Although first identified as the product of sour grapes (acetum, vinegar), acetate recently has proven to be a sweet harvest for biologists interested in transcriptional regulation. Over the past few years, a congruence of many separate lines of investigation has revealed that acetylation of chromatin is likely to play a key role in gene regulation in eukaryotes, from yeast to mammals (reviewed in refs. 1–4). Indeed, acetate appears to serve as a molecular fulcrum by which chromatin structure is levered open and closed by transcription factors and thereby may contribute to the changes in gene expression that distinguish neoplastic from normal cells. Intriguingly, the crucial role of acetate in gene regulation may have been exploited by microorganisms long before these processes were evident to researchers: a number of microbial products appear to target the acetylation/deacetylation pathways that are critical for proper gene regulation and normal cell function (5).
These themes are underscored in the paper in this issue of the Proceedings by Kwon and colleagues (6). This work grew out of an approach rooted in the beginnings of pharmacology: the mass screening of natural compounds for medically useful functions. One product of this screening approach was depudecin, a compound isolated from the fungus Alternaria brassicicola and identified by its ability to induce the morphological reversion of oncogenically transformed NIH 3T3 cells (7). Kwon et al. sought to identify the molecular target(s) of this potential anti-tumor drug; they now provide evidence that depudecin belongs to an expanding group of pharmaceuticals that operate, at least in part, by inhibiting histone deacetylases. Several lines of evidence are provided: (i) depudecin can compete for binding to the same cellular targets as trapoxin, a known inhibitor of histone deacetylases, (ii) depudecin inhibits histone deacetylases in vitro, and (iii) treatment of cells with depudecin leads to histone hyperacetylation in vivo. This work, taken as a whole, provides a plausible molecular basis for the actions of depudecin, and suggests, if somewhat more tentatively, that changes in gene expression lie behind the morphological reversion induced in transformed fibroblasts by depudecin.
These observations on the actions of depudecin coincide with a recent revival of interest in histone modification as an effector of eukaryotic transcriptional regulation. It has long been recognized that histones in transcriptionally active chromatin tend to be hyperacetylated, whereas silent chromatin generally is associated with histone hypoacetylation (1–4). However, these observations have gained new immediacy from revelations that many transcriptional activators can physically interact with cofactors that are histone acetyltransferases and that the ability to recruit these histone modifying enzymes is closely intertwined with the ability of the transcription factor to activate gene expression (reviewed in refs. 1–4). Transcriptional cofactors that possess histone acetyltransferase activity include the p300/CBP transcriptional integrator, the SRC-1 family of nuclear hormone receptor coactivators, the p/CAF cofactor, and the TAFII250 component of general transcription factor TF-IID (8–12). This apparent camaraderie between transcriptional activators and histone acetyltransferases can approach an obsession: the nuclear hormone receptors, for example, are potentially able to recruit not just one, but all four, of the histone acetyltransferases listed above. Providing a pleasing conceptual symmetry is the observation that many transcriptional repressors are associated with histone deacetylases (reviewed in refs. 2, 4, and 13). Thus, gene expression may be regulated through a balance between histone acetylation and deacetylation, with inhibitors of histone deacetylases, such as depudecin, able to shift the chromatin equilibrium toward acetylation.
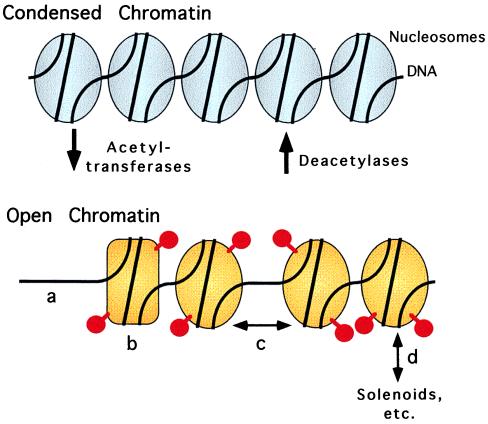
How might histone acetylation and deacetylation actually influence transcription? Prevailing wisdom suggests that acetylation of the N termini of certain core histones can lead to an “opening” of the chromatin structure, thereby increasing access of the DNA to the general transcriptional machinery and increasing transcriptional initiation rate (Fig. 1; refs. 1–4 and 13–18). Many predictions of this conceptual view are consistent with actual experimental observations. Hyperacetylated chromatin generally is more accessible to probes of DNA structure than is hypoacetylated chromatin, and transcriptionally active genes often are associated with areas of this more “open” chromatin. However, the devil is in the details, and it is the molecular details of the term “open” that remain demonically resistant to experimental dissection. The “open” state of chromatin induced by acetylation is not inevitably manifested as an overt change in nucleosome occupancy, but is often an amalgamation of more subtle changes in nucleosome structure and higher order packing; these delicate changes in chromatin structure can be at the borderline of resolution of the techniques currently available to dissect these phenomena. Adding to the complexity is the fact that acetylation can occur on different histones and at different sites within the same histone, presumably each with different consequences in terms of transcriptional regulation (1–4, 19). Clearly much work must still be done to elucidate the manner in which these changes in histone acetylation are manifested in the chromatin architecture and how these changes are interpreted by the transcriptional machinery.
Figure 1.
Schematic diagram of the effects of histone acetylation and deacetylation on chromatin structure (red spheres on stalks represent acetyl groups linked to histones). Gold tone depicts a more “open” structure for acetylated chromatin; this open structure could conceivably be manifested as changes in nucleosome occupancy (a), changes in the nucleosome conformation (b), changes in internucleosome interactions (c), and/or changes in higher-order chromatin packaging and structure (d).
It also must be noted that acetylation of chromatin is unlikely to be the sole determinant of transcription regulation. Many transcription activators interact not only with histone acetyltransferases, but also with components of the general transcriptional machinery itself, and thus may contribute to transcriptional activation by direct recruitment of a preinitiation complex to the target promoter (reviewed in ref. 20). Indeed, histone acetyltransferases and deacetylases themselves may have physiological targets beyond the limits implied by their nomenclature; CREBP binding protein, for example, can acetylate the p53 transcription factor, not just histones, in an manner that significantly enhances the DNA binding properties of p53 (21). It is also far from established that acetylation is invariably associated with transcriptional activation and that deacetylation is inevitably inhibitory; inhibition of histone deacetylase, for example, has different effects on different target genes, with defects observed in both transcriptional repression and activation (1–4). In summary, it appears likely that both activation and repression operate through multifaceted mechanisms involving alterations in the chromatin template and interactions with the general transcriptional machinery itself. Different mechanisms likely will predominate on different promoters and in different cellular contexts.
To return to the depudecin story: how might inhibition of histone deacetylase by depudecin lead to morphological reversion of transformed fibroblasts? The canonical view would be that a change of gene expression is involved, perhaps through activation of a set of previously repressed genes. This notion is consistent with indications that the anti-transformation effects of depudecin require de novo mRNA and protein synthesis. Kwon et al. (6) go further to propose a specific candidate as a possible target of depudecin action: the gene for gelsolin, a calcium-dependent actin filament severing and capping protein. Histone deacetylase inhibitors induce gelsolin expression (22), and perhaps more provocative, Kwon et al. allude to data indicating that microinjection of antibodies to gelsolin can prevent the morphological effects of these histone deacetylases. Much remains to be established, of course. Are changes in gene regulation sufficient, as well as necessary, for depudecin-mediated morphological reversion? Might there be targets of depudecin in addition to histone deacetylase, particularly given the relatively high concentrations of depudecin necessary for deacetylase inhibition, relative to the effective concentrations of trapoxin or trichostatin A? What other aspects of the transformed phenotype are influenced by depudecin treatment? Do changes in expression of known oncogenes contribute to the depudecin phenotype? We can expect interesting answers to these questions in the future.
In light of the actions of depudecin on v-ras/v-src transformed NIH 3T3 cells, it is perhaps not surprising that histone deacetylases also have been implicated in other neoplasias. For example, the v-erb A oncogene is derived from a nuclear hormone receptor and acts in retrovirus-induced erythroleukemia by blocking differentiation. The actions of v-erb A in neoplasia are tightly linked to the ability of this oncoprotein to recruit a corepressor complex that is believed to include histone deacetylase activity (23, 24). Analogous links between corepressor/deacetylase recruitment and oncogenesis, some tenuous and some more firmly established, also have been elucidated for the chimeric retinoic acid receptors implicated in human acute promyelocytic leukemias, and for the BCL-6 oncogene involved in follicular and large-cell lymphomas (25–28). Inhibitors of histone deacetylases also have been reported to revert the transformed morphology of a number of established tumor cell lines (5, 29).
Depudecin’s addition to the pharmacopoeia potentially contributes to both basic and clinical science. Depudecin possesses a chemical structure distinct from that of the previously identified inhibitors of histone deacetylase (trapoxin, trichostatin A, and sodium butyrate), and therefore depudecin may exhibit a selectivity toward histone deacetylases distinct from that of these previously described drugs. If true, depudecin, in combination with these other reagents, may be of significant value for dissecting the roles of specific histone acetylations in gene regulation. The novel structure of depudecin also is likely to contribute to understanding the enzymatic mechanisms of action of the deacetylases and may have unique advantages for therapeutic purposes.
In summary, both researchers and physicians owe a significant debt to the alchemy of microorganisms, which has provided an extensive inventory of pharmacologically active compounds for study of, and for medical intervention in, basic biological processes. Depudecin joins this inventory of useful biological tools, and we look forward to the knowledge that will be discovered in the future with its help.
References
- 1.Grunstein M. Nature (London) 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 2.Wu C. J Biol Chem. 1997;272:28171–28174. doi: 10.1074/jbc.272.45.28171. [DOI] [PubMed] [Google Scholar]
- 3.Wolffe A P, Pruss D. Cell. 1996;84:817–819. doi: 10.1016/s0092-8674(00)81059-4. [DOI] [PubMed] [Google Scholar]
- 4.Pazin M J, Kadonaga J T. Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida M, Horinouchi S, Beppu T. BioEssays. 1995;17:423–430. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]
- 6.Kwon H J, Owa T, Hassig C A, Shimada J, Schreiber S L. Proc Natl Acad Sci USA. 1998;95:3356–3361. doi: 10.1073/pnas.95.7.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsumoto M, Matsutani S, Sugita K, Yoshida H, Hayashi F, Terui Y, Nakai H, Uotani N, Kawamura Y, Matsumoto K, et al. J Antibiot. 1992;45:879–885. doi: 10.7164/antibiotics.45.879. [DOI] [PubMed] [Google Scholar]
- 8.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 9.Bannister A J, Kouzarides T. Nature (London) 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 10.Mizzen C A, Yang X-J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, et al. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 11.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O’Malley B O. Nature (London) 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 12.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 13.Wolffe A P. Nature (London) 1997;387:16–17. doi: 10.1038/387016a0. [DOI] [PubMed] [Google Scholar]
- 14.Brownell J E, Allis C D. Curr Opin Genet Dev. 1996;6:176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 15.Turner B M, O’Neil L P. Semin Cell Biol. 1995;6:229–236. doi: 10.1006/scel.1995.0031. [DOI] [PubMed] [Google Scholar]
- 16.Hong L, Schroth G P, Matthews H R, Yau P, Bradbury E M. J Biol Chem. 1993;268:305–314. [PubMed] [Google Scholar]
- 17.Lee D Y, Hayes J J, Pruss D, Wolffe A P. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Rameriz M, Rocchini C, Ausio J. J Biol Chem. 1995;270:17923–17928. doi: 10.1074/jbc.270.30.17923. [DOI] [PubMed] [Google Scholar]
- 19.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Nature (London) 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 20.Goodrich J A, Cutler G, Tjian R. Cell. 1996;84:825–830. doi: 10.1016/s0092-8674(00)81061-2. [DOI] [PubMed] [Google Scholar]
- 21.Gu W, Roeder R G. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 22.Hoshikawa Y, Kwon H J, Yoshida M, Horinouchi S, Beppu T. Exp Cell Res. 1994;214:189–197. doi: 10.1006/excr.1994.1248. [DOI] [PubMed] [Google Scholar]
- 23.Chen J D, Evans R M. Nature (London) 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 24.Sande S, Privalsky M L. Mol Endocrinol. 1996;10:813–825. doi: 10.1210/mend.10.7.8813722. [DOI] [PubMed] [Google Scholar]
- 25.Dhordain P, Albagli O, Lin R J, Ansieau S, Quief S, Leutz A, Kerckaert J P, Evans R M, Leprince D. Proc Natl Acad Sci USA. 1997;94:10762–10767. doi: 10.1073/pnas.94.20.10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong S-H, David G, Wong C W, Dejean A, Privalsky M L. Proc Natl Acad Sci USA. 1997;94:9028–9033. doi: 10.1073/pnas.94.17.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin R J, Nagy L, Inoue S, Shao W, Miller W H, Jr, Evans R M. Nature (London) 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 28.Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara F F, Zamir I, et al. Nature (London) 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- 29.Futamura M, Monden Y, Okabe T, Fujita-Yoshigaki J, Yokoyama S, Nishimura S. Oncogene. 1995;10:1119–1123. [PubMed] [Google Scholar]