Abstract
Metformin has been reported to lower cancer incidence among type II diabetics. Metformin exhibits antiproliferative and antineoplastic effects associated with inhibition of mammalian target of rapamycin complex 1 (mTORC1), but the mechanisms are poorly understood. We provide a unique genome-wide analysis of translational targets of canonical mTOR inhibitors (rapamycin and PP242) compared with metformin, revealing that metformin controls gene expression at the level of mRNA translation to an extent comparable to that of canonical mTOR inhibitors. Importantly, metformin's antiproliferative activity can be explained by selective translational suppression of mRNAs encoding cell-cycle regulators via the mTORC1/eukaryotic translation initiation factor 4E-binding protein pathway. Thus, metformin selectively inhibits translation of mRNAs encoding proteins that promote neoplastic proliferation, which should facilitate studies on metformin and related biguanides in cancer prevention and treatment.
Keywords: polysome-microarray, posttranscriptional regulation, translatomics
The mechanistic/mammalian target of rapamycin complex 1 (mTORC1) is a key component of the PI3K/Akt anabolic pathway, which is frequently up-regulated in cancer (1). Consequently, there is heightened interest in developing therapeutic strategies to target aberrant mTORC1 signaling in cancer patients (2). Rapamycin, a naturally occurring allosteric inhibitor of mTORC1 and its analogs (rapalogs), are used in the clinic to treat advanced renal cell carcinoma, albeit with partial success (3). This partial success has been explained by the inability of rapalogs to completely inhibit the phosphorylation of a family of translational repressors, the eukaryotic translation initiation factor 4E (eIF4E)-binding proteins (4E-BPs) (4–6). In contrast, newly developed mTOR inhibitors, which target the active site of mTOR (asTORi; e.g., PP242) abolish the phosphorylation of 4E-BPs and induce more potent antiproliferative and antitumorigenic effects than rapamycin (4–7).
A widely used biguanide antidiabetic drug, metformin, exhibits antiproliferative properties, inhibits mTORC1, and is linked to decreased incidence of cancer in diabetic patients (8–10). However, the mechanisms underlying metformin’s antiproliferative and anticancer activities are not known. Metformin activates AMP-kinase (AMPK), most probably by interfering with mitochondrial respiratory complex I (11, 12) and altering cellular energy balance (13, 14). AMPK down-regulates anabolic processes, including protein synthesis, in large part via inhibition of the mTORC1 pathway (15–17). It has also been suggested that metformin suppresses mTORC1 signaling independently of AMPK by inhibiting Rag GTPases or activating REDD1 (18, 19). Herein, we compared quantitative and qualitative effects of “classic” mTOR inhibitors and metformin on mRNA translation. We show that the antiproliferative effect of metformin, similarly to asTORi, is in part mediated by selective suppression of translation of a subset of mRNAs encoding cell-cycle factors via the mTORC1/4E-BP pathway.
Results
Metformin Primarily Regulates Gene Expression at the Level of mRNA Translation.
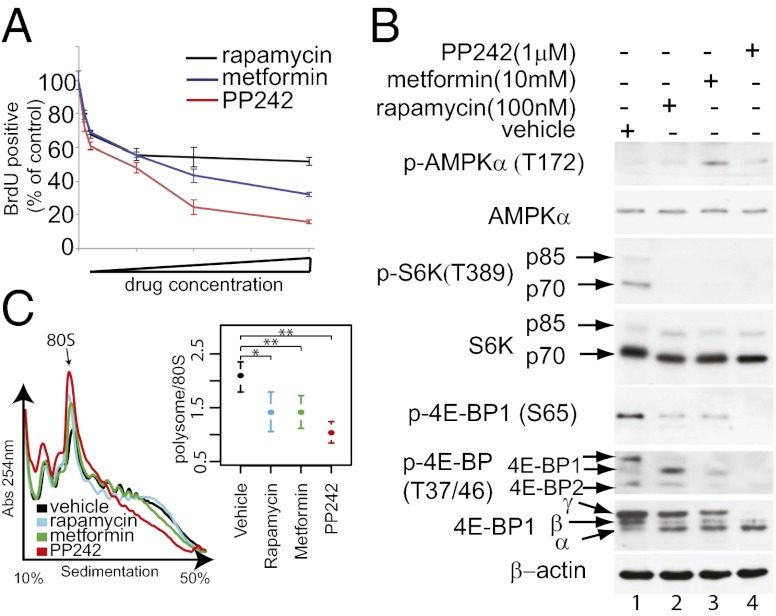
mTORC1 stimulates cell growth and proliferation by activating mRNA translation (20). The effects of metformin on the translatome [genome-wide pool of translated mRNAs (21)] and their functional consequences are, however, unknown. To address this important question, we compared the impact of metformin and established mTOR inhibitors (i.e., rapamycin and PP242) on translation and steady-state mRNA levels by monitoring polysome-associated and total cytoplasmic mRNAs, respectively. Because the comparison of effects of metformin and mTOR inhibitors on gene expression in vivo is likely confounded by pharmacokinetic factors, we investigated cell-autonomous effects of the drugs in cells. MCF7 cells were used previously to demonstrate that metformin reduces global protein synthesis and cell proliferation via AMPK-dependent inhibition of mTORC1 signaling (15, 16). To rigorously compare the impact of metformin, rapamycin, and PP242 on gene expression, we first determined the concentration of each compound that inhibits proliferation of MCF7 cells to 50% of vehicle-treated cells (absolute IC50; aIC50) (Fig. 1A). Although PP242 or metformin reduced proliferation >50% compared with control, rapamycin reached a plateau (∼50%) where increase in its concentration reduced proliferation only marginally. Next, we assessed the effects of the drugs at aIC50 concentrations on mTORC1 signaling by monitoring the phosphorylation of ribosomal protein S6 kinases (S6Ks) and 4E-BP1. In agreement with previous reports, rapamycin and PP242 inhibited mTORC1-dependent phosphorylation of S6Ks to the same extent, whereas PP242 inhibited more the phosphorylation of 4E-BP1 (Fig. 1B) (4–7). Metformin inhibited the phosphorylation of S6Ks to a similar extent as rapamycin and PP242, but its effect on the phosphorylation of 4E-BP1, on Thr-37/46, was intermediate between that of rapamycin and PP242 (Fig. 1B, compare lanes 2, 3, and 4). Thus, metformin inhibits phosphorylation of key residues on 4E-BP1 more strongly than rapamycin. This finding can be explained by the differences in the mechanisms of action of metformin and rapamycin, whereby metformin inhibits mTORC1 via modulation of its upstream regulators but rapamycin acts as an allosteric inhibitor (3, 8).
Fig. 1.
Antiproliferative effects of mTOR inhibitors and metformin correlate with inhibition of mTORC1 signaling and mRNA translation. (A) MCF7 cells were incubated with vehicle or increasing concentrations of metformin, PP242, or rapamycin for 24 h. Proliferation was determined by BrdU incorporation and expressed as a percentage of the inhibition of BrdU incorporation relative to control (vehicle-treated cells). Results are presented as mean values ± SD (n = 4). Concentrations of rapamycin, PP242, and metformin that inhibit proliferation by ∼50% of the control were 100 nm, 1 μM, and 10 mM, respectively. (B) Western blot analysis of the levels and the phosphorylation status of indicated proteins in MCF7 cells treated with vehicle, rapamycin (100 nM), PP242 (1 μM), and metformin (10 mM) for 12 h. β-actin served as a loading control. α, β, and γ indicate hypo-, intermediate-, and hyperphosphorylated forms of 4E-BP1. (C) Polysome profiles from cells treated with vehicle, rapamycin (100 nM), PP242 (1 μM), and metformin (10 mM) for 12 h. Absorbance at 254 nm is shown as a function of sedimentation and the 80S monosome is indicated. The area under the curve for polysomes and the 80S peak were calculated and the ratio is shown (mean ± SD from four experiments; P values from paired Student t tests are indicated: *P < 0.05, **P < 0.01)
We next examined the impact of drug treatments on overall mRNA translation by studying polysome formation. The number of ribosomes engaged in polysomes is directly proportional to the translation initiation rate (22). All drug treatments led to a reduction in the fraction of ribosomes that are engaged in polysomes compared with control (Fig. 1C). Taken together, these findings demonstrate that the antiproliferative effects of metformin, rapamycin, and PP242 are paralleled by inhibition of mTORC1 signaling and reduction in mRNA translation.
Strikingly, because none of the compounds led to a complete dissociation of polysomes (Fig. 1C), a sizable number of mRNAs must be translated in the presence of these agents. It is thus highly likely that metformin, like PP242, exerts its antiproliferative effect by selectively impairing the translation of a subset of mRNAs. We therefore studied the quantitative and qualitative effects of each drug on the total cytoplasmic (alterations in cytoplasmic mRNA levels reflect changes in transcription, mRNA export, and stability) and polysome-associated mRNAs on a genome-wide scale. MCF7 cells were treated with aIC50 drug concentrations (Fig. 1A) for 12 h, thus allowing determination of the early effect of the drugs on gene expression. Total cytoplasmic mRNA and mRNAs associated with four or more ribosomes (hereafter referred to as polysome-associated mRNAs) from four biological replicates were isolated, and their genome-wide expression profiles were determined using DNA-microarrays.
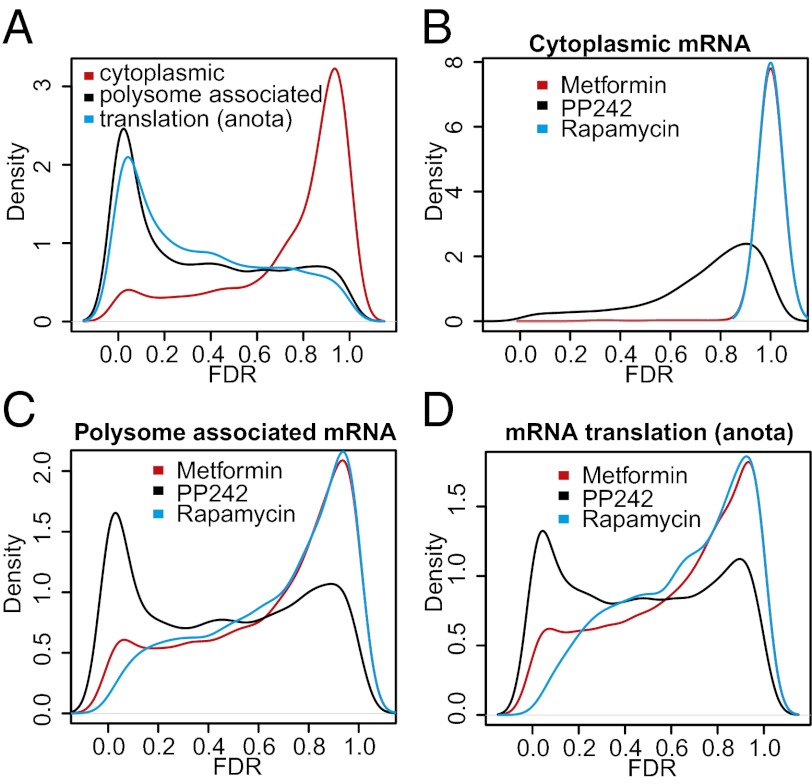
First, we analyzed at which step the drugs affect gene expression by comparing levels of cytoplasmic or polysome-associated mRNA between all conditions using ANOVA, incorporating a variance shrinkage method to reduce false-positive rates [the random variance model (RVM) (23)] and adjusting the P values for multiple testing using the Benjamini and Hochberg’s false-discovery rate (FDR) method. The drugs induced dramatic changes in polysome-associated mRNAs (1,254 of 18,185 genes changed with FDR < 0.001), but not in total cytoplasmic mRNA levels (63 of 18,185 genes changed with FDR < 0.001). Accordingly, the distributions of FDRs differed significantly between the analysis of total cytoplasmic and polysome-associated mRNAs, as illustrated by the Kolmogorov–Smirnov (KS) test (P < 2.2e-16) (Fig. 2A; histograms in Fig. S1A). To further establish the extent to which the differences in polysome-associated mRNAs can be attributed to differential translation of individual mRNAs, we used the recently developed “anota” algorithm (24, 25). Anota specifically captures differences in translational activity of individual mRNAs on a genome-wide scale by comparing polysome-associated mRNA levels to cytoplasmic mRNA levels. The density of the obtained anota FDRs demonstrated substantial qualitative and quantitative differences in the translational activity across the conditions (Fig. 2A and Fig. S1A).
Fig. 2.
mTOR inhibitors and metformin suppress gene expression at the level of mRNA translation. (A) Kernel densities of adjusted P values (FDRs) for all assessed genes from ANOVAs comparing all conditions using data obtained from cytoplasmic or polysome-associated mRNA and from analysis of translational activity using anota. Kernel densities of adjusted P values (FDRs) for all assessed genes from treatment (metformin, PP242, or rapamycin) compared with vehicle using data obtained from cytoplasmic (B), polysome-associated mRNA (C), or translation as analyzed using anota (D).
Next, we determined individual effects of drugs on the total cytoplasmic versus polysome-associated mRNA. PP242 induced a modest perturbation of cytoplasmic mRNA levels (15 mRNAs with FDR < 0.01), whereas the effects of metformin or rapamycin were minimal (0 genes with FDR < 0.01) (Fig. 2B and Fig. S1B). In sharp contrast, all drugs altered the pools of polysome-associated mRNAs (1,676, 177, and 5 mRNAs with an FDR < 0.01 for PP242, metformin, and rapamycin, respectively). Notably, PP242 induced a larger shift of the FDRs compared with rapamycin and metformin (Fig. 2C and Fig. S1C) (KS P < 2.2e-16 for both comparisons), whereas metformin was more potent than rapamycin (KS P = 4.4e-16). Next, we deployed anota to establish genome-wide effects of each drug on the translation of individual mRNAs. PP242 treatment resulted in a shift of the FDRs obtained from anota congruent with a stronger perturbation of translational activity compared with metformin or rapamycin (Fig. 2D and Fig. S1D) (KS P < 2.2e-16 for comparisons to both distributions), whereas the effects of metformin were stronger than those of rapamycin (KS P < 2.2e-16). Thus, it is striking that metformin, which was not previously recognized as a qualitative modulator of translation of specific mRNAs, perturbs the translatome to an extent comparable to that of the canonical mTOR inhibitors.
Effects of Metformin and mTOR Inhibitors on the Translatome Partially Overlap.
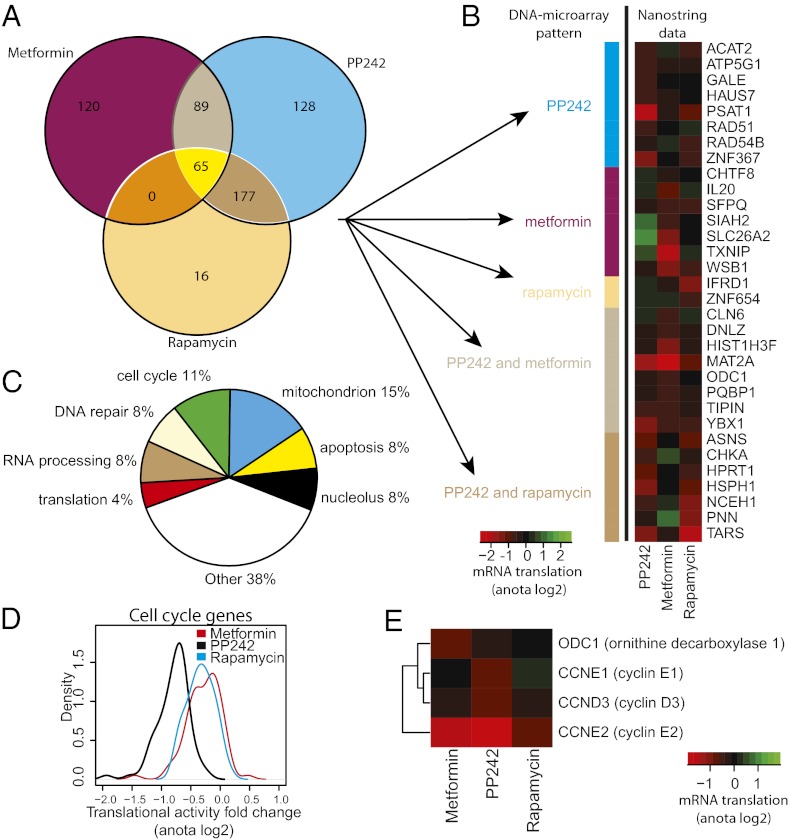
Because each drug induces substantial perturbations in the translatome, it was pertinent to determine the extent to which the effects of the drugs on the translatome overlap. A total of 595 mRNAs were translationally suppressed by at least one of the drugs with a fold-change >1.5 and FDR < 0.15 (using anota analysis). Instead of using list comparisons (i.e., comparing overlaps between lists of mRNAs that show differential translation under each condition), we compared translational activity across the different treatments. The advantage of this approach is that mRNAs will not appear to be selectively targeted by a single drug because of a small difference in variance. Therefore, the 595 mRNAs were subjected to k-means clustering and each cluster was manually annotated according to drug-specificity (Fig. S2). This analysis suggested that the three drugs suppress both distinct and shared translational targets, as summarized in a Venn diagram (Fig. 3A).
Fig. 3.
Rapamycin, PP242, or metformin engender overlapping and disparate effects on the translatome. (A) A Venn diagram comparing translatomes following treatment with rapamycin, PP242, or metformin. (B) Validation of translational profiles using nanostring technology. The heatmap indicates differences in mRNA translation (log2 scale) compared with vehicle for each treatment using data from nanostring that were analyzed using anota (n = 2). The color bar to the left indicates the regulation pattern of the mRNAs according to DNA-microarray analysis (i.e., A). (C) A pie chart of cellular functions of encoded proteins whose translation was suppressed by all drugs. (D) Kernel densities of fold-changes (calculated with anota) for cell cycle-related genes (shown in Fig. S4) after treatment with metformin, PP242, or rapamycin compared with vehicle. (E) Translational activity for selected cell-cycle genes after treatment with metformin, PP242, or rapamycin compared with vehicle (calculated with anota, log2 scale).
To validate these results we used nanostring, a targeted sequencing technology allowing parallel validation of multiple mRNAs (26), on duplicate samples from each condition in conjunction with anota analysis. We obtained quantitative data for 34 of 45 mRNAs that were selected by random sampling for representative mRNAs (see Materials and Methods) showing specific drug-sensitivity patterns (Fig. 3A and Fig. S2) and validated 32 as translationally suppressed (anota FDR < 0.15). Moreover, similar drug-sensitivity patterns that were identified using DNA-microarrays (Fig. 3A) were observed by nanostring (Fig. 3B). These results demonstrate that in addition to the shared targets, metformin, rapamycin, and PP242 induce disparate alterations in the translatome. Moreover, the considerable overlap of translational targets of metformin and PP242 strongly suggests that metformin acts as an mTORC1-dependent modulator of mRNA translation.
Metformin and mTOR Inhibitors Suppress Translation of a Specific Subset of Functionally Related mRNAs.
To determine the functional consequences of the drug-induced perturbations in the translatome, we grouped mRNAs whose translation was suppressed based on the drug-specific regulation patterns shown in the Venn diagram (Fig. 3A). We then searched for enrichment of mRNAs, which encode proteins with shared biological function according to annotations from the gene ontology (GO) consortium (27) (Fig. S3). Transcripts encoding cell-cycle regulating proteins were enriched [enriched categories define >5 mRNAs in the Venn diagram group (Fig. 3A), and show a >twofold enrichment compared with the background with an FDR < 0.05] in the pool of mRNAs, which are translationally suppressed by all three drugs (e.g., “cell cycle” showed a 3.4-fold enrichment). In addition, mRNAs implicated in cell-cycle control were also enriched in the “PP242 and rapamycin” or “PP242 and metformin” groups (e.g., the cell cycle category showed a 3.3- and 2.1-fold enrichment, respectively). mRNAs that were translationally suppressed by PP242, but not metformin or rapamycin, showed strong enrichment for several additional cell cycle-related functions (e.g., “DNA strand elongation” showed a 27-fold enrichment), mitochondrial transport (9.8-fold enrichment), and mRNA translation (4.1-fold enrichment). In turn, mRNAs, the translation of which was inhibited by metformin, but not PP242 or rapamycin, are enriched for those encoding proteins that are implicated in RNA processing and noncoding RNA processing (2.4- and 4.2-fold enrichment, respectively). Thus, metformin, rapamycin, and PP242 suppress translation of limited subsets of functionally related mRNAs that encode proteins involved in processes, such as cell-cycle control, metabolism, mRNA translation, and RNA processing.
Metformin Inhibits Proliferation by Downregulating Translation of mRNAs Encoding Cell-Cycle Regulators via the mTORC1/4E-BP Pathway.
It is thought that suppression of the mTOR pathway inhibits cell proliferation largely by attenuating cell-cycle progression (1, 28). Consistent with this theory, mRNAs, the translation of which is suppressed by all three drugs, include a sizable number of those encoding cell cycle-related factors (11% of the targets) (Fig. 3C). Surprisingly, inhibition of translation of some of the cell cycle-regulating mRNAs was, however, specific for each drug (Fig. 3A and Fig. S3). We therefore examined the effects of each drug on the translation of mRNAs implicated in the cell cycle. In accordance with the functional analysis, most of the mRNAs encoding cell cycle-related factors were suppressed by PP242, some of which overlapped with mRNAs whose translation was inhibited by metformin or rapamycin (Fig. S4). To compare the magnitude of the effects of each drug on mRNA translation, we evaluated distributions of fold-changes (Fig. 3D). PP242 induced stronger translational suppression of mRNAs encoding cell-cycle factors compared with rapamycin and metformin (Fig. 3D) (KS P < 2.2e-16 for comparisons to both distributions). Differences in the magnitude of inhibition for a subset of mRNAs were, however, observed (Fig. S4). For example, translation of cyclin E2 and ODC1 mRNAs was strongly inhibited by PP242 and metformin, but only marginally by rapamycin, whereas translation of cyclin E1 and cyclin D3 mRNAs was suppressed by PP242, but not by rapamycin or metformin (Fig. 3E). Possible reasons for these differences are addressed in the Discussion.
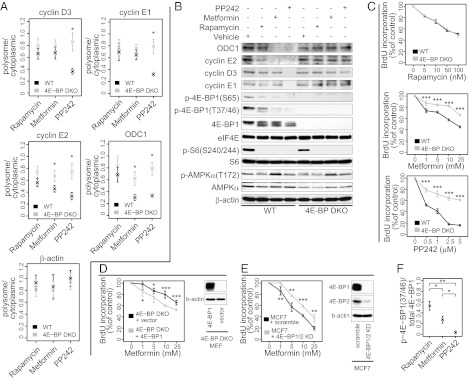
Translational efficiency of D- and E-type cyclins and ODC1 mRNAs is dictated by eIF4E activity (29). 4E-BPs bind to eIF4E and inhibit eIF4E association with eIF4G, and consequently the assembly of the eIF4F initiation complex and recruitment of mRNA to the ribosome (30). When activated, mTORC1 phosphorylates and inactivates 4E-BPs (31, 32), thereby promoting eIF4E activity, cell-cycle progression, and proliferation (20). We therefore determined the role of 4E-BPs in mediating the effects of metformin, rapamycin, and PP242 on translation of cyclin E1, -E2, -D3, and ODC1 mRNAs in WT and 4E-BP1 and 4E-BP2 double knock-out (DKO) mouse embryonic fibroblasts (MEFs) using quantitative RT-PCR (qRT-PCR). The use of 4E-BP DKO MEFs provided a means to study the translational inhibition caused by suppression of mTORC1 signaling to 4E-BPs, thus by-passing off-target effects of the drugs and the multitude of potential mediators of metformin’s actions on mTORC1 signaling [e.g., AMPK, Rags, REDD1 (15, 16, 18, 19)]. Consistent with the microarray data from MCF7 cells (Fig. 3E), PP242, but not metformin or rapamycin, suppressed translation of cyclin D3 and -E1 mRNAs in WT compared with 4E-BP DKO MEFs. In turn, metformin and PP242, but not rapamycin, suppressed translation of ODC1 and cyclin E2 mRNAs in WT compared with 4E-BP DKO MEFs. Thus, akin to PP242, metformin inhibits mRNA translation of cell cycle regulators via inhibition of mTORC1 signaling to 4E-BPs (Fig. 4 A and B).
Fig. 4.
Rapamycin, PP242, and metformin differentially decrease translation in a 4E-BP–dependent manner. (A) qRT-PCR of polysome-associated and total cytoplasmic mRNA in WT and 4E-BP DKO MEFs treated with drugs or vehicle for 12 h. Levels of cyclin E1, ODC1, cyclin E2, and cyclin D3 were assessed. β-actin was used as a control. The ratio between polysome-associated mRNA and total cytoplasmic mRNA was normalized to vehicle treated cells. The results are presented as mean values ± SD (n = 3). (B) Western blotting of whole-cell protein extract from WT and 4E-BP DKO MEFs treated with drugs or vehicle for 12 h. eIF4E and β-actin were used as loading controls. (C) WT and 4E-BP DKO MEFs were incubated with vehicle (DMSO+PBS), or the indicated concentrations of drugs for 48 h. Proliferation was determined by BrdU incorporation and expressed as a percentage of inhibition of BrdU incorporation relative to control (vehicle-treated cells). Results are presented as mean values ± SD (n = 3). 4E-BP DKO MEFs expressing vector-control or 4E-BP1 (D) and MCF7 cells expressing scrambled or 4E-BP1/2 shRNA (E) were incubated with vehicle (DMSO+PBS), or the indicated concentrations of metformin for 48 h. Proliferation was determined by BrdU incorporation and expressed as a percentage of inhibition of BrdU incorporation relative to control (vehicle-treated cells). Results are presented as mean values ± SD (n = 5). Shown is also Western blotting of whole-cell protein extracts (D and E: β-actin was used as a loading control). (F) Protein quantification using densitometry for p-4E-BP1 (Thr-37/46) and total 4E-BP1. Shown are mean of ratios ± SD after normalization to vehicle (n = 3). KD, knockdown; P values from two-tailed Student t tests are indicated: *P < 0.05, **P < 0.01, ***P < 0.001.
To ascertain that inhibition of mRNA translation of cell-cycle regulators by the drugs mirror their antiproliferative effects, we treated WT and 4E-BP DKO MEFs with increasing drug concentrations for 48 h. PP242 induced the strongest inhibition of proliferation of WT MEFs, whereas the effects of rapamycin and metformin were comparable (Fig. 4C). As reported previously (5), 4E-BP DKO MEFs were less sensitive to the antiproliferative effects of PP242 compared with WT MEFs, whereas rapamycin inhibited proliferation of WT and 4E-BP DKO MEFs to the same extent (Fig. 4C). Importantly, the proliferation of 4E-BP DKO MEFs was partially resistant to metformin compared with WT MEFs. Moreover, the sensitivity of 4E-BP DKO MEFs to metformin was restored by expressing 4E-BP1 (Fig. 4D), and MCF7 cells in which 4E-BP1/2 were silenced showed reduced sensitivity to metformin (Fig. 4E). These results are in agreement with the findings that 4E-BP DKO MEFs were relatively resistant to inhibition of translation of cyclin E2 and ODC1 mRNAs by metformin compared with WT MEFs (Fig. 4 A and B). Nonetheless, the resistance of 4E-BP DKO MEFs to the antiproliferative activity of PP242 is stronger than that observed for metformin treated cells. Indeed, metformin inhibits the phosphorylation of 4E-BP1 (at Thr-37/46) to a lesser extent than PP242 (Fig. 4F), and only suppresses translation of some of the tested mRNAs, which encode cell-cycle regulators (e.g., ODC1 and cyclin E2 but not cyclin E1 or cyclin D3) (Fig. 4 A and B). As we reported previously, rapamycin inhibited proliferation of WT and 4E-BP DKO MEFs to a similar extent (5). This result has been explained by the finding that rapamycin only weakly inhibited phosphorylation of 4E-BPs (at Thr-37/46) (Fig. 4F), and failed to suppress translation of mRNAs encoding key cell-cycle proteins, including cyclin D3, -E1, -E2, and ODC1 (Fig. 4 A and B). In contrast to 4E-BPs, all three drugs equipotently suppressed phosphorylation of ribosomal protein S6 (rpS6) and this effect was similar in WT and 4E-BP DKO MEFs (Fig. 4B). These results demonstrate that the effects of metformin and PP242 on proliferation are mediated by 4E-BPs, but the effect of rapamycin is not. In addition, they show that the biological effects of metformin are exerted via suppression of translation of a subset of mRNAs in a 4E-BP–dependent manner.
Discussion
Metformin suppresses cellular anabolism and thus proliferation via multiple mechanisms, including activation of AMPK (33). Our findings reveal a previously unrecognized role of metformin in regulation of translation of a specific subset of functionally related mRNAs, and demonstrate that its antiproliferative effects are in large part a consequence of translational suppression of mRNAs encoding cell-cycle functions via the mTORC1/4E-BP pathway. As mRNA translation is the most energy-consuming process in the cell (34), it is thought that under conditions of energy stress induced by metformin, global mRNA translation is down-regulated to conserve energy. However, our findings reveal that although this occurs, metformin preferentially inhibits the translation of a unique subset of mRNAs. These mRNAs include those encoding proliferation and tumor-promoting proteins, such as cyclin E2 and ODC1. In contrast, translation of mRNAs encoding other cell-cycle regulators (e.g., cyclin E1 and -D3), is suppressed by PP242 but not by metformin. PP242 inhibits 4E-BP1 phosphorylation and thus reduces eIF4E availability more strongly than metformin (Fig. 4F). eIF4E drives tumorigenesis by selectively activating translation of a subset of tumor-promoting mRNAs (including those encoding cyclins and ODC) commonly referred to as “eIF4E-sensitive” (29). Our findings, that translation of cyclin E2 and ODC1 mRNAs is more sensitive to inhibition of 4E-BP1 phosphorylation than that of cyclin E1 and -D3 mRNAs, suggest previously unrecognized differences in the relative dependence on eIF4E availability among eIF4E-sensitive mRNAs. Importantly, it is highly unlikely that these observations are a result of off-target effects of the drugs or downstream effectors of mTORC1 other than 4E-BPs (e.g., S6Ks), inasmuch as metformin and PP242 failed to inhibit translation of cyclin E1, -E2, -D3, and ODC1 mRNAs in 4E-BP DKO MEFs (Fig. 4 A and B). In addition, rapamycin, which inhibits phosphorylation of S6Ks/rpS6—but not 4E-BP1—to an extent comparable to PP242 and metformin, did not dramatically inhibit translation of the aforementioned transcripts (Figs. 1B and 4B). Intriguingly, we also observed that mRNAs encoding mitochondrial proteins are enriched in the cohort of mRNAs suppressed by metformin. Nevertheless, because metformin inhibits complex I in isolated mitochondria (12), which synthesize only a limited number of proteins, these effects do not appear to be necessary for metformin’s primary effects on the mitochondria.
During the revision of this article, a study examining the effects of rapamycin and PP242 on mRNA translation in PC3 cells was published (35). Although there is an overlap for a number of PP242 mRNA targets [e.g., YBX1 (YB1)], there are apparent differences between mRNAs identified to be PP242-sensitive in the aforementioned and present study. These discrepancies are likely a result of differences in methodology, selected time points, and cell types used.
It is well established that up-regulated translation of proliferation and tumor-promoting mRNAs critically contributes to malignant transformation and tumor progression (36). Our findings therefore reveal previously unrecognized aspects of the action of metformin that may contribute to its antineoplastic activity in experimental models (37). Moreover, the similarities and differences between the effects of metformin and classic mTOR inhibitors raise the possibility that biguanides may be well-tolerated alternatives to mTOR inhibitors currently in use. It is of particular interest that our data were obtained using MCF-7 cells, which model estrogen receptor positive breast cancer, a disease where mTOR inhibition has recently been shown to be effective in a pivotal clinical trial (38). Our data also suggest that use of metformin should be considered for cancer prevention in syndromes involving hyperactivation of the mTORC1 pathway, such as tuberous sclerosis and Cowden disease.
Materials and Methods
Cell Culture, Cell Proliferation, and Western Blot Analysis.
MCF7 cells were obtained from ATCC and grown in RPMI (Invitrogen), supplemented with 10% (vol/vol) FBS (Invitrogen), 2 mM l-glutamine, and 100 units/mL penicillin/streptomycin (all from Invitrogen) at 37 °C and 5% CO2. MCF7 cells with silenced 4E-BP1/2 were generated by transducing MCF-7 cells with shRNA for human 4E-BP1 (Sigma; TRCN0000040203) and human 4E-BP2 (Sigma; TRCN0000117814). WT (p53−/−) and 4E-BP1/4E-BP2−/− (p53−/−) DKO MEFs and 4E-BP DKO MEFs expressing 4E-BP1 were described previously (5, 39). All MEFs were maintained in DMEM (Invitrogen) supplemented with 10% (vol/vol) FBS (Invitrogen), 2 mM l-glutamine, and 100 units/mL penicillin/streptomycin (all from Invitrogen) at 37 °C and 5% CO2. Cells were seeded at ∼50% confluency, grown overnight, and treated using concentrations of rapamycin (stock in DMSO), metformin (stock in PBS), and PP242 (stock in DMSO) indicated in the text. As a control, cells were incubated in the presence of the vehicle (DMSO+PBS). Cell proliferation assays and Western blot conditions are described in SI Materials and Methods.
Polysome Analysis, RNA Isolation, Microarray, Nanostring Analysis, and qRT-PCR.
Polysome RNA preparations are described in the SI Materials and Methods. Polysome fractions with mRNA associated with >3 ribosomes were pooled (polysome-associated mRNA) and RNA was isolated using TRIzol. A parallel sample was collected from the postnuclear lysates that were loaded onto the sucrose gradient (cytoplasmic mRNA) and RNA was isolated using TRIzol. For microarrays, 500 μg cytoplasmic or polysome-associated RNA (n = 4 from each condition) was used as starting material for the 3′ IVT Express Kit (Affymetrix). The resulting labeled samples were probed with the Human Genome U133 Plus 2.0 gene arrays from Affymetrix according the instructions of the manufacturer and scanned using the GeneArray Scanner 3000. For NanoString, a codeset targeting 50 mRNAs was designed by the manufacturer. Next, 150 ng RNA was used as input for the NanoString nCounter assay (n = 2 from each condition). Data were generated as previously described (26). For RT-PCR, the RNA was treated with DNaseTurbo (Ambion) according to the manufacturer’s instructions. qRT-PCR reactions were carried out using SuperScript III First-Strand Synthesis System (Invitrogen) and iQ SYBR Green Supermix (Bio-Rad) according to the manufacturer’s instructions. The list of primers is provided in Table S1.
Data Analysis.
For microarray-data we used updated probe-set definitions (40, 41) and robust mutiarray averaging to normalize gene expression data. Differential polysome or cytosolic mRNA levels were identified using the RVM modified t test (23) and differential translation was identified using anota (24, 25). For nanostring-data, differential translation was identified using anota. Enriched biological processes as defined by the gene ontology consortium (27) were identified using GO::Termfinder (42). For more details on data analysis, see SI Materials and Methods. The data has been deposited at the Gene Expression Omnibus (accession no. GSE36847). Genes that showed differential translation are shown in Table S2.
Note Added in Proof.
A relevant paper on the translational program of mTORC1 was recently published by Thoreen et al. (2012) Nature 485:109–113.
Supplementary Material
Acknowledgments
We thank C. Rommel for providing PP242, and C. Lister and P. Kirk for assistance. This work was supported by a grant from the Canadian Cancer Society Research Institute (CCSR) (to M.P.); grants from the Canadian Institute of Health Research (CIHR) and CCSR (to N.S.); the Swedish Research Council, the Swedish Cancer Foundation, the Cancer Society in Stockholm, the Jeansson Foundation, and the Åke Wiberg Foundation (O.L); CIHR and The Terry Fox Research Institute (I.T.); the Uehara Memorial foundation postdoctoral fellowship and the Japan Society for the Promotion of Science Excellent Young Researcher Overseas Visit Program (M.M); and the Brain Tumour Foundation of Canada (T.A.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE36847).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201689109/-/DCSupplemental.
References
- 1.Zoncu R, Efeyan A, Sabatini DM. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guertin DA, Sabatini DM. The pharmacology of mTOR inhibition. Sci Signal. 2009;2:pe24. doi: 10.1126/scisignal.267pe24. [DOI] [PubMed] [Google Scholar]
- 4.Thoreen CC, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dowling RJ, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328:1172–1176. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feldman ME, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7(2):e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.García-Martínez JM, et al. Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR) Biochem J. 2009;421:29–42. doi: 10.1042/BJ20090489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben Sahra I, Le Marchand-Brustel Y, Tanti JF, Bost F. Metformin in cancer therapy: A new perspective for an old antidiabetic drug? Mol Cancer Ther. 2010;9:1092–1099. doi: 10.1158/1535-7163.MCT-09-1186. [DOI] [PubMed] [Google Scholar]
- 9.Larsson SC, Mantzoros CS, Wolk A. Diabetes mellitus and risk of breast cancer: A meta-analysis. Int J Cancer. 2007;121:856–862. doi: 10.1002/ijc.22717. [DOI] [PubMed] [Google Scholar]
- 10.Larsson SC, Orsini N, Wolk A. Diabetes mellitus and risk of colorectal cancer: A meta-analysis. J Natl Cancer Inst. 2005;97:1679–1687. doi: 10.1093/jnci/dji375. [DOI] [PubMed] [Google Scholar]
- 11.El-Mir MY, et al. Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. J Biol Chem. 2000;275:223–228. doi: 10.1074/jbc.275.1.223. [DOI] [PubMed] [Google Scholar]
- 12.Owen MR, Doran E, Halestrap AP. Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem J. 2000;348:607–614. [PMC free article] [PubMed] [Google Scholar]
- 13.Gwinn DM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 15.Dowling RJ, Zakikhani M, Fantus IG, Pollak M, Sonenberg N. Metformin inhibits mammalian target of rapamycin-dependent translation initiation in breast cancer cells. Cancer Res. 2007;67:10804–10812. doi: 10.1158/0008-5472.CAN-07-2310. [DOI] [PubMed] [Google Scholar]
- 16.Zakikhani M, Dowling R, Fantus IG, Sonenberg N, Pollak M. Metformin is an AMP kinase-dependent growth inhibitor for breast cancer cells. Cancer Res. 2006;66:10269–10273. doi: 10.1158/0008-5472.CAN-06-1500. [DOI] [PubMed] [Google Scholar]
- 17.Shaw RJ. LKB1 and AMP-activated protein kinase control of mTOR signalling and growth. Acta Physiol (Oxf) 2009;196:65–80. doi: 10.1111/j.1748-1716.2009.01972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ben Sahra I, et al. Metformin, independent of AMPK, induces mTOR inhibition and cell-cycle arrest through REDD1. Cancer Res. 2011;71:4366–4372. doi: 10.1158/0008-5472.CAN-10-1769. [DOI] [PubMed] [Google Scholar]
- 19.Kalender A, et al. Metformin, independent of AMPK, inhibits mTORC1 in a rag GTPase-dependent manner. Cell Metab. 2010;11:390–401. doi: 10.1016/j.cmet.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 21.Greenbaum D, Luscombe NM, Jansen R, Qian J, Gerstein M. Interrelating different types of genomic data, from proteome to secretome: ’Oming in on function. Genome Res. 2001;11:1463–1468. doi: 10.1101/gr.207401. [DOI] [PubMed] [Google Scholar]
- 22.Warner JR, Knopf PM, Rich A. A multiple ribosomal structure in protein synthesis. Proc Natl Acad Sci USA. 1963;49:122–129. doi: 10.1073/pnas.49.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19:2448–2455. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- 24.Larsson O, Sonenberg N, Nadon R. anota: Analysis of differential translation in genome-wide studies. Bioinformatics. 2011;27:1440–1441. doi: 10.1093/bioinformatics/btr146. [DOI] [PubMed] [Google Scholar]
- 25.Larsson O, Sonenberg N, Nadon R. Identification of differential translation in genome wide studies. Proc Natl Acad Sci USA. 2010;107:21487–21492. doi: 10.1073/pnas.1006821107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geiss GK, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 27.Anonymous Gene Ontology Consortium The Gene Ontology project in 2008. Nucleic Acids Res. 2008;36(Database issue):D440–D444. doi: 10.1093/nar/gkm883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pollak M. Insulin and insulin-like growth factor signalling in neoplasia. Nat Rev Cancer. 2008;8:915–928. doi: 10.1038/nrc2536. [DOI] [PubMed] [Google Scholar]
- 29.Graff JR, Konicek BW, Carter JH, Marcusson EG. Targeting the eukaryotic translation initiation factor 4E for cancer therapy. Cancer Res. 2008;68:631–634. doi: 10.1158/0008-5472.CAN-07-5635. [DOI] [PubMed] [Google Scholar]
- 30.Pause A, et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 31.Gingras AC, et al. Regulation of 4E-BP1 phosphorylation: A novel two-step mechanism. Genes Dev. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gingras AC, et al. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15:2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pollak M. Metformin and other biguanides in oncology: Advancing the research agenda. Cancer Prev Res (Phila) 2010;3:1060–1065. doi: 10.1158/1940-6207.CAPR-10-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buttgereit F, Brand MD. A hierarchy of ATP-consuming processes in mammalian cells. Biochem J. 1995;312:163–167. doi: 10.1042/bj3120163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsieh AC, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer. 2010;10:254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 37.Pollak M. The insulin and insulin-like growth factor receptor family in neoplasia: An update. Nat Rev Cancer. 2012;12:159–169. doi: 10.1038/nrc3215. [DOI] [PubMed] [Google Scholar]
- 38.Baselga J, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012;366(6):520–529. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petroulakis E, et al. p53-dependent translational control of senescence and transformation via 4E-BPs. Cancer Cell. 2009;16:439–446. doi: 10.1016/j.ccr.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 40.Dai M, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33(20):e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandberg R, Larsson O. Improved precision and accuracy for microarrays using updated probe set definitions. BMC Bioinformatics. 2007;8(1):48. doi: 10.1186/1471-2105-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyle EI, et al. GO:TermFinder—Open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.