Abstract
Nonsense-mediated RNA decay (NMD) is an evolutionarily conserved RNA quality control mechanism that eliminates transcripts containing nonsense mutations. NMD has also been shown to affect the expression of numerous genes, and inactivation of this pathway is lethal in higher eukaryotes. However, despite relatively detailed knowledge of the molecular basis of NMD, our understanding of its physiological functions is still limited and the underlying causes of lethality are unknown. In this study, we examined the importance of NMD in plants by analyzing an allelic series of Arabidopsis thaliana mutants impaired in the core NMD components SMG7 and UPF1. We found that impaired NMD elicits a pathogen defense response which appears to be proportional to the extent of NMD deficiency. We also demonstrate that developmental aberrations and lethality of the strong smg7 and upf1 alleles are caused by constitutive pathogen response upregulation. Disruption of pathogen signaling suppresses the lethality of the upf1-3 null allele and growth defects associated with SMG7 dysfunction. Interestingly, infertility and abortive meiosis observed in smg7 mutants is not coupled with impaired NMD suggesting a broader function of SMG7 in cellular metabolism. Taken together, our results uncover a major physiological consequence of NMD deficiency in Arabidopsis and revealed multifaceted roles of SMG7 in plant growth and development.
INTRODUCTION
Nonsense-mediated RNA decay (NMD) is an important surveillance mechanism that detects and targets aberrant RNA transcripts for degradation (1,2). NMD substrates are typically transcripts containing a premature translation termination codon (PTC) that can arise by mutation and gene rearrangements, transcription errors or alternative splicing.
Research from various model organisms has provided reasonably detailed molecular insights into how aberrant RNAs are recognized and processed by the NMD machinery. One of the most defining features of NMD substrates is a PTC that gives rise to a long 3′ UTR, which is sensed, in a translation-dependent manner, by the evolutionarily conserved RNA helicase UPF1 (3). The current NMD model predicts that the decision of whether RNA will be degraded or not is determined by competition between cytoplasmic poly(A)-binding protein 1 and UPF1 for binding to translation release factors eRF1 and eRF3 at the terminating ribosome (1). This interaction may be modulated by numerous structural features within mRNA. For example, the presence of splicing boundaries downstream of a stop codon acts as a strong enhancer of NMD. This is due to the activity of two other conserved NMD components, UPF2 and UPF3, which associate with the exon junction complex that is deposited at the exon–exon boundaries and are thought to enhance interaction between UPF1 and the release factors (1,4). The UPF1-eRF binding stimulates UPF1 phosphorylation by SMG1 kinase and promotes RNA degradation. In many eukaryotic organisms, this process relies on the conserved SMG5–7 phosphoserine binding proteins that interact with phosphorylated UPF1 and promote its dephosphorylation by the protein phosphatase PP2A (5,6). In metazoans, these proteins are suggested to determine two distinct pathways of RNA degradation. One mechanism relies on SMG5 and SMG7, which are thought to direct aberrant RNAs destined for exonucleolytic degradation to cytoplasmic P-bodies (7,8). The alternative pathway initiates mRNA degradation by endonucleolytic cleavage in the vicinity of the PTC via the PilT N-terminus (PIN) nuclease domain of SMG6 (9,10).
Although NMD is primarily described as a quality control mechanism, studies in a number of organisms have shown that it affects the stability of many physiological mRNAs as well as non-coding transcripts and pseudogenes, indicating a broader role in gene regulation (11–14). Nevertheless, the biological function of NMD is still not fully understood, partially because NMD null mutations are lethal in higher eukaryotes. While NMD is dispensable for viability in yeast and worms (15–18), inactivation of the core NMD components such as UPF1, UPF2 or members of the SMG5–7 protein family confer embryonic lethality in flies, zebra fish and mice (19–23). However, the primary cause of the lethality in these organisms is unknown. Transcriptome analysis in aborted embryos of SMG1 mouse knock-outs revealed massive mis-expression of a number of genes, suggesting that defective embryogenesis in NMD mutants is underpinned by deregulation of multiple cellular pathways (24). Alternatively, NMD may be critical for a specific process, deregulation of which has fatal consequences. It is also possible that the essential functions of the above-mentioned proteins are linked to their roles in processes unrelated to NMD (2).
Nonsense-mediated RNA decay is also conserved in plants and several of its components have been functionally characterized in Arabidopsis (25–29). These studies demonstrated that NMD is essential for plant viability as null mutations in SMG7 and UPF1 cause embryo and seedling lethality, respectively. In our previous work, we took advantage of viable hypomorphic smg7 alleles, which allowed us to study the role of SMG7 in plants. We showed that these mutants are infertile and that the infertility is caused by a specific defect in meiotic cell cycle progression that prevents formation of functional gametes (29,30). The smg7 mutants also exhibited a range of vegetative phenotypes suggesting a pleiotropic role of SMG7 in plant development. Interestingly, these phenotypes were modulated by environmental conditions as the growth defects were largely suppressed by cultivation at higher temperature (27°C) or elevated humidity (80–100%). In this study, we demonstrate that the vegetative growth defects in hypomorphic smg7 mutants are caused by deregulation of a specific pathway involved in pathogen response. Furthermore, we show that lethality of the UPF1 deficient plants is also caused by massive upregulation of pathogen response. Nevertheless, meiotic defects observed in smg7 mutants do not appear to be a consequence of aberrant NMD and indicate a broader role of SMG7 in cellular metabolism.
MATERIALS AND METHODS
Plant growth conditions and treatments
All mutant strains used, in this study, were obtained from the Arabidopsis Stock Center (Supplementary Table S1). Arabidopsis thaliana ecotype Col-0 was used as a control line. Seeds were either directly sown on soil or were germinated on 0.5 × MS medium supplemented with 1% sucrose and 0.6% plant agar (Duchefa, Netherlands). In most experiments, plants were grown at 21°C and 60–70% humidity under long day conditions (16 h light/8 h dark). To examine phenotypic difference among upf1 and upf3 alleles, plants were grown at 60–70% humidity and 19°C during the photoperiod (16 h) and at 16°C in the dark (8 h). Plants used for infiltration with P. syringae were grown at 21°C and 60–70% humidity under short day conditions (8 h light/16 h dark). Infection of Arabidopsis plants with P. syringae was performed according to (31). In brief, an inoculum of P. syringae pv. maculicula ES4326 containing the LuxCDABE operon (32) was prepared by growing the bacterial culture at 28°C in King’s B medium, followed by sedimentation and two washing steps. The bacterial pellet was diluted in water supplemented with 0.004% Silvet L-77 (Lehle Seeds) to an OD600 of 0.005 or 0.002. Six-week-old plants grown in short day conditions at 21°C were pre-conditioned for 3 days in a grow chamber under a 19/16°C temperature regime and then vacuum infiltrated with either the bacterial inoculum or with distilled water. Imaging of luciferase activity was performed using the VisiLuxx imaging system (Visitron System). For determination of the bacterial titer, discs of leaf tissue were excised from infiltrated leaves 48 h post-infection, washed and ground. The extract was then plated in a serial dilution on plates containing King’s medium B and bacteria were allowed to grow at 28°C for 2 days, followed by quantification of colony forming units.
RNA analyses
Total RNA was isolated from plant tissues using TriReagent (Sigma). For Northern blot analysis, 10 µg of total RNA was fractionated by electrophoresis on a 1.2% formaldehyde agarose gel, blotted onto a nylon membrane (Hybond-NX, Amersham) and probed with 32P-labeled probes specific for PR1 (At2g14610), PR5 (At1g75040) and PDF1.2 (At1g55010). The autoradiogram was recorded on a Kodak Phospor Screen (Biorad) and scanned with the Molecular Imager FX (Biorad) imaging system. Equal loading of RNA samples was verified with ethidium bromide staining of the RNA gel. For analysis of alternatively spliced At2g45670 transcripts by RT-PCR, cDNA was reverse transcribed from 2 µg of total RNA using oligo-dT primers followed by PCR with primers listed in Supplementary Table S2. PCR products were separated on an 8% polyacrylamide gel, DNA was stained with SybrGreen I, and fluorescence signal was scanned using a Molecular Imager FX (Biorad). Cordycepin and cycloheximide treatments were carried out as previously described (33,34). RNA for qPCR analysis was pretreated with TURBO DNA-free DNAse (Ambion) and reverse transcribed with qScript Flex cDNA synthesis kit (Quanta Biosciences) and oligo dT in a 20 µl reaction. Two microlitres of the cDNA diluted in a 1:5 ratio were then used as a template in qPCR using the SensiMix SYBR & Fluorescein kit (Bioline) and the iQ5 cycler (BioRad) with PR1 and At2g45670 specific primers (Supplementary Table S2). The At4g26410 gene, expression of which does not alter after biotic stress, was used for normalization (35). All presented data are derived from three biological replicas, each of which represents an average of three technical replicas.
Histology and cytology
Necrotic lesions on leaves were detected by trypan blue staining according to (36). The stained leaves were examined under a phase-contrast stereo microscope (Leica). Meiosis in pollen mother cells and pollen viability were analyzed as described (29,37).
Salicylic acid quantification
Leaves of three 5-week-old plants were pooled and ground in liquid nitrogen to a fine powder. Free and total levels of salicylic acid were measured via high-performance anion-exchange chromatography as previously described (38).
RESULTS
Vegetative defects in smg7 mutants can be genetically uncoupled from infertility
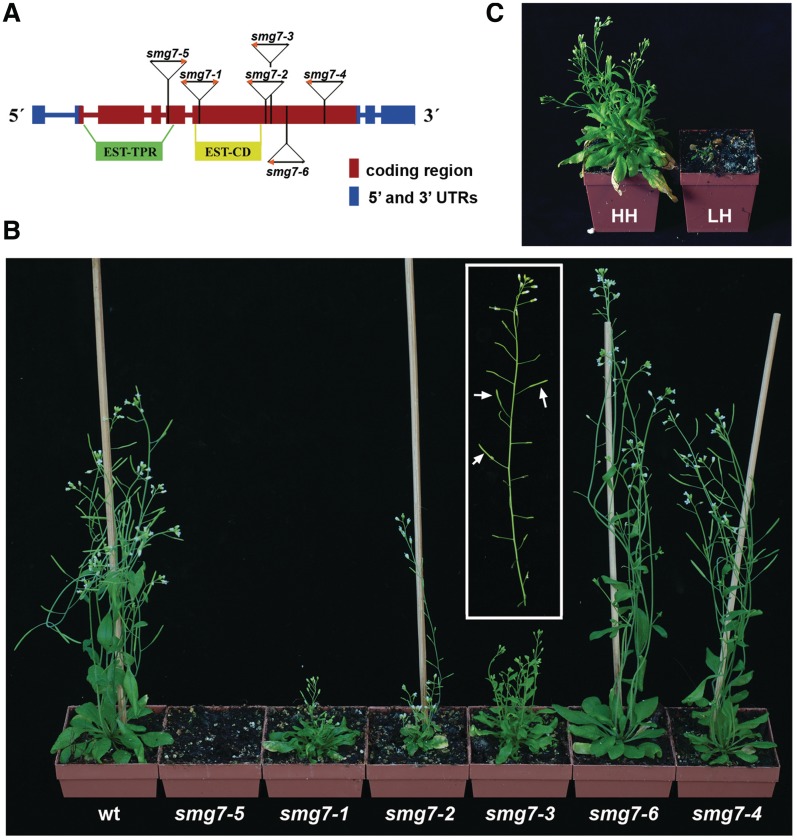
SMG7 is a member of the SMG5–7 protein family, which is characterized by an N-terminal TPR (tetratrico peptide repeat)-domain and a conserved central domain. SMG7 lacks the C-terminal PIN domain that is present in SMG5 and SMG6. SMG7 proteins are present in most metazoans and in plants, which appear to lack the SMG5 and SMG6 homologues (29). In our previous study, we analyzed three Arabidopsis smg7 mutant lines carrying T-DNA insertions in the conserved N-terminal and central domains. While disruption of the N-terminal TPR-domain in smg7-5 mutants is embryonic lethal, mutants with more distal insertions (smg7-1 and smg7-3, Figure 1A) were viable. The smg7-1 and smg7-3 plants were infertile and exhibited pleiotropic growth phenotypes that included dwarf stature, narrow serrated leaves and suppressed apical dominance (29). To gain further insight into the role of SMG7 in plant growth and development, we analyzed three additional smg7 T-DNA insertion alleles (Figure 1). The smg7-2 plants, which harbour a disruption in the vicinity of the insertion site in the smg7-3 allele disrupting the gene at the end of the central domain, show a similar set of phenotypes to smg7-1 and smg7-3 mutants and are completely sterile. Interestingly, the smg7-6 and smg7-4 alleles, which have insertions in the non-conserved C-terminal domain of SMG7, do not cause vegetative growth defects and are indistinguishable from wild-type. However, while smg7-4 plants are fully fertile, the smg7-6 mutants have significantly reduced fertility and only some late developing flowers give rise to seeds (Figure 1B).
Figure 1.
An allelic series of Arabidopsis smg7 mutants. (A) Diagram of the SMG7 gene that includes positions of T-DNA insertions with indicated orientation of T-DNA left borders (orange arrowhead). Boxes represent exons, regions coding for the conserved TPR (EST-TPR) and central (EST-CD) domains are also shown. Structure of T-DNA insertions in smg7-4 and smg7-6 alleles is shown in Supplementary Figure 1. (B) Six-week-old smg7 mutant plants grown at 60–70% humidity. In the inset is an older inflorescence from smg7-6 mutant carrying partially fertile siliques that are indicated by arrows. (C) The smg7-1 mutant plants grown at 60–70% (HH, high) and 40–50% (LH; low) humidity at 21°C.
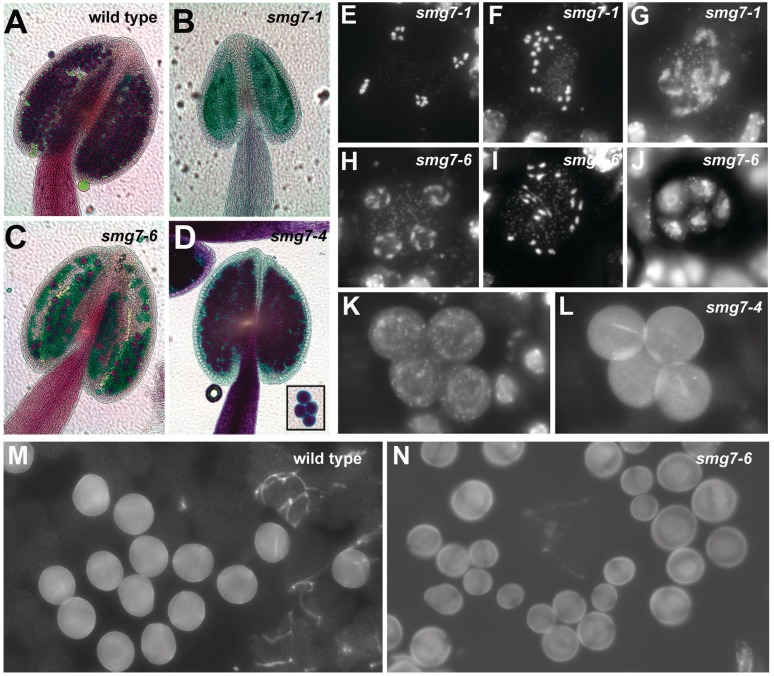
To more fully understand the cause of impaired fertility in smg7-6 mutants, we undertook a closer inspection of anthers and meiosis in pollen mother cells (PMC). Anthers of infertile smg7-1 mutants are devoid of any pollen, because meiotic progression arrests in anaphase II. This arrest results in aberrant meiocytes that contain separated condensed chromatids that do not undergo cytokinesis [Figure 2; (29)]. Although analysis of meiosis in smg7-6 PMCs revealed figures typical for meiocytes arrested in anaphase II (Figure 2I), we also detected normal telophase II stages with four haploid nuclei (Figure 2H). In addition, we often observed polyads containing a variable number of nuclei of different size (Figure 2J). These polyads underwent cytokinesis producing microspores of unequal size (Figure 2N). We conclude from these data that meiosis in smg7-6 PMCs is delayed in anaphase II, but in contrast to the situation smg7-1 mutants, it eventually proceeds to cytokinesis, even producing a small number of viable pollen (Figure 2C). Although smg7-4 plants are fully fertile, we noticed that all four meiotic microspores tend to remain attached to each other, and this attachment is also preserved in mature pollen (Figure 2D, K, L) indicating that SMG7 function in the smg7-4 germ line is still partially compromised. These data suggest that while the non-conserved SMG7 C-terminal domain is dispensable for normal vegetative growth, it is essential for proper germ-line development.
Figure 2.
Development of male germ-line in smg7-6 and smg7-4 mutants. (A–D) Anthers of different smg7 mutants after Alexander staining. Viable pollen stain red, no pollen is present in smg7-1 anthers. Inset in (D) shows four joint pollen grains in smg7-4 mutants. (E–J) End of second meiotic division in smg7-1 and smg7-6 mutants. Meiotic chromosomes are stained with DAPI: (E) regular anaphase II, (F, I) irregular anaphase II, (G) slowly decondensing chromosomes after anaphase II, (H) regular telophase II, (J) a polyad consisting of five separated cells with irregular nuclei. (K, L) Microspores typical for smg7-4 mutants remain attached after meiotic division. Nuclei are stained with DAPI (K), cell walls detected due to their auto-fluorescence using FITC filters (L). (M, N) Fields of microspores in wild-type and smg7-6 mutants.
Vegetative defects in smg7 mutants are caused by constitutive pathogen signaling
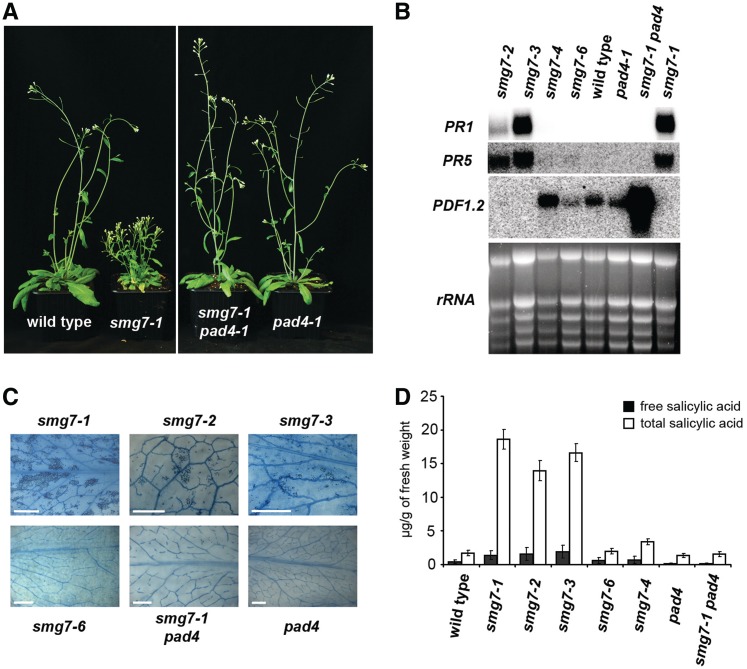
We next decided to decipher the cause of the severe vegetative phenotypes. We noticed that growth performance of smg7-1 mutants is strongly influenced by environmental conditions. Plants grown at high humidity were much stronger and bigger than plants grown at low humidity (Figure 1C). We also observed similar modulation of the phenotype by temperature as smg7-1 mutants grown at 27°C were almost indistinguishable from wild-type plants (data not shown). Under suboptimal conditions (either 19°C/16°C, 60–70% humidity or 21°C, 40–50% humidity), smg7-1 plants often succumbed to massive necrosis within the first 3 weeks after germination [Supplementary Figure S4; (29)]. Necrotic lesions were apparent in all mutants with vegetative growth defects (smg7-1,-2,-3), but not in smg7-6 plants (Figure 3C). Leaf necrosis and improved survival at high humidity or temperature are hallmarks of lesion mimic mutants (39–41). Such mutants have constitutively up-regulated pathogen signaling, characterized by high levels of salicylic acid (SA) and pathogen related (PR) transcripts, which leads to an enhanced hypersensitive response. Indeed, all strong smg7 alleles (smg7-1,-2,-3) show elevated expression of PR1 and PR5 and up to ∼10-fold increase in concentration of SA (Figure 3). In contrast, levels of PR transcripts and SA in the mild smg7 mutants (smg7-6,-4) were comparable to wild-type plants.
Figure 3.
Aberrant growth of smg7 mutants is associated with elevated pathogen response. (A) Six-week-old plants carrying different mutant combinations grown at 60–70% humidity. (B) Northern blot analysis of expression of pathogen related transcripts (PR1 and PR5) and the defensin PDF1.2 in leaves of smg7 mutants. Ethidium bromide stained gel with rRNA species is shown as a RNA-loading control. (C) Necrotic lesions in leaves visualized by staining with trypan blue. Scale bar = 2 mm. (D) Concentrations of SA in leaves of smg7 mutants. Each data point represents three biological replicates.
A strong hypersensitive response is usually caused by effector triggered immunity, which is initiated by activation of R-gene(s) in response to pathogen avirulence factors, and is further transduced, depending on the type of R-gene, through either the NDR1 protein or the PAD4/EDS1 complex (42–44). To determine whether inactivation of these pathways attenuates the constitutive pathogen signaling in SMG7 deficient plants, we generated smg7-1 ndr1 and smg7-1 pad4 double mutants. Whereas growth performance and PR1 transcription were not affected by the ndr1 mutation (Supplementary Figure S2), inactivation of PAD4 in smg7-1 mutants led to full suppression of all vegetative defects. Furthermore, levels of SA and PR transcripts were restored to normal (Figure 3). Nevertheless, smg7-1 pad4 still exhibited increased expression of the defensin PDF1.2, which is regulated by the jasmonic acid signaling pathway. Jasmonic acid mediates a subset of pathogen responses, but this pathway is usually antagonized by SA (45). Accordingly, expression of PDF1.2 was suppressed in smg7 mutants with high levels of SA (Figure 3B). The active jasmonic acid pathway in smg7-1 pad4 mutants argues that PAD4 inactivation only aborts the SA mediated response, but there is still ongoing defense signaling.
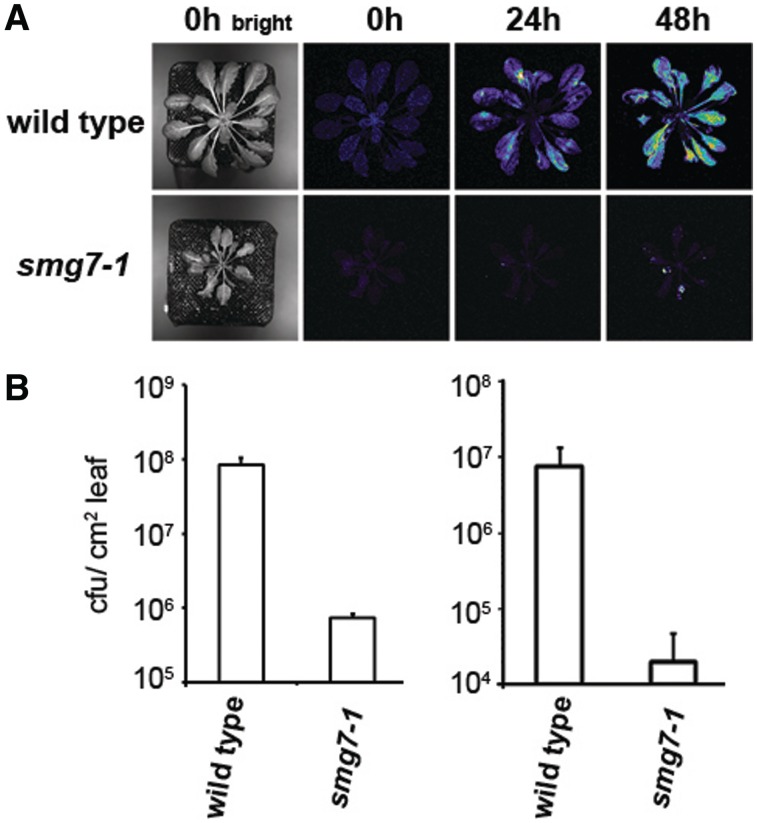
Enhanced SA signaling confers increased resistance to pathogens (46). To analyze the susceptibility of SMG7 deficient plants with up-regulated SA signaling to a bacterial pathogen, we compared growth of a virulent strain of Pseudomonas syringae pv. maculicola ES4326 containing the luxCDAEB operon (32) in leaves of wild-type and smg7-1 mutants. By measuring luminescence and titer of P. syringae, we observed that bacterial growth was inhibited in smg7-1 mutants by more than two orders of magnitude (Figure 4). In conclusion, upregulation of molecular markers, as well as increased resistance to P. syringae, demonstrate that SMG7 deficiency in Arabidopsis leads to auto-activation of the immune response.
Figure 4.
Resistance of smg7-1 mutants to P. syringae. (A) Approximately six-week-old plants grown under short day conditions were infiltrated with P. syringae pv. maculicola ES4326 harbouring the LuxCDABE operon. Growth of bacteria in leaves was monitored at 24 and 48 h after inoculation by measuring the amount of light produced by bacteria within a 5 min interval. Pseudo-colouring of the images from blue through white and yellow to red reflects increasing light intensity. The left panel shows pictures of the same plants taken under bright light. (B) Concentration of bacteria in leaves determined by plating method in two independent experiments. Left chart shows concentration of bacteria 48 h after infection with higher concentration of bacteria (OD600 = 0.005). The right chart shows concentration of bacteria 72 h after infection with a lower concentration of bacteria (OD600 = 0.002). Standard deviations are derived from three independent biological replicates.
Activation of pathogen response is caused by NMD deficiency
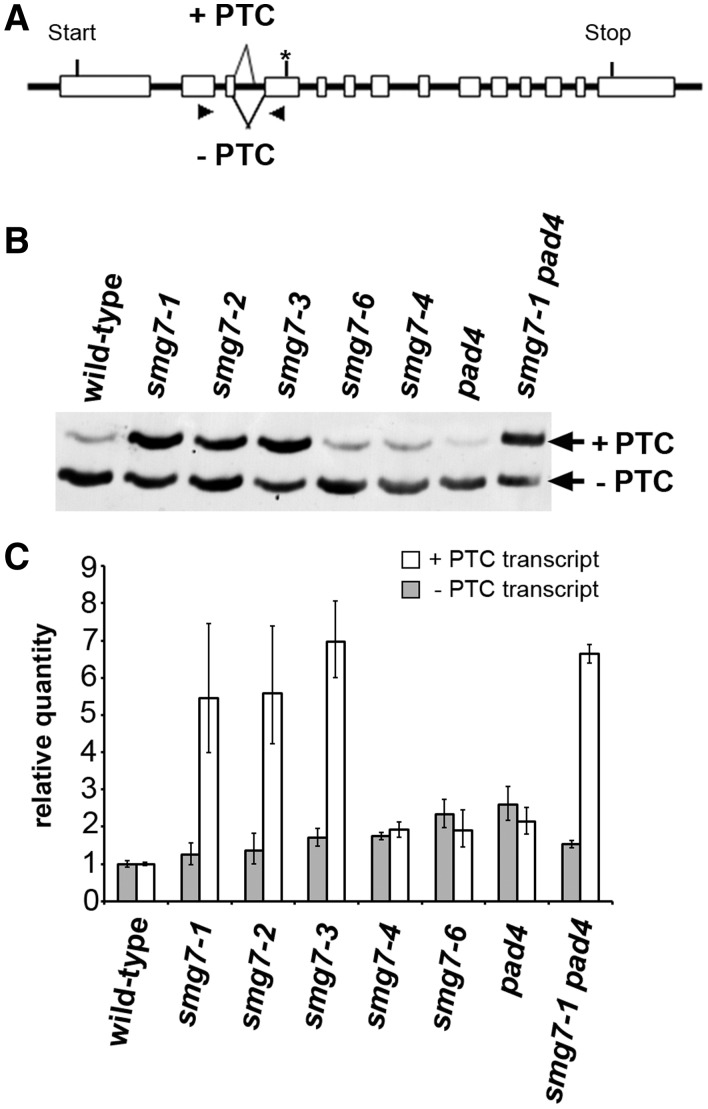
Although the pad4 mutation rescues the vegetative growth phenotypes of smg7 mutants, smg7-1 pad4 mutants are still infertile. Furthermore, the mild smg7-6 and smg7-4 truncated alleles do not show pathogen response activation, but are still partially impaired in reproductive development. These observations indicate that SMG7 may participate in these processes through different molecular mechanisms. Because SMG7 is primarily known for its function in NMD, we assayed whether there was a correlation between NMD deficiency and any of the observed phenotypes. We used an alternatively spliced transcript of the At2g45670 gene harbouring a PTC as an endogenous NMD reporter (Figure 5A). The PTC-containing spliced variant is rapidly degraded after inhibition of transcription by cordycepin, and its amount is elevated after cycloheximide treatment that blocks translation (Supplementary Figure S3). This confirms that the alternatively spliced At2g45670 variant is a genuine NMD substrate (34). Quantitative PCR and RT-PCR analyses showed that the PTC-containing transcript is strongly increased relative to the transcript without PTC in smg7-1, -2 and -3 mutants confirming impaired NMD (Figure 5B and C). As expected, the pad4 mutation did not rescue NMD deficiency in smg7-1 plants. However, the ratios of +PTC and −PTC transcript levels in smg7-4 and smg7-6 mutants are comparable with wild-type suggesting that these plants are NMD proficient (Figure 5).
Figure 5.
NMD efficiency in smg7 mutants. (A) Alternative splicing of the At2g45670 results in a PTC in the 4th exon (indicated by an asterisk). Primers used to amplify the cDNA region spanning the alternatively spliced sites are indicated by arrowheads. (B, C) Effect of smg7 alleles on the abundance of alternatively spliced At2g45670 transcripts analyzed by (B) RT-PCR and polyacrylamide gel electrophoresis (C) or qPCR. Error bars represent standard deviations from three independent RNA samples.
The detection of increased +PTC transcripts only in the smg7 mutants with vegetative growth defects indicated that auto-activation of pathogen response is caused by aberrant NMD. In this case, deregulation of pathogen response should also occur in plants deficient in other NMD genes. In support of this prediction, a recent study showed slightly elevated levels of SA and PR1 transcripts in Arabidopsis upf1-5 and upf3-1 mutants that were not exposed to a pathogen (47). These mutants were also reported to exhibit abnormal growth phenotypes and altered response to P. syringae infection (47,48). We have also detected ∼2- to 3-fold increase in SA accumulation in the upf1-5 and upf3-1 mutants along with mild growth defects, such as narrower and slightly smaller rosette leaves, when the plants were cultivated at 16°C. However, smg7-1 plants grown at the same conditions exhibited much stronger growth retardation and more than 20-times higher concentration of SA in leaves than upf1-5 and upf3-1 mutants (Supplementary Figure S4).
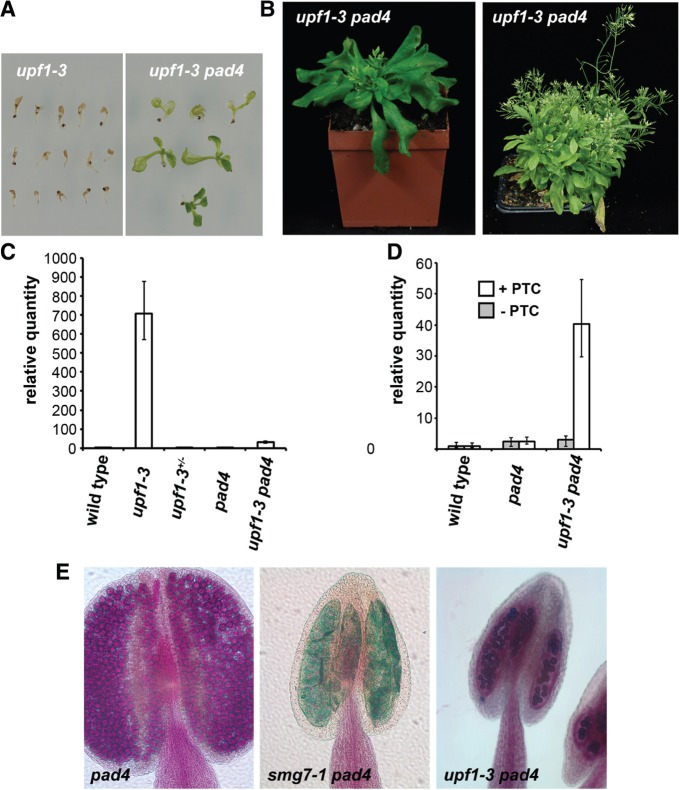
The upf1-5 allele is a hypomorphic mutation with a T-DNA insertion in the 3′UTR that leads to a reduction of UPF1 expression (26). In contrast, the upf1-3 allele harbours a T-DNA disruption in the conserved RNA helicase domain and is lethal (26,27). Seedlings homozygous for the upf1-3 allele succumb to massive necrosis before development of the first true leaves and qRT-PCR analysis revealed that the upf1-3 seedlings have highly elevated expression of PR1 (Figure 6A and C). To determine whether the excessive hypersensitive response causes seedling lethality, we attenuated the pathogen response pathway by inactivating PAD4. Surprisingly, upf1-3 pad4 mutant seedlings not only remained green, with most developing true leaves, many even survived to produce flowering adult plants (Figure 6A,B). The upf1-3 pad4 plants grew more slowly than wild-type plants and also flowered much later, producing a large number of leaves before forming inflorescence bolts (Figure 6B). These data demonstrate that deregulation of pathogen response is the primary cause of lethality in UPF1 deficient plants. Analysis of At2g45670 expression revealed that while the relative quantity of the +PTC transcript is increased six times in smg7-1 pad4 mutants (Figure 5C), the +PTC transcript is more than 40 times elevated in upf1-3 pad-4 plants (Figure 6D). This data indicates that the extent of pathogen response upregulation is proportional to the decreased efficiency of NMD.
Figure 6.
Pathogen response signaling causes lethality of upf1-3 mutants. (A) Seedlings of upf1-3 and upf1-3 pad4 mutants grown for 10 days on agar plates. (B) Six- and 12-week-old upf1-3 pad4 plants. (C) Levels of the PR1 transcript measured by qPCR in seedlings of upf1-3 plants and corresponding controls. (D) Levels of alternatively spliced At2g45670 transcripts in upf1-3 pad4 mutants. Error bars in (C) and (D) indicate standard deviations from three independent RNA samples. (E) Anthers of pad4, smg7-1 pad4 and upf1-3 pad4 mutants analyzed by Alexander staining. While smg7-1 pad4 anthers are empty, a reduced number of viable pollen can be detected in upf1-3 pad4 mutants.
Interestingly, despite the more severe vegetative phenotypes, the upf1-3 pad4 mutants are, in contrast to smg7-1 pad4 plants, semisterile and produced viable pollen (Figure 6E). This observation further supports the notion that aberrant NMD does not underlie the meiotic defects observed in smg7 mutants.
DISCUSSION
Nonsense-mediated RNA decay is an mRNA quality control mechanism that is highly conserved from yeast to humans. While the consequences of NMD dysfunction have a rather uniform molecular manifestation across all tested organisms, namely the increased stability of aberrant RNA transcripts, phenotypic outcomes differ significantly between species. In budding and fission yeast, as well as in Caenorhabditis elegans, NMD is dispensable for viability, although NMD deficiency in worms causes mild morphological defects in reproductive organs (15–17). In contrast, inactivation of UPF1 and UPF2 genes in Drosophila, zebra fish and mice is embryonic lethal, although the underlying causes of lethality are not well understood (19–23). Based on genome-wide expression analysis in mammalian cell lines impaired in NMD, an estimated 6–9% of alternatively spliced genes may be subject to regulation by NMD (12,24,49). Similarly, up to 13% of Arabidopsis intron containing genes were predicted to be affected by NMD (50). Deregulation of such a large number of genes may interfere with essential cellular and developmental processes, thus causing lethality. However, some of the core components of the NMD machinery are implicated in diverse molecular pathways, such as DNA replication, telomere maintenance and cell cycle progression (2,51). Therefore, there is the possibility that the essential function of these genes is associated with a role outside of NMD.
In this study, we were able to pinpoint a specific pathway whose deregulation is responsible for the lethal consequences of NMD dysfunction in Arabidopsis. We show that aberrant NMD leads to constitutive auto-activation of the immune response. Importantly, disruption of the PAD4/EDS1 pathogen signaling pathway suppressed the vegetative defects and lethality of the strong smg7 and upf1 alleles, arguing that activation of pathogen signaling pathways is a major physiological response to declined NMD in Arabidopsis. Activation of an immune response upon NMD attenuation may not be limited to plants. A recent study showed that siRNA knock-down of SMG1 or UPF2 in tumour cells led to their immune mediated rejection in mice (52). The key question that remains is how NMD deficiency triggers an immune response. A strong hypersensitive response along with signaling through the PAD4/EDS1 complex indicates that impaired NMD leads to activation of effector triggered immunity in Arabidopsis. This defense mechanism relies on a set of R-proteins that act as intracellular sensors to recognize effector proteins, which are injected by an invading pathogen into plant cells to inhibit other pathogen signaling pathways. An R-protein can recognize an effector either through direct interaction, or can sense its presence indirectly by monitoring host proteins that are targeted by the effectors (43,44,53). Two scenarios can be envisioned for triggering an autoimmune response by impaired NMD. One possibility is that activity of R-genes is unleashed by truncated or novel proteins that are expected to be produced under relaxed mRNA surveillance from transcripts carrying a PTC or a codon frame shift. A similar mechanism was proposed to be responsible for immune rejection of NMD deficient tumours in mice (52) and NMD-resistant transcripts with frame shift mutations were shown to yield novel protein epitopes in human tumour cells (54).
Another possibility is that NMD regulates expression of genes directly involved in pathogen perception or signaling. Indeed, expression of a number of pathogen response genes is altered in Arabidopsis upf1-3 and upf1-5 mutants. These include several WRKY transcription factors, PAD4, EDS1 and SNC1, an R-gene whose mis-regulation leads to a lesion mimic phenotype (13,47,55). However, it is currently unclear whether these genes are direct targets of NMD, or whether their deregulation is a secondary consequence of elevated pathogen signaling. A promising group of putative NMD targets with a regulatory function in pathogen response are R-genes. There are about 150 NBS-LRR R-genes in the Arabidopsis genome. Their global expression analysis showed that at least 12 NBS-LRR R-genes produce alternatively spliced variants and 15 genes contain introns in 5′ or 3′ UTRs (56). Thus, a significant subset of R-gene transcripts carries features typical for NMD substrates.
Our data show that pathogen response upregulation has more detrimental consequences in upf1-3 mutants than in smg7-1 plants. Accordingly, the upf1-3 allele has a greater effect on the +PTC transcript than smg7-1,-2 and -3 alleles. Thus, the extent of pathogen response deregulation appears to be proportional to the extent of NMD deficiency. It is likely that the upf1-3 are NMD-null due to a T-DNA disruption in the highly conserved RNA helicase domain (27), while the smg7-1, -2 and -3 mutants still retain some NMD activity due to expression of the conserved TPR-domain. Alternatively, the role of UPF1 in NMD may be more important than the function of SMG7. Remarkably, the smg7-4 and -6 alleles are fully proficient in degrading the At2g45670 +PTC transcript indicating that, in contrast to mammals (7), the non-conserved C-terminus of SMG7 is dispensable for NMD in Arabidopsis.
Further phenotypic analysis of the SMG7 allelic series demonstrates that the function of this protein goes beyond traditional NMD. Hypomorphic smg7 alleles give rise to seemingly pleiotropic defects, which we were able to dissect into two specific pathways, namely pathogen response and meiotic progression (this study) (29,30). However, while the autoimmune response is caused by NMD deficiency, abnormalities in reproductive development also occur in smg7 alleles, which are NMD proficient. Furthermore, NMD null upf1-3 pad4 mutants do not exhibit the meiotic defects typical of smg7 plants. These data argue that SMG7 function in meiosis is not mediated through NMD. A broader role for SMG7 in cellular metabolism is also inferred from the observation that, in contrast to the upf1-3 mutation, we were not able to rescue embryonic lethality of the smg7-5 allele by inactivating PAD4 (data not shown). An additional function, distinct from the other NMD factors, was also suggested for SMG7 in zebra fish (21).
In conclusion, our detailed functional and phenotypic analysis of SMG7 in Arabidopsis has revealed a multifaceted role for this protein in cellular metabolism. Identification of specific pathways that are affected by SMG7 dysfunction in Arabidopsis (e.g. pathogen signaling and meiosis) will facilitate more detailed understanding of molecular mechanisms that underlie the function of this evolutionary conserved gene and provide important research directions for elucidating the biological function of NMD in an organism with complex RNA regulatory networks.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–4, Supplementary Tables 1–2 and Supplementary References [57–59].
FUNDING
Austrian Science Fund (FWF; grant P19256-B03); Austrian Academy of Sciences. Funding for open access charge: Austrian Science Fund.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Svetlana Akimcheva for excellent technical assistance.
REFERENCES
- 1.Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Muhlemann O. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol. Life Sci. 2010;67:677–700. doi: 10.1007/s00018-009-0177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Isken O, Maquat LE. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat. Rev. Genet. 2008;9:699–712. doi: 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hogg JR, Goff SP. Upf1 senses 3′ UTR length to potentiate mRNA decay. Cell. 2010;143:379–389. doi: 10.1016/j.cell.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conti E, Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr. Opin. Cell. Biol. 2005;17:316–325. doi: 10.1016/j.ceb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, Ohno S. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol. Cell. 2003;12:1187–1200. doi: 10.1016/s1097-2765(03)00443-x. [DOI] [PubMed] [Google Scholar]
- 6.Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol. Cell. 2005;17:537–547. doi: 10.1016/j.molcel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 7.Unterholzner L, Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol. Cell. 2004;16:587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 8.Nicholson P, Muhlemann O. Cutting the nonsense: the degradation of PTC-containing mRNAs. Biochem. Soc. Trans. 2010;38:1615–1620. doi: 10.1042/BST0381615. [DOI] [PubMed] [Google Scholar]
- 9.Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat. Struct. Mol. Biol. 2009;16:49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- 10.Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA. 2008;14:2609–2617. doi: 10.1261/rna.1386208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Monch K, Thoren LA, Nielsen FC, Jacobsen SE, Nerlov C, Porse BT. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 2008;22:1381–1396. doi: 10.1101/gad.468808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 13.Kurihara Y, Matsui A, Hanada K, Kawashima M, Ishida J, Morosawa T, Tanaka M, Kaminuma E, Mochizuki Y, Matsushima A, et al. Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis. Proc. Natl Acad. Sci. USA. 2009;106:2453–2458. doi: 10.1073/pnas.0808902106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitrovich QM, Anderson P. mRNA surveillance of expressed pseudogenes in C. elegans. Curr. Biol. 2005;15:963–967. doi: 10.1016/j.cub.2005.04.055. [DOI] [PubMed] [Google Scholar]
- 15.Hodgkin J, Papp A, Pulak R, Ambros V, Anderson P. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics. 1989;123:301–313. doi: 10.1093/genetics/123.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui Y, Hagan KW, Zhang S, Peltz SW. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 1995;9:423–436. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- 17.Mendell JT, Medghalchi SM, Lake RG, Noensie EN, Dietz HC. Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol. Cell. Biol. 2000;20:8944–8957. doi: 10.1128/mcb.20.23.8944-8957.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cali BM, Kuchma SL, Latham J, Anderson P. smg-7 is required for mRNA surveillance in Caenorhabditis elegans. Genetics. 1999;151:605–616. doi: 10.1093/genetics/151.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metzstein MM, Krasnow MA. Functions of the nonsense-mediated mRNA decay pathway in Drosophila development. PLoS Genet. 2006;2:e180. doi: 10.1371/journal.pgen.0020180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avery P, Vicente-Crespo M, Francis D, Nashchekina O, Alonso CR, Palacios IM. Drosophila Upf1 and Upf2 loss of function inhibits cell growth and causes animal death in a Upf3-independent manner. RNA. 2011;17:624–638. doi: 10.1261/rna.2404211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wittkopp N, Huntzinger E, Weiler C, Sauliere J, Schmidt S, Sonawane M, Izaurralde E. Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol. Cell Biol. 2009;29:3517–3528. doi: 10.1128/MCB.00177-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum. Mol. Genet. 2001;10:99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- 23.Hwang J, Maquat LE. Nonsense-mediated mRNA decay (NMD) in animal embryogenesis: to die or not to die, that is the question. Curr. Opin. Genet. Dev. 2011;21:422–430. doi: 10.1016/j.gde.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McIlwain DR, Pan Q, Reilly PT, Elia AJ, McCracken S, Wakeham AC, Itie-Youten A, Blencowe BJ, Mak TW. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc. Natl Acad. Sci. USA. 2010;107:12186–12191. doi: 10.1073/pnas.1007336107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hori K, Watanabe Y. UPF3 suppresses aberrant spliced mRNA in Arabidopsis. Plant J. 2005;43:530–540. doi: 10.1111/j.1365-313X.2005.02473.x. [DOI] [PubMed] [Google Scholar]
- 26.Arciga-Reyes L, Wootton L, Kieffer M, Davies B. UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 2006;47:480–489. doi: 10.1111/j.1365-313X.2006.02802.x. [DOI] [PubMed] [Google Scholar]
- 27.Yoine M, Nishii T, Nakamura K. Arabidopsis UPF1 RNA helicase for nonsense-mediated mRNA decay is involved in seed size control and is essential for growth. Plant Cell Physiol. 2006;47:572–580. doi: 10.1093/pcp/pcj035. [DOI] [PubMed] [Google Scholar]
- 28.Yoine M, Ohto MA, Onai K, Mita S, Nakamura K. The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis. Plant J. 2006;47:49–62. doi: 10.1111/j.1365-313X.2006.02771.x. [DOI] [PubMed] [Google Scholar]
- 29.Riehs N, Akimcheva S, Puizina J, Bulankova P, Idol RA, Siroky J, Schleiffer A, Schweizer D, Shippen DE, Riha K. Arabidopsis SMG7 protein is required for exit from meiosis. J. Cell Sci. 2008;121:2208–2216. doi: 10.1242/jcs.027862. [DOI] [PubMed] [Google Scholar]
- 30.Bulankova P, Riehs-Kearnan N, Nowack MK, Schnittger A, Riha K. Meiotic progression in Arabidopsis is governed by complex regulatory interactions between SMG7, TDM1, and the meiosis I-specific cyclin TAM. Plant Cell. 2010;22:3791–3803. doi: 10.1105/tpc.110.078378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Katagiri,F., Thilmony,R. and He,S.Y. (2002) The Arabidopsis thaliana-Pseudomonas syringae interaction. In: Somerville,C.R. and Meyerowitz,E.M. (eds), The Arabidopsis Book. America Society of Plant Biologists, Rockville, MD. [DOI] [PMC free article] [PubMed]
- 32.Fan J, Crooks C, Lamb C. High-throughput quantitative luminescence assay of the growth in planta of Pseudomonas syringae chromosomally tagged with Photorhabdus luminescens luxCDABE. Plant J. 2008;53:393–399. doi: 10.1111/j.1365-313X.2007.03303.x. [DOI] [PubMed] [Google Scholar]
- 33.Gutierrez RA, Ewing RM, Cherry JM, Green PJ. Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc. Natl Acad. Sci. USA. 2002;99:11513–11518. doi: 10.1073/pnas.152204099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hori K, Watanabe Y. Chapter 8. In vivo analysis of plant nonsense-mediated mRNA decay. Methods Enzymol. 2008;449:165–176. doi: 10.1016/S0076-6879(08)02408-7. [DOI] [PubMed] [Google Scholar]
- 35.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koch E, Slusarenko A. Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell. 1990;2:437–445. doi: 10.1105/tpc.2.5.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alexander MP. Differential staining of aborted and nonaborted pollen. Stain Technol. 1969;44:117–122. doi: 10.3109/10520296909063335. [DOI] [PubMed] [Google Scholar]
- 38.Rozhon W, Petutschnig E, Wrzaczek M, Jonak C. Quantification of free and total salicylic acid in plants by solid-phase extraction and isocratic high-performance anion-exchange chromatography. Anal. Bioanal. Chem. 2005;382:1620–1627. doi: 10.1007/s00216-005-3326-x. [DOI] [PubMed] [Google Scholar]
- 39.Noutoshi Y, Ito T, Seki M, Nakashita H, Yoshida S, Marco Y, Shirasu K, Shinozaki K. A single amino acid insertion in the WRKY domain of the Arabidopsis TIR-NBS-LRR-WRKY-type disease resistance protein SLH1 (sensitive to low humidity 1) causes activation of defense responses and hypersensitive cell death. Plant J. 2005;43:873–888. doi: 10.1111/j.1365-313X.2005.02500.x. [DOI] [PubMed] [Google Scholar]
- 40.Yang S, Hua J. A haplotype-specific Resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell. 2004;16:1060–1071. doi: 10.1105/tpc.020479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jambunathan N, Siani JM, McNellis TW. A humidity-sensitive Arabidopsis copine mutant exhibits precocious cell death and increased disease resistance. Plant Cell. 2001;13:2225–2240. doi: 10.1105/tpc.010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 43.Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 44.Eitas TK, Dangl JL. NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr. Opin. Plant Biol. 2010;13:472–477. doi: 10.1016/j.pbi.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spoel SH, Dong X. Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe. 2008;3:348–351. doi: 10.1016/j.chom.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 46.Vlot AC, Dempsey DA, Klessig DF. Salicylic Acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009;47:177–206. doi: 10.1146/annurev.phyto.050908.135202. [DOI] [PubMed] [Google Scholar]
- 47.Jeong HJ, Kim YJ, Kim SH, Kim YH, Lee IJ, Kim YK, Shin JS. Nonsense-mediated mRNA decay factors, UPF1 and UPF3, contribute to plant defense. Plant Cell Physiol. 2011;52:2147–2156. doi: 10.1093/pcp/pcr144. [DOI] [PubMed] [Google Scholar]
- 48.Shi C, Baldwin IT, Wu J. Arabidopsis plants having defects in nonsense-mediated mRNA decay factors UPF1, UPF2, and UPF3 show photoperiod-dependent phenotypes in development and stress responses. J. Integr. Plant Biol. 2012;54:99–114. doi: 10.1111/j.1744-7909.2012.01093.x. [DOI] [PubMed] [Google Scholar]
- 49.Pan Q, Saltzman AL, Kim YK, Misquitta C, Shai O, Maquat LE, Frey BJ, Blencowe BJ. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes Dev. 2006;20:153–158. doi: 10.1101/gad.1382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalyna M, Simpson CG, Syed NH, Lewandowska D, Marquez Y, Kusenda B, Marshall J, Fuller J, Cardle L, McNicol J, et al. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 2012;40:2454–2469. doi: 10.1093/nar/gkr932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feuerhahn S, Iglesias N, Panza A, Porro A, Lingner J. TERRA biogenesis, turnover and implications for function. FEBS Lett. 2010;584:3812–3818. doi: 10.1016/j.febslet.2010.07.032. [DOI] [PubMed] [Google Scholar]
- 52.Pastor F, Kolonias D, Giangrande PH, Gilboa E. Induction of tumour immunity by targeted inhibition of nonsense-mediated mRNA decay. Nature. 2010;465:227–230. doi: 10.1038/nature08999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 54.Williams DS, Bird MJ, Jorissen RN, Yu YL, Walker F, Zhang HH, Nice EC, Burgess AW. Nonsense mediated decay resistant mutations are a source of expressed mutant proteins in colon cancer cell lines with microsatellite instability. PLoS One. 2010;5:e16012. doi: 10.1371/journal.pone.0016012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yi H, Richards EJ. A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. Plant Cell. 2007;19:2929–2939. doi: 10.1105/tpc.107.051821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tan X, Meyers BC, Kozik A, West MA, Morgante M, St Clair DA, Bent AF, Michelmore RW. Global expression analysis of nucleotide binding site-leucine rich repeat-encoding and related genes in Arabidopsis. BMC Plant Biol. 2007;7:56. doi: 10.1186/1471-2229-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Glazebrook J, Zook M, Mert F, Kagan I, Rogers EE, Crute IR, Holub EB, Hammerschmidt R, Ausubel FM. Phytoalexin-deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics. 1997;146:381–392. doi: 10.1093/genetics/146.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Century KS, Shapiro AD, Repetti PP, Dahlbeck D, Holub E, Staskawicz BJ. NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science. 1997;278:1963–1965. doi: 10.1126/science.278.5345.1963. [DOI] [PubMed] [Google Scholar]
- 59.Shapiro AD, Zhang C. The role of NDR1 in avirulence gene-directed signaling and control of programmed cell death in Arabidopsis. Plant Physiol. 2001;127:1089–1101. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.