Abstract
Vibrio cholerae is the waterborne bacterium responsible for worldwide outbreaks of the acute, potentially fatal cholera diarrhea. The primary factors this human pathogen uses to cause the disease are controlled by a complex regulatory program linking extracellular signaling inputs to changes in expression of several critical virulence genes. Recently it has been uncovered that many non-coding regulatory sRNAs are important components of the V. cholerae virulence regulon. Most of these sRNAs appear to require the RNA-binding protein, Hfq, to interact with and alter the expression of target genes, while a few sRNAs appear to function by an Hfq-independent mechanism. Direct base-pairing between the sRNAs and putative target mRNAs has been shown in a few cases but the extent of each sRNAs regulon is not fully known. Genetic and biochemical methods, coupled with computational and genomics approaches, are being used to validate known sRNAs and also to identify many additional putative sRNAs that may play a role in the pathogenic lifestyle of V. cholerae.
Keywords: Hfq, Quorum sensing, Vibrio cholerae, sRNA, virulence
Introduction
The bacterium Vibrio cholerae causes over 3,000,000 cases of the potentially fatal diarrheal disease cholera, resulting in approximately 100,000 deaths annually.1 The lifestyle of this human pathogen includes an aquatic niche where it survives as either a free-living bacterium or in biofilm communities often associated with the surface of plankton. Once ingested via contaminated food or water, V. cholerae can gain access to the human intestine and replicate to large numbers while producing the cholera symptoms that aid in transmission back to the aquatic environment. The relative contribution to epidemics of person-to-person transmission, vs. naturally occurring environmental V. cholerae bacteria, remains a contentious issue, recently debated following the 2010 cholera outbreak in Haiti.2
The pathogenesis of V. cholerae within the human host is controlled by a complex signaling cascade of regulatory factors and virulence genes, which have been the subject of several decades of intense study.3-5 During the last two decades, the identification of non-coding small RNAs (sRNAs) has revealed a new layer in bacterial gene regulation. Work in the model organism Escherichia coli led to the discovery of sRNAs as ubiquitous regulators of numerous cellular processes in many bacteria. In V. cholerae, several trans-acting sRNAs have recently been identified that regulate signaling pathways important in environmental settings; as well as in pathways that alter expression of genes affecting the ability of this pathogen to cause disease, the focus of this review. This field of research is likely to be a fecund area for future study with potential biomedical implications.
Bacteria encode both cis-acting and trans-acting non-coding sRNAs. Cis-acting sRNAs are perfectly complementary to an mRNA target expressed on the opposite strand of the DNA. In contrast, the trans-acting sRNAs described here are only partially complementary to one or more mRNA targets encoded elsewhere in the genome. In E. coli and V. cholerae, the RNA binding protein Hfq typically binds to both a trans-acting sRNA and its cognate mRNA, to lower the apparent dissociation constant between a sRNA and target mRNA,6-11 to stabilize sRNAs,12 and in some cases to facilitate degradation of the RNAs by recruiting RNase E.13 As Hfq is involved in many different sRNA/mRNA interactions, bacteria with an hfq gene deletion usually display pleiotropic defects.14,15 However, V. cholerae also encodes several trans-acting sRNAs, described below, that appear to interact with mRNAs without the necessity of Hfq,16,17 and also that act by titration of a protein rather than an Hfq-dependent base-pairing mechanism.18
Most bacterial trans-acting sRNAs regulate cognate target mRNAs through formation of a sRNA/mRNA duplex that negatively or positively alters the translation of the target gene. Repression is generally accomplished by base-pairing of the sRNA to nucleotides within the 5′untranslated region (5′ UTR) of an mRNA that are adjacent to or within the ribosome binding site (RBS), thus preventing access by the translation machinery,19 or in some cases recruiting an RNase.13 In contrast, fewer examples of sRNA-dependent activation have been described, and typically occur as a consequence of sRNA pairing in an mRNA region distinct from the RBS. Activation results from an alteration of the secondary structure within a 5′ UTR, that relieves an inhibitory structure in the mRNA, such as a hairpin, that blocks initiation or elongation of translation.20
Initial discoveries of bacterial sRNAs grew out of genetic studies in which signaling pathways with missing components were shown to be controlled by non-coding regulatory sRNAs. More recently, algorithms have been developed to predict putative sRNAs and their potential targets based on DNA sequence features, coupled with current genomics methods that have likewise identified numerous putative regulatory sRNAs in V. cholerae and other bacteria.21 However, robust experimental validation of sRNA/mRNA base-pairing is required for a definitive demonstration of sRNA/mRNA base-pairing interactions. The benchmark for experimental evidence is obtained by the construction of a sRNA and an mRNA that both contain one or more nucleotide substitutions within the predicted pairing region. Often a native nucleotide(s) within the sRNA and the mRNA is replaced by the complementary base. This allows demonstration that pairing does not occur between a mutated sRNA and its native mRNA target, and vice versa; but that pairing is restored between a mutated sRNA and the mRNA target carrying a compensatory mutation. Computational predictions coupled with both in vivo and in vitro experimental methods have identified and validated in bacteria numerous sRNA/mRNA interactions and elegant regulatory mechanisms, with new discoveries invariably to follow. This review focuses on the relatively small group of trans-acting sRNAs currently known to play a role in the pathogenesis of the waterborne bacterial pathogen, V. cholerae.
V. cholerae ecology and pathogenesis
V. cholerae in marine environments exists in a free living state and in association with surfaces (such as chitinous zooplankton and crab exoskeletons) as biofilms composed of bacterial cells, extracellular matrix material, and DNA.22-25 Biofilm formation protects bacteria against environmental insults,26 and biofilm-derived V. cholerae cells are also enhanced for virulence compared with free-living cells.27-29 Biofilm growth in standard nutrient-rich broth requires the production of exopolysaccharides, which are synthesized by the gene products of two VPS (Vibriopolysaccharide) operons, vpsA-L and vpsK-Q; though VPS production is not required for biofilms in sea water.23,30,31 Growth of V. cholerae in association with chitin also induces the bacteria to become naturally competent to take up extracellular DNA, which can be incorporated onto the chromosome allowing acquisition of genes encoding virulence factors and in antigenicity evolution.32,33 Thus, the environment may serve as a reservoir for potentially pathogenic V. cholerae, and also as a source of genetic diversity and an arsenal of virulence factors.
Following ingestion of bacteria in contaminated food or water, a subset of ingested V. cholerae cells survive passage through the acidic conditions of the stomach and gain access to the small intestine.34 The two primary virulence factors responsible for colonization and subsequent disease are the cholera toxin (CT), encoded by ctxAB, and the toxin-co-regulated pilus (TCP), encoded by tcpA-F. CT is the canonical AB-type toxin, composed of five B subunits that bind as a pentameric ring to the GM1 ganglioside of intestinal epithelial cells to deliver the A subunit of the toxin into the cell. Within intestinal cells, the A subunit of CT ADP-ribosylates the Gαs protein leading to activation of adenylate cyclase, resulting in a massive increase in cyclic AMP (cAMP) production. Accumulation of cAMP leads to the activation of the CFTR ion channel via phosphorylation, causing an efflux of chloride ions and water into the lumen of the small intestine, creating the diarrheal disease.35
The second major virulence factor of V. cholerae, the type IV pilus TCP, is required for efficient colonization of the small intestine.36,37 The thread-like TCP pili promote cell-cell aggregation that aids in intestinal colonization,4 and also serve as a receptor for the CTXφ phage, which encodes the ctxAB genes for CT.38 TCP is encoded by the tcpA-F operon located in a region of the genome called the Vibrio pathogenicity island (VPI). The VPI also contains other genes, including acfA-D, which are required for full colonization of a mouse intestine,39 and toxT, the primary regulator of tcpA-F and ctxAB, the expression of which is coordinately regulated with TCP.
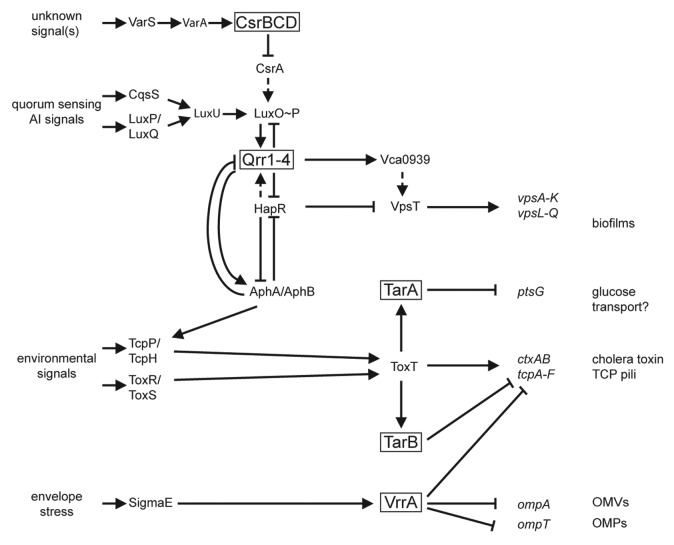
ToxT is the primary regulatory protein controlling transcription of both ctxAB and tcpA-F and a member of the AraC/XylS family of transcriptional activators (Fig. 1).40 Degenerate 13 nucleotide (nt) sequences upstream of the -35 promoter element of ToxT-regulated genes, called “toxboxes,” serve as binding sites for the transcription factor.41 Transcription of ToxT is activated by ToxR, a transmembrane protein that contains a C-terminal DNA binding domain. ToxR stability requires ToxS, another inner membrane associated protein.42 TcpP, a second transmembrane protein, along with its partner protein TcpH, also activates ToxT transcription. Several environmental cues, including temperature, pH, osmolarity, and bile salts alter expression of the virulence factors controlled by ToxT, via mechanisms both dependent and independent of TcpP/H and ToxR/S.43,44 Both ToxR and TcpP bind to the promoter of toxT to activate transcription.45 While transcription of the toxRS operon is constitutively active, tcpPH transcription is controlled by two activator proteins, AphA and AphB, which both bind to the promoter of tcpPH to directly activate its transcription.46,47 This complex AphA regulon maintains control of ToxT, which is absolutely required for expression of ctxAB and tcpA-F, and thus virulence. With one exception, to date, the V. cholerae sRNAs described below that participate in virulence regulation either impinge on toxT expression, or are in turn controlled by ToxT (Fig. 1).
Figure 1. Regulatory network linking sRNAs to virulence in V. cholerae. Extracellular signal inputs coordinate regulatory cascades that connect the transcription of multiple sRNAs to the expression of genes that play a role in V. cholerae virulence. Sensory information is perceived by cognate receptors in or at the bacterial membrane (left), which transduce information into the regulatory network of the cell. Transcription of multiple sRNAs (boxed) is altered, resulting in changes in the expression of numerous genes involved in virulence and pathogenesis (right). The signal(s) sensed by VarS remains unclear. Dashed lines indicate indirect regulation. Solid lines indicate direct regulation. See text for additional details.
The Qrr sRNAs and the V. cholerae Quorum Sensing system
In 2002, collaborative studies by the Bassler group demonstrated that a cell-cell communication system, first described in bioluminescent marine bacterium Vibrio harveyi, termed quorum sensing (QS), impinges on the ToxT virulence cascade of V. cholerae.48-50 The V. cholerae QS system described below allows for synchronization of gene expression in response to small chemical signal molecules (autoinducers, or AIs) that are produced by V. cholerae and released into the extracellular environment. Importantly, multiple sRNAs are critical components of the V. cholerae QS system and provided the first examples of non-coding sRNAs that regulate virulence gene expression in this pathogen (Fig. 1).
V. cholerae produces and responds to two QS autoinducers, CAI-1 (cholera autoinducer-1) and AI-2 (autoinducer-2), which are produced by CqsA and LuxS, respectively. CAI-1 is (S)-3-hydroxytridecan-4-one, produced and used by numerous Vibrios.51 AI-2 is 4,5-dihydroxy-2,3-pentanedione (DPD), which is produced by many bacteria and described as a “universal” signaling molecule for inter-species communication.52,53V. cholerae AIs are produced at a constant rate and thus serve as a proxy measure of bacterial number; with low AI levels at low cell density and high AI levels at high cell density.
Each AI has a cognate inner membrane receptor: CqsS for CAI-1 and the LuxP/Q heterodimer for AI-2. When V. cholerae experiences low cell density (LCD) conditions, and AI signals are scarce, the cognate AI receptors function as histidine kinases and transfer phosphate, via LuxU, to a response regulator, LuxO, which is a member of the NtrC family of transcription factors that activate transcription in conjunction with the alternate sigma factor, σ54.54 Based on parallel genetic studies of the related QS system in bioluminescent V. harveyi, it was hypothesized that phosphorylated LuxO (LuxO~P) repressed the activator of bioluminescence, LuxR (homologous to HapR in V. cholerae). This hypothesis was validated with a genetic screen that identified gene disruptions in luxO and rpoN (encoding σ54), as expected, and also the previously described hfq gene.55 A bioinformatics study followed, which identified the four V. cholerae sRNAs, Qrr1–4 (quorum-regulated RNA), that are transcriptionally activated at low cell density by LuxO~P in concert with σ54.55
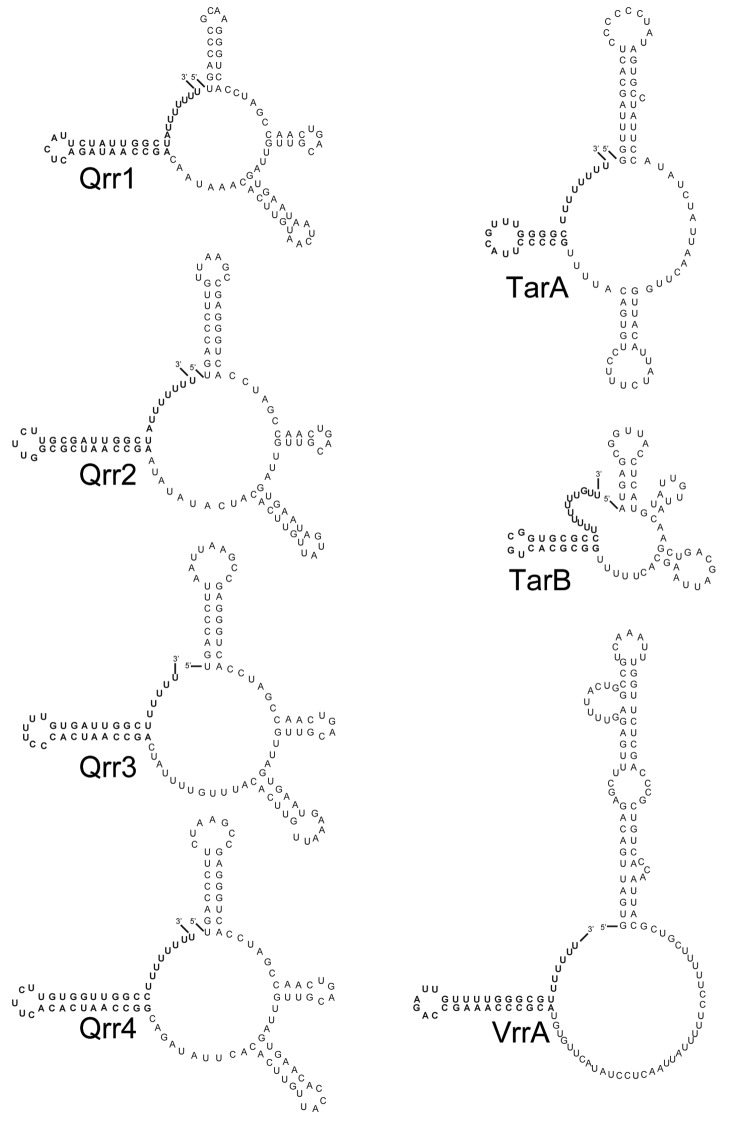
The Qrr sRNAs act in concert with Hfq to regulate several different mRNAs, both positively and negatively. In V. cholerae, a strain carrying any single Qrr (with deletion of the other three qrr genes) exhibits WT QS behavior, thus the Qrr sRNAs were deemed functionally redundant for hapR regulation, which controls most QS phenotypes.55 This is due to several feedback mechanisms that control Qrr transcription and maintain total Qrr levels even when one, two, or three qrr genes are deleted.56 The Qrr sRNAs vary in length from 96 to 108 nucleotides and are predicted by minimal free energy (MFE) calculations to form similar secondary structures to one another (Fig. 2).55,57 All sequenced pathogenic Vibrios, encode multiple Qrr sRNAs that each contain an absolutely conserved 32 nt sequence, which was predicted to be the site of interaction with mRNA targets.55 When V. cholerae reaches high cell density (HCD), and thus high AI levels, the AI-bound receptors switch to phosphatases, and as a result, unphosphorylated LuxO cannot activate Qrr transcription, initiating the QS response.
Figure 2. Predicted secondary structures of V. cholerae Hfq-dependent sRNAs that play a role in virulence gene expression. The lowest free energy structures of each sRNA are shown as predicted by mfold analysis (http://mfold.rna.albany.edu/?q=mfold). Qrr1 and Qrr4 structures appear different than described in 2004 in ref. 55 using current mfold analysis tools. A predicted TarB secondary structure was not described in ref. 87. To generate a single lowest free energy TarB structure, restrictions were imposed to prevent long range base-pairs by setting a maximum distance between paired bases at 30. The Rho-independent terminator of each sRNA is shown in bold.
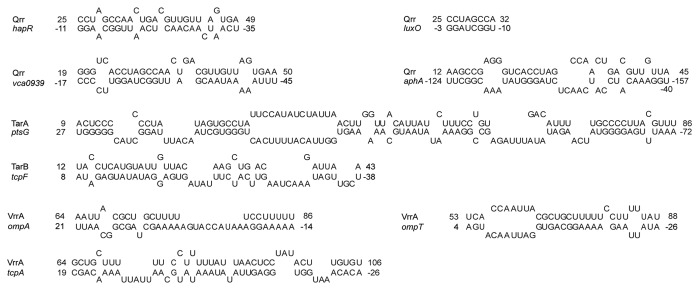
Several members of the Qrr regulon play a role in V. cholerae virulence, most prominently the transcriptional regulator, HapR,58 which controls a variety of behaviors of V. cholerae, including virulence and biofilm formation.28,48,49,59,60 We have recently shown by in vivo and in vitro experiments with defined mutations in each RNA, that the Qrr sRNAs base-pair with hapR mRNA at the ribosome binding site (RBS), resulting in repression of hapR translation and a halt in HapR protein production (Fig. 3).11,55 The Qrr sRNAs may also facilitate destruction of the hapR mRNA by cellular RNases as increased levels of the sRNAs correlate with decreased levels of hapR mRNA.61 Repression of hapR translation by the Qrr sRNAs requires the RNA chaperone Hfq,55 which also facilitates sRNA/mRNA interactions in E. coli as described above.6-10 V. cholerae Hfq serves a similar function by decreasing the apparent dissociation constant of hapR mRNA and Qrr sRNA in vitro.11 Furthermore, the V. cholerae hfq gene is required for colonization of a suckling mouse intestine by the bacterium, a virulence defect presumably caused by a loss of function of many sRNAs.15 Indeed, microarray analysis indicated several important classes of genes with altered expression in a strain lacking Hfq, including genes encoding RpoE (stress response sigma factor), chemotaxis components, several histidine kinases, and multiple outer membrane proteins and periplasmic stress response proteins.15
Figure 3. Predicted base pairing interactions between V. cholerae sRNAs and mRNA targets. Each sRNA/mRNA pairing shown was predicted with RNAhybrid program (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/submission.html) analysis. The predictions shown were previously described for Qrr interactions with hapR, luxO and vca0939 in refs. 55, 56 and 72; TarA/ptsG in ref. 86; VrrA/ompA and VrrA/tcpA in ref. 17; and VrrA/ompT in ref. 94. The Qrr/aphA and TarB/tcpF pairing predictions were performed for this review. For each predicted pairing shown, unpaired nucleotides were omitted from the ends for simplicity.
QS regulation of the V. cholerae Qrr sRNAs plays a role in regulation of the virulence cascade described above. At HCD, in the absence of Qrr sRNAs, HapR directly represses transcription of aphA by binding to its promoter to block transcription.50 Strains of V. cholerae engineered to produce no Qrr sRNAs, do not make TcpP or CTX, and are severely impaired for colonization in an infant mouse model of V. cholerae pathogenesis.48,49 Virulence factor repression depends on AIs, as addition of synthetic AIs to a strain that cannot synthesize its own QS signals also causes a dramatic decrease in TCP production, indicating that production of the Qrr sRNAs is critical for negative regulation of HapR that enables CT and TCP production.62
HapR accumulation at HCD controls additional phenotypes that may also play a role in virulence and/or biofilm formation. Notably, the AI-induced termination of Qrr synthesis and the resulting accumulation of HapR at HCD positively regulates expression of multiple competence genes, including comEA, required for V. cholerae to become capable of horizontally acquiring genetic material in the form of extracellular DNA.11,32,63 Natural competence in several other bacteria results in the acquisition of virulence factors and antibiotic resistance genes.64,65 QS-induced natural competence in V. cholerae can promote acquisition of the CTXφ prophage carrying ctxAB,66 and alteration of genetic determinants of antigen specificity.67 Indeed, the “mosaic structured” genome of V. cholerae is believed to be the result of numerous horizontal gene transfer events mediated by activities including chitin-induced natural competence.68
V. cholerae biofilm formation is also controlled by Qrr/hapR interactions, as HapR serves as a repressor of vps transcription.28,59,60,69 HapR repression of vps transcription at HCD is directly mediated by binding of HapR to the promoter of vpsT, which encodes a positive regulator of the vpsA-K and vpsL-Q loci. QS, via HapR, also indirectly controls vps transcription and biofilm formation by modulating the accumulation of a secondary signaling molecule, cyclic di-GMP (c-di-GMP).69-71 Interestingly, the Qrr sRNAs can directly activate translation of the mRNA of vca0939, a protein that contains an amino acid motif (GGDEF) indicative of proteins that directly synthesize c-di-GMP.72,73 Genetic evidence supports a model that at LCD, Hfq-dependent base-pairing of the 32 nt interaction region of the Qrr sRNAs to the 5′UTR of vca0939 mRNA prevents formation of an inhibitory secondary structure at the RBS (Fig. 3), thereby promoting translation in a manner similar to interactions in E. coli that promote translation of sigma factor RpoS by pairing with the RprA and DsrA sRNAs.74,75 Thus, the Qrr sRNAs appear to link QS to biofilm formation by multiple pathways: direct activation of vca0939 translation, negative regulation of HapR resulting in high c-di-GMP and enhanced VPS-dependent biofilm formation at LCD, and low levels of c-di-GMP and biofilm repression at HCD.
Previous studies established that quorum sensing and the Qrr sRNAs play a role in V. cholerae pathogenesis through HapR-mediated effects on aphA transcription.76 However, recently it was shown that V. cholerae Qrr sRNAs also directly regulate aphA mRNA translation independent of HapR.77 Specifically, aphA expression is 3-fold increased in a hapR- V. cholerae strain constitutively expressing the Qrr sRNAs. The Qrr sRNAs base-pair with the 5′UTR of aphA over 100 nt upstream of the ATG start codon and perhaps alter a putative secondary structure within the leader sequence that hinders translation (Fig. 3).78 Interestingly, several Qrr nucleotides predicted to participate in aphA base-pairing lie outside of the absolutely conserved 32 nt sequence predicted to be important in the regulation of other direct Qrr targets (Fig. Two and 3). Because Vibrio Qrr sRNAs show some sequence divergence outside of the conserved 32 nt region, it has been proposed that individual Qrr sRNAs may possess unique regulons not controlled by the other Qrr sRNAs. Indeed, in both V.harveyi and V. cholerae, individual Qrr sRNAs show different abilities to activate AphA translation due to sequence divergence that influences the ability of the Qrr sRNAs to bind aphA mRNA.77,78
Finally, several regulatory feedback loops involving the Vibrio Qrrs also appear to maintain levels of the QS sRNAs in the cell. First, in V. cholerae, AphA represses transcription of both the qrr sRNAs and hapR, creating two feedback loops that refine the control of the quorum sensing system. In V. harveyi, AphA binds to the promoter of qrr4 in a region that potentially occludes binding of the activator protein LuxO.77 AphA appears to control several hundred genes at LCD in V. harveyi, however, it is unclear whether AphA controls those same genes in V. cholerae as the experiments to determine the AphA regulon were performed at HCD. Second, Qrr/luxO base-pairing establishes an additional feedback loop that is sufficient to downregulate luxO transcription by occluding the RBS of luxOU mRNA (Fig. 3).56 Predicted Qrr/luxO pairing only represses luxO transcription several fold, compared with the more extensive Qrr/hapR pairing that halts HapR production (Fig. 3); presumably because full LuxO repression would in turn prevent Qrr transcription. Third, HapR itself activates Qrr transcription by an indirect mechanism that requires LuxO~P but does not involve direct interaction of HapR protein with the Qrr promoters.61 Presumably this feed-back loop is important in a transition from HCD to LCD, since dense HCD cultures (devoid of Qrr sRNAs) rapidly accumulate the QS sRNA when transitioned to LCD conditions, but isogenic HapR- strains incubated under similar conditions do not rapidly accumulate the sRNAs.61 Thus the levels of three proteins serve as inputs to control the levels of Qrr sRNAs in the V. cholerae cell: LuxO~P, HapR, and AphA. Interestingly, each of these regulators participate in auto-inhibitory feedback loops, repressing their own transcription.56,61,77
CsrBCD sRNAs and the VarA/S signaling pathway
The V. cholerae QS system described above controls multiple Qrr sRNAs in response to secreted AI signals. Remarkably, an additional second set of sRNAs also contributes to the V. cholerae QS system by altering the activity of the QS LuxO response regulator. A genetic screen to identify an additional regulator(s) of V. cholerae QS revealed that a two component regulatory system composed of a sensor kinase, VarS, and a response regulator, VarA, contributes to the regulation of hapR (Fig. 1).79 A strain lacking VarA shows decreased expression of hapR, as measured by a luciferase-based reporter gene fusion that is activated by HapR. VarA had previously been identified and implicated in virulence; as a strain lacking varA show reduced production of CT and TcpA, the main structural pilus component of TCP, with a corresponding defect in colonization of infant mouse intestines.80 Oddly, these experiments were performed in a HapR- V. cholerae strain, suggesting that VarA has both QS-dependent and QS-independent roles in virulence regulation.
The V. cholerae VarS/VarA system is homologous to the GacS/GacA two-component system from Pseudomonas species that controls secondary metabolite production,81,82 and to the BarA/UvrA system in E. coli.18,83 The sensory information these signaling systems respond to is poorly understood. In E. coli, UvrA, activates transcription of two sRNAs (CsrB and CsrC) that control the activity of CsrA, an RNA binding protein, which regulates a variety of processes in E. coli by destabilizing RNA.84 The sRNAs control CsrA by directly binding to and titrating out CsrA protein, preventing CsrA from destabilizing its target RNAs.18,83 Since, the Csr sRNAs in V. cholerae, E. coli and other bacteria do not function by base-pairing interactions with other RNAs, these Csr sRNAs act by an Hfq-independent mechanism.
In V. cholerae, the VarS/VarA sensory system activates the expression of three sRNAs, CsrB-D, which are approximately 350 nt in length and functionally homologous to the CsrB sRNA from E. coli.79 Predicted secondary structures indicate that each of the sRNAs contains numerous hairpins with the loop regions often containing the sequences AGGA/AGGGA, which are thought to facilitate CsrA binding.18,83 In V. cholerae, CsrA is required for the effect of Vars/VarA on HapR expression, consistent with the interpretation that the CsrB-D sRNAs function similarly in V. cholerae and E. coli by titrating CsrA. V. cholerae strains with a deletion in the varS or varA gene, or a triple mutant lacking all three CsrBCD sRNAs display an increase in Qrr levels and lower HapR production.79 In contrast, a csrA mutation in each of these genetic backgrounds results in an opposite QS phenotype. CsrA appears to have a positive effect on LuxO activity, because a strain carrying an allele of luxO (luxO D47E) that mimics the phosphorylated active form of the protein shows low HapR levels and these levels are unaltered when the strain also carries the csrA mutation. Transcription of luxO is not altered by the VarS.VarA/CsrBCD/A system, therefore it is believed an unidentified factor links CsrA to LuxO.79,85
ToxT-controlled TarA and TarB sRNAs
The seven V. cholerae sRNAs discussed above all regulate virulence by reducing the levels of HapR, a repressor of the AphA regulon that controls production of ToxT. Recently, two additional sRNAs, TarA and TarB (for ToxT activated RNAs) have been discovered in V. cholerae that are regulated by ToxT and appear to have effects on virulence in animal models. Both of these sRNAs are encoded within the VPI and are activated by ToxT.86,87 The 99 nt TarA sRNA (Fig. 2) is encoded upstream of tcpI and contains two toxboxes for ToxT binding within its promoter.86 To date, the only target gene known to be controlled by TarA is ptsG, which is involved in regulating the uptake of glucose. Like the other Hfq-dependent sRNAs described here, TarA is a trans-acting sRNA. Although trans-encoded, TarA shows remarkable sequence complementarity with the 5′UTR of ptsG, with almost two thirds of its nucleotides predicted to base-pair with ptsG mRNA by RNAhybrid (Fig. 3). Repression of ptsG translation by the TarA sRNA is based in part on the observation that deletion of the tarA gene leads to increased levels of ptsG mRNA.86 The reciprocal is also observed; tarA overexpression causes growth defects in glucose minimal medium that are similar to the phenotype of a V. cholerae ptsG deletion mutant.86 This regulation appears to be dependent on Hfq, as TarA is unstable in a V. cholerae hfq mutant. However, the role of TarA in V. cholerae virulence is not entirely clear as different studies have documented conflicting results. The group that initially discovered TarA reported a slight disadvantage for a tarA mutant when competed against wild type in an infant mouse colonization model.86 However, a more recent study using a different V. cholerae strain documented no virulence defect for a tarA mutant.87 It is also unclear how or if control of glucose transport is responsible for the modest virulence defect in the original study, or whether ctxAB or tcpA-F are also under TarA (and/or ptsG) control. Demonstration of TarA/ptsG pairing by mutational analysis of the sRNA/mRNA interaction and in vitro RNA interaction, as well as an analysis of the ToxT regulon in strains altered in TarA/ptsG pairing will likely shed light on the role of this new sRNA in the in vivo ecology of this pathogen.
An additional ToxT-regulated sRNA was recently identified that also appears to play a role a role in V. cholerae virulence. TarB, which is also encoded within the VPI, along with tcpA-F, is a relatively small 60 nt sRNA (Fig. 2) that is positively regulated by ToxT.87 Discovered using a high-throughput sequencing approach to identify sRNAs regulated by ToxT, the TarB promoter contains two toxboxes shown to be bound by ToxT protein in gel-shift experiments, consistent with a model of direct transcriptional activation of tarB by ToxT.87 Interestingly, V. cholerae strains lacking TarB outcompete wild type strains in an infant mouse model, suggesting that TarB has a negative effect on V. cholerae virulence. Specifically TarB targets the 5′UTR of tcpF (Fig. 3), the final gene in the tcpA-F operon, which encodes the secreted TcpF colonization factor. Surprisingly, TarB/tcpF interactions do not require Hfq, as levels of TarB sRNA and tcpF mRNA are unaltered in an Hfq- compared with an Hfq+ genetic background.87,88 The regulation of TcpF appears to be responsible for the increased ability of TarB- strain to colonize the mouse small intestine, as a strain carrying a tcpF allele with a mutation in the 5′UTR rendering it TarB-independent displays a colonization phenotype identical to a strain lacking tarB.87 It remains unclear why a sRNA that is activated by ToxT, a positive regulator of the major V. cholerae virulence factors (CTX and TCP), would repress TcpF expression. It is proposed that TarB may be expressed only in certain microenvironments as a mechanism for fine-tuning expression of virulence factors, but no studies have yet addressed this hypothesis.
The VrrA sRNA and Tcp
Many sRNAs in other gram-negative species have been shown to directly target mRNAs encoding outer membrane proteins (OMPs).89 In V. cholerae the sRNA, MicX, which was identified by bioinformatics methods, appears to interact with the mRNA of an uncharacterized OMP, vc0972, although this presumptive pairing does not have an effect on virulence.16 In contrast, another V. cholerae sRNA, VrrA, has been experimentally shown to interact with the mRNA of an OMP and in doing so alters virulence expression.17 VrrA is an approximately 140 nt sRNA (Fig. 2) that was identified following a transposon mutagenesis screen. Like the Qrr sRNAs, transcription of vrrA depends on an alternative sigma factor, in this case σE, indicating it may be expressed in response to stress.17 In both E. coli and Salmonella typhimurium, σE activates the MicA and RybB sRNAs, which regulate OMPs, consistent with the σE function of monitoring envelope stress.90-92 Deletion of vrrA causes an increase in the levels of OmpA, an outer membrane porin. So too, loss of OmpA due to overexpression of vrrA increases outer membrane vesicle production, which has been hypothesized to be involved in a variety of functions in gram-negative bacteria including pathogenesis and biofilm development.93 VrrA binds to the 5′UTR of ompA mRNA in a region overlapping the translation initiation region (Fig. 3), as determined by in vitro toeprinting analysis. However, VrrA-mediated repression of ompA translation does not strictly require Hfq as overexpression of VrrA represses OmpA production even in a ∆hfq mutant.17 Interestingly, VrrA requires Hfq to bind to and repress the mRNA encoding another outer membrane porin, OmpT (Fig. 3).94 Like TarB, VrrA may also negatively regulate virulence in V. cholerae, as a vrrA deletion strain out-competes WT V. cholerae for infant mouse colonization.17 An ompA deficient strain also shows a 10-fold defect in infant mouse colonization, consistent with the in vivo effect of the vrrA deletion.
The role of the VrrA sRNA in V. cholerae virulence may be more complex, however, as VrrA, like the Qrr sRNAs, may regulate a large set of mRNAs. Specifically, VrrA was shown to be complementarity to the 5′ UTR of a third mRNA, namely, tcpA, and thus may also directly alter its translation by a base-pairing mechanism (Fig. 3), although this has not yet been demonstrated.17 That VrrA negatively regulates tcpA, was demonstrated by overexpression of VrrA, which causes a decrease in TCP levels, while deletion of the VrrA sRNA results in a modest increase in TCP pilin levels.17 It is unclear what the relative contributions are of OmpA, OmpT and TcpA regulation by the VrrA sRNA to V. cholerae pathogenesis, as the regulation of each target has not been genetically separated.
Additional sRNAs
While a relatively small number of sRNAs have been identified that play a role in V. cholerae pathogenesis, it is likely that many more regulatory RNAs remain to be discovered and characterized in this pathogen. Suggestions of more sRNAs have come from both bioinformatic and high-throughput sequencing studies.87,95,96 Experimental validation of these new putative regulators will likely identify a role for many of these sRNAs in both in vivo and environmental settings.
A bioinformatic study relying on genomic DNA features such as Rho-independent terminators within intergenic regions of DNA identified 32 novel putative sRNAs.95 Validation of expression by northern blot analysis showed that 6 of 9 predicted sRNAs tested were expressed under the conditions used. Bioinformatic approaches are still limited however, by a reliance on known sequence motifs and a need for validation of the expression of the sRNA.
Deep-sequencing studies have revealed a wealth of potential sRNAs in V. cholerae. Previously, direct cloning and sequencing of the bacterial transcriptome to search for sRNAs had been technically difficult due to the challenges of removing the housekeeping tRNA and rRNA that are similar in size to potential sRNAs. To resolve this dilemma, the Camilli group devised a technique termed sRNA-Seq.96 Briefly, 10–200 nt RNAs gel-extracted from total RNAs are bound to linkers and then annealed to DNA oligonucleotides that can be used to differentiate and exclusively degrade DNA complexes with rRNA and tRNA using RNase H. The remaining linkered RNAs can be reverse transcribed and subjected to high throughput sequencing. This novel study identified all of the sRNAs experimentally known at that time in V. cholerae, as well as more than 600 potentially novel sRNAs, from both antisense strands of coding sequences and putative intergenic regions.96 A second sRNA-Seq study sought to specifically identify sRNAs involved in pathogenesis using a V. cholerae strain that overexpressed ToxT to activate the major V. cholerae virulence factors.87 By cross-referencing with a set of sequence data of ToxT binding sequences, a putative list of sRNAs likely to be directly activated by ToxT was established. This method uncovered the previously discovered TarA and led to the identification of TarB, both of which appear to affect V. cholerae virulence, as described above. In addition, 16 other putative ToxT-activated sRNAs were discovered. Whether one or more of these sRNAs play a role in the virulence of V. cholerae remains to be determined. A third study examining the transcriptome of V. cholerae in animal models of infection detected the presence of several known sRNAs, including the previously mentioned Qrr sRNAs, TarA, and CsrC, as well as over 70 potentially novel sRNAs.97 Thus it is increasingly apparent that sRNAs are important, newly recognized regulatory, not only in V. cholerae virulence, but in the virulence of many other pathogens as well.
Concluding Remarks
The sRNAs discovered in the bioinformatic and deep sequencing studies described above could be involved in many aspects of bacterial physiology important for V. cholerae during infection or in the environment. Several V. cholerae sRNAs have been discovered, such as RyhB, which is potentially involved in iron acquisition, though is not required for mouse colonization.98 MicX, like VrrA, regulates an OMP,16 but does not appear to play a role in virulence, as described above. The MtlX sRNA regulates carbon metabolism through post-transcriptional regulation of the mRNA of a mannitol tranporter, MtlA.96,99 Finally, the TfoR sRNA activates translation of tfoX mRNA, which encodes the TfoX regulator required for natural competence in response to chitin.100 Thus, regulatory networks are being uncovered that link multiple sensory inputs, such as chitin and QS AIs, via small RNAs, to the control of genes for behaviors important in the environment and in the human host.32,63,100,101
It is increasingly appreciated that non-coding sRNAs are critical regulatory factors that play fundamental roles in bacteria. While the full extent of sRNA-based regulation in V. cholerae is not yet known, several sRNAs have already been identified that alter the production of the two main virulence factors of V. cholerae critical for causing the cholera disease CTX and TCP. Some of the sRNAs, such as the Qrr sRNAs, control both in vivo and environmental behaviors, while other sRNAs appear to have a role in fundamental bacterial physiology. The Qrr sRNAs, and VrrA, also appear to directly interact with many mRNA target genes that participate in virulence gene expression, and it remains possible that multiple target genes will be found for each of the other sRNAs described here. Determining the specific signaling inputs that activate the transcription of these sRNAs will also aid in defining the molecular mechanisms by which these regulatory pathways are controlled. Ultimately, interdisciplinary methods that incorporate genetic, bioinformatic, genomic and biochemical approaches will continue to be essential for a comprehensive understanding of the contribution of sRNAs to virulence in V. cholerae.
Acknowledgments
BK Hammer acknowledges financial support from a National Science Foundation grant (MCB-0919821).
Glossary
Abbreviations:
- QS
quorum sensing
- Qrr
Quorum regulatory RNA
- sRNA
small RNA
- LCD
low cell density
- HCD
high cell density
- 5′ UTR
5′ untranslated region
- RBS
ribosome binding site
- nt
nucleotide
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/19975
References
- 1.Mutreja A, Kim DW, Thomson NR, Connor TR, Lee JH, Kariuki S, et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature. 2011;477:462–5. doi: 10.1038/nature10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hendriksen RS, Price LB, Schupp JM, Gillece JD, Kaas RS, Engelthaler DM, et al. Population genetics of Vibrio cholerae from Nepal in 2010: evidence on the origin of the Haitian outbreak. MBio. 2011;2:e00157–11. doi: 10.1128/mBio.00157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matson JS, Withey JH, DiRita VJ. Regulatory networks controlling Vibrio cholerae virulence gene expression. Infect Immun. 2007;75:5542–9. doi: 10.1128/IAI.01094-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis BM, Waldor MK. Filamentous phages linked to virulence of Vibrio cholerae. Curr Opin Microbiol. 2003;6:35–42. doi: 10.1016/S1369-5274(02)00005-X. [DOI] [PubMed] [Google Scholar]
- 5.Nelson EJ, Harris JB, Morris JG, Jr., Calderwood SB, Camilli A. Cholera transmission: the host, pathogen and bacteriophage dynamic. Nat Rev Microbiol. 2009;7:693–702. doi: 10.1038/nrmicro2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sledjeski DD, Whitman C, Zhang A. Hfq is necessary for regulation by the untranslated RNA DsrA. J Bacteriol. 2001;183:1997–2005. doi: 10.1128/JB.183.6.1997-2005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lease RA, Woodson SA. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J Mol Biol. 2004;344:1211–23. doi: 10.1016/j.jmb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. Positive regulation by small RNAs and the role of Hfq. Proc Natl Acad Sci U S A. 2010;107:9602–7. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Udekwu KI, Darfeuille F, Vogel J, Reimegård J, Holmqvist E, Wagner EG. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 2005;19:2355–66. doi: 10.1101/gad.354405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Updegrove T, Wilf N, Sun X, Wartell RM. Effect of Hfq on RprA-rpoS mRNA pairing: Hfq-RNA binding and the influence of the 5′ rpoS mRNA leader region. Biochemistry. 2008;47:11184–95. doi: 10.1021/bi800479p. [DOI] [PubMed] [Google Scholar]
- 11.Bardill JP, Zhao X, Hammer BK. The Vibrio cholerae quorum sensing response is mediated by Hfq-dependent sRNA/mRNA base pairing interactions. Mol Microbiol. 2011;80:1381–94. doi: 10.1111/j.1365-2958.2011.07655.x. [DOI] [PubMed] [Google Scholar]
- 12.Folichon M, Arluison V, Pellegrini O, Huntzinger E, Régnier P, Hajnsdorf E. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res. 2003;31:7302–10. doi: 10.1093/nar/gkg915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Massé E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–83. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsui HC, Leung HC, Winkler ME. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 15.Ding Y, Davis BM, Waldor MK. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol Microbiol. 2004;53:345–54. doi: 10.1111/j.1365-2958.2004.04142.x. [DOI] [PubMed] [Google Scholar]
- 16.Davis BM, Waldor MK. RNase E-dependent processing stabilizes MicX, a Vibrio cholerae sRNA. Mol Microbiol. 2007;65:373–85. doi: 10.1111/j.1365-2958.2007.05796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song T, Mika F, Lindmark B, Liu Z, Schild S, Bishop A, et al. A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol Microbiol. 2008;70:100–11. doi: 10.1111/j.1365-2958.2008.06392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu MY, Gui G, Wei B, Preston JF, 3rd, Oakford L, Yüksel U, et al. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J Biol Chem. 1997;272:17502–10. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- 19.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–89. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fröhlich KS, Vogel J. Activation of gene expression by small RNA. Curr Opin Microbiol. 2009;12:674–82. doi: 10.1016/j.mib.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 21.Vogel J, Wagner EG. Target identification of small noncoding RNAs in bacteria. Curr Opin Microbiol. 2007;10:262–70. doi: 10.1016/j.mib.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Huq A, Small EB, West PA, Huq MI, Rahman R, Colwell RR. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl Environ Microbiol. 1983;45:275–83. doi: 10.1128/aem.45.1.275-283.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watnick PI, Kolter R. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol Microbiol. 1999;34:586–95. doi: 10.1046/j.1365-2958.1999.01624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meibom KL, Li XB, Nielsen AT, Wu CY, Roseman S, Schoolnik GK. The Vibrio cholerae chitin utilization program. Proc Natl Acad Sci U S A. 2004;101:2524–9. doi: 10.1073/pnas.0308707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seper A, Fengler VH, Roier S, Wolinski H, Kohlwein SD, Bishop AL, et al. Extracellular nucleases and extracellular DNA play important roles in Vibrio cholerae biofilm formation. Mol Microbiol. 2011;82:1015–37. doi: 10.1111/j.1365-2958.2011.07867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15:167–93. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faruque SM, Biswas K, Udden SM, Ahmad QS, Sack DA, Nair GB, et al. Transmissibility of cholera: in vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc Natl Acad Sci U S A. 2006;103:6350–5. doi: 10.1073/pnas.0601277103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu J, Mekalanos JJ. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev Cell. 2003;5:647–56. doi: 10.1016/S1534-5807(03)00295-8. [DOI] [PubMed] [Google Scholar]
- 29.Tamayo R, Patimalla B, Camilli A. Growth in a biofilm induces a hyperinfectious phenotype in Vibrio cholerae. Infect Immun. 2010;78:3560–9. doi: 10.1128/IAI.00048-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yildiz FH, Schoolnik GK. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci U S A. 1999;96:4028–33. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kierek K, Watnick PI. Environmental determinants of Vibrio cholerae biofilm development. Appl Environ Microbiol. 2003;69:5079–88. doi: 10.1128/AEM.69.9.5079-5088.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. Chitin induces natural competence in Vibrio cholerae. Science. 2005;310:1824–7. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- 33.Blokesch M, Schoolnik GK. The extracellular nuclease Dns and its role in natural transformation of Vibrio cholerae. J Bacteriol. 2008;190:7232–40. doi: 10.1128/JB.00959-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sack DA, Sack RB, Nair GB, Siddique AK. Cholera. Lancet. 2004;363:223–33. doi: 10.1016/S0140-6736(03)15328-7. [DOI] [PubMed] [Google Scholar]
- 35.Sánchez J, Holmgren J. Virulence factors, pathogenesis and vaccine protection in cholera and ETEC diarrhea. Curr Opin Immunol. 2005;17:388–98. doi: 10.1016/j.coi.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 36.Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci U S A. 1987;84:2833–7. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herrington DA, Hall RH, Losonsky G, Mekalanos JJ, Taylor RK, Levine MM. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J Exp Med. 1988;168:1487–92. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–4. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 39.Brown RC, Taylor RK. Organization of tcp, acf, and toxT genes within a ToxT-dependent operon. Mol Microbiol. 1995;16:425–39. doi: 10.1111/j.1365-2958.1995.tb02408.x. [DOI] [PubMed] [Google Scholar]
- 40.Higgins DE, Nazareno E, DiRita VJ. The virulence gene activator ToxT from Vibrio cholerae is a member of the AraC family of transcriptional activators. J Bacteriol. 1992;174:6974–80. doi: 10.1128/jb.174.21.6974-6980.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Withey JH, DiRita VJ. The toxbox: specific DNA sequence requirements for activation of Vibrio cholerae virulence genes by ToxT. Mol Microbiol. 2006;59:1779–89. doi: 10.1111/j.1365-2958.2006.05053.x. [DOI] [PubMed] [Google Scholar]
- 42.DiRita VJ, Mekalanos JJ. Periplasmic interaction between two membrane regulatory proteins, ToxR and ToxS, results in signal transduction and transcriptional activation. Cell. 1991;64:29–37. doi: 10.1016/0092-8674(91)90206-E. [DOI] [PubMed] [Google Scholar]
- 43.Skorupski K, Taylor RK. Cyclic AMP and its receptor protein negatively regulate the coordinate expression of cholera toxin and toxin-coregulated pilus in Vibrio cholerae. Proc Natl Acad Sci U S A. 1997;94:265–70. doi: 10.1073/pnas.94.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skorupski K, Taylor RK. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol Microbiol. 1997;25:1003–9. doi: 10.1046/j.1365-2958.1997.5481909.x. [DOI] [PubMed] [Google Scholar]
- 45.Krukonis ES, Yu RR, Dirita VJ. The Vibrio cholerae ToxR/TcpP/ToxT virulence cascade: distinct roles for two membrane-localized transcriptional activators on a single promoter. Mol Microbiol. 2000;38:67–84. doi: 10.1046/j.1365-2958.2000.02111.x. [DOI] [PubMed] [Google Scholar]
- 46.Kovacikova G, Skorupski K. Overlapping binding sites for the virulence gene regulators AphA, AphB and cAMP-CRP at the Vibrio cholerae tcpPH promoter. Mol Microbiol. 2001;41:393–407. doi: 10.1046/j.1365-2958.2001.02518.x. [DOI] [PubMed] [Google Scholar]
- 47.Kovacikova G, Lin W, Skorupski K. Vibrio cholerae AphA uses a novel mechanism for virulence gene activation that involves interaction with the LysR-type regulator AphB at the tcpPH promoter. Mol Microbiol. 2004;53:129–42. doi: 10.1111/j.1365-2958.2004.04121.x. [DOI] [PubMed] [Google Scholar]
- 48.Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell. 2002;110:303–14. doi: 10.1016/S0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- 49.Zhu J, Miller MB, Vance RE, Dziejman M, Bassler BL, Mekalanos JJ. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci U S A. 2002;99:3129–34. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kovacikova G, Skorupski K. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol Microbiol. 2002;46:1135–47. doi: 10.1046/j.1365-2958.2002.03229.x. [DOI] [PubMed] [Google Scholar]
- 51.Ng WL, Perez LJ, Wei Y, Kraml C, Semmelhack MF, Bassler BL. Signal production and detection specificity in Vibrio CqsA/CqsS quorum-sensing systems. Mol Microbiol. 2011;79:1407–17. doi: 10.1111/j.1365-2958.2011.07548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller ST, Xavier KB, Campagna SR, Taga ME, Semmelhack MF, Bassler BL, et al. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol Cell. 2004;15:677–87. doi: 10.1016/j.molcel.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 53.Xavier KB, Bassler BL. Interference with AI-2-mediated bacterial cell-cell communication. Nature. 2005;437:750–3. doi: 10.1038/nature03960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lilley BN, Bassler BL. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol Microbiol. 2000;36:940–54. doi: 10.1046/j.1365-2958.2000.01913.x. [DOI] [PubMed] [Google Scholar]
- 55.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 56.Svenningsen SL, Tu KC, Bassler BL. Gene dosage compensation calibrates four regulatory RNAs to control Vibrio cholerae quorum sensing. EMBO J. 2009;28:429–39. doi: 10.1038/emboj.2008.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jobling MG, Holmes RK. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol Microbiol. 1997;26:1023–34. doi: 10.1046/j.1365-2958.1997.6402011.x. [DOI] [PubMed] [Google Scholar]
- 59.Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101–4. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- 60.Yildiz FH, Liu XS, Heydorn A, Schoolnik GK. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol Microbiol. 2004;53:497–515. doi: 10.1111/j.1365-2958.2004.04154.x. [DOI] [PubMed] [Google Scholar]
- 61.Svenningsen SL, Waters CM, Bassler BL. A negative feedback loop involving small RNAs accelerates Vibrio cholerae’s transition out of quorum-sensing mode. Genes Dev. 2008;22:226–38. doi: 10.1101/gad.1629908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Higgins DA, Pomianek ME, Kraml CM, Taylor RK, Semmelhack MF, Bassler BL. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature. 2007;450:883–6. doi: 10.1038/nature06284. [DOI] [PubMed] [Google Scholar]
- 63.Antonova ES, Hammer BK. Quorum-sensing autoinducer molecules produced by members of a multispecies biofilm promote horizontal gene transfer to Vibrio cholerae. FEMS Microbiol Lett. 2011;322:68–76. doi: 10.1111/j.1574-6968.2011.02328.x. [DOI] [PubMed] [Google Scholar]
- 64.Johnsborg O, Håvarstein LS. Regulation of natural genetic transformation and acquisition of transforming DNA in Streptococcus pneumoniae. FEMS Microbiol Rev. 2009;33:627–42. doi: 10.1111/j.1574-6976.2009.00167.x. [DOI] [PubMed] [Google Scholar]
- 65.Carroll IM, Khan AA, Ahmed N. Revisiting the pestilence of Helicobacter pylori: insights into geographical genomics and pathogen evolution. Infect Genet Evol. 2004;4:81–90. doi: 10.1016/j.meegid.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 66.Udden SM, Zahid MS, Biswas K, Ahmad QS, Cravioto A, Nair GB, et al. Acquisition of classical CTX prophage from Vibrio cholerae O141 by El Tor strains aided by lytic phages and chitin-induced competence. Proc Natl Acad Sci U S A. 2008;105:11951–6. doi: 10.1073/pnas.0805560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blokesch M, Schoolnik GK. Serogroup conversion of Vibrio cholerae in aquatic reservoirs. PLoS Pathog. 2007;3:e81. doi: 10.1371/journal.ppat.0030081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chun J, Grim CJ, Hasan NA, Lee JH, Choi SY, Haley BJ, et al. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci U S A. 2009;106:15442–7. doi: 10.1073/pnas.0907787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Waters CM, Lu W, Rabinowitz JD, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J Bacteriol. 2008;190:2527–36. doi: 10.1128/JB.01756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Srivastava D, Harris RC, Waters CM. Integration of cyclic di-GMP and quorum sensing in the control of vpsT and aphA in Vibrio cholerae. J Bacteriol. 2011;193:6331–41. doi: 10.1128/JB.05167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yildiz FH, Visick KL. Vibrio biofilms: so much the same yet so different. Trends Microbiol. 2009;17:109–18. doi: 10.1016/j.tim.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hammer BK, Bassler BL. Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae. Proc Natl Acad Sci U S A. 2007;104:11145–9. doi: 10.1073/pnas.0703860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Camilli A, Bassler BL. Bacterial small-molecule signaling pathways. Science. 2006;311:1113–6. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci U S A. 1998;95:12462–7. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Majdalani N, Hernandez D, Gottesman S. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol Microbiol. 2002;46:813–26. doi: 10.1046/j.1365-2958.2002.03203.x. [DOI] [PubMed] [Google Scholar]
- 76.Lin W, Kovacikova G, Skorupski K. The quorum sensing regulator HapR downregulates the expression of the virulence gene transcription factor AphA in Vibrio cholerae by antagonizing Lrp- and VpsR-mediated activation. Mol Microbiol. 2007;64:953–67. doi: 10.1111/j.1365-2958.2007.05693.x. [DOI] [PubMed] [Google Scholar]
- 77.Rutherford ST, van Kessel JC, Shao Y, Bassler BL. AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev. 2011;25:397–408. doi: 10.1101/gad.2015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shao Y, Bassler BL. Quorum-sensing non-coding small RNAs use unique pairing regions to differentially control mRNA targets. Mol Microbiol. 2012;83:599–611. doi: 10.1111/j.1365-2958.2011.07959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lenz DH, Miller MB, Zhu J, Kulkarni RV, Bassler BL. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol Microbiol. 2005;58:1186–202. doi: 10.1111/j.1365-2958.2005.04902.x. [DOI] [PubMed] [Google Scholar]
- 80.Wong SM, Carroll PA, Rahme LG, Ausubel FM, Calderwood SB. Modulation of expression of the ToxR regulon in Vibrio cholerae by a member of the two-component family of response regulators. Infect Immun. 1998;66:5854–61. doi: 10.1128/iai.66.12.5854-5861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hrabak EM, Willis DK. The lemA gene required for pathogenicity of Pseudomonas syringae pv. syringae on bean is a member of a family of two-component regulators. J Bacteriol. 1992;174:3011–20. doi: 10.1128/jb.174.9.3011-3020.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Laville J, Voisard C, Keel C, Maurhofer M, Défago G, Haas D. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc Natl Acad Sci U S A. 1992;89:1562–6. doi: 10.1073/pnas.89.5.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weilbacher T, Suzuki K, Dubey AK, Wang X, Gudapaty S, Morozov I, et al. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol Microbiol. 2003;48:657–70. doi: 10.1046/j.1365-2958.2003.03459.x. [DOI] [PubMed] [Google Scholar]
- 84.Romeo T. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol Microbiol. 1998;29:1321–30. doi: 10.1046/j.1365-2958.1998.01021.x. [DOI] [PubMed] [Google Scholar]
- 85.Lenz DH, Bassler BL. The small nucleoid protein Fis is involved in Vibrio cholerae quorum sensing. Mol Microbiol. 2007;63:859–71. doi: 10.1111/j.1365-2958.2006.05545.x. [DOI] [PubMed] [Google Scholar]
- 86.Richard AL, Withey JH, Beyhan S, Yildiz F, DiRita VJ. The Vibrio cholerae virulence regulatory cascade controls glucose uptake through activation of TarA, a small regulatory RNA. Mol Microbiol. 2010;78:1171–81. doi: 10.1111/j.1365-2958.2010.07397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bradley ES, Bodi K, Ismail AM, Camilli A. A genome-wide approach to discovery of small RNAs involved in regulation of virulence in Vibrio cholerae. PLoS Pathog. 2011;7:e1002126. doi: 10.1371/journal.ppat.1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kirn TJ, Bose N, Taylor RK. Secretion of a soluble colonization factor by the TCP type 4 pilus biogenesis pathway in Vibrio cholerae. Mol Microbiol. 2003;49:81–92. doi: 10.1046/j.1365-2958.2003.03546.x. [DOI] [PubMed] [Google Scholar]
- 89.Vogel J, Papenfort K. Small non-coding RNAs and the bacterial outer membrane. Curr Opin Microbiol. 2006;9:605–11. doi: 10.1016/j.mib.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 90.Papenfort K, Pfeiffer V, Mika F, Lucchini S, Hinton JC, Vogel J. SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol Microbiol. 2006;62:1674–88. doi: 10.1111/j.1365-2958.2006.05524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gogol EB, Rhodius VA, Papenfort K, Vogel J, Gross CA. Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon. Proc Natl Acad Sci U S A. 2011;108:12875–80. doi: 10.1073/pnas.1109379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Udekwu KI, Wagner EG. Sigma E controls biogenesis of the antisense RNA MicA. Nucleic Acids Res. 2007;35:1279–88. doi: 10.1093/nar/gkl1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol. 2010;64:163–84. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Song T, Sabharwal D, Wai SN. VrrA mediates Hfq-dependent regulation of OmpT synthesis in Vibrio cholerae. J Mol Biol. 2010;400:682–8. doi: 10.1016/j.jmb.2010.05.061. [DOI] [PubMed] [Google Scholar]
- 95.Livny J, Fogel MA, Davis BM, Waldor MK. sRNAPredict: an integrative computational approach to identify sRNAs in bacterial genomes. Nucleic Acids Res. 2005;33:4096–105. doi: 10.1093/nar/gki715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu JM, Livny J, Lawrence MS, Kimball MD, Waldor MK, Camilli A. Experimental discovery of sRNAs in Vibrio cholerae by direct cloning, 5S/tRNA depletion and parallel sequencing. Nucleic Acids Res. 2009;37:e46. doi: 10.1093/nar/gkp080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mandlik A, Livny J, Robins WP, Ritchie JM, Mekalanos JJ, Waldor MK. RNA-Seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe. 2011;10:165–74. doi: 10.1016/j.chom.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Davis BM, Quinones M, Pratt J, Ding Y, Waldor MK. Characterization of the small untranslated RNA RyhB and its regulon in Vibrio cholerae. J Bacteriol. 2005;187:4005–14. doi: 10.1128/JB.187.12.4005-4014.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mustachio LM, Aksit S, Mistry RH, Scheffler R, Yamada A, Liu JM. The Vibrio cholerae mannitol transporter is regulated posttranscriptionally by the MtlS small regulatory RNA. J Bacteriol. 2012;194:598–606. doi: 10.1128/JB.06153-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yamamoto S, Izumiya H, Mitobe J, Morita M, Arakawa E, Ohnishi M, et al. Identification of a chitin-induced small RNA that regulates translation of the tfoX gene, encoding a positive regulator of natural competence in Vibrio cholerae. J Bacteriol. 2011;193:1953–65. doi: 10.1128/JB.01340-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Suckow G, Seitz P, Blokesch M. Quorum sensing contributes to natural transformation of Vibrio cholerae in a species-specific manner. J Bacteriol. 2011;193:4914–24. doi: 10.1128/JB.05396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]