/bi901020d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chervitz SA, Falke JJ. Lock on/off disulfides identify the transmembrane signaling helix of the aspartate receptor. J Biol Chem. 1995;270:24043–24053. doi: 10.1074/jbc.270.41.24043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Draheim RR, Bormans AF, Lai RZ, Manson MD. Tuning a bacterial chemoreceptor with protein-membrane interactions. Biochemistry. 2006;45:14655–14664. doi: 10.1021/bi061259i. [DOI] [PubMed] [Google Scholar]
- 50.Ames P, Parkinson JS. Transmembrane signaling by bacterial chemoreceptors: E. coli transducers with locked signal output. Cell. 1988;55:817–826. doi: 10.1016/0092-8674(88)90137-7. [DOI] [PubMed] [Google Scholar]
- 51.Zhou Q, Ames P, Parkinson JS. Biphasic control logic of HAMP domain signalling in the Escherichia coli serine chemoreceptor. Mol Microbiol. 2011;80:596–611. doi: 10.1111/j.1365-2958.2011.07577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Briegel A, et al. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc Natl Acad Sci USA. 2012;109:3766–3771. doi: 10.1073/pnas.1115719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vu A, Wang X, Zhou H, Dahlquist FW. The receptor-CheW binding interface in bacterial chemotaxis. J Mol Biol. 2012;415:759–767. doi: 10.1016/j.jmb.2011.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 56.Amat F, et al. Markov random field based automatic image alignment for electron tomography. J Struct Biol. 2008;161:260–275. doi: 10.1016/j.jsb.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 57.Winkler H, et al. Tomographic subvolume alignment and subvolume classification applied to myosin V and SIV envelope spikes. J Struct Biol. 2009;165:64–77. doi: 10.1016/j.jsb.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Winkler H. 3D reconstruction and processing of volumetric data in cryo-electron tomography. J Struct Biol. 2007;157:126–137. doi: 10.1016/j.jsb.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 59.Liu J, Wright ER, Winkler H. 3D visualization of HIV virions by cryoelectron tomography. Methods Enzymol. 2010;483:267–290. doi: 10.1016/S0076-6879(10)83014-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu J, et al. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: Evidence for stator ring curvature and rotor/C-ring assembly flexion. J Bacteriol. 2009;191:5026–5036. doi: 10.1128/JB.00340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Förster F, Medalia O, Zauberman N, Baumeister W, Fass D. Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc Natl Acad Sci USA. 2005;102:4729–4734. doi: 10.1073/pnas.0409178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 64.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]