SUMMARY
Non-coding RNAs form unique three-dimensional structures, which perform many biochemical and regulatory functions. To understand how RNAs fold uniquely despite a small number of tertiary interaction motifs, we mutated the major tertiary interactions in a group I ribozyme. The resulting perturbations to the folding energy landscape were measured using SAXS, ribozyme activity, hydroxyl radical footprinting and native PAGE. Double and triple mutant cycles show that most tertiary interactions have a small effect on native state stability. Instead, formation of core and peripheral structural motifs are cooperatively linked in near-native folding intermediates, and this cooperativity depends on the native topology of the ribozyme. The emergence of a cooperative interaction network at an early stage of folding suppresses non-native structures and makes the search for the native state more efficient. We suggest that cooperativity in non-coding RNAs arose from natural selection of architectures that promote a unique fold despite a rugged energy landscape.
INTRODUCTION
Many non-coding RNAs fold into unique three-dimensional structures, which allow them to catalyze biochemical reactions or act as regulators of gene expression. Recent genomic sequence analyses indicate that non-coding RNAs with complex structures are widespread in nature (Weinberg et al., 2009; Westhof, 2010) and may encode a greater variety of biochemical functions than currently known. Structural and computational studies of ribozymes and other non-coding RNAs have revealed tertiary motifs and organizational principles (Holbrook, 2008; Laing and Schlick, 2011; Lescoute and Westhof, 2006), but how these sequence motifs encode the self-assembly of unique three-dimensional RNA structures remains an open question with implications for the design and evolution of new RNA functions.
Unlike globular proteins, non-coding RNAs lack extensive hydrophobic cores through which distant side chains interact (Creighton, 1990; Thirumalai and Hyeon, 2005). Instead, RNA double helices are pinned together by a smaller number of tertiary interaction motifs that encode the native structure (Leontis et al., 2006). The orientation or topology of helices in the core of the RNA is important for biological function and invariably conserved within RNA families (Michel and Westhof, 1990; Montange and Batey, 2008). By contrast, the stability of the folded RNA often depends on peripheral tertiary structure modules that vary during molecular evolution (Baird et al., 2005; Fujita et al., 2009; Lehnert et al., 1996). Owing to their stable secondary structure and modular construction, non-coding RNAs have the potential to form incorrect structures that compete with the biologically active structure (Thirumalai and Hyeon, 2005). Despite these “rough” energy landscapes for folding, many RNAs nevertheless achieve remarkable folding specificity (Fang et al., 1999; Rangan et al., 2003; Su et al., 2003).
Here, we use energy perturbation theory and double mutant cycles to show that tertiary interactions are strongly coupled in the folding intermediates of a natural group I ribozyme. This cooperativity depends strongly on the native architecture of the ribozyme. Small perturbations to the ribozyme structure allowed the entire folding landscape to be determined. By separately monitoring the stability of the native state and near-native folding intermediates, our results unexpectedly reveal that thermodynamic cooperativity emerges early in the RNA folding process, suppressing incorrect structures and simplifying the search for the native state. We discuss these findings and previous observations to suggest that folding cooperativity arose during evolution by the natural selection of RNA topologies conducive to specific folding.
RESULTS
Rationally designed mutations disrupt tertiary interactions motifs
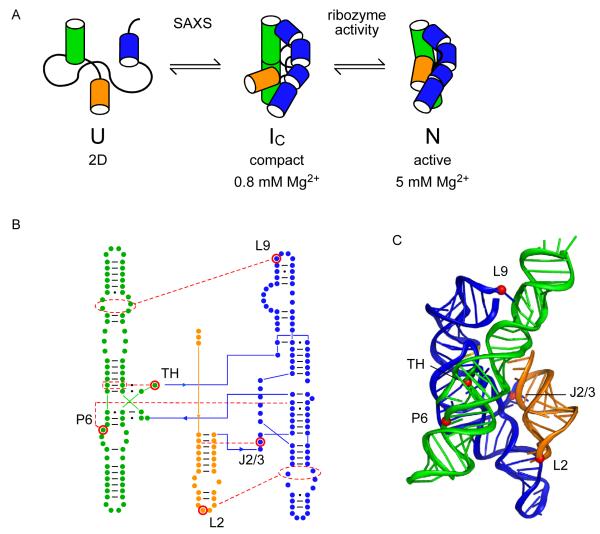
For our analysis, we used the 195 nt. Azoarcus sp. bacterial group I ribozyme because it has a stable tertiary structure (Tanner and Cech, 1996) and well-characterized equilibrium folding pathway (Fig. 1A) (Rangan et al., 2003). In 0.2-0.4 mM Mg2+, assembly of the core helices produces native-like intermediates (IC) that are almost as compact as the native ribozyme (Perez-Salas et al., 2004). The IC intermediates are stabilized by tertiary interactions (Chauhan et al., 2005; Chauhan et al., 2009), but the tertiary contacts between double helical domains are not fully stable, as judged by hydroxyl radical footprinting of the RNA backbone (Rangan et al., 2003). Local reorganization of the tertiary structure in 2-4 mM Mg2+ yields the fully folded and catalytically active native state (N) (Rangan and Woodson, 2003). These folding transitions are well separated and can be distinguished by different experimental assays.
Figure 1. Cooperative folding of the Azoarcus group I ribozyme.
(A) Compact, native-like intermediates (IC) form in low Mg2+ (Perez-Salas et al., 2004; Rangan et al., 2003) and are detected by SAXS or native PAGE. Formation of the native structure (N) is reported by ribozyme activity and the solvent accessibility of the RNA backbone. See also Figure S1. (B,C) Tertiary interaction motifs indicated by red dots were perturbed by single base substitutions: loop L2, A25U; joining region J2/3, A39U; paired region P6, A97U; triple helix TH, G125A; loop L9, A190U (see Table S1). J8/7, cyan ribbon. Cooperative interactions indicated by red (IC) or blue (N) arrowheads (positive, pointed; negative, flat). Thickness indicates relative strength. (C) Ribbon drawn with PyMOL; 1u6b (Adams et al., 2004b).
We mutated five tertiary interaction motifs singly and in combination, avoiding active site residues and mutations likely to change the secondary structure of the RNA. Single base substitutions were designed to disrupt the joining region J2/3, the central triple helix (TH), and paired region P6 within the ribozyme core, and two peripheral tetraloops (TL) L2 and L9 that dock with helical receptors (TR) in P8 and P5, respectively (Fig. 1B-C; see Supplementary Table S1 for details). The single base substitutions were combined in six double mutants, four triple mutants and one quadruple mutant to measure the thermodynamic cooperativity between tertiary interaction motifs in different regions of the RNA.
Mutant ribozymes are active and fold correctly
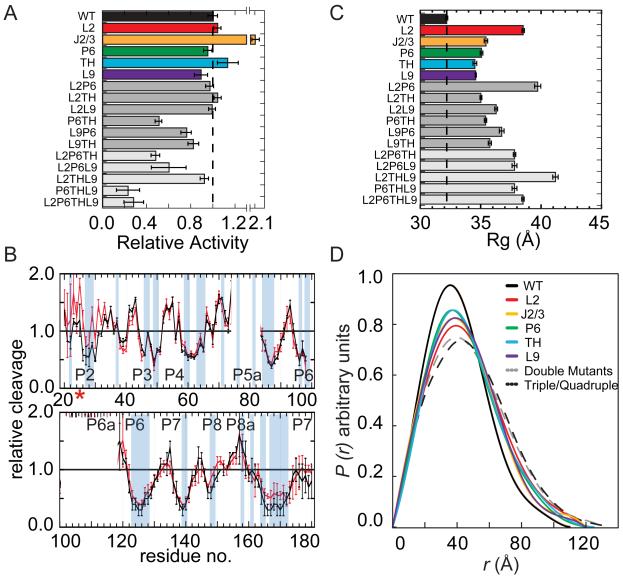
All of the mutant ribozymes, including the quadruple mutant, were active in a single-turnover oligonucleotide substrate cleavage assay (Herschlag and Cech, 1990), with single mutants attaining 80 – 200% of the wild type (WT) activity in 15 mM MgCl2 (Fig. 2A; see Experimental Procedures and Fig. S1 for details). To determine whether the mutants have the same structure as the WT RNA, we probed the solvent accessibility of the RNA backbone in several mutant ribozymes using hydroxyl radical footprinting under native conditions (Fig. 2B and Fig. S2). A similar pattern of backbone solvent accessibility in the WT and mutant RNAs indicated that they form similar native-state ensembles. However, residues near the mutation site and in the catalytic core (J8/7 residues 167-172) were slightly more exposed in 15 mM Mg2+ than the corresponding regions in the wild type ribozyme, consistent with increased unfolding due to the loss of a tertiary contact.
Figure 2. Mutant and wild type ribozymes fold similarly.
(A) Mutations do not abolish the catalytic activity of the Azoarcus ribozyme in 15 mM MgCl2 Product formed in the first turnover (fproduct) reports the fraction native RNA. Dashed line, WT activity. (B) Hydroxyl radical footprinting of wild type (black) and mutant L2 (A25U) ribozymes (red) in 15 mM Mg2+. Blue bars, buried riboses predicted by the crystal structure (Adams et al., 2004b). Residues 72-83 and 100-118 were not quantified due to gel compressions. See Figure S2 for further data. (C) Mutant ribozymes are less compact than the WT ribozyme. Radii of gyration (Rg) from SAXS in ≥ 2.6 mM MgCl2 at 37 °C; dashed line shows WT Rg = 32 Å. (D) Pair distribution functions P(r) normalized to equal area and colored as in (C).
The hydroxyl radical footprinting results indicate that although the mutant ribozymes form an active structure, they are more likely than the wild type RNA to populate extended or locally unfolded conformations. To evaluate this possibility, we obtained information about the average compactness and shape of each RNA under native conditions from small angle X-ray scattering (SAXS) experiments (Fig. S1). All of the mutant ribozymes formed globular structures above 3 mM MgCl2 similar to that of the WT RNA. However, the mutants were less compact than the WT RNA, as revealed by their larger radii of gyration (Rg) (Fig. 2C) and longer maximum distances (Rmax) in the pair distribution function P(r) (Fig. 2D). A triple mutant lacking both tetraloop-tetraloop receptor (TL-TR) interactions and the central triple helix (L2THL9) was the least tightly folded (gray; Fig. 2C, D), but was still catalytically active (Fig. 2A). Thus, the mutant ribozymes form compact, functional structures, although they are more likely than the WT ribozyme to explore extended conformations under native conditions.
Free energies of Mg2+-dependent folding transitions
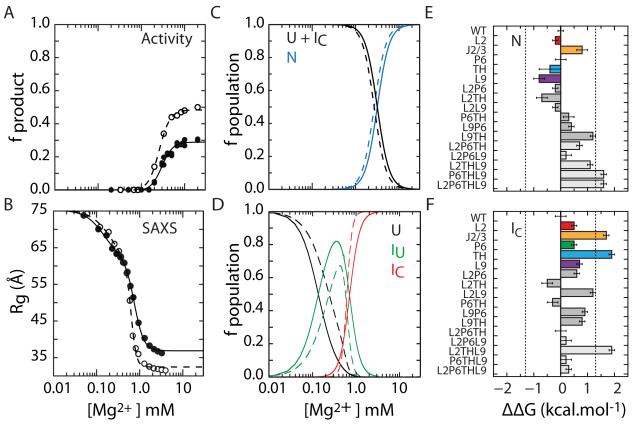
The contribution of each tertiary interaction to the folding free energy of the Azoarcus ribozyme was measured by titrating the WT and mutant ribozymes with Mg2+ (Fig. 3). In general, stable RNA tertiary structures require less Mg2+ to form, and the steepness of the folding transition correlates with the free energy gap between the unfolded and folded states (see Methods) (Fang et al., 1999; Pan et al., 1999). The tolerance of the Azoarcus ribozyme toward destabilizing mutations validated our energy perturbation approach, and allowed us to measure the complete free energy landscape for folding and coupling energies without having to assume that mutations ablate a tertiary interaction motif.
Figure 3. Mutations perturb folding free energies of IC and N.
(A) Single-turnover activity assay (fproduct) of IC to N folding transition (eq. 1). The maximum product is 50% due to slow product release and religation (Tanner and Cech, 1996). (B) SAXS measurement of U to IC folding transition under the conditions in (A). The change in R 2g was fit to a three-state model (eq. 2). Filled symbols, L9P6 mutant; open symbols, WT. (C,D) Populations calculated from data in A and B, respectively. U, unfolded state; IC, compact intermediate; N, native state; IU, extended non-native intermediate. (E,F) Differences in folding free energies ΔΔG = ΔGmut – ΔG evaluated at 5 mM MgCl2 (N) and 0.8 mM MgCl2 (IC). These Mg2+ concentrations (Cref) corresponded to 90% saturation of the respective WT folding transitions. Error bars depict standard deviations calculated from 10,000 resamplings of fit residuals. Dashed lines at ±1.3 kcal mol−1 indicate the magnitude of WT ΔG. See Figure S3 and Table S2 for details.
The free energy change associated with forming the native state ( ΔGI→N ) was obtained from the increase in pre-steady-state ribozyme activity with Mg2+ (Fig. 3A, Fig. S3). The free energy change between U and IC ( ΔGU→I ) was obtained from the decrease in Rg2 measured by SAXS, under the same conditions as the activity assays (Fig. 3B, Fig. S3). Using previously validated folding models (Moghaddam et al., 2009; Rangan et al., 2003) to extract populations of U, IC and N (Fig. 3C,D), free energies for each folding transition were evaluated at 0.8 mM Mg2+ for IC, 5 mM Mg2+ for N (Fang et al., 1999). These reference conditions correspond to 90% saturation of each transition, and were chosen to be near the midpoint of each transition where the assumptions underlying the free energy calculations are most valid (Leipply and Draper, 2010). Full details of the analysis are given in Experimental Procedures, Fig. S3 and Table S2.
Loss of tertiary interactions perturbs both steps of folding
All of the single mutations destabilized both the IC intermediate and the native state N, demonstrating the importance of each tertiary interaction to folding stability (colored bars; Fig. 3E, F). Surprisingly, IC was perturbed more than N, amounting to 0.5 – 2 kcal/mol or 35-150% of the total free energy change of –1.3 kcal/mol for the WT U → IC transition. Among the single base substitutions, only A39U in the J2/3 linker affected N more than IC, narrowing the energy gap for the second folding step (ΔΔGN > 0, Fig. 3E). The other single mutations raised the free energy of IC more than N, and ΔΔGN ≤0.
The large effect of each mutation on the IC state generalized previous conclusions that native tertiary contacts are present in compact folding intermediates of the Azoarcus ribozyme (Chauhan et al., 2005) and other other ribozymes (Bokinsky et al., 2003; Buchmueller and Weeks, 2003; Das et al., 2003; Fang et al., 1999). As IC has considerable native character, the free energy surfaces for IC and N are similar (Fig. 4 and Fig. S4). In other words, the degree to which a mutation perturbs IC predicts how much that mutation destabilizes the native state. In the WT and all of the mutants, the energy gap between U and IC is much larger than the gap between IC and N. This observation and the large effects of the mutations on IC suggests that formation of the compact intermediate is the dominant step of tertiary folding, rather than formation of the native state as previously assumed.
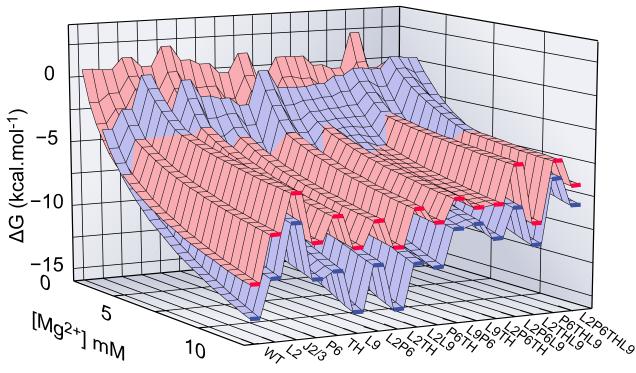
Figure 4. Free energy landscape for folding.
Energetic perturbations in the Azoarcus ribozyme due to mutations in IC (red) and N (blue) are shown versus Mg2+ concentration (back to front). Mutations that destabilize N more than IC, e.g. J2/3 and most of triplets, shift the crossover of surfaces to higher Mg2+ compared to WT. Red and blue surfaces are similar because IC is similar in structure to N. Single mutations located in helical junctions (J2/3 and TH) are particularly destabilizing. See also Figure S4.
Free energy landscape of folding
The free energy perturbations arising from the loss of each tertiary interaction motif show how the folding landscape is defined by the architecture of the ribozyme (Fig. 1B and Fig. 4). Mutations in joining segments J2/3 and J6/7 (triple helix) were most destabilizing, reflecting their role in aligning double helices in the ribozyme core (Adams et al., 2004b). J2/3 helps dock the P1 substrate helix in the active site (Strauss-Soukup and Strobel, 2000), while the triple helix between J6/7 and P4 aligns the P4-P6 and P3-P9 domains (Doudna and Cech, 1995; Zarrinkar and Williamson, 1996) and is conserved in all group I ribozymes (Michel and Westhof, 1990). Mutations that weaken these interactions hinder the ribozyme from achieving a compact fold.
Second in importance were the peripheral TL-TR interactions (L2 and L9; Fig. 4A) previously identified as stability elements (Ikawa et al., 2000; Jaeger et al., 1994). The reduced steepness of the folding transitions (Table S2, Fig. S6) as well as a larger Rg in 1 mM Mg2+ indicates that a higher fraction of ribozymes containing L2 and L9 mutations form non-native structures that are more extended than native-like intermediates.
Lastly, the A-minor interaction between A97 in P6 and the P3 double helix (Rangan et al., 2003) contributed least to the stability of IC, but had a comparable effect on the native state as TL-TR interactions. The perturbation (ΔΔGN) from the single mutations were consistent with ~0.5 to 1 kcal/mol associated with individual tertiary interactions in other RNA molecules (Battle and Doudna, 2002; Fiore et al., 2009; Silverman and Cech, 1999), when the data were analyzed as in this study.
Cooperativity of tertiary interaction motifs
In addition to their individual contributions to stability, tertiary interactions in different regions of the RNA may cooperatively stabilize the native structure when both interactions form simultaneously. When formation of one tertiary interaction favors formation of another, the two are thermodynamically coupled (or linked), and stabilize the structure by more than the sum of individual interactions. Conversely, if a tertiary interaction stabilizes a local structure that is incompatible with another interaction, they are antagonistic and reduce the overall contribution to the stability.
We used the well-established framework of double mutant cycles (di Cera, 1998) to measure thermodynamic coupling (ΔΔGlink) between tertiary interactions in different regions of the RNA as a function of Mg2+ concentration (Fig. 5). In this method, ΔΔGlink between each pair of tertiary interactions is calculated by comparing the perturbation from introducing mutation i in the WT or in a background containing a mutation at site j (see thermodynamic box in Fig. S5-A) and measuring the folding free energy of single and double mutants (see Experimental Procedures). We used activity assays to measure linkage in the native state, and SAXS to detect the formation of compact (native-like) folding intermediates as described above.
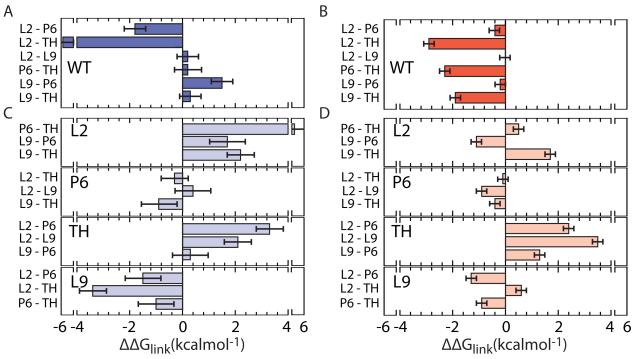
Figure 5. Thermodynamic cooperativity in folding intermediates.
Linking free energies between tertiary interactions, ΔΔGlink, calculated from double mutant cycles (see Extended Experimental Procedures and Figure S5A for details). ΔΔGlink < 0 indicates cooperativity between tertiary interactions. (A) Pairwise linking energies in WT RNA in native state (U to N; 5 mM Mg2+). (B) As in (A), for the intermediate state (U to IC; 0.8 mM Mg2+). (C,D) Pairwise linking energies in the background of a third mutation (large letters). See Extended Experimental Procedures for details.
Linkage analysis for all combinations of the L2, TH, P6, and L9 mutations revealed strong interactions between the central triple helix (TH) and the other tertiary interactions (negative ΔGlink for pairs of contacts on left of Fig. 5A). This strong cooperativity explains why single mutations were nearly as destabilizing as double mutations (Fig. 4): the first mutation destroys the favorable interaction between sites, in addition to any local effect of the perturbed contact. Unexpectedly, however, linkage with TH was strongest in the U to IC transition (-2 to −3 kcal/mol) (combinations with TH; Fig. 5B). Thus, tertiary interactions not only stabilize the “I” state, but also become energetically coupled with each other during assembly of the core helices and before the native state is fully achieved.
These results are easily rationalized by the architecture of the ribozyme, and show further how this architecture encodes cooperativity between tertiary interaction motifs. Although the A-minor interaction in P6 and the two peripheral TL-TR motifs (L2 and L9) are each coupled to the triple helix, they form independently of each other in IC (ΔΔGlink ~ 0) (Fig. 5B). Each TL-TR motif bridges a different domain interface, and can be expected to form independently. By contrast, docking either tetraloop with its receptor depends greatly on the orientation of the P4-P6 and P3-P9 domains established by the triple helix (Adams et al., 2004a; Michel and Westhof, 1990), which is in turn stabilized by the TL-TR interactions (Chauhan and Woodson, 2008). Therefore, the peripheral and core tertiary interactions mutually reinforce each other, producing the thermodynamic cooperativity that we observe.
This cooperativity network reorganizes in the second folding step leading to the native state. First, the L2 tetraloop interaction becomes more strongly coupled to the triple helix (-6 kcal/mol) and the P6 A-minor interactions (Fig. 5A). This likely reflects the need to dock the P1-P2 helices with the ribozyme core for catalysis (Pyle and Cech, 1991). Second, the L9 TR-TL interaction is decoupled from other tertiary contacts and antagonizes formation of the P6 A-minor contact in the native state (Fig. 5A), consistent with a structural rearrangement in P9/P9.0 during the transition from the intermediates to the native state (Bevilacqua et al., 1996; Chauhan et al., 2009). Antagonism between L9 and P6 interactions intensifies with higher Mg2+ (Fig S5B), as the native state becomes more stable, causing the energetic cost of structural reorganizations to rise. Thus, peripheral tertiary contacts are cooperatively linked with assembly of the core helices during the first stage of folding, but these linkages change in importance as the ribozyme progresses toward its native structure.
Cooperativity requires a native architecture
As the pattern of thermodynamic coupling reflects the ribozyme architecture, we asked whether assembly of a native-like structure is needed to link tertiary interactions in different parts of the RNA. When triple mutants were used to measure the linking free energies between pairs of interactions in the absence of the triple helix (Fig 5C,D), cooperativity was lost, corresponding to a rise in ΔΔGlink of 4.4 ± 0.3 kcal/mol in IC (0.8 mM Mg2+) and up to 5.0 ± 0.5 kcal/mol in N (5 mM Mg2+). Thus, the loss of one tertiary interaction decouples the others, underlining the importance of native topology for concerted folding.
Destabilization of the triple helix (TH) had the greatest effect on cooperative relationships with the other interactions in both IC and N (Fig. 5C,D, third panel), consistent with its role in coordinating the formation of tertiary interactions in different regions of the structure. Similarly, destabilization of the L2 TL-TR interactions weakened coupling between other pairs of interactions in the native state (Fig. 5C, first panel). The apparent gain in cooperativity when L9 is mutated reflects the energetic penalty imposed by reorganization of P6 and L9 late in the folding pathway of the WT ribozyme (Fig. 5C, fourth panel). It is possible that the topological stress stored in the native RNA by this rearrangement contributes to catalysis.
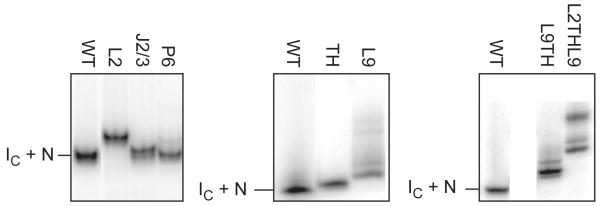
Mutants populate non-native intermediates
The decoupling of tertiary interactions in the triple mutant could be explained if non-native structures are populated when core tertiary interactions are weakened, as previously suggested by time-resolved footprinting studies (Chauhan and Woodson, 2008). Our SAXS data also suggested this is the case, as extended (non-native) intermediates (IU) are more heavily populated by the mutants than the WT ribozyme (Fig. S3 and Table S2). As a further test, native-like and non-native RNAs were separated using non-denaturing polyacrylamide gel electrophoresis (PAGE). As expected, mutants which appeared less compact by SAXS also had a higher probability of forming alternative, non-native structures (Fig. 6). Qualitatively similar free energies for the U to IC transition were obtained using native PAGE, indicating that non-native intermediates (IU) did not noticeably contribute to the stability of IC measured by SAXS (data not shown). From these results, we conclude that perturbations to tertiary interactions allow more frequent excursions into non-native folds below 2 mM Mg2+, at the expense of native-like I states.
Figure 6. Loss of tertiary interactions increases misfolding.
Native 8% polyacrylamide gel electrophoresis (PAGE) resolves native-like (IC+N) and non-native conformers (lower mobility). WT and mutant ribozymes were refolded in 15 mM MgCl2 before loading. Gels were run at 4 °C in 3 mM MgCl2 to stabilize intermediates and prevent unfolding during electrophoresis (Rangan et al., 2003). Incubation at 37 °C or 50 °C before electrophoresis had no effect on the population of alternative structures, confirming they are equilibrium states.
DISCUSSION
Our results show that the energetic coupling between RNA tertiary interactions (-1.8 to −5.5 kcal/mol) is larger than the energies associated with individual tertiary contacts (~ −0.5-1 kcal/mol), and larger than the cooperativity typically observed between residues in globular proteins (−0.5 to −2 kcal/mol) (Wells, 1990). Cooperativity in RNA folding unexpectedly emerges during the initial assembly of the core helices (IC), and not in the formation of the native state. Although the tertiary interaction sites we probed are 25-35 Å apart in the folded ribozyme, the stiffness of RNA double helices can transmit local perturbations to helix packing over long distances. This may explain why a small number of tertiary interaction motifs is sufficient to guide helix assembly and specific a unique 3D fold.
The coupling free energies between pairs of tertiary interactions can be expressed as the fraction of molecules in the folded ensemble that have formed both interactions coincidently, which we call linking frequencies or probabilities (see Extended Experimental Procedures). As folding is not perfectly cooperative, these probabilities are less than 100%. However, the magnitude of thermodynamic linkage in the Azoarcus ribozyme (ΔΔGlink ~ −1.8 to −5.5 kcal/mol) implies that 80% to 99% of folded molecules contain both interactions at the transition midpoint, compared to 50% for independent interactions (ΔΔGlink ~ 0). These values are similar to ΔGcoop obtained from ablating two tertiary motifs in the Tetrahymena ribozyme P4-P6 RNA (Sattin et al., 2008), but much greater than the additive stabilization previously obtained for ribose zipper motifs (Silverman and Cech, 1999). Earlier studies did not resolve the I and N states nor the Mg2+ dependence of cooperativity.
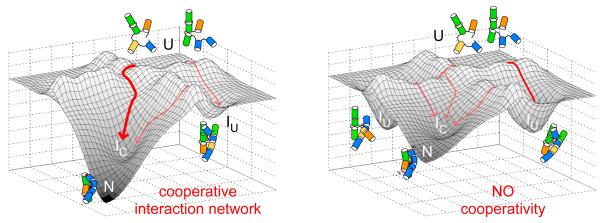
Coupling between tertiary contacts in different regions of the RNA suppresses non-native structures (Fig. 6), and preferentially favors the active RNA structure by widening the free energy gap between the native state and the next most stable structure on the free energy landscape (Brion et al., 1999; Fang et al., 2001) (Fig. 7). The free energy gap between N and other stable intermediates was previously used to explain the differences between RNase P from thermostable and mesostable bacteria (Baird et al., 2006), and the in vitro selection of a thermostable variant of the Tetrahymena ribozyme (Wan and Russell, 2011). Non-native folding intermediates in RNA typically have extended conformations, are stabilized by additional base pairing and base stacking interactions which compensate lost tertiary contacts (Baird et al., 2010; Szewczak and Cech, 1997; Walter et al., 1999).
Figure 7. Coupling core and peripheral tertiary interactions ensures a unique fold.
A cooperative network of tertiary interaction motifs in the WT ribozyme (left) favors proper helix assembly and lower the free energies of the native-like IC intermediate and the active native state (N), improving fitness. Mutations that perturb one or more tertiary interaction motifs (right) disrupt this cooperative network, increasing populations of extended, non-native intermediates (IU). Natural selection of the native state is expected to favor RNA architectures that link peripheral and core tertiary interactions.
Since thermodynamic cooperativity emerges early in RNA folding, the collapse transition becomes more specific, simplifying the search for the native fold. Our results provide a foundation for understanding how tertiary interactions in RNA are linked through its helical architecture, driving cooperative self-assembly even in the face of a rugged folding landscape and individually stable domains. As native-like intermediates are observed in many large RNAs (Bokinsky et al., 2003; Buchmueller and Weeks, 2003; Chauhan et al., 2005; Das et al., 2003; Fang et al., 1999), we posit that early emergence of cooperativity is a general property of RNA folding.
In the Azoarcus ribozyme, peripheral tertiary interactions are coupled with each other via the central triple helix, underscoring the importance of core topology in self-assembly. The triple helix is one of the most conserved elements of group I ribozymes (Cannone et al., 2002; Michel and Westhof, 1990), is necessary for correct folding (Zarrinkar and Williamson, 1996), and orients adjacent double helices during folding along with other single-stranded “junctions” such as J2/3 (Adams et al., 2004a). Here, we observe that peripheral tertiary interaction motifs produce cooperative folding when coupled to assembly of the core architecture. Our observations support the idea that junctions greatly constrain the available conformational space by restricting helix orientations (Bailor et al., 2010; Butcher and Pyle, 2011).
Interestingly, triple helices are ubiquitous in RNA structures, including group II introns (Toor et al., 2008), riboswitches (Gilbert et al., 2008), and telomerase RNA (Theimer et al., 2005). The frequent occurrence of such motifs in conserved regions of structured RNAs may reflect their ability to correctly orient double helices early in folding. A conserved triple helix in the catalytic core of a group II intron not only stabilizes the native fold by merging two functionally important domains, but directly contributes to catalysis (Toor et al., 2008). As the core topology is often stabilized by peripheral tertiary interactions, we believe the relationship between native topology and folding cooperativity is universal in structured RNAs.
Random RNA sequences lack the folding specificity of evolved sequences with same size and base composition and generally form simple, extended structures (Schultes et al., 2005). This observation supports the idea that highly cooperative (specific) folding is a product of natural selection, and not a general property of RNA chains. Likewise, comparing natural and de novo designed small polypeptides, Watters et al. (Watters et al., 2007) suggested that the highly cooperative folding and smooth energy landscapes are products of evolution. Moreover, single molecule studies indicated that native topology is necessary for coupling between domains in a large multi-domain protein (Shank et al., 2010). Here, by elucidating the connection between folding specificity and early-stage cooperativity in a large natural ribozyme, we complete this argument by suggesting that cooperativity in RNA arose from natural selection of topologies that efficiently couple core and peripheral tertiary interactions and thus promote specific folding (Fig. 7).
Despite partial loss of folding specificity in ribozymes with weakened tertiary interactions (see Fig. 2), a fraction of RNAs always folds to the active structure. This observation underscores the robustness of RNA structure, and hence, its capacity for further evolution. Finally, our results suggest that the prediction and design of RNA structures can be improved by considering the potential for energetic cooperativity. As individual tertiary interactions provide only small contributions to overall stability, they may stabilize native and non-native structures equally well, thereby reducing folding specificity (Treiber et al., 1998; Wan and Russell, 2011). By contrast, concerted changes in more than one helical domain are necessary to increase the folding cooperativity and specificity (Baird et al., 2006). Screening simulated or real sequence libraries for early folding cooperativity may increase the efficiency of rational design algorithms or in vitro evolution experiments.
EXPERIMENTAL PROCEDURES
RNA preparation and site-directed mutagenesis
The L-9 Azoarcus ribozyme (195 nucleotides) was prepared by run-off transcription of pAz-IVS DNA digested with EarI and purified by PAGE as previously described (Kilburn et al., 2010; Rangan et al., 2003). Purified RNA was resuspended in TE (10 mM Tris-HCl, pH 7.5, 0.1 mM EDTA) or CE (10 mM sodium cacodylate, pH 7.5, 0.1 mM EDTA) and incubated 5 minutes at 50°C before use. Mutations that disrupt conserved tertiary interactions in the ribozyme were introduced into pAZ-IVS by QuikChange (Stratagene) or by PCR amplification of transcription templates. Mutation sites were selected based on crystal structures (Adams et al., 2004b), Nucleotide Analogue Interference Mapping (NAIM) (Strauss-Soukup and Strobel, 2000) and other biochemical studies (Ikawa et al., 2000; Jaeger et al., 1994; Tanner and Cech, 1997). See Table S1 for details.
Ribozyme activity assays
Pre-steady-state cleavage of 5′-32P-labeled rCAUAUCGCC substrate was performed as described in 20 mM Tris-HCl containing 0 – 30 mM MgCl2 using L-3 ribozyme(Chauhan et al., 2005; Herschlag and Cech, 1990; Kuo et al., 1999) except that folding and substrate cleavage were done at 37°C (Fig. S1B) and the end of the burst phase (first turnover) was determined to be 20 s (Fig. S1C). To verify that the RNA was fully equilibrated, another set of assays were done in which the ribozyme was folded 10 – 15 minutes at 50°C, then cooled to 37°C for at least 15 minutes before addition of the substrate oligonucleotide. Folding transitions under these two conditions were identical for all tested samples within experimental uncertainty (Fig. S1E).
Equilibrium hydroxyl radical footprinting
Fe(II)-EDTA-dependent hydroxyl radical cleavage reactions were carried out as previously described (Chauhan and Woodson, 2008), except that 5′-32P-labeled RNA was folded 30-40 min at 37 °C and incubated 5 min on ice, before the addition of Fenton reagents. The intensities of the cleavage products were corrected and quantified as described previously (Chauhan et al., 2009). To ensure that protections indicate the equilibrium population distribution, experiments were repeated with ribozymes folded 20 minutes in 50 °C and cooled down to 37 °C for at least 20 minutes before footprinting. The protection profiles were identical.
Small Angle X-ray Scattering (SAXS) measurements
SAXS measurements were conducted at BioCAT (ID18; Advanced Photon Source, Argonne National Laboratory) as described previously (Chauhan et al., 2005; Kilburn et al., 2010). RNA solutions (0.4 mg/mL in 20 mM Tris-HCl) with 0 −10 mM MgCl2 were equilibrated at 37 °C before each measurement (four 2-s exposures), over Q = 0.008 to 0.2 Å−1 (Fig. S1A). Data were corrected for background scattering of the buffer after radial averaging. Distance distribution functions, P(r), were obtained by indirect Fourier transform of scattering intensity data using GNOM (SVERGUN, 1992). The best estimate of rmax was obtained by fitting the data without constraining P(rmax). Next, rmax was held constant while varying the regularization parameter (α) to obtain the best least-squares fit to the data that also maximized smoothness constraints. Values of Rg obtained from the indirect transform method were more precise and less sensitive to experimental noise than those obtained from the Guinier method. Errors in Rg were less than ± 1 Å in low [Mg2+] and less than ± 0.5 Å in high [Mg2+] in the titration curves.
Equilibrium folding transitions
The amount of product formed after 20 s at each Mg2+ concentration relative to the maximum activity in 15 mM MgCl2 was taken to be proportional to the amount of native ribozyme (Herschlag and Cech, 1990), and was fit to a two-state cooperative transition (Fig. S3):
| (1) |
in which C is the molar Mg2+ concentration, Cm is the midpoint and n is the steepness of the transition near the midpoint.
Data from two or more independent trials (25 to 80 data points) were globally fit to obtain the midpoints and slopes of the transition. To determine the confidence intervals, residuals were randomly sampled with replacement 10,000 times and redistributed over the data points. The resulting datasets with added noise were fitted with the same model. Distributions of fit parameters were unimodal and non-skewed. After removing outliers (Hogg and Tanis, 2006), the median and standard deviation of each parameter were recorded.
The change in Rg2 of the wild type and mutant ribozymes were fit with a three state model as described previously (Moghaddam et al., 2009), in which the midpoint and slope of each transition defines the statistical weight of each conformational state (Fig. S3):
| (2) |
Rg(U) and Rg(IC) are the experimentally determined values for U and N, respectively, and Rg(IU) of the extended intermediate is obtained from the fits. Fitted parameter values were used to obtain apparent two-state Cm and n for IC (Table S2). Confidence intervals for fitted parameters were obtained as explained above. Medians and standard deviations are reported.
Calculation of folding free energy
The steepness of the Mg2+-dependent folding transition, n, is directly proportional to the change in folding free energy with lnC. Fractional populations of IC and N were calculated from fits to experimental folding transitions (Moghaddam et al., 2009); the average parameters and two state approximation were used to determine the free energy changes associated with the U to I and I 2+ C C to N folding steps at a common [Mg ], Cref, using ΔG(C) = –nRT ln(Cref /Cm) (Fang et al., 1999). Values of Cref were 0.8 mM for U to IC (SAXS) and 5 mM for IC to N (activity), and were chosen to represent the same point (90% saturation) along U to I and I to N equilibria for the wild type ribozyme. In both cases, Cref is close to the transition midpoint, where the assumption of constant n is likely valid (Lambert et al., 2010).
Thermodynamic linkage
Thermodynamic linkage (coupling) between two tertiary interactions i and j were calculated from the folding energies of single and double mutants by the use of a thermodynamic cycle (Fig. S9) (di Cera, 1998; Weber, 1975). A derivation of the linkage equations is given in Extended Experimental Procedures. The experimentally observed linkage was calculated from the folding energies using
| (3a) |
| (3b) |
in which and designate the free energy perturbations caused by mutation x (i, j, or i+j) in the IC or N states at the reference Mg2+ concentration. We assumed tertiary interactions are uncoupled in U. Accounting for the contributions of tertiary interactions to the IC state when calculating linkage in N yields and . The linkage between pairs of tertiary contacts in the presence of a third site mutation was determined using a triple mutant (Siegfried and Bevilacqua, 2009) (see Extended Experimental Procedures).
Supplementary Material
HIGHLIGHTS.
RNA tertiary interactions are present in native-like folding intermediates.
RNA tertiary interactions become cooperatively linked in folding intermediates.
Cooperativity depends on the native architecture of the ribozyme.
We suggest cooperativity arose during natural selection of unique RNA structures.
ACKNOWLEDGMENTS
The authors thank L. Guo and T. Irving for assistance with SAXS experiments, and D. Barrick, D. Draper, E. Westhof, J. Lecomte, D. Zappulla and members of Woodson laboratory for discussion and comments. This work was supported by grants from the NIH (GM60819) and NIST. BioCAT is an NIH-supported Research Center (RR-08630). Access to the APS was made possible by support from the US DOE.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: R. B. designed and performed experiments, analyzed the data and wrote the paper, J.-H. R and D.K. performed SAXS experiments, R.M.B. designed SAXS experiments and edited the paper, S.A.W. designed the experiments and wrote the paper.
REFERENCES
- Adams PL, Stahley MR, Gill ML, Kosek AB, Wang J, Strobel SA. Crystal structure of a group I intron splicing intermediate. RNA. 2004a;10:1867–1887. doi: 10.1261/rna.7140504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004b;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- Bailor MH, Sun X, Al-Hashimi HM. Topology links RNA secondary structure with global conformation, dynamics, and adaptation. Science. 2010;327:202–206. doi: 10.1126/science.1181085. [DOI] [PubMed] [Google Scholar]
- Baird NJ, Gong H, Zaheer SS, Freed KF, Pan T, Sosnick TR. Extended structures in RNA folding intermediates are due to nonnative interactions rather than electrostatic repulsion. J. Mol. Biol. 2010;397:1298–1306. doi: 10.1016/j.jmb.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NJ, Srividya N, Krasilnikov AS, Mondragon A, Sosnick TR, Pan T. Structural basis for altering the stability of homologous RNAs from a mesophilic and a thermophilic bacterium. RNA. 2006;12:598–606. doi: 10.1261/rna.2186506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NJ, Westhof E, Qin H, Pan T, Sosnick TR. Structure of a folding intermediate reveals the interplay between core and peripheral elements in RNA folding. J. Mol. Biol. 2005;352:712–722. doi: 10.1016/j.jmb.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Battle DJ, Doudna JA. Specificity of RNA-RNA helix recognition. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11676–11681. doi: 10.1073/pnas.182221799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua PC, Sugimoto N, Turner DH. A mechanistic framework for the second step of splicing catalyzed by the Tetrahymena ribozyme. Biochemistry. 1996;35:648–658. doi: 10.1021/bi951962z. [DOI] [PubMed] [Google Scholar]
- Bokinsky G, Rueda D, Misra VK, Rhodes MM, Gordus A, Babcock HP, Walter NG, Zhuang X. Single-molecule transition-state analysis of RNA folding. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9302–9307. doi: 10.1073/pnas.1133280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brion P, Schroeder R, Michel F, Westhof E. Influence of specific mutations on the thermal stability of the td group I intron in vitro and on its splicing efficiency in vivo: a comparative study. RNA. 1999;5:947–958. doi: 10.1017/s1355838299990477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmueller KL, Weeks KM. Near native structure in an RNA collapsed state. Biochemistry. 2003;42:13869–13878. doi: 10.1021/bi035476k. [DOI] [PubMed] [Google Scholar]
- Butcher SE, Pyle AM. The Molecular Interactions That Stabilize RNA Tertiary Structure: RNA Motifs, Patterns, and Networks. Acc. Chem. Res. 2011 doi: 10.1021/ar200098t. [DOI] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D’Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S, Behrouzi R, Rangan P, Woodson SA. Structural rearrangements linked to global folding pathways of the Azoarcus group I ribozyme. J. Mol. Biol. 2009;386:1167–1178. doi: 10.1016/j.jmb.2008.12.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S, Caliskan G, Briber RM, Perez-Salas U, Rangan P, Thirumalai D, Woodson SA. RNA tertiary interactions mediate native collapse of a bacterial group I ribozyme. J. Mol. Biol. 2005;353:1199–1209. doi: 10.1016/j.jmb.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Chauhan S, Woodson SA. Tertiary interactions determine the accuracy of RNA folding. J. Am. Chem. Soc. 2008;130:1296–1303. doi: 10.1021/ja076166i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton TE. Protein folding. Biochem. J. 1990;270:1–16. doi: 10.1042/bj2700001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Kwok LW, Millett IS, Bai Y, Mills TT, Jacob J, Maskel GS, Seifert S, Mochrie SG, Thiyagarajan P, et al. The fastest global events in RNA folding: electrostatic relaxation and tertiary collapse of the Tetrahymena ribozyme. J. Mol. Biol. 2003;332:311–319. doi: 10.1016/s0022-2836(03)00854-4. [DOI] [PubMed] [Google Scholar]
- di Cera E. Site-specific analysis of mutational effects in proteins. Adv. Protein Chem. 1998;51:59–119. doi: 10.1016/s0065-3233(08)60651-8. [DOI] [PubMed] [Google Scholar]
- Doudna JA, Cech TR. Self-assembly of a group I intron active site from its component tertiary structural domains. RNA. 1995;1:36–45. [PMC free article] [PubMed] [Google Scholar]
- Fang X, Pan T, Sosnick TR. A thermodynamic framework and cooperativity in the tertiary folding of a Mg2+-dependent ribozyme. Biochemistry. 1999;38:16840–16846. doi: 10.1021/bi991700n. [DOI] [PubMed] [Google Scholar]
- Fang XW, Golden BL, Littrell K, Shelton V, Thiyagarajan P, Pan T, Sosnick TR. The thermodynamic origin of the stability of a thermophilic ribozyme. Proc. Natl. Acad. Sci. U. S. A. 2001;98:4355–4360. doi: 10.1073/pnas.071050698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang XW, Pan T, Sosnick TR. Mg2+-dependent folding of a large ribozyme without kinetic traps. Nat. Struct. Biol. 1999;6:1091–1095. doi: 10.1038/70016. [DOI] [PubMed] [Google Scholar]
- Fiore JL, Kraemer B, Koberling F, Edmann R, Nesbitt DJ. Enthalpy-driven RNA folding: single-molecule thermodynamics of tetraloop-receptor tertiary interaction. Biochemistry. 2009;48:2550–2558. doi: 10.1021/bi8019788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Furuta H, Ikawa Y. Tailoring RNA modular units on a common scaffold: a modular ribozyme with a catalytic unit for beta-nicotinamide mononucleotide-activated RNA ligation. RNA. 2009;15:877–888. doi: 10.1261/rna.1461309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SD, Rambo RP, Van Tyne D, Batey RT. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat. Struct. Mol. Biol. 2008;15:177–182. doi: 10.1038/nsmb.1371. [DOI] [PubMed] [Google Scholar]
- Herschlag D, Cech TR. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry. 1990;29:10159–10171. doi: 10.1021/bi00496a003. [DOI] [PubMed] [Google Scholar]
- Hogg RV, Tanis EA. Probability and statistical inference. Prentice Hall; Upper Saddle River, NJ: 2006. [Google Scholar]
- Holbrook SR. Structural principles from large RNAs. Annu. Rev. Biophys. 2008;37:445–464. doi: 10.1146/annurev.biophys.36.040306.132755. [DOI] [PubMed] [Google Scholar]
- Ikawa Y, Naito D, Shiraishi H, Inoue T. Structure-function relationships of two closely related group IC3 intron ribozymes from Azoarcus and Synechococcus pre-tRNA. Nucleic Acids Res. 2000;28:3269–3277. doi: 10.1093/nar/28.17.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger L, Michel F, Westhof E. Involvement of a GNRA tetraloop in long-range RNA tertiary interactions. J. Mol. Biol. 1994;236:1271–1276. doi: 10.1016/0022-2836(94)90055-8. [DOI] [PubMed] [Google Scholar]
- Kilburn D, Roh JH, Guo L, Briber RM, Woodson SA. Molecular crowding stabilizes folded RNA structure by the excluded volume effect. J. Am. Chem. Soc. 2010;132:8690–8696. doi: 10.1021/ja101500g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo LY, Davidson LA, Pico S. Characterization of the Azoarcus ribozyme: tight binding to guanosine and substrate by an unusually small group I ribozyme. Biochim. Biophys. Acta. 1999;1489:281–292. doi: 10.1016/s0167-4781(99)00200-6. [DOI] [PubMed] [Google Scholar]
- Laing C, Schlick T. Computational approaches to RNA structure prediction, analysis, and design. Curr. Opin. Struct. Biol. 2011;21:306–318. doi: 10.1016/j.sbi.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert D, Leipply D, Draper DE. The osmolyte TMAO stabilizes native RNA tertiary structures in the absence of Mg2+: evidence for a large barrier to folding from phosphate dehydration. J. Mol. Biol. 2010;404:138–157. doi: 10.1016/j.jmb.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnert V, Jaeger L, Michel F, Westhof E. New loop-loop tertiary interactions in self-splicing introns of subgroup IC and ID: a complete 3D model of the Tetrahymena thermophila ribozyme. Chem. Biol. 1996;3:993–1009. doi: 10.1016/s1074-5521(96)90166-0. [DOI] [PubMed] [Google Scholar]
- Leipply D, Draper DE. Dependence of RNA tertiary structural stability on Mg2+ concentration: interpretation of the Hill equation and coefficient. Biochemistry. 2010;49:1843–1853. doi: 10.1021/bi902036j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Lescoute A, Westhof E. The building blocks and motifs of RNA architecture. Curr. Opin. Struct. Biol. 2006;16:279–287. doi: 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescoute A, Westhof E. The interaction networks of structured RNAs. Nucleic Acids Res. 2006;34:6587–6604. doi: 10.1093/nar/gkl963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Moghaddam S, Caliskan G, Chauhan S, Hyeon C, Briber RM, Thirumalai D, Woodson SA. Metal ion dependence of cooperative collapse transitions in RNA. J. Mol. Biol. 2009;393:753–764. doi: 10.1016/j.jmb.2009.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annu. Rev. Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- Pan J, Thirumalai D, Woodson SA. Magnesium-dependent folding of self-splicing RNA: exploring the link between cooperativity, thermodynamics, and kinetics. Proc. Natl. Acad. Sci. U. S. A. 1999;96:6149–6154. doi: 10.1073/pnas.96.11.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Salas UA, Rangan P, Krueger S, Briber RM, Thirumalai D, Woodson SA. Compaction of a bacterial group I ribozyme coincides with the assembly of core helices. Biochemistry. 2004;43:1746–1753. doi: 10.1021/bi035642o. [DOI] [PubMed] [Google Scholar]
- Pyle AM, Cech TR. Ribozyme recognition of RNA by tertiary interactions with specific ribose 2′-OH groups. Nature. 1991;350:628–631. doi: 10.1038/350628a0. [DOI] [PubMed] [Google Scholar]
- Rangan P, Masquida B, Westhof E, Woodson SA. Assembly of core helices and rapid tertiary folding of a small bacterial group I ribozyme. Proc. Natl. Acad. Sci. U. S. A. 2003;100:1574–1579. doi: 10.1073/pnas.0337743100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan P, Woodson SA. Structural requirement for Mg2+ binding in the group I intron core. J. Mol. Biol. 2003;329:229–238. doi: 10.1016/s0022-2836(03)00430-3. [DOI] [PubMed] [Google Scholar]
- Sattin BD, Zhao W, Travers K, Chu S, Herschlag D. Direct measurement of tertiary contact cooperativity in RNA folding. J. Am. Chem. Soc. 2008;130:6085–6087. doi: 10.1021/ja800919q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultes EA, Spasic A, Mohanty U, Bartel DP. Compact and ordered collapse of randomly generated RNA sequences. Nat. Struct. Mol. Biol. 2005;12:1130–1136. doi: 10.1038/nsmb1014. [DOI] [PubMed] [Google Scholar]
- Shank EA, Cecconi C, Dill JW, Marqusee S, Bustamante C. The folding cooperativity of a protein is controlled by its chain topology. Nature. 2010;465:637–640. doi: 10.1038/nature09021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried NA, Bevilacqua PC. Thinking inside the box: designing, implementing, and interpreting thermodynamic cycles to dissect cooperativity in RNA and DNA folding. Methods Enzymol. 2009;455:365–393. doi: 10.1016/S0076-6879(08)04213-4. [DOI] [PubMed] [Google Scholar]
- Silverman SK, Cech TR. Energetics and cooperativity of tertiary hydrogen bonds in RNA structure. Biochemistry. 1999;38:8691–8702. doi: 10.1021/bi9906118. [DOI] [PubMed] [Google Scholar]
- Strauss-Soukup JK, Strobel SA. A chemical phylogeny of group I introns based upon interference mapping of a bacterial ribozyme. J. Mol. Biol. 2000;302:339–358. doi: 10.1006/jmbi.2000.4056. [DOI] [PubMed] [Google Scholar]
- Su LJ, Brenowitz M, Pyle AM. An alternative route for the folding of large RNAs: apparent two-state folding by a group II intron ribozyme. J. Mol. Biol. 2003;334:639–652. doi: 10.1016/j.jmb.2003.09.071. [DOI] [PubMed] [Google Scholar]
- Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 1992;25:495–503. [Google Scholar]
- Szewczak AA, Cech TR. An RNA internal loop acts as a hinge to facilitate ribozyme folding and catalysis. RNA. 1997;3:838–849. [PMC free article] [PubMed] [Google Scholar]
- Tanner M, Cech T. Activity and thermostability of the small self-splicing group I intron in the pre-tRNA(lle) of the purple bacterium Azoarcus. RNA. 1996;2:74–83. [PMC free article] [PubMed] [Google Scholar]
- Tanner MA, Cech TR. Joining the two domains of a group I ribozyme to form the catalytic core. Science. 1997;275:847–849. doi: 10.1126/science.275.5301.847. [DOI] [PubMed] [Google Scholar]
- Theimer CA, Blois CA, Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol. Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Thirumalai D, Hyeon C. RNA and protein folding: common themes and variations. Biochemistry. 2005;44:4957–4970. doi: 10.1021/bi047314+. [DOI] [PubMed] [Google Scholar]
- Toor N, Rajashankar K, Keating KS, Pyle AM. Structural basis for exon recognition by a group II intron. Nat. Struct. Mol. Biol. 2008;15:1221–1222. doi: 10.1038/nsmb.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber DK, Rook MS, Zarrinkar PP, Williamson JR. Kinetic intermediates trapped by native interactions in RNA folding. Science. 1998;279:1943–1946. doi: 10.1126/science.279.5358.1943. [DOI] [PubMed] [Google Scholar]
- Walter NG, Burke JM, Millar DP. Stability of hairpin ribozyme tertiary structure is governed by the interdomain junction. Nat. Struct. Biol. 1999;6:544–549. doi: 10.1038/9316. [DOI] [PubMed] [Google Scholar]
- Wan Y, Russell R. Enhanced specificity against misfolding in a thermostable mutant of the Tetrahymena ribozyme. Biochemistry. 2011;50:864–874. doi: 10.1021/bi101467q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters AL, Deka P, Corrent C, Callender D, Varani G, Sosnick T, Baker D. The highly cooperative folding of small naturally occurring proteins is likely the result of natural selection. Cell. 2007;128:613–624. doi: 10.1016/j.cell.2006.12.042. [DOI] [PubMed] [Google Scholar]
- Weber G. Energetics of ligand binding to proteins. Adv. Protein Chem. 1975;29:1–83. doi: 10.1016/s0065-3233(08)60410-6. [DOI] [PubMed] [Google Scholar]
- Weinberg Z, Perreault J, Meyer MM, Breaker RR. Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis. Nature. 2009;462:656–659. doi: 10.1038/nature08586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JA. Additivity of mutational effects in proteins. Biochemistry. 1990;29:8509–8517. doi: 10.1021/bi00489a001. [DOI] [PubMed] [Google Scholar]
- Westhof E. The amazing world of bacterial structured RNAs. Genome Biol. 2010;11:108. doi: 10.1186/gb-2010-11-3-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinkar PP, Williamson JR. The kinetic folding pathway of the Tetrahymena ribozyme reveals possible similarities between RNA and protein folding. Nat. Struct. Biol. 1996;3:432–438. doi: 10.1038/nsb0596-432. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.