Abstract
We assert that it is possible to trap and identify proteins, and even (conceivably) manipulate proteins secreted from a single cell (i.e. the secretome) through transfection via electroporation by exploiting the exquisite control over the electrostatic potential available in a nanopore. These capabilities may be leveraged for single cell analysis and transfection with single molecule resolution, ultimately enabling a careful scrutiny of tissue heterogeneity.
Introduction
Biological function emerges from a cell through a hierarchical network of molecules, which generally involves large protein complexes, that can affect gene expression.1 This hierarchy senses the internal state or external environment of the cell and reconfigures itself both spatially and functionally, while in the process transducing signals such as: internal damage, metabolites, mechanical force, signaling molecules from nearby and distant cells, nutrients, and poisons. Like any hierarchical organization, the elements at the top exert the most influence, and so both the internal state and external environment of the cell influence the interconnected control machinery, ultimately affecting gene expression. Thus, the cellular microenvironment is just as important as the genes in determining biological function.
Here we explore the prospects for using a nanopore to both detect aspects of the cellular microenvironment and transfect genes into single cells. While detecting and analyzing analytes, such as protein, originating either from a lysed cell or in the microenvironment of a single cell is a daunting task, a merger of nanopore sensing with optical trapping for the manipulation of single molecules2-4 and single cells5,6 have made it feasible.
Nanopore detection represents the logical conclusion in the development of chemical analysis, which extracts the maximum amount of information from a minimum amount of material. Both proteinaceous and solid-state pores are effective analytical tools for single molecule analysis of DNA, RNA, peptides and proteins.2-4,7 Though protein pores can show high selectivity for a ligand,8,9 they are fragile—they do not share the robustness of solid-state pores, nor do they easily allow for tailored selectivity of ligands.2,4,8 In particular, solid-state nanopores have been used to inform on the chemical, thermal, and electric field effects on protein folding.10-14 However, most of the strategies that have been pursued so far for detecting proteins rely on using the pore as a stochastic sensor.4 Although it has not been demonstrated unequivocally, it has been asserted that a statistically representative sample of current blockades could be used to discriminate between different molecules in a pore that is comparable to the size of the protein.10
Proteins inherently have a very specific distribution of surface charge, which is used to attract different targets to different parts of the protein. The exquisite control over the electrostatic potential available in a nanometer-diameter pore could be used to exploit the distinctive surface charge on a protein to trap and identify it. The promise of a nanopore as a non-optical, molecular sensor relies on the electric signal that develops when an analyte translocates across a membrane through a pore immerse in electrolyte. If the size of the pore is such that only one analyte is admitted at a time and each has a characteristic signature, a pore can be used to detect analytes with single molecule sensitivity and discriminate single molecules against a complex chemical background.2-4
While the sensitivity of a nanopore is incontrovertible, it has a drawback for detection of dilute concentrations of molecules that is related to the diffusion equivalent capacitance.15,16 The response time affects the minimum detectable concentration17 but the molecule must first diffuse within range of the pore to be driven through it by the electric field.18 Thus, the specifications for detecting proteins with a nanopore are very stringent—the geometry and electric field in the pore are determined by the molecular size and surface charge, while the capture rate increases only with proximity to the molecular source.
To improve the capture rate, we show that it is possible to place a cell immediately above a pore using optical tweezers. In doing so, we discovered serendipitously that the same stringent specifications for detecting and selectively analyzing single molecules in a pore in close proximity to a cell also offers the prospect for transfection of a single cell with the same pore. Provided that the cell is in close proximity, the control of the electric field in and beyond the pore lumen enables transfection of genes via electroporation.
In this report, we test the efficacy of using a nanopore for detecting single proteins and transfecting single cells. We assert that by using a nanopore as a stochastic sensor, proteins can be discriminated based on size and charge.19 We offer preliminary measurements showing distinctive and dramatic changes in the transmembrane current when a protein translocates across the membrane. We interpret blockades in the current as changes in the volume accessible to electrolytic ions in the pore, and show a dependence on both the pH and the pore size. Furthermore, we show that by controlling the electric field, a single protein can be trapped in a pore smaller than the molecule, which facilitates analysis and increases the likelihood of correctly identifying a single molecule. Control of the electric field in and beyond the pore lumen also enables transfection of a cell in close proximity via electroporation. To demonstrate this, we position a single breast cancer cell over a nanopore using optical tweezers, and then use electroporation to transfect it with fluorescent DNA. These results extend the frontier of biology towards single cell analysis with single molecule resolution, offering the potential for an unprecedented probe of tissue heterogeneity.
Methods and Materials
Nanopores
Silicon nitride membranes ≤ 30nm thick on a silicon handle wafer (Protochips) were first prepared using a 10-30 s O2 plasma clean. The silicon chips were then loaded into a FEI Titan 80-300 transmission electron microscope (TEM) for sputtering. The thickness of the relieved nitride membrane was measured in-situ using Electron Energy Loss Spectroscopy; the thicknesses ranged from 11 to 31 nm, depending on the deposition and etch conditions used to define the film. A nanometer-size pore was sputtered into the membrane using spot-sizes ranging from 0.2-2 nm. From TEM images taken at different tilt angles, we infer that the pore geometry is bi-conical, each with >10° cone angle.20
Subsequently, the chip containing the membrane was plasma bonded to a multi-level microfluidic device. The plasma bond sealed the chip into the microfluidic between two vias, leaving the pore as the only connection between the cis and trans channels as illustrated in Figures 1(d,e). The trans reservoir (red) has a volume of ~6 nL, while the cis reservoir (blue) has a volume of ~100 μL. To characterize the pore, a transmembrane voltage was applied using a program written in LabView (National Instrument, ver. 2010) and the current was measured at 23±1°C with an Axopatch 200B amplifier (Molecular Devices) using Ag/AgCl electrodes embedded in each fluidic channel. The signal was filtered at 10 kHz by a low-pass 8-pole Bessel filter before being digitized at a rate of 100 kHz. Current blockades were detected automatically using an algorithm in Clampfit 10.2 (Molecular Devices) with a threshold >5% of the open pore current.
FIGURE 1. Nanopores sputtered with sub-nanometer precision in small area membranes embedded in microfluidics.
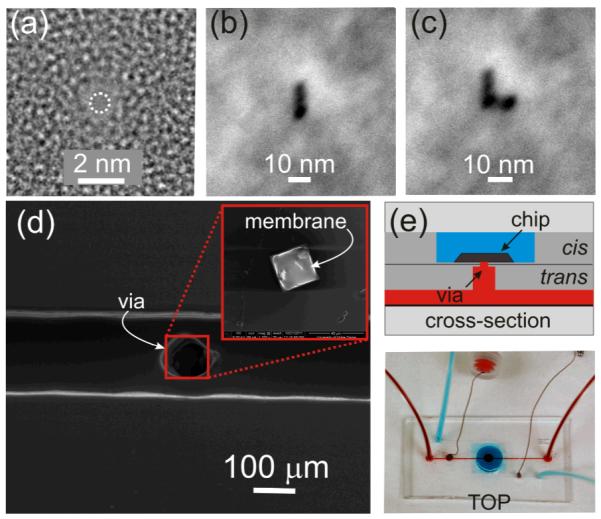
(a) TEM micrograph of a nanopore 0.7×0.9 nm in cross-section. (b,c) Scanning TEM (STEM) micrograph of a slit and an “L” pattern respectively, indicating stability of the beam (<1 nm drift/30 min.) (d) Scanning Electron Microscopy (SEM) of a silicon chip with a Si3N4 membrane on it (inset) bonded to a PDMS microfluidic at the top of a 75 μm via in the middle of a 150 μm microchannel.(e) (top) Schematic showing a magnified view of a horizontal cross-section through the device and (bottom) optical micrograph showing the top view of a PDMS microfluidic with two channels: one above (blue) and the other below (red) the silicon membrane.
Microfluidic
The microfluidic devices consist of two microchannels configured to independently address the cis and trans-sides of the membrane through vias. The entry-channels were all 250 μm wide and 100 μm high, but the vias (see Figure 1(d)) ranged in size from 250×250 μm to 75×75 μm square and 200 μm to 50 μm thick. A master mold of the design was generated by stereo-lithography (FineLine Prototyping) and made of DSM Somos ProtoTherm 12120, a strong, high temperature tolerant plastic. The microfluidic device was formed from poly dimethylsiloxane (PDMS) using a mold-casting technique. The PDMS silicone polymer used to create the chips is commercially available as Sylgard 184 (Dow Corning), a two-part polymer mix. The two parts were mixed thoroughly at a 1:10 ratio of curing agent to base. The mixture was degassed in house vacuum for 30 min. and then poured into a master mold where it was cured at 75°C for ~12 hrs. After cooling to room temperature, the plastic, which hardened to a rubber-like consistency, was peeled away from the mold yielding a piece of silicone with the inverse pattern of the master mold.
The silicon chip containing the silicon nitride membrane with the pore in it was tightly sealed to the PDMS trans-microfluidic channel with a plasma bonding process (Harrick PDS-001) that exposed the chip and PDMS to an oxygen plasma at a power of 30 W for 180 s. The plasma generates silanol (Si-OH) groups on the surface of PDMS, which reacts with silanol groups on the nitride surface to form an Si-O-Si bond. After exposure to the plasma, the PDMS was gripped by the sides, placed in contact with the chip and a uniform pressure was applied for 10 s to form the bond. At the same time, to provide optical access, we likewise sealed the trans-channel in the PDMS to a #1 cover glass (Corning) using the same bonding strategy described above. We subsequently tested the seal against a 30 nm thick silicon nitride membrane without a pore in 100 mM KCl pH 8 for > 3 weeks without failure; the leakage current was < 8 pA at 1V.
Finally, the microfluidic channels were connected to external pressure and fluid reservoirs through polyethylene tubing at the input and output ports. Through these ports the cis-microfluidic was used to convey dsDNA and various proteins to the pore, but not necessarily through the small diameter pore without an applied transmembrane bias voltage. On the other hand, the trans-microchannel was used to convey cells to the membrane where optical tweezers were used to precisely position them over the pore.
Live Cell Lithography
MDA-MB-231 human breast adenocarcinoma cells (ATCC) were cultured in DMEM high glucose medium (Invitrogen, #11995) supplemented with 10% FBS at 37°C with 5% CO2. A confluent cell culture was first trypsinized using a solution of 0.25% trypsin/EDTA (Gibco) and then centrifuged at 200g. The supernatant was discarded and the cell pellet re-suspended in 500 μL of DMEM. Immediately before injection into the microfluidic, the cell suspension was combined 1:1 with a pre-polymer mixture made of 3.4kDa polyethylene glycol diacrylate (PEGDA, Laysan Bio) dissolved at 16% (w/v) in phosphate buffered saline along with a photoinitiator, 2-hydroxy-2-methyl-propiophenone, at a concentration of 1.5% (v/v).
A single cell was extracted using optical tweezers from a laminar flow in the microfluidic channel and positioned over the nanopore. Optical tweezers were formed by focusing light (λ=900 nm) from a tunable CW Ti:sapphire laser (Spectra Physics) pumped at 532 nm by a 15 W Nd:YVO4 diode-pumped solid state laser (Spectra Physics) using a Zeiss Fluar 100× oil immersion objective (1.3NA). Acousto-optic deflectors (AOD) (AA-Optoelectronic) were used to steer the optical trap over the pore. We have previously established that under optimum trapping conditions the maximum radiation dose limits the cell’s exposure to the laser.21 Nevertheless, the trapping conditions can be optimized to allow for very long duration (>hr) exposures.22 To facilitate our preliminary work and minimize exposure to the laser beam, the cells were encapsulated in hydrogel.21,23 The pre-polymer solution was photopolymerized over the membrane using a metal halide light source (X-CITE 120Q, Lumen Dynamics) and a 360±15 nm bandpass UV filter (Semrock). A square mask (~5.0 mm × 5.0 mm) placed in front of the UV source was used to control the shape and size of the hydrogel spot. The light generated by the lamp is focused using a Köhler light train described elsewhere.23
Biomolecules
To explore the feasibility of using a nanopore to discriminate between different proteins, we compared two proteins of similar molecular weight at concentrations in the range 10-15 pM (New England Biolabs): bovine serum albumin (BSA) with molecular weight of 66.5 kDa, 5.7 pI and an approximate size24 of 750nm3=14×7×8 nm to streptavidin (STR) with MW 60 kDa, 6.5 pI, and a size25 of 230 nm3 =7×6×6 nm. To demonstrate transfection via electroporation we used double-stranded DNA (dsDNA) intercalated with YOYO-1 iodide dye (Invitrogen). A 20 kbp dsDNA was added to YOYO-1 (40 mM Tris-acetate, 2 mM EDTA, pH 8.0) for a final ratio of 5:1 nucleotides to dye molecules. This mixture was incubated for 60 min. at room temperature then stored at 4°C. A 25 pM solution of DNA-YOYO was made in 100 mM KCl (pH 8.0).
Fluorescence data was collected using a Leica TCS SP5 II (Leica Microsystems) confocal microscope with enhanced, hybrid GaAsP detectors for improved sensitivity to fluorescence. All confocal images were acquired using an HCX PL APO lambda blue 63× 1.2 NA (Leica) water immersion objective.
Results and Discussion
It is possible to detect a single molecule by measuring the electric signal from its translocation through a nanopore. The molecular configuration, the pore geometry and charge determine how the electrolytic ions passing through the pore interact with the potential presented by the molecule. Therefore, the differences in surface charge associated with each molecule should have a substantial effect on the current-voltage relationship and facilitate discrimination. However, to control the electric field and the forces in a pore, the geometry must be defined with high precision due to the molecular size.
The technique used prevalently to create nanometer-diameter pores in silicon nitride membranes is stimulated beam decomposition and sputtering from the tightly focused electron beam of a transmission electron microscope (TEM).20 An elaboration of that same method allows us to produce nanopores with a bi-conical geometry and sub-nanometer precision, such as the pore illustrated in Figure 1(a); this pore has a diameter comparable to an α-helix. Leveraging this exquisite spatial resolution in combination with minimal drift in the microscope, it is possible to form larger pores or merge them together to form slits (see Figure 1(b)), or even an “L”-shape (Figure 1(c)) for example. Fashioned this way—with high precision and minimal drift—nanopores can be used to selectively address higher order structure in protein.
To provide direct fluidic and optical access to the nanopore, while at the same time reducing the concentration of analyte (10-15 pM) required for detection, we embedded the silicon chips containing the nanopores in a multi-level microfluidic device. As shown in Figures 1(d,e), this microfluidic has two separate channels addressing the cis/trans (cathode/anode) sides of the membrane. Ag/AgCl electrodes are embedded in the channel via ports built into the microfluidic device. The cis/trans channels are connected to the nitride membrane supporting the pore through narrow access vias as shown in Figure 1(d). The vias have the added benefit of mitigating the effect of parasitic membrane capacitances (~100pF) on the dielectric component of the electrolytic noise.26,27
Nanopores as Stochastic Sensors for Protein
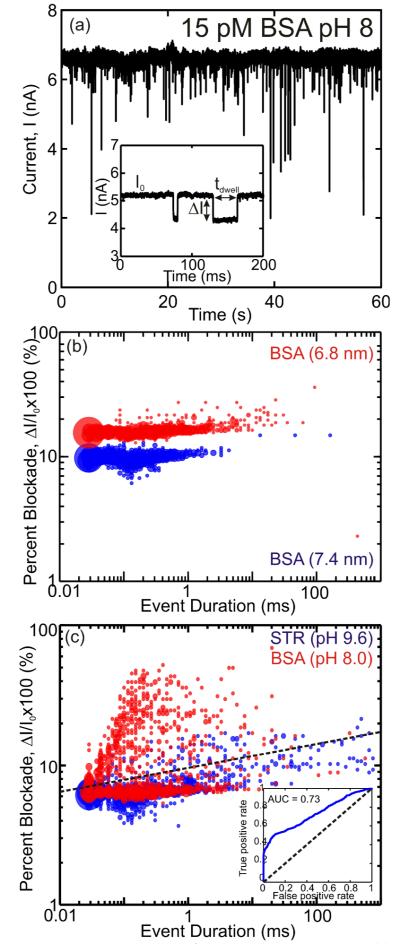
We tested the prospects for using a pore to detect and analyze proteins in two different ways: first, using the pore as a stochastic sensor we tried to discriminate between two proteins BSA and STR; and second, we trapped single BSA proteins in a pore smaller than the molecule to scrutinize unfolding. Figure 2(a) shows a typical 60 s current trace of 15 pM BSA in 100 mM KCl stabilized by 10 mM TRIS-HCl at pH 8 interacting with a 7.4 nm diameter pore at a 1 V transmembrane bias. We observed that, despite the dilute concentration of protein, the response time measured by the interarrival time between current transients in the pore current is only 820±14 μs. This facility we have for detecting dilute concentrations of protein (15 pM) with a short response time is superior to past efforts17 and a consequence of embedding the nanopore in a microfluidic, which localizes the analyte near the pore reducing the diffusion equivalent capacitance. Generally, data like that in Figure 2(a) can be modeled by a log-normal distribution with a mean translocation time, tdwell, and a mean blockade to open pore current ratio, ΔI/I0, (as defined in the inset to Figure 2(a)) of 140±10 μs and 0.145±0.004 respectively.
FIGURE 2. Proteins translocating through a nanopore.
(a) Pore current at 1 V bias through a 7.4 nm pore after 15 pM BSA in 100 mM KCl, 10 mM TRIS-HCl at pH 8 is introduced on the cis-side of the membrane. The frequent blockades observed in the current are indicative of the BSA interacting with the pore. The inset shows two typical events; the arrows indicate the dwell time, tdwell, the mean blockade amplitude, ΔI, and the open pore current, I0. (b) Event distribution of ΔI/I0 vs tdwell for the first 1000 recorded events of BSA at pH 8 through 6.8-nm-diameter (red) and 7.4-nm-diameter pores (blue). (c) Event distribution of ΔI/I0 vs tdwell for the first 1000 recorded events of BSA at pH 8 (red) and STR at pH 9.6 (blue) through the 7.4-nm-diameter pore. Black line shows group demarcation when the events are classified with LDA. (inset) ROC of the protein discriminator with an AUC of 0.73.
To test the effect of pore geometry on the distribution, we collected data on the same protein, BSA, at pH 8 in two different pores, 6.8 nm and 7.4 nm in diameter. Applying a t-test, we found a statistically significant difference between the resulting pore blockades with a p-value of <10−6. Thus, the result shown Figure 2(b) provides compelling evidence that the pore geometry can be used to manipulate the distribution—the smaller pore seems to broaden the event duration and increase the blockade percentage—which could possibly be used to isolate a specific protein from a complex chemical background.
Using an appropriately designed pore it should be possible to discriminate between different proteins this way. To demonstrate this strategy, we tried to force STR, which has about the same molecular weight as BSA, through the same 7.4 nm pore at pH 8, but could not detect any current transients indicative of STR interacting with the pore. We attribute this observation to the relatively neutral charge on the protein: at pH 8, BSA supposedly has a charge of −25e, while STR has a charge of only −4e. According to this interpretation, BSA can be discriminated by charge from STR perfectly at pH 8.
The charge of the protein can be adjusted by controlling the pH of the solution. However, the pH can also affect the secondary structure of the protein.28 For example, it has been shown that BSA transitions from an α-helix to a β-sheet when the pH is either below 5 or above 10.29 Since the current measures the molecular volume occluding the pore, the pH has to be maintained in a range where the secondary structure of the protein is not affected.
Lending further support to our interpretation of the STR data obtained at pH 8, at pH 9.6, STR has an overall charge of −12e and current blockades are observed in the 7.4 nm pore with a distribution defined by tdwell=150±50 μs and ΔI/I0=0.066±0.005. Figure 2(c) represents a compilation of the two distributions: one corresponding to BSA at pH 8 and the other to STR at pH 9.6. Both distributions of dwell time are highlighted by a large number of events of very short tdwell < 30 μs duration, which are likely due to collisions between the pore and the protein that may not result in a translocation. These events are outside of the log-normal distribution used to calculate the mean dwell time and blockade magnitude. These very short duration events are not specific to any one protein, but are more common with STR occurring in ~19% of all events. The distribution of dwell times indicates that, for the most part, BSA and streptavidin have similar translocation velocities; with roughly ~9% of the events lasting longer than 10 ms for STR while for BSA, only ~2% of the events lasted as long.
To discriminate a specific protein, each event is classified by the distribution that defines it. In multivariate analysis a common technique used to classify observations is linear discriminant analysis (LDA), which identifies linear combinations of features between different events. When we apply LDA to the event duration and blockade amplitude we find a distinct separation of events, indicated by the dashed line in Figure 2(b). Events occurring above the line have a high probability of belonging to BSA while events occurring below the line are likely due to STR. To further quantify the ability to discriminate between the two proteins, we generated a receiver operating characteristic curve (ROC),30 which measures the ratio of true positives (an event that correctly identifies the protein) to false positives (an event that identifies the wrong protein) in the LDA model. The ROC for the distribution of events, shown in the inset of Figure 2(c), indicates that the probability to correctly identify a protein, typically measured by the area under curve (AUC) is AUC=0.73, which is significantly better than random (AUC=0.5).
Nanopores for Trapping a Single Protein
It may be possible to discriminate between two different proteins by manipulating the selectivity of the pore by either varying the applied potential,31 changing the pH14,32 or the electrolyte concentration of the solution.33 However, the classification is imperfect due in part to the stochastic nature of the single molecule measurement. Noise associated with different molecular conformations in the pore likely contributes to the distribution of translocation events. In addition, translocation noise in the motion of the molecule relative to the detector is invariably introduced when a continuous molecular strand is driven electrophoretically through a pore.34,35 Translocation noise is captured succinctly in a one-dimensional transport model by the ratio of the drift to diffusion velocities, vdrift/vdiff=V/(kT/q). From this relation we infer that voltages larger than kT/q are desirable to offset diffusion of the molecule through the pore. Unfortunately, large voltages can also adversely affect membrane reliability, affect the molecular conformation, and increase translocation velocity, forcing high frequency operation to electrically detect a molecule.
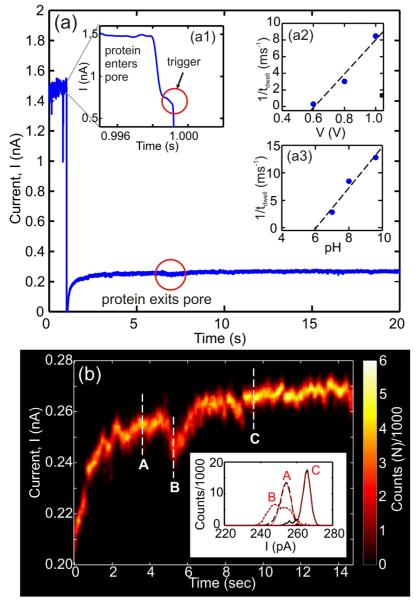
Instead of stochastic sensing on a large sample of proteins, we tested the feasibility of detecting and analyzing a single protein trapped in a nanopore. This strategy is motivated by recent work on dsDNA, which demonstrated the possibility of trapping a single molecule in a pore smaller than the double helix by rapidly switching the electric field below the threshold for stretching the molecule.36 The conditions required for trapping a protein like BSA can be inferred from the dependence of the reciprocal of the dwell time as a function of voltage and pH. The insets to Figure 3 (a2,a3) indicate that for a 5 nm pore at pH 8, 1/tdwell vanishes below ~0.6 V or below pH 6 at 1V. Accordingly, we forced BSA at pH 8 from the cis-side of the membrane into a pore smaller than the protein using 0.6 V and then trapped it there by lowering the voltage while simultaneously measuring the blockade current until the trapped molecule escaped. A typical event is shown in Figure 3(a). Initially, the transmembrane voltage bias is set to 0.6 V, but when a dramatic change in the current is observed—see inset (a1)—corresponding to BSA entering the pore lumen, it triggers a change in the transmembrane bias from 0.6 V to 0.1 V resulting in a substantial reduction in the translocation velocity— the molecule exits this pore after about 6 s.
FIGURE 3. A single protein trapped in a nanopore.
(a) Current trace of a protein molecule trapped in a pore. When the protein enters the pore at V=0.6V, Inset (a1), it triggers a signal that switches the voltage bias to V=0.1V. After ~6 s the protein exits the pore. Inset (a2) shows the inverse dwell time versus bias voltage, for BSA passing through a 5×5nm pore in 100 mM KCl at pH8. The linear fit to the data, indicated by the dashed line, extrapolates to a threshold of 0.59V. Inset (a3) shows the inverse dwell time versus pH of BSA in the same pore as (a2) at 1 V bias. (b) Intensity map showing current fluctuations within a moving 250 ms window associated with a trapped protein. Inset: Histograms of 1 s of current data at 3.6 (A), 5.25 (B) and 9.5 s (C) after the protein is trapped. Solid lines (red) are fits to the data.
Figure 3(b) is a compilation of intensity plots that capture the amplitude of current fluctuations observed while a single protein is in the trap. It is produced by generating a histogram of the current in consecutive 250 ms windows. Because of the long dwell time in the trap, the current trace can be averaged to eliminate noise using an 8-pole Bessel low-pass filter with a cut-off at 1 kHz. Representative histograms encompassing 1 second of current data obtained at 3.6, 5.25 and 9.5 s after the molecule was trapped are shown in the respective insets to Figure 3(b). The open pore current is represented by position C, while two supposedly different protein conformations are represented by histograms A and B.
We assert that the fluctuations in the blockade current inform on the molecular configuration in the pore. Specifically, we claim that current fluctuations reflect the change in the electrolytic volume in the pore as the protein denatures under force associated with the electric field in the undersized pore. Optical tweezer37 and atomic force microscopy38 studies on titin have shown that the muscle protein undergoes forced unfolding with applied forces as low as 20-30 pN. The electric field in a nanopore, which is approximately E≈V/L, where V is the transmembrane voltage and L is the effective thickness of the nitride membrane, is E~3.2 pN/e at 0.1 V with a length of 10 nm, indicating that the effective force on BSA with a charge of −25e at pH 8 is ~75 pN. While this force is large enough to unfold the protein, the charge distribution of BSA is not uniform, however. The amino acid sequence is such that positively charged residues are adjacent to negatively charged ones. Therefore, it is feasable to have strong opposing forces on localized regions of the protein within the pore, which may also affect how the protein unfolds. While the forced unfolding of BSA has not been reported, structural studies of BSA have shown that it undergoes α-helix to β-sheet conformational change above >58°C.39 Thus, we assert that the force associated with the electric field in the pore can have a similar effect as the application of heat to the protein.
The relatively narrow histogram associated with the open pore current in C reflects only noise associated with the ionic current through the pore and is well fit by a single Gaussian distribution with mean μ=265.65±0.03 pA, standard deviation σ=3.06±0.04 pA and coefficient of determination r2=0.991. On the other hand, the histogram represented in B is better modeled by two Gaussian distributions with μ1=247.1±0.2 pA and μ2=255.2±0.2 pA; σ1=5.1±0.2 pA and σ2=4.9±0.2 pA; and r2=0.9865 rather than a single Gaussian distribution (r2=0.9762), which indicates a change in the molecular configuration while the molecule resides in the trap. Lastly, although it is broader than C, A can also be fit to a single Gaussian distribution (μ= 253.94±0.04 pA σ=4.11±0.05 pA with r2=0.989).
The width of the current distribution near A is consistent with the protein partially blocking the entrance of the pore. In contrast, the smaller pore current at B indicates that the protein is occluding a larger volume of the pore. The current distribution becomes broader due to flucuations in the occluded volume. According to this interpretation, the protein samples various configurations till it can transit through the smaller pore. At C, the current in the pore has returned to the open pore value and the narrow width current distribution indicates that the protein has escaped.
Trapping a protein obviates the requirement for continuous high voltage operation to suppress translocation noise and even conformational noise might be mitigated through repeated measurements. Following this strategy, single proteins in the microenvironment of a cell can be analyzed over an extended interval, enabling an accurate classification and quantitation. In principle, a protein trapped by the pore, can be analyzed and re-analyzed, pushing the molecule back and forth until it is identified. So, by using a combination of specific ligands or markers and by manipulating the pH and the electric field around the pore, a trapped protein may be unambiguously identified. This, of course, is only possible provided that the protein is within the capture radius defined by the electric field emanating from the pore.
A Nanopore Juxtaposed with a Single Cell
Another advantage of this microfluidic device is that we can use optical tweezers23 to position a cell directly over the nanopore, effectively eliminating the diffusion capacitance. To position a single cell over the nanopore, first the cell was conveyed via the microfluidics to the trans-side of the membrane where 9 time-shared optical tweezers (35 mW total power at λ=900 nm) were used to precisely position it. The cell was then encapsulated via photopolymerization in a 3.4 kDa PEGDA hydrogel, holding it in a fixed position relative to the pore. In separate experiments (not shown), we have observed that MDA-MB-231 cells remain viable in hydrogel for >12 hrs. The optical micrograph shown in Figure 4(a) shows the cell just prior to photopolymerization of the hydrogel. Using the same microfluidic device, we then conveyed 20 kbp YOYO intercalated dsDNA to the cis-side of the membrane. By applying a 2 V transmembrane bias through Ag/AgCl electrodes embedded directly in the microfluidic, it is possible to force the dsDNA through a 3.4 nm pore,36 a pore that is too narrow to allow for translocation of YOYO intercalated dsDNA at low voltage.40 At the same time, the voltage in the vicinity of the pore is apparently sufficient (>100 mV) to induce electroporation of the cell membrane, allowing the DNA to enter the cell. Figures 4(b,c) show x-z cross-sectional confocal images taken at different times showing the translocation of DNA through the pore and the concomitant transfection of the cell via electroporation. As time progresses and more DNA transfects the cell, the fluorescence intensifies, eventually showing the outline of the cell in Figure 4(c). Eventually, the fluorescence migrates to the nucleus of the cell as shown in Figure 4(d) by the false-color perspective of iso-surfaces reconstructed from confocal images of the same cell obtained 18 min after the start of transfection. Figure 4(d) starkly illustrates the concentration of fluorescence in the nucleus. DNA seems to be retained in the cell, as no extracellular fluorescence was observed, suggesting that the cell membrane remains intact >15 min. after electroporation.
FIGURE 4. Using a nanopore to transfect a breast cancer cell via electroporation.

(a) Optical micrograph showing an MDA-MB-231 cell held by optical tweezers over a 3.4-nm-diameter pore in a 30-nm-thick silicon nitride membrane, 35 μm on edge. The nitride membrane is the light gray square, transparent compared to the silicon handle wafer around it. (b,c) Confocal (x-z) cuts showing the translocation of 20 kbp YOYO intercalated dsDNA and the subsequent transfection of a 231 cell positioned over the 3.4 nm pore. As time progresses from t=3 to 16 min. more DNA enters the cell and the fluorescence increases, eventually showing the outline of the cell. (d) False-color perspective iso-surfaces reconstructed from volumetric data obtained from confocal images of the same cell as (a,b). At this late time (t=18 min), the fluorescent DNA appears to concentrate in the nucleus. Scattering from the membrane as well as fluorescent DNA in the pore are both indicated.
Although we have not established the ability to produce new biological function, we have demonstrated the potential for transfecting a single cell by electroporation using a nanopore. While many other single cell transfection approaches have been developed41,42 that are reportedly efficient and suitable for the transfection of virtually any type of cell, the strategy that we have developed employing a nanopore is the only one that offers the possibility of manipulating the cell’s genes and detecting proteins secreted from it at the same time. Moreover, this may all be accomplished with dose-controlled transfection and single molecule precision.
Conclusions
Using a nanopore embedded in a microfluidic as a stochastic sensor we have shown that it is possible to discriminate between different proteins of similar molecular weight by size and pH. Using the same device it is also possible to trap a single protein molecule and follow, through measurements of the blockade current, changes in its conformation as it unfolds in the pore. The microfluidic platform provides, not only an improvement in the minimum detectable concentration, but also optical access enabling the use of optical tweezers for positioning cells over the nanopore, which could possibly eliminate the diffusion capacitance altogether. Serendipitously, we discovered that cells positioned in this way can be transfected via electroporation. We demonstrated this by transfecting a breast cancer cell, encapsulated in hydrogel next to a nanopore, with YOYO intercalated dsDNA and observing fluorescence in the cell.
Thus, a nanopore embedded in a microfluidic used in combination with optical tweezers can be used to both transfect genes and discriminate the resulting secretome of a single cell, conceivably with single molecule resolution. The proteins secreted from cells comprise a complex and scarce set of molecules referred to as the ‘secretome’.43 About 10-20% of human genes encode (~2000) secreted proteins44 with an average molecular weight of 41.9 kDa.45 Most secreted proteins add complexity to higher eukaryotic systems and are vital for understanding cell-cell communications for tissue development and differentiation, among other processes. Some secreted proteins are involved in human disease, including metastasis and tumorigenesis.46,47 Thus, the secretome offers new biomarkers for the diagnosis of disease, a promising approach to drug discovery, and a deeper understanding of the mechanisms for tissue development. But these proteins are secreted in only minute quantities, and then diluted in body fluid or cell culture medium, making the secretome very difficult to detect and analyze. Moreover, stochastic gene expression produces a heterogeneous population of cells in tissue forcing analysis of the secretome with single cell resolution.
Using a nanopore with optical tweezers allows for the deconvolution of a single cell secretome from the bulk populations in which they are usually observed. With such a system, we can not only study the output, i.e. secreted proteins and small molecules, but also provide input to individual cells, in the form of plasmids, siRNAs, aptamers, or even prions. This will enable individual cell responses to be parsed and modulated to understand the stochastic behaviors driving biological function.
Acknowledgement
We gratefully acknowledge support from NSF CCF-1129098 and NIH F32CA138111.
Notes
This article is part of a themed issue highlighting the targeted study of single units, such as molecules, cells, organelles and pores – The “Single” Issue, guest edited by Henry White.
References
- 1.Alon U. An Introduction to Systems Biology. CRC Press; Baton Rouge, FL: 2007. [Google Scholar]
- 2.Iqbal SM, Bashir R, editors. Nanopores. Springer; New York, NY: 2011. [Google Scholar]
- 3.Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, Jovanovich SB, Krstic PS, Lindsay S, Ling XS, Mastrangelo CH, Meller A, Oliver JS, Pershin YV, Ramsey JM, Riehn R, Soni GV, Tabard-Cossa V, Wanunu M, Wiggin M, Schloss JA. Nat. Biotech. 2008;26:1146. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayley H, Cremer PS. Nature. 2001;413:226–230. doi: 10.1038/35093038. [DOI] [PubMed] [Google Scholar]
- 5.Wang D, Bodovitz S. Trends Biotech. 2010;28:281–290. doi: 10.1016/j.tibtech.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zare RN, Kim S. Annu. Rev. Biomed. Eng. 2010;12:187–201. doi: 10.1146/annurev-bioeng-070909-105238. [DOI] [PubMed] [Google Scholar]
- 7.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Proc. Natl. Acad. Sci. (USA) 1996;93:13770. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kullman L, Winterhalter M, Bezrukov SM. Biophys. J. 2002;82:803–812. doi: 10.1016/S0006-3495(02)75442-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kasianowicz JJ, et al. Anal. Chem. 2001;73:2268–2272. doi: 10.1021/ac000958c. [DOI] [PubMed] [Google Scholar]
- 10.Talaga DS, Li J. J. Am. Chem. Soc. 2009;131:9287. doi: 10.1021/ja901088b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oukhaled A, Cressiot B, Bacri L, Pastoriza-Gallego M, Betton J-M, Bourhis E, Gierak R, Auvray L, Pelta J. ACS Nano. 2011;5:3628. doi: 10.1021/nn1034795. [DOI] [PubMed] [Google Scholar]
- 12.Freedman KJ, Prabhu M, Ahn CW, Jemth P, Edel JB, Kim MJ. Anal. Chem, 2011;83:5137. doi: 10.1021/ac2001725. [DOI] [PubMed] [Google Scholar]
- 13.Fologea D, Ledden B, McNabb DS, Li J. Appl. Phys. Lett. 2007;91:053901. doi: 10.1063/1.2767206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Movileanu L. Trends Biotechnol. 2009;27:333. doi: 10.1016/j.tibtech.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 15.Berg HC. Random walks in biology. Princeton University Press; Princeton, NJ: 1993. [Google Scholar]
- 16.Nair PR, Alam MA. Appl. Phys. Let. 2006;88:3. [Google Scholar]
- 17.Halverson KM, et al. J. Biol. Chem. 2005;280:34056–34062. doi: 10.1074/jbc.M507928200. [DOI] [PubMed] [Google Scholar]
- 18.Nakane J, Akeson M, Marziali A. Electrophoresis. 2002;23:2592–2601. doi: 10.1002/1522-2683(200208)23:16<2592::AID-ELPS2592>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 19.Robertson JWF, et al. Proc. Natl. Acad. Sci. (USA) 2007;104:8207–8211. doi: 10.1073/pnas.0611085104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho C, Qiao R, Heng JB, Chatterjee A, Timp R, Aluru NR, Timp G. Proc. Nat. Acad. Sci. (USA) 2005;102:10445. doi: 10.1073/pnas.0500796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mirsaidov U, Timp W, Timp K, Mir M, Matsudaira P, Timp G. Phys. Rev. E. 2008;78:021910. doi: 10.1103/PhysRevE.78.021910. [DOI] [PubMed] [Google Scholar]
- 22.Min TL, Mears PJ, Chubiz LM, Rao CV, Golding I, Chemla YR. Nat. Methods. 2009;6(11):831. doi: 10.1038/nmeth.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mirsaidov U, Scrimgeour J, Timp W, Beck K, Mir M, Matsudaira P, Timp G. Lab Chip. 2008;8:2174. doi: 10.1039/b807987k. [DOI] [PubMed] [Google Scholar]
- 24.Wright AK, Thompson MR. Biophys. J. 1975;15(2 Pt 1):137–141. doi: 10.1016/s0006-3495(75)85797-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Trong I, Wang Z, Hyre DE, Lybrand TP, Stayton PS, Stenkamp RE. Acta Crystallogr., Sect D. 2011;67:813. doi: 10.1107/S0907444911027806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smeets RMM, Keyser UF, Dekker NH, Dekker C. Proc. Natl. Acad. Sci. (U SA) 2008;105:417–21. doi: 10.1073/pnas.0705349105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dimitrov V, Mirsaidov U, Wang D, Sorsch T, Mansfield W, Miner J, Klemens F, Cirelli R, Yemenicioglu S, Timp G. Nanotechnology. 2010;21:065502. doi: 10.1088/0957-4484/21/6/065502. [DOI] [PubMed] [Google Scholar]
- 28.Carter DC, Ho JX. Adv. Protein Chem. 1994;45:153. doi: 10.1016/s0065-3233(08)60640-3. [DOI] [PubMed] [Google Scholar]
- 29.Lin VJ, Koenig JL. Biopolymers. 1976;15:203. doi: 10.1002/bip.1976.360150114. [DOI] [PubMed] [Google Scholar]
- 30.Fawcett T. Pattern Recognition Letters. 2006;27:861. [Google Scholar]
- 31.Fologea D, Uplinger J, Thomas B, McNabb DS, Li J. Nano Letters. 2005;5:1734. doi: 10.1021/nl051063o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hooderheide DP, Garaj S, Golovchenko JA. Phys. Rev. Lett. 2009;102:256804. doi: 10.1103/PhysRevLett.102.256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smeets RMM, Keyser UF, Krapf D, Wu M-Y, Dekker NH, Dekker C. Nano Letters. 2006;6:89. doi: 10.1021/nl052107w. [DOI] [PubMed] [Google Scholar]
- 34.Golovchenko JA. private communication.
- 35.Timp W, Mirsaidov U, Wang D, Comer J, Aksimentiev O, Timp G. IEEE Trans. Nanotechnology. 2010;9(3):281–294. doi: 10.1109/TNANO.2010.2044418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mirsaidov U, Dimitrov V, Comer J, Aksimentiev A, Timp G. Nanotechnology. 2010;21:395501. doi: 10.1088/0957-4484/21/39/395501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kellermayer MS, Smith SB, Granzier HL, Bustamante C. Science. 1997;276:1112. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- 38.Brockwell DJ, Beddard GS, Clarkson J, Zinober RC, Blake AW, Trinick J, Olmsted PD, Smith DA, Radford SE. Biophys. J. 2002;83:458. doi: 10.1016/S0006-3495(02)75182-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poole S, West SI, Fry JC. Food Hydrocolloids. 1987;1:301. [Google Scholar]
- 40.Zhang C, Zhang F, van Kan JA, van der Maarel JRC. J. Chem. Phys. 2008;128:225109. doi: 10.1063/1.2937441. [DOI] [PubMed] [Google Scholar]
- 41.Ghafar-Zadeh E, Yeh E-C, Fu C-C, Lee LP. Biophys. J. 2011;100:522a. [Google Scholar]
- 42.Boukany PE, Morss A, Liao W-C, Henslee B, Jung HC, Zhang X, Yu B, Wang X, Wu Y, Li L, Gao K, Hu X, Zhao X, Hemminger O, Lu W, Lafyatis GP, Lee LJ. Nat. Nanotech. 2011;6:747. doi: 10.1038/nnano.2011.164. [DOI] [PubMed] [Google Scholar]
- 43.Tjalsma H, Bolhuis A, Jongbloed JD, van Dijl JM. Microbiol. Mol. Biol. Rev. 2000;64:515. doi: 10.1128/mmbr.64.3.515-547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hathout Y. Expert Rev Proteomics. 2007;4:239–248. doi: 10.1586/14789450.4.2.239. [DOI] [PubMed] [Google Scholar]
- 45.Klee EW, Carlson DF, Fahrenkrug SC, Ekker SC, Ellis BM. Nucl. Acids Res. 2004;32:1414. doi: 10.1093/nar/gkh286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Makridakis M, Vlahou A. J. Proteomics. 2010;73:2291. doi: 10.1016/j.jprot.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 47.Karagiannis GS, Pavlou MP, Diamandis EP. Molecular Oncology. 2010;4:496. doi: 10.1016/j.molonc.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]