Abstract
Analyses of the DNA replication-associated proteins of hyperthermophilic archaea have yielded considerable insight into the structure and biochemical function of these evolutionarily conserved factors. However, little is known about the regulation and progression of DNA replication in the context of archaeal cells. In the current work, we describe the generation of strains of Sulfolobus solfataricus and Sulfolobus acidocaldarius that allow the incorporation of nucleoside analogues during DNA replication. We employ this technology, in conjunction with immunolocalization analyses of replisomes, to investigate the sub-cellular localization of nascent DNA and replisomes. Our data reveal a peripheral localization of replisomes in the cell. Furthermore, while the two replication forks emerging from any one of the three replication origins in the Sulfolobus chromosome remain in close proximity, the three origin loci are separated.
INTRODUCTION
As in other archaea, the DNA replication machinery of members of the genus Sulfolobus resembles that of eukaryotes (1). In general, the archaeal machinery is a simplified version of that found in eukarya (2). This relationship between archaeal and eukaryotic replication-associated proteins extends across the replication process, from initiator proteins to factors involved in Okazaki fragment maturation. Furthermore, some archaea, including Sulfolobus species, possess multiple origins of replication per chromosome; a situation reminiscent of that in eukaryotes and clearly distinct from the single origin systems found in bacterial chromosomes (3–7). Thus, we, and others, have been studying Sulfolobus species as model organisms to investigate the fundamental, conserved processes of DNA replication.
Neutral–neutral 2D gel analyses coupled with marker frequency analysis (MFA) in synchronized S. acidocaldarius cells have revealed that all three replication origins fire in every cell and in every cell cycle (8). Two of the origins fire within a narrow temporal window in the 3-h cell cycle, at the onset of S-phase, while the third appears to have slightly more relaxed temporal constraints. Nevertheless, origin firing can be readily detected in some cells at all three origins within 2 min following the preceding cell division event, and by 30-min initiation of replication at all three origins has taken place in the majority of cells. Replication proceeds bi-directionally from all three origins and S-phase is completed by 90-min post-initiation (8). However, while 2D agarose gels and MFA have been of some utility in following replication fork progression, to date no other tools have been developed to accurately detect and map the progression of archaeal replication forks at the single cell level.
Numerous studies over the last few decades have taken advantage of the ability of thymidine salvage pathways to direct incorporation of radiolabelled thymidine or thymidine analogues into newly synthesized DNA (9,10). The key enzyme in this process is thymidine kinase (TK), which converts thymidine to thymidylate. Thymidylate then enters the cellular nucleotide pools. Importantly, TK can also direct the phosphorylation of halogenated analogues of thymidine such as 5-bromo-, 5-chloro- or 5-iodo-2-deoxyuridine (where the 5-methyl group of the thymine base is substituted by the halogen). Incorporation of the halogenated base into DNA can be visualized by detection with specific antisera. More recently, with the development of ‘click chemistry’ it has been possible to direct the incorporation of the synthetic base analogue 5-ethynyl-2-deoxyuridine, EdU, into DNA via the TK-mediated salvage pathway (11). EdU has an alkyne group in place of thymine’s 5-methyl group that, in a copper-catalysed manner, will react with an azide. Thus, incorporation of EdU into DNA can be monitored by conjugation to an azide-containing fluorophore. One potential advantage of EdU over halogenated (halo-dU) base incorporation lies in the ability of the click chemistry to be performed on intact duplex DNA. In contrast, the antibody-mediated detection of 5-halogen deoxyuridine is dependent on the prior denaturation of the DNA to expose single-stranded regions.
Here, we describe the development of strains of Sulfolobus that have the capacity to incorporate base analogues via the thymidine salvage pathway. Utilizing the ability to incorporate EdU, we apply this technology to examine the sub-cellular distribution of nascent DNA in Sulfolobus. These data, in conjunction with immunolocalization analyses of the distribution of the DNA sliding clamp PCNA, support a model in which there is no obligate spatial linkage of the replisomes emerging from the three replication origins in the cell, indicating that replication is not confined to a single replication cluster. However, there does appear to be a co-association of sister replisomes emerging from a given replication origin. Interestingly, we also detect a significant association of replisome foci with the cell periphery.
MATERIALS AND METHODS
Cell growth, synchronization and flow cytometry
Sulfolobus acidocaldarius MW001, a pyrEF− strain derived from S. acidocaldarius DSM639, and S. solfataricus PBL2025 were used as the parental strains (12,13). Unless otherwise stated, cells were grown at 75°C in Brock’s medium pH 3.2 containing 0.1% (w/v) tryptone, 0.2% (w/v) xylose and 0.025 mg/ml uracil. Following transformations, MW001 cultures contained 0.1% NZ-amine as an alternative to tryptone. Following transformations, PBL2025 cultures contained 0.4% lactose without tryptone or xylose. When indicated, l-arabinose was added to 0.4% (w/v). When required, Brock’s medium was solidified with 0.7% Gelrite (GmbH).
SacTK cells were synchronized using the baby machine method as described previously (8) except cells were collected after 5-h elution. Cell synchronization was assessed by flow cytometry as described previously (8).
Construction of plasmids for integration of TK into S. acidocaldarius and S. solfataricus
The S. solfataricus P2 pyrEF genes (SSO0615 and SSO0616) were amplified using primers EF5: CTACGTGTCTTAATCTCACAAAGCCC and TG1: TTAATGTTAGTTTATAAAGACCG. The resulting product was cloned into the SrfI site of pPCR-Script Amp SK(+) to generate pEF. Fragments of ∼1 kb upstream and 1 kb downstream of the S. acidocaldarius amyA gene (Saci1162) were amplified using primers TG2: TTTGAATTCTGTTATTATGTTCAGATACTCG and TG3: CTGCAGCTTAGCACTAGGACTACTTTC, and TG4: GTCCTAGTGCTAAGCTGCAGAAGGAGGTTCCTGAAATAGC and TG5: GTTGGATCCATAAAGGAGGAGCTACTGC, respectively. The resulting fragments were then joined by overlap PCR using primers TG2 and TG5, generating a fusion product containing a central PstI site. This product was cloned into the EcoRI and BamHI sites of pEF to generate pEFamyA. Primers igd254: CCCCCGCGGTAAGCCATCAAGA and igd287: CCCCCAAAAAGCGGCCGCCCGGGGGCCATGGCCATACCTCACAACACCGTTCCG were used to amplify the S. solfataricus P2 tf55α promoter and the resulting product was inserted between the SacII and NotI sites in pC (14), generating pIDSB19. The open reading frame for TK (PAE2453) was amplified from Pyrobaculum aerophilum genomic DNA using primers igd244: GAGGTGAGAATTCCATGGGACTTGTTGTTATTGTGGGGC, and igd245: GATCGCGGCCGCTTATTTGTCGTCGTCGTCTTTGTAGTCTGGAGGAATTACATAATGACGTC, which incorporated a FLAG-tag sequence at the C-terminus of the encoded protein (underlined). The fragment was inserted between the NcoI and NotI sites of pIDSB19, generating pIDSB20. Finally, the Ptf55α-TK-FLAG product was amplified from pIDSB20 using primers TG6: GTTCTGCAGTAAGCCATCAAGAAATCTGC and TG7: GTTCTGCAGTTATTTGTCGTCGTCGTCTTT and the resulting product cloned into the PstI site of pEFamyA to generate pTK3.
To produce vectors for integration into the S. solfataricus chromosome, a 5′-region of the S. solfataricus amyA gene (SSO1172) was amplified from S. solfataricus P2 genomic DNA using primers amyAFront-F: CCCGGTACCTTAGCCATGGGTAATTTGCC and amyAFront-R: CCCCTCGAGCGATCTGAATCGAAATTGAGG. The PCR product was inserted into the KpnI and XhoI sites of pPCR-Script Amp SK(+) to generate pIDSB11. Similarly, a 3′-region of amyA amplified with primers amyABack-F: CCCCGCGGAACATTTCACCATTAGTCTATCTACC and amyABack-R: CCCGAGCTCAATGTGCCCCCTCTTCATTAC, was then cloned between the SacI and SacII sites of pIDSB11, resulting in pIDSB12 (which thus contains a deletion mutant of the amyA gene). The lacS* marker from pET2268 (15) was amplified with primers lacS-F: CCCCTGCAGCTCCTCTTATTATTAGAATTGTACGC and lacS-R: CCCCATCGATCCCTAGTGTTGCAAGGCAG. The product was then cloned between the PstI and ClaI sites of pIDSB12, so that lacS* was located between the ‘front’ and ‘back’ portions of the amyA gene, to give pIDSB13. To place the TK gene under the control of the S. solfataricus arabinose-inducible araS promoter [Para (16)], the TK PCR product (primers igd244 and igd245, as shown above) was first cloned between the NcoI and NotI sites of pRYS_Sso0909 (17) so as to place TK-FLAG under the control of Para. The Para-TK-FLAG fragment from the resulting plasmid (pIDSB5) was then sub-cloned with SacII and NotI into pIDSB13, yielding pIDSB16.
Construction of strains SacTK and SsoTK
Sulfolobus acidocaldarius MW001 was transformed as described previously (12) with pTK3, methylated as described in (14). Strains containing integrated plasmids were isolated on solid NZ-amine media, followed by liquid NZ-amine media. Unmarked mutants were then isolated by second selection using solid NZ-amine media containing 0.25 mg/ml 5-fluoroorotic acid and 0.025 mg/ml uracil. PCR amplification across the amyA locus followed by DNA sequencing was used to confirm the TK-FLAG insertion. S. solfataricus PBL2025 was transformed as described previously (13), using DraIII-linearized pIDSB16 and with initial selection in lactose liquid media (18). Single colonies were isolated on solid media and the expected integration at the amyA locus was confirmed by PCR amplification across the amyA locus followed by DNA sequencing. Western blotting was also used to detect production of the TK-FLAG protein, using an anti-FLAG primary antibody (1:2000 dilution; Sigma, F3165) and anti-IgG (mouse) horseradish peroxidase-coupled secondary antibody followed by standard chemiluminescence detection.
Assessing bromodeoxyuridine/EdU incorporation into DNA
Fifty millilitre cultures, containing the indicated amount of bromodeoxyuridine (BrdU)/EdU, were inoculated with Sulfolobus to a starting OD600 of 0.05 and growth was monitored over time by measuring OD600. S. acidocaldarius genomic DNA labelled with BrdU for 6 h was extracted using a Nucleospin Tissue kit (Macherey-Nagel), digested with PstI, and 750 ng DNA was separated by agarose gel electrophoresis. Sulfolobus solfataricus cells were labelled with BrdU for 12 h and then prepared for pulsed-field gel electrophoresis (PFGE) using the agarose plug method described previously to give a cell density equivalent of A600 = 1 in each plug (8). The cells were lysed, the DNA was purified and then digested with EagI in the plug, as described (8). PFGE was carried out with 1% agarose (SeaKem Gold; FMC BioProducts) in 90 mM Tris–borate, 0.2 mM EDTA (pH 8.2) for 37 h at 5 V/cm at 14°C, with the switching time ramped linearly from 20 to 60 s with included angle of 120°. In both cases, after electrophoresis DNA was transferred to Protran nitrocellulose membrane in 0.5 M Tris–Cl, pH 7.0, 1.5 M NaCl after standard acid/base treatments of the gel. The membrane was blocked with a standard Tris-buffered saline (TBS), 0.05% (v/v) Tween-20, 5% (w/v) skim milk powder solution and then probed using an anti-BrdU primary antibody (2.6 µg/ml; Abcam) and anti-IgG (sheep) HRP-coupled secondary antibody, followed by standard chemiluminescence detection. To detect EdU labelled S. acidocaldarius DNA, 450 ng genomic DNA was extracted using a Nucleospin Tissue kit, digested with PstI, cleaned-up using a Nucleospin Extract II kit (Macherey-Nagel) and eluted in 25 µl water. The PstI digestion step was used simply to generate smaller DNA fragments for increased solubility and ease of handling. DNA was then incubated for 30 min with 20 µl Click-iT reaction mix (Click-iT EdU Imaging Kit, Invitrogen), prepared as described by the manufacturer before cleaning-up using a Nucleospin Extract II kit and eluting in 25 µl water. Ten microlitres of this processed DNA was spotted onto Protran nitrocellulose membrane and visualized using a FLA5000 fluorescence imager.
Fluorescence microscopy
Cells collected from the baby machine were fixed for microscopy as described in (17). Where indicated, prior to fixing, cell grow-out was performed at 75°C for the indicated time period, with or without addition of 200 µM EdU. Immunolocalization of PCNA1 was performed as described previously (17) using a 1:4000 dilution of anti-SSO0391 (S. solfataricus PCNA1) primary antibody, which cross-reacts specifically with the S. acidocaldarius PCNA1 protein. Coverslips were then incubated with Alexa 488-conjugated anti-rabbit secondary antibody diluted 1:1000. When detecting EdU, this was performed using the Click-iT EdU Imaging Kit (Invitrogen), as per the manufacturer’s instructions. A Metamorph-driven Olympus BX-61 microscope was used to capture 30 image steps of 0.1 µM step size and processed using AutoQuant X to generate deconvolved images. The number of EdU/PCNA1 foci observed in 3D reconstructions was comparable to those observed using central 2D slices (20 cells were assessed), thus deconvolved 2D slices were used for foci counting. Cells in which a single EdU focus comprised more than one quarter of the cell area, or where the shape of the foci were irregular and the number of distinct foci could not be determined, were classified as fully labelled. Similar to the method described by Hediger et al. (19), in order to assess cellular localization of replication foci we first generated a template circle to encompass 100% of the cell area (using a central 2D slices). Based on this, we then generated an inner central circle of 50% area and determined whether EdU or PCNA1 foci were located in the inner or outer 50% of the cell area. A binomial test was performed using Excel to determine the probability of obtaining the observed number of foci in the outer 50% cell area if the localization was random i.e. if the probability of being in the outer 50% of the cell was 50%.
RESULTS
Construction of Sulfolobus strains capable of incorporating thymidine analogues
Genes for TK orthologues show a very restricted distribution in the archaeal domain of life. Of 97 fully sequenced archaeal genomes, belonging to the phyla Euryarchaeota, Crenarchaeota, Thaumarchaeota and Korarchaeota, TK superfamily members were found in only 16 species and those were restricted to subsets of the euryarchaeal orders Thermoplasmatales, Halobacteriales and Thermococcales and to three crenarchaeal species, all members of the Thermoproteales; P. aerophilum, P. arsenaticus and Thermoproteus neutrophilus (Supplementary Figure S1). Notably, the Sulfolobales, including S. acidocaldarius and S. solfataricus lack genes for TK. Previous studies in fungi, which also lack endogenous tk genes, have shown that introduction of heterologous tk genes can generate a thymidine salvage pathway in these organisms (20–22).
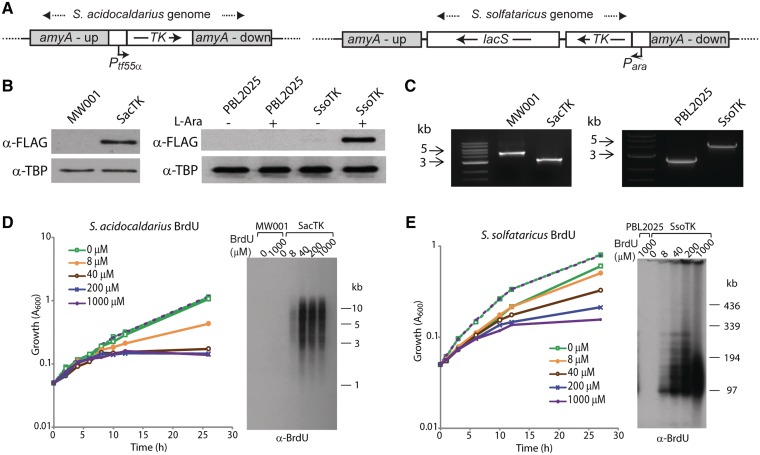
As Sulfolobus species are hyperthermophiles, growing at 80°C, we wished to identify a tk gene from a similarly thermostable organism and to use it to attempt to confer thymidine salvage capability on Sulfolobus cells. Accordingly, we amplified the tk gene (Gene Locus PAE2453) from P. aerophilum (23) and cloned it downstream of the strong arabinose-inducible promoter (Para) or TF55 promoter (Ptf55α) for expression in S. solfataricus or S. acidocaldarius, respectively (16,24). We then used homologous recombination to direct integration of the relevant cassette into the non-essential α-amylase genes of S. solfataricus PBL2025 and S. acidocaldarius MW001 (Figure 1A) (12,25). Integrants were confirmed by PCR amplification across the amyA locus followed by sequencing of the resultant product and TK expression was verified by western blotting, employing a FLAG tag incorporated at the C-terminus of the TK protein (Figure 1B and C). Next, tk+ cells were incubated in the presence of various concentrations of BrdU and the effect on growth monitored. As can be seen in Figure 1D and E, the tk+ cells show a dose-dependent inhibition of growth by BrdU. In contrast, the parental strains show no inhibition even at the highest concentration of BrdU tested.
Figure 1.
Sulfolobus acidocaldarius and S. solfataricus strains expressing FLAG-tagged TK from P. aerophilum incorporate BrdU into genomic DNA. (A) The TK-FLAG gene, expressed from a tf55α promoter, was integrated into the amyA locus of S. acidocaldarius MW001 using a ‘pop-in pop-out’ approach. Double crossover homologous recombination was used to integrate the TK-FLAG gene, expressed from an arabinose-inducible promoter, into the amyA locus of S. solfataricus PBL2025. The lacS marker confers growth to PBL2025 in lactose minimal media. The regions flanking the amyA gene are labelled amyA-up and amyA-down. (B) Western blot analysis of whole cell extracts from exponential cultures of parental strains (MW001 or PBL2025) and TK-FLAG integration strains (SacTK and SsoTK) using an anti-FLAG antibody, and anti-TBP antibody as a control. S. solfataricus cultures were grown in the absence or presence of 0.4% l-arabinose (l-Ara). (C) PCR amplification across the amyA locus using genomic DNA template generated 4.8 and 2.7 kb products in the parental strains MW001 and PBL2025, respectively, and 3.2 and 4.1 kb products in the mutant strains SacTK and SsoTK, respectively, indicating replacement of amyA with the tf55α-TK-FLAG constructs. (D) MW001 (dashed lines) and SacTK (solid lines) cells were grown in medium containing the indicated concentrations of BrdU and growth was monitored by A600. Genomic DNA was extracted from 6 h samples, digested with PstI and separated by electrophoresis. DNA was transferred to a nitrocellulose membrane and probed with an anti-BrdU antibody. (E) PBL2025 (dashed lines) and SsoTK (solid lines) cells were grown in medium containing 0.4% l-Ara and the indicated concentrations of BrdU, and growth was monitored by A600. Genomic DNA was extracted from 12 h samples, digested with EagI and separated by PFGE. DNA was transferred to a nitrocellulose membrane and probed with an anti-BrdU antibody.
The incorporation of BrdU into DNA was confirmed by Southern blotting of DNA recovered from cells incubated with BrdU, followed by detection with an anti-BrdU antibody. As can be seen in the right-hand panels of Figure 1D and E, incubation of tk+ strains with BrdU resulted in a dose-dependent incorporation of BrdU into genomic DNA, whereas there was no detectable incorporation of BrdU in the parental strains.
Visualization of labelled DNA in Sulfolobus cells
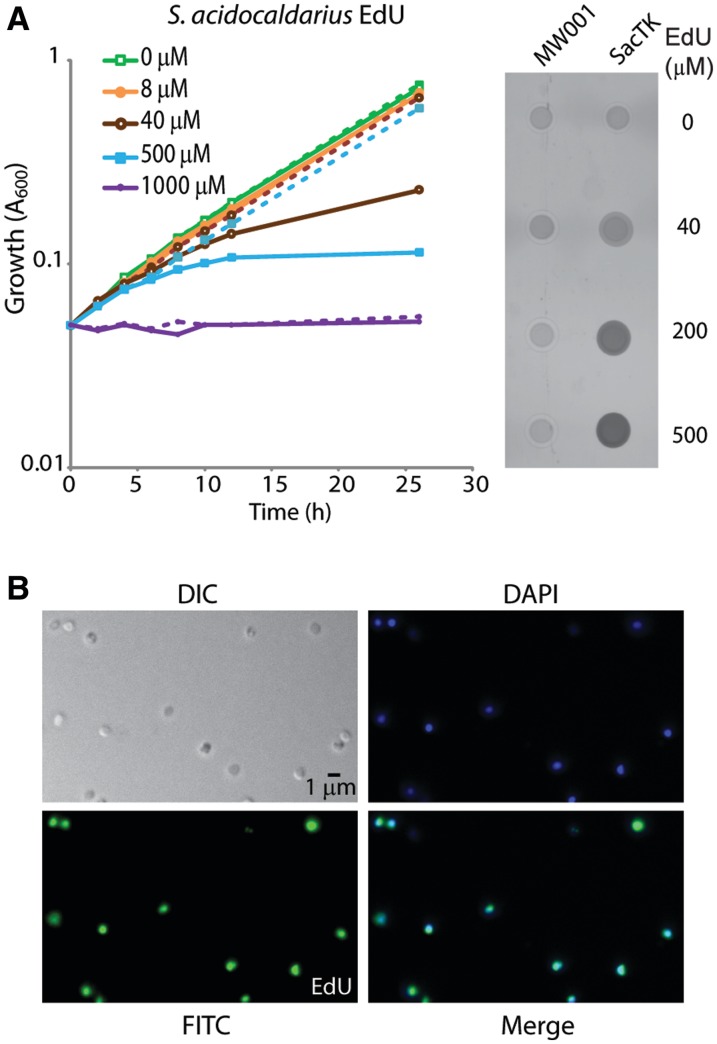
We initially attempted to visualize BrdU incorporation using the available antisera against this moiety. However, the treatments required to generate the single-stranded DNA recognized by these antibodies appeared to grossly perturb the morphology of the nucleoid, resulting in compaction and often resulting in disruption of cells (not shown). We therefore sought to utilize the EdU labelling technology, which allows visualization of the nucleotide analogue in the context of non-denatured double-stranded DNA. As can be seen in Figure 2A and B, cells expressing TK readily incorporated EdU and incubation with 100 µM EdU for 180 min resulted in >70% of cells showing strong labelling throughout the nucleoid (Figure 2B).
Figure 2.
Sulfolobus acidocaldarius cells expressing FLAG-TK incorporate EdU into genomic DNA. (A) MW001 (dashed lines) and SacTK (solid lines) cells were grown in medium containing the indicated concentrations of EdU and growth was monitored by A600. Genomic DNA was extracted from 24 h samples and digested with PstI. Incorporated EdU was fluorescently labelled using ‘click’ chemistry. Three hundred nanograms of each DNA sample were spotted onto a nitrocellulose membrane and visualized using a FLA5000 Phosphoimager. (B) Fluorescent microscopy of SacTK cells grown for 3 h with 100 µM EdU. After fixing with 2.5% paraformaldehyde, cellular DNA was fluorescently labelled using ‘click’ chemistry. Images show DIC, DAPI staining of DNA (blue), EdU-AlexaFluor488 (green) and merged images.
Assessing the impact of BrdU/EdU on cell-cycle progression
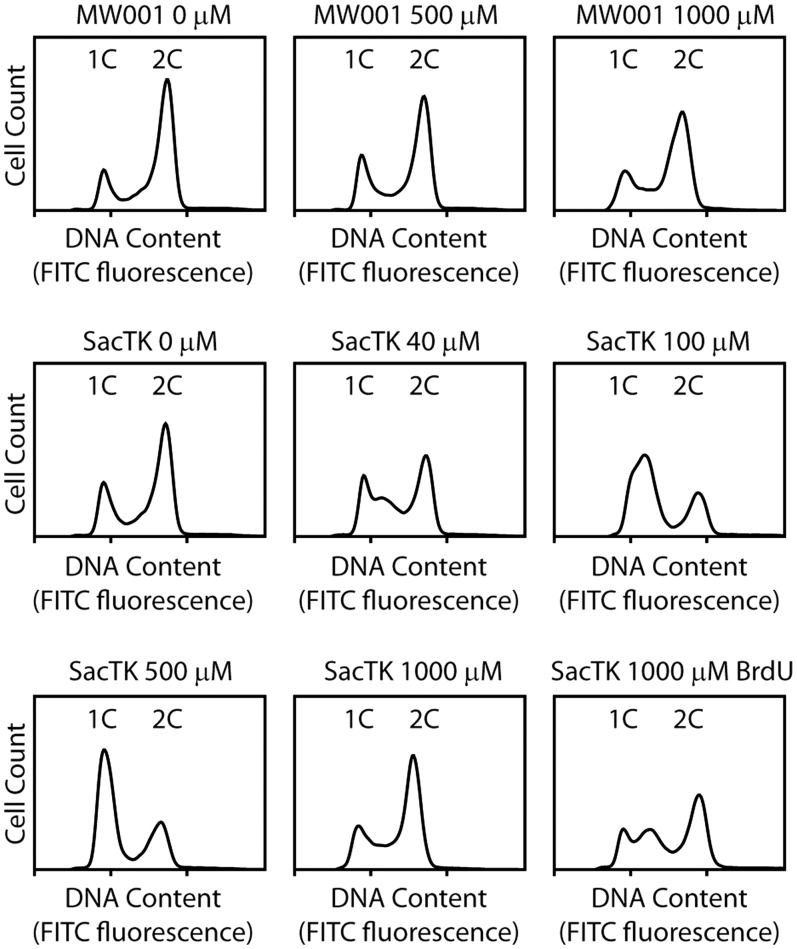
As growth of the tk+ cells was inhibited after 4-h growth (approximately one doubling time) in the presence of BrdU or EdU (Figures 1 and 2), we used flow cytometry to assess the impact of the nucleoside analogues on the cell cycle. As shown in Figure 3, addition of EdU did not significantly alter the cell-cycle profile of the parental strain, MW001 (top three panels). In contrast, treatment of SacTK with EdU or BrdU resulted in an increased S-phase population, indicating a slowed progression through S-phase or an S-phase arrest. This effect was not visible after 1-h incubation with BrdU/EdU (data not shown), but was clearly detected after 3 h, by which time there was an accumulation of S-phase cells (containing between 1n and 2n DNA content) (Figure 3). Strikingly, as EdU concentration was increased, the peak of accumulated cells occurred at lower fluorescence, indicating an increasingly early S-phase arrest (Figure 3). Following release from EdU-mediated cell-cycle arrest, SacTK cells retained a limited, dose-dependent capacity to resume progression through the cell cycle (Supplementary Figure S2).
Figure 3.
Flow cytometry of S. acidocaldarius grown in the presence of nucleoside analogues. Fifty millilitre cultures containing the indicated amount of EdU (or BrdU where shown) were inoculated with S. acidocaldarius SacTK or MW001 to an OD600 of 0.05 and grown for 3 h before performing flow cytometry.
At the highest concentration of EdU tested (1000 µM), there was no significant change in the cell-cycle profile of either MW001 or SacTK (Figure 3). Under these conditions, cell growth was previously shown to be severely inhibited in both MW001 and SacTK (Figure 2A). There is no evidence that EdU is incorporated into the DNA of MW001, suggesting that the growth impact at very high EdU concentrations may occur via a mechanism that is independent of DNA incorporation. It should be noted that, in our hands, the absolute concentration of EdU required to reach the threshold at which cell growth was completely arrested, and cell-cycle profile was unperturbed, was variable between experiments (1000–2000 µM), potentially due to differences between batches of EdU supplied by the manufacturer. Finally, BrdU addition resulted in a less marked effect on the cell cycle, with 1000 µM BrdU eliciting an approximately similar effect to 40 µM EdU (Figure 3).
Visualization of DNA replication in synchronized Sulfolobus cells
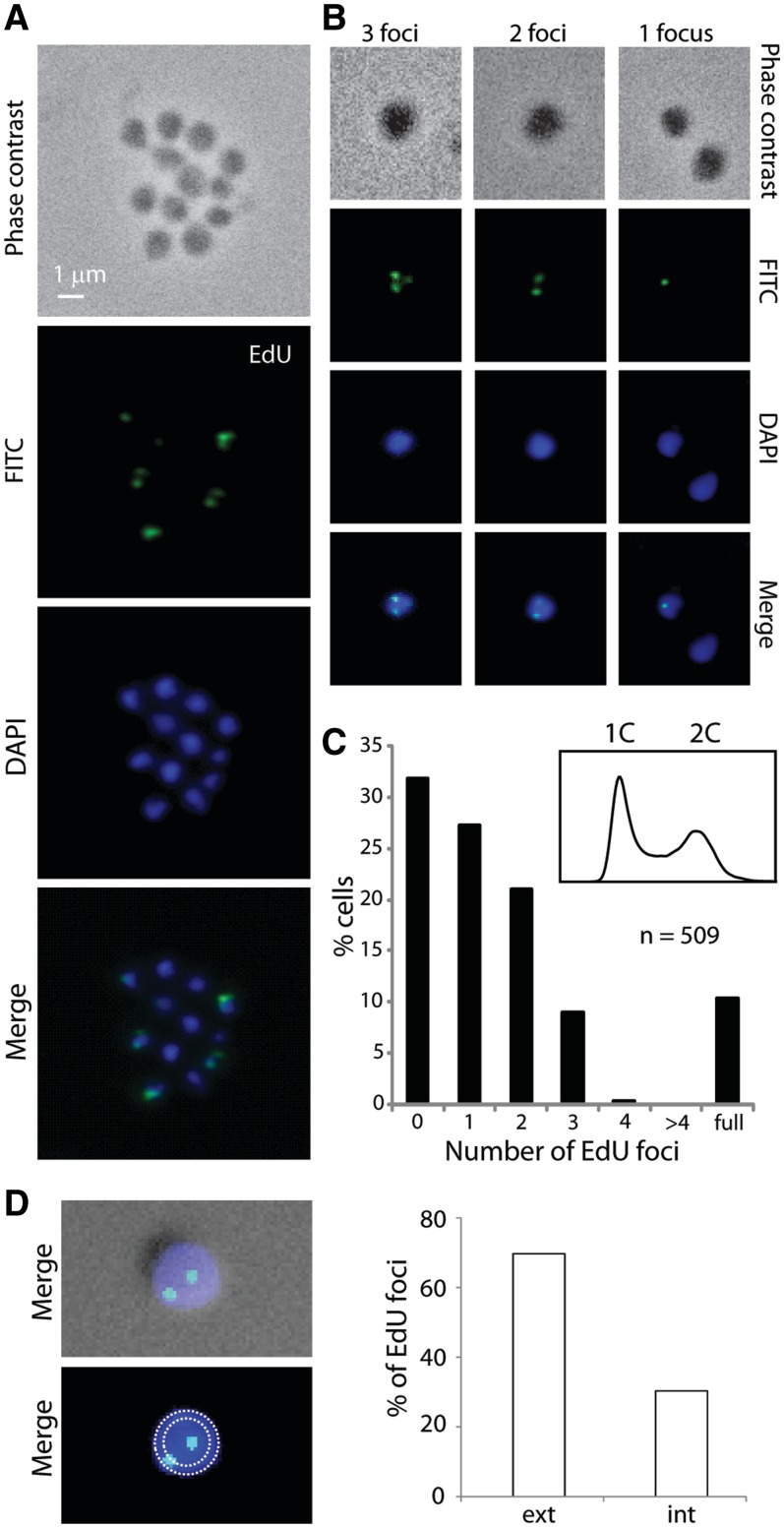
Next we sought to label a synchronized cell population taking advantage of the previously described ‘Baby Machine’ technique (8). This method generates an enriched G1 population of cells that then undergoes synchronous progression through S-phase and the later stages of the cell cycle. Synchronized populations were grown for 20 min with 200 µM EdU to allow uptake of the nucleoside analogue and its incorporation into nascent DNA. The localization of this newly incorporated alkyne-containing DNA was then monitored by in situ detection with an azide-containing fluorescent probe. We observed cells with either a single fluorescent focus or with two or three foci (Figure 4A and B). We only very rarely detected cells with four foci (0.4%; two cells out of 509 cells counted) and did not observe any cells with more than four foci (Figure 4C). Notably, in many instances the foci appeared to be at the periphery of the cell (Figure 4D). Indeed, quantitation revealed that 70% of the foci were fully contained in the outer 50% of the 2D image’s area, demonstrating a statistically significant localization to the outer region (n = 145; P = 7.2 × 10−7). This analysis is complicated by the small size (∼1 µm) of Sulfolobus cells coupled with their coccoidal shape and lack of cellular reference points. We then carried out image deconvolution and 3D reconstruction with a Z-series of images; this revealed that some central foci on the 2D images were located on the top or bottom surface of the cell as viewed and thus were located at the cell periphery (see Supplementary Movies S1–S5).
Figure 4.
Localization of replicating DNA in synchronized S. acidocaldarius cells. SacTK cells were synchronized using the baby machine method and then incubated at 75°C for 20 min with 200 µM EdU to label newly synthesized DNA. Images show phase contrast, DAPI staining of DNA (blue), EdU-AlexaFluor488 (green) and merged images. (A) A representative image of cells and (B) representative images of cells scored as showing 1, 2 or 3 EdU foci are shown. (C) The number of EdU foci detected was counted (509 cells total) and the percentage of cells in each category is shown. Cell synchronization was assessed using flow cytometry, shown inset. (D) Using 2D slices, template circles encompassing 100 and 50% of the cell area were generated and EdU foci were assigned to the inner or outer 50% of the cell area (n = 145).
Localization of replisomes in synchronized Sulfolobus cells
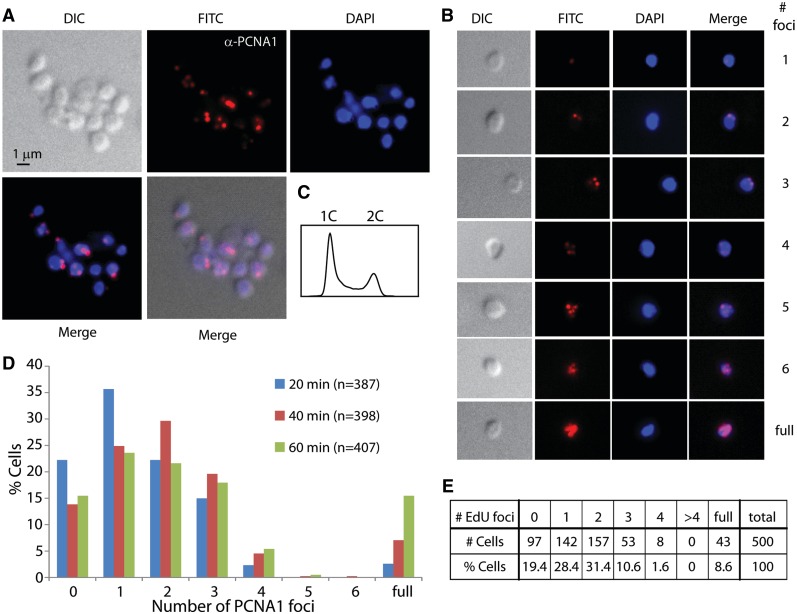
Next, we sought to visualize the location of replisomes within the cell. We accomplished this by using immunolocalization with anti-sera generated against the PCNA1 protein. PCNA is widely used as a marker for replisome position in eukaryotic cells, and biochemical and genetic experiments have demonstrated that this protein also functions in Sulfolobus DNA replication (26,27). Cells were synchronized in G1 with the baby machine and grown in the absence of EdU for 20, 40 or 60 min prior to harvesting and use in immunolocalization. As may be seen in Figure 5 and Supplementary Movies S6–S8, the majority of labelled cells contained one, two or three foci. From a total of 1192 cells counted, we observed 49 cells (4%) with four foci, three cells (0.25%) with five foci and one cell (0.08%) with six foci (Figure 5B and D). Many cells had peripheral localization of the PCNA foci; at each time-point, a statistically significant proportion of cells possessed foci in the peripheral 50% of the cell’s image area: 80.1% at 20 min (n = 216; P = 4.3 × 10−20), 72.3% at 40 min (n = 202; P =6.0 × 10−11), 74.7% at 60 min (n = 178; P = 9.1 × 10−12). We also detected similar levels of peripheral localization when considering the total number of foci within the cell: 76.2% of foci in single-focus cells (n = 181; P = 2.8 × 10−13), 77.0% in two-foci cells (n = 230; P =3.0 × 10−17), 76.0% in three-foci cells (n = 129; P = 9.4 × 10−10) and 69.6% in four-foci cells (n = 56; P = 1.4 × 10−3). In addition, we did not observe a consistent relative positioning of, or relative distance between, foci in multi-foci cells (Supplementary Figure S3).
Figure 5.
Localization of replisomes in synchronized S. acidocaldarius cells. SacTK cells were synchronized and then grown at 75°C for 20, 40 or 60 min before fixing. Images show DIC, DAPI staining of DNA (blue), antibody labelling of PCNA1 (red; images were obtained in the FITC channel and false coloured red) and merged images. (A) A representative image of cells and (B) representative images of cells scored as showing 1–6 PCNA1 foci, or fully labelled, are shown. (C) Cell synchronization was assessed using flow cytometry. (D) The number of PCNA1 foci detected was counted from cells fixed at the indicated time after release, and the percentage of cells in each category is shown. (E) Synchronized SacTK cells were grown at 75°C for 15 min and then labelled with 200 µM EdU for 10 min before fixing. The number of EdU foci detected was counted (500 cells total) and the percentage of cells in each category is shown.
Finally, we performed an EdU labelling experiment in which cells were synchronized and then grown in the absence of label for 15 min followed by a 10-min pulse with EdU prior to collection and processing for microscopy (Figure 5E). Under these conditions, where replication forks have the potential to separate prior to labelling, 28.4% of cells contained a single focus, 31.4% had two foci and 10.6% of cells had three foci. Only 1.6% of cells contained more than three foci.
DISCUSSION
Here we have described the application of nucleoside analogue-based DNA labelling technologies to perform the first examination of the localization of DNA replication within individual archaeal cells. In our engineered strains the artificial nucleosides BrdU and EdU were incorporated rapidly and quantitatively into genomic DNA of growing cells. The strains and plasmids developed here should have broader application, and they significantly expand the genetic toolbox for Sulfolobus research. For example, it will now be possible to perform chromosome combing experiments using synchronized pulse-labelled cells, or cells incubated with BrdU/EdU following DNA damage, to investigate the in vivo dynamics of DNA replication and repair on single chromosomes for the first time in archaea.
Extended growth of Sulfolobus in the presence of the artificial nucleosides resulted in obvious growth retardation after >4 h (approximately one doubling time), and flow cytometry indicated that, by 3 h, incorporation of BrdU/EdU into S. acidocaldarius DNA had resulted in slowed or arrested progression through S-phase in a significant fraction of cells. This dose-depended effect was more pronounced in the presence of EdU than BrdU. These observations suggest that BrdU and EdU are incorporated efficiently during DNA replication, but cells later encounter difficulty in DNA replication with a BrdU- or EdU-containing DNA template. EdU has been shown to have similar effects on growth of fission yeast, where it was found to cause cell arrest via activation of the rad3-dependent checkpoint after one cell cycle (22). Toxicity of EdU has also been reported in mammalian cells (28). Therefore, while EdU is suitable for pulse-labelling DNA, it may not be suitable for studies requiring continuous longer-term growth, and this should be considered when designing future experiments utilizing EdU in archaea or other systems. In our hands, BrdU is not suitable for Sulfolobus studies that require downstream microscopy as the conditions required to generate single-stranded DNA, essential for BrdU detection, grossly altered the structure of the nucleoid.
A previous study in our laboratory used MFA to assess origin firing within a population of synchronized S. acidocaldarius cells. That work revealed that all three origins fired in every cell in each cell cycle. MFA also indicated that two of the origins (oriC1 and oriC3) fire within a temporal window of ∼ 20 min from the onset of S-phase, while the third (oriC2) appears to have slightly more relaxed temporal constraints, with the vast majority of cells having fired all three origins after 30 min from the beginning of S-phase (8). Those studies were performed on whole populations of cells and so the relative timing of firing of the three origins in a single cell was unknown. In the current study, pulse-labelling of G1-synchronized S. acidocaldarius cells with EdU for 20 min during initiation of replication resulted in cells containing one (27.3%), two (21.0%) or three (9.0%) foci. We interpret these foci as the locations of DNA synthesis arising from the three known replication origins in Sulfolobus. The number of cells classified as containing three foci may be slightly under-represented due to the difficulties in resolving multiple foci within the small Sulfolobus cells. In addition, although origin firing is detected in some cells immediately following collection of the synchronized population, it takes ∼30 min for all three origins to have fired in every cell in the culture (8) which may contribute to an under-representation of cells containing three foci. Labelling of Sulfolobus cells for longer than 20 min resulted in essentially global labelling of the nucleoid.
Cells containing fewer than three EdU foci may represent cells in which two or more replication foci are co-localized, forming replication clusters reminiscent of those observed in eukaryotes. [In the following discussion we shall refer to paired sister replisomes emerging from a single replication origin as replication factories; higher-order associations of replication factories will be termed replication clusters (29)]. However, we did not observe a consistent pattern of labelling intensity; for example, cells containing two EdU foci did not consistently show one focus with double the intensity of the other, and we regularly saw cells in which the three foci were of different intensities. Therefore, although we do not rule out formation of some replication clusters, we believe that the variation in focus number is likely to represent a partial asynchrony of origin firing within the more general initiation stage of replication. Furthermore, our data strongly suggest that there is no requirement for all replisomes to be maintained in a single higher-order replication cluster. This conclusion is supported by our immunolocalization studies of the replisome component PCNA. In agreement with our EdU labelling studies, we observed cells in which variable numbers of replisomes were visible, supporting a model in which there is no obligate spatial linkage of the replisomes emerging from the three replication origins in the cell. Further, the peripheral bias in the localization of PCNA foci correlates with the position of EdU foci. The PCNA localization experiments were performed in the absence of EdU. This, therefore, indicates that, despite the growth effects of prolonged incubation with high concentrations of EdU, in this study EdU incorporation has not perturbed the localization of the Sulfolobus replisomes. This is consistent with a recent study in Escherichia coli in which origin localization as determined by EdU pulse labelling was identical to data obtained using live-cell imaging of fluorescently labelled replisomes (30).
When assessing the number of PCNA1 foci using immunolocalization, we observed a trend towards higher numbers of foci over time, in agreement with our previous data revealing that all three origins fire in each cell cycle. However, even after 60 min growth, we observed relatively few examples of >3 PCNA1 foci. This suggests that the two sister replication forks arising from initiation at a single origin may remain directly associated in a replication factory or within a confined region that makes them irresolvable by light microscopy. Furthermore, in G1-synchronized cells grown for 15 min into S-phase prior to EdU labelling we still observed the majority of cells with only three or fewer foci. Thus these data also suggest that sister replisomes do not rapidly segregate after initiation of replication in Sulfolobus. This contrasts with the situation in the model bacterium E. coli where the replisomes move around the chromosome independently in opposite cell halves, before converging near mid-cell where termination occurs (31). Thus, Sulfolobus appears more similar to budding yeast, in which sister replisomes remain associated during replication (32). Replication factories or close spatial positioning of replisomes have also been observed in the bacteria Bacillus subtilis and Caulobacter crescentus (24,27). In this regard, we note that an analysis of the genomes of seven distinct strains of Sulfolobus islandicus, revealed high levels of synteny between the genomes. However, one strain, Y.N.15.51 possesses a clear inversion of around 600 kbp symmetrically centered on oriC2 (33 and Supplementary Figure S4). The existence of such a symmetrically positioned inversion has been proposed to be indicative of recombination in the vicinity of paired replisomes derived from the central replication origin (34).
Finally, we observed that EdU and PCNA1 foci were preferentially located at the periphery of S. acidocaldarius cells and this localization was still evident 60 min after release from synchronization, when the replisomes are expected to have moved away from the origins of replication. In E. coli, oriC is located at mid-cell during initiation of replication, although specific proteins anchoring the region at mid-cell have not been found (35). In contrast, in Caulobacter crescentus, which also acts as a model for bacterial DNA replication, the origin region is anchored to the cellular envelope at the cell pole via the PopZ and ParB proteins (36). In yeast, some correlation between nuclear localization and timing of origin firing has been observed, with late-firing origins preferentially localizing to the nuclear periphery. However, a variety of factors affecting chromatin structure appear to be involved, and sub-nuclear localization per se does not appear to determine replication timing (33). In light of our observations, it seems possible that components of the Sulfolobus replication apparatus may be tethered to the cell membrane and this is an area that will be explored in future studies. In this regard it may be significant that we have previously observed that perturbation of cell division results in mis-regulation of DNA replication in Sulfolobus cells (17).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Materials and Methods, Supplementary Figures 1–4, Supplementary Movies 1–8 and Supplementary References [36–38].
FUNDING
Biotechnology and Biological Sciences Research Council, UK (to T.G., I.G.D. and S.D.B.); intramural funds of the Max Planck Society (to M.W. and S.V.A.). Funding for open access charge: BBSRC.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are indebted to Peter Cook for valuable comments on this work and would like to thank Bill Wicksted for help with PFGE.
REFERENCES
- 1.Duggin IG, Bell SD. The chromosome replication machinery of the archaeon Sulfolobus solfataricus. J. Biol. Chem. 2006;281:15029–15032. doi: 10.1074/jbc.R500029200. [DOI] [PubMed] [Google Scholar]
- 2.Edgell DR, Doolittle WF. Archaea and the origin(s) of DNA replication proteins. Cell. 1997;89:995–998. doi: 10.1016/s0092-8674(00)80285-8. [DOI] [PubMed] [Google Scholar]
- 3.Robinson NP, Bell SD. Origins of DNA replication in the three domains of life. FEBS J. 2005;272:3757–3766. doi: 10.1111/j.1742-4658.2005.04768.x. [DOI] [PubMed] [Google Scholar]
- 4.Robinson NP, Bell SD. Extrachromosomal element capture and the evolution of multiple replication origins in archaeal chromosomes. Proc. Natl Acad. Sci. USA. 2007;104:5806–5811. doi: 10.1073/pnas.0700206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson NP, Blood KA, McCallum SA, Edwards PA, Bell SD. Sister chromatid junctions in the hyperthermophilic archaeon Sulfolobus solfataricus. EMBO J. 2007;26:816–824. doi: 10.1038/sj.emboj.7601529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson NP, Dionne I, Lundgren M, Marsh VL, Bernander R, Bell SD. Identification of two origins of replication in the single chromosome of the Archaeon Sulfolobus solfataricus. Cell. 2004;116:25–38. doi: 10.1016/s0092-8674(03)01034-1. [DOI] [PubMed] [Google Scholar]
- 7.Lundgren M, Andersson A, Chen L, Nilsson P, Bernander R. Three replication origins in Sulfolobus species: synchronous initiation of chromosome replication and asynchronous termination. Proc. Natl Acad. Sci. USA. 2004;101:7046–7051. doi: 10.1073/pnas.0400656101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duggin IG, McCallum SA, Bell SD. Chromosome replication dynamics in the archaeon Sulfolobus acidocaldarius. Proc. Natl Acad. Sci. USA. 2008;105:16737–16742. doi: 10.1073/pnas.0806414105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bick MD, Davidson RL. Total substitution of bromodeoxyuridine for thymidine in dna of a bromodeoxyuridine-dependent cell line. Proc. Natl Acad. Sci. USA. 1974;71:2082–2086. doi: 10.1073/pnas.71.5.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gratzner HG. Monoclonal-antibody to 5-bromodeoxyuridine and 5-iododeoxyuridine - a new reagent for detection of dna-replication. Science. 1982;218:474–475. doi: 10.1126/science.7123245. [DOI] [PubMed] [Google Scholar]
- 11.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl Acad. Sci. USA. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellen AF, Rohulya OV, Fusetti F, Wagner M, Albers SV, Driessen AJM. The sulfolobicin genes of Sulfolobus acidocaldarius encode novel antimicrobial proteins. J. Bacteriol. 2011;193:4380–4387. doi: 10.1128/JB.05028-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albers S-V, Driessen AJM. Conditions for gene disruption by homologous recombination of exogenous DNA into the Sulfolobus solfataricus genome. Archaea. 2008;2:145–149. doi: 10.1155/2008/948014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berkner S, Grogan D, Albers S-V, Lipps G. Small multicopy, non-integrative shuttle vectors based on the plasmid pRN1 for Sulfolobus acidocaldarius and Sulfolobus solfataricus, model organisms of the (cren-)archaea. Nucleic Acids Res. 2007;35:e88. doi: 10.1093/nar/gkm449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szabó Z, Sani M, Groeneveld M, Zolghadr B, Schelert J, Albers SV, Blum P, Boekema EJ, Driessen AJM. Flagellar motility and structure in the hyperthermoacidophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 2007;189:4305–4309. doi: 10.1128/JB.00042-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Albers SV, Jonuscheit M, Dinkelaker S, Urich T, Kletzin A, Tampé R, Driessen AJ, Schleper C. Production of recombinant and tagged proteins in the hyperthermophilic archaeon Sulfolobus solfataricus. Appl. Environ. Microb. 2006;72:102–111. doi: 10.1128/AEM.72.1.102-111.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in Archaea. Science. 2008;322:1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duggin IG, Dubarry N, Bell SD. Replication termination and chromosome dimer resolution in the archaeon Sulfolobus solfataricus. EMBO J. 2011;30:145–153. doi: 10.1038/emboj.2010.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hediger F, Neumann FR, Van Houwe G, Dubrana K, Gasser SM. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr. Biol. 2002;12:2076–2089. doi: 10.1016/s0960-9822(02)01338-6. [DOI] [PubMed] [Google Scholar]
- 20.Sivakumar S, Porter-Goff M, Patel PK, Benoit K, Rhind N. In vivo labeling of fission yeast DNA with thymidine and thymidine analogs. Methods. 2004;33:213–219. doi: 10.1016/j.ymeth.2003.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hodson JA, Bailis JM, Forsburg SL. Efficient labeling of fission yeast Schizosaccharomyces pombe with thymidine and BUdR. Nucleic Acids Res. 2003;31:E134. doi: 10.1093/nar/gng134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lengronne A, Pasero P, Bensimon A, Schwob E. Monitoring S phase progression globally and locally using BrdU incorporation in TK+ yeast strains. Nucleic Acids Res. 2001;29:1433–1442. doi: 10.1093/nar/29.7.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fitz-Gibbon ST, Ladner H, Kim UJ, Stetter KO, Simon MI, Miller JH. Genome sequence of the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Proc. Natl Acad. Sci. USA. 2002;99:984–989. doi: 10.1073/pnas.241636498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lubelska JM, Jonuscheit M, Schleper C, Albers S-V, Driessen AJM. Regulation of expression of the arabinose and glucose transporter genes in the thermophilic archaeon Sulfolobus solfataricus. Extremophiles. 2006;10:383–391. doi: 10.1007/s00792-006-0510-7. [DOI] [PubMed] [Google Scholar]
- 25.Worthington P, Hoang V, Perez-Pomares F, Blum P. Targeted disruption of the alpha-amylase gene in the hyperthermophilic archaeon Sulfolobus solfataficus. J. Bacteriol. 2003;185:482–488. doi: 10.1128/JB.185.2.482-488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dionne I, Nookala RK, Jackson SP, Doherty AJ, Bell SD. A heterotrimeric PCNA in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol. Cell. 2003;11:275–282. doi: 10.1016/s1097-2765(02)00824-9. [DOI] [PubMed] [Google Scholar]
- 27.Zhang C, Guo L, Deng L, Wu Y, Liang Y, Huang L, She Q. Revealing the essentiality of multiple archaeal pcna genes using a mutant propagation assay based on an improved knockout method. Microbiology (SGM) 2010;156:3386–3397. doi: 10.1099/mic.0.042523-0. [DOI] [PubMed] [Google Scholar]
- 28.Diermeier-Daucher S, Clarke ST, Hill D, Vollmann-Zwerenz A, Bradford JA, Brockhoff G. Cell type specific applicability of 5-ethynyl-2'-deoxyuridine (EdU) for dynamic proliferation assessment in flow cytometry. Cytometry A. 2009;75:535–546. doi: 10.1002/cyto.a.20712. [DOI] [PubMed] [Google Scholar]
- 29.Bates D. The bacterial replisome: back on track? Mol. Microbiol. 2008;69:1341–1348. doi: 10.1111/j.1365-2958.2008.06378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Lesterlin C, Reyes-Lamothe R, Ball G, Sherratt DJ. Replication and segregation of an Escherichia coli chromosome with two replication origins. Proc. Natl Acad. Sci. USA. 2011;108:E243–E250. doi: 10.1073/pnas.1100874108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reyes-Lamothe R, Possoz C, Danilova O, Sherratt DJ. Independent positioning and action of Escherichia coli replisomes in live cells. Cell. 2008;133:90–102. doi: 10.1016/j.cell.2008.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitamura E, Blow JJ, Tanaka TU. Live-cell imaging reveals replication of individual replicons in eukaryotic replication factories. Cell. 2006;125:1297–1308. doi: 10.1016/j.cell.2006.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ebrahimi H, Robertson ED, Taddei A, Gasser SM, Donaldson AD, Hiraga S-i. Early initiation of a replication origin tethered at the nuclear periphery. J. Cell Sci. 2010;123:1015–1019. doi: 10.1242/jcs.060392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard J-F, Guindon S, Lefort V, Lescot M, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acid Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sherratt DJ. Bacterial chromosome dynamics. Science. 2003;301:780–785. doi: 10.1126/science.1084780. [DOI] [PubMed] [Google Scholar]
- 36.Bowman GR, Comolli LR, Zhu J, Eckart M, Koenig M, Downing KH, Moerner WE, Earnest T, Shapiro L. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell. 2008;134:945–955. doi: 10.1016/j.cell.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reno ML, Held NL, Fields CJ, Burke PV, Whitaker RJ. Biogeography of the pan-genome of Sulfolobus islandicus. Proc. Natl Acad. Sci. USA. 2009;106:8605–8610. doi: 10.1073/pnas.0808945106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahadevan P, Seto D. Rapid pair-wise synteny analysis of large bacterial genomes using web-based GeneOrder4.0. BMC Res. 2010;3:41. doi: 10.1186/1756-0500-3-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.