Abstract
Point mutations in the gene encoding copper-zinc superoxide dismutase (SOD1) impart a gain-of-function to this protein that underlies 20-25% of all familial amyotrophic lateral sclerosis (FALS) cases. However, the specific mechanism of mutant SOD1 toxicity has remained elusive. Using the complementary techniques of atomic force microscopy (AFM), electrophysiology, and cell and molecular biology, here we examine the structure and activity of A4VSOD1, a mutant SOD1. AFM of A4VSOD1 reconstituted in lipid membrane shows discrete tetrameric pore-like structure with outer and inner diameters 12.2 and 3.0 nm respectively. Electrophysiological recordings show distinct ionic conductances across bilayer for A4VSOD1 and none for wild-type SOD1. Mouse neuroblastoma cells exposed to A4VSOD1 undergo membrane depolarization and increases in intracellular calcium. These results provide compelling new evidence that a mutant SOD1 is capable of disrupting cellular homeostasis via an unregulated ion channel mechanism. Such a “toxic channel” mechanism presents a new therapeutic direction for ALS research.
Keywords: amyotrophic lateral sclerosis, superoxide dismutase, A4V, protein misfolding, atomic force microscopy, Fluo-4 calcium assay, DiBAC4(3) membrane potential measurement
INTRODUCTION
Amyotrophic lateral sclerosis (ALS) is a rapidly progressive neurodegenerative disease characterized by selective loss of motor neurons (MNs) in the brain and spinal cord leading to muscle inactivity and degeneration. At present, no viable treatment exists to alter the course of the disease, with death usually occurring within 1-5 years of the disease onset (Cleveland and Rothstein, 2001). Approximately 10% of ALS cases are familial (FALS), most typically with an autosomal dominant inheritance pattern (Bruijn et al., 2004). FALS cases are remarkably similar in clinical manifestations to sporadic ALS (SALS) (Bruijn et al., 2004). Genetic studies have linked 20-25% of FALS cases to >100 single, missense mutations in the copper-zinc superoxide dismutase type 1 (SOD1) gene located on chromosome 21 (Boillee et al., 2006; Bruijn et al., 2004; Rosen et al., 1993). These point mutations in the SOD1 locus are reported to induce ALS pathophysiology not by “loss of function” as supported by the three lines of evidence: i) some of the mutant forms of SOD1 that induce FALS have full dismutase activity; ii) an SOD-/- mouse does not develop ALS; and iii) mice that carry mutant SOD1 as a transgene develop ALS, despite a normal endogenous dismutase activity (reviewed in (Boillee et al., 2006)). These findings suggests an alternate competing notion that mutated SOD1 acquires a toxic “gain of function” to induce the disease.
Despite the identification of mutant SOD1 as a cause of ALS for almost 20 years, the specific toxic function gained by mutant SOD1 that causes the selective loss of MNs has remained elusive. High molecular weight aggregates of mutant SOD1 have been detected in cell extracts from FALS patients, FALS transgenic mice and cultured cells (Bruening et al., 1999; Bruijn et al., 1998; Johnston et al., 2000) and they have been linked to ALS disease progression, however, the structural and functional details of these aggregates are not fully characterized. Significantly, mutant SOD1 forms misfolded aggregates and kills MNs because of its acquired toxicity rather than its diminished dismutase activity (Ghadge et al., 2006; Gurney et al., 1994; Reaume et al., 1996). Several findings, including a reduction in mutual repulsive charge and increase in hydrophobicity, have been reported for the genetic mutants of SOD1 and are likely to contribute to the misfolding and aggregation of mutant SOD1 peptides (Sandelin et al., 2007; Tiwari et al., 2005). Recent studies also suggest that SALS may also be caused by a similar SOD1 neurotoxicity related to misfolding (Ezzi et al., 2007; Gruzman et al., 2007; Kabashi et al., 2007). Therefore understanding the mechanistic details underlying ALS pathology is of paramount importance for developing specific diagnostic markers and/or developing effective treatment modalities.
Growing evidence in support of a toxic channel hypothesis for Alzheimer's disease and other neurodegenerative diseases lends credence to a common disease mechanism in protein misfolding diseases (Jang et al., 2010a; Quist et al., 2005). Consistent with this scenario, altered levels of intracellular calcium are reported to contribute to the motor neuron degeneration and mitochondrial dysfunction associated with ALS (Arimura et al., 2002; Grosskreutz et al., 2007; Ionov, 2007; Kuwabara and Kanai, 2007; Menzies et al., 2002; Pieri et al., 2003; Urushitani et al., 2001). In addition, abnormal sodium currents have also been implicated in ALS (Camerino et al., 2007). Recently, Israelson et al. (Israelson et al., 2010), reported a very important study underpinning mitochondrial dysfunction in ALS pathology. They showed direct inhibition of VDAC1, a key mitochondrial membrane protein, by mutant SOD1G37R leading to mitochondrial dysfunction. However, this study failed to conclude that toxic interaction of mutant SOD1 with VDAC1 is intrinsic to the disease mechanism, since deletion of VDAC1 also diminished the survival in the SODG37R/VDAC-/- mice.
The focus of the present study is to understand the structure-function relationship of mutant SOD1 aggregates using complementary techniques of atomic force microscopy (AFM), ion channel conductance, fluorescence microscopy, cell and molecular biology. In this study, we investigate the A4VSOD1 mutant, which has a single amino acid difference from WTSOD1 and causes the most aggressive and prevalent form of ALS in North America with a mean survival time of ~1 year from disease onset (Boillee et al., 2006). We report the ability of A4VSOD1 to incorporate into lipid membranes as discrete pores and render the membrane permeable to ions. We also show that these pores facilitate cellular calcium uptake and cell membrane depolarization in neuroblastoma cells.
MATERIALS AND METHODS
Expression and purification of SOD1 proteins
Human wildtype SOD1 and a control, the Src homology 3 (SH3) domain of c-Src kinase, were cloned in a prokaryotic expression vector that has been previously described (Scholle et al., 2004); the expressed SOD1 protein contained 6XHis tags on the C-terminus while the expressed SH3 contained the tags on the N-terminus. A4VSOD1 cDNA was cloned in the same manner as that for the wildtype SOD1. These vectors were used to express the fusion proteins in E. coli BL21 (DE3) pBirA by inducing with isopropyl-b-D-thiogalactopyranoside (IPTG) and purification using a Ni-affinity column (Qiagen, MD) as described (Scholle et al., 2004). The proteins were dialyzed against phosphate buffered saline (PBS) containing 10% glycerol and stored at -80° C. The purified proteins were then analyzed on SDS-PAGE gels by Coomassie blue staining and by western blot using rabbit anti-SOD1 polyclonal antibody (Stressgen, Canada, 1:1000 dilution) and horseradish peroxidase (HRP)-linked anti-rabbit IgG (1:2000 dilution) followed by detection with an ECL-Plus detection kit (Amersham, NJ).
Analytical ultracentrifugation
A Beckman Optima XL-A (Beckman Coulter, Fullerton, CA) analytical ultracentrifuge (AUC) equipped with a Ti60 rotor was used to determine the molecular weights and molar percentages of the oligomers present in each of the two purified protein fractions (wildtype and mutant SOD1). Fifty µM preparations of the proteins were analyzed as dissolved in PBS under non-denaturing, non-reducing conditions, the results of which are shown in Figure 1. Sedimentation velocity data was obtained for each protein at 40,000 rpm using an Epon two-channel centerpiece. Absorbance of the samples at 280 nm was monitored in a continuous-mode time interval of 360-480 sec and a step size of 0.003 cm.
Fig. 1.
In vitro analyses of the A4VSOD1 protein fraction show the presence of higher order oligomers, including high levels of a tetrameric species, compared to WTSOD1. A. Coomassie blue stained SDS-PAGE of purified WTSOD1 (WT) and A4VSOD1 (A4V) . The WT and A4V have an electrophoretic mobility at ~25 kDa, the expected molecular weight of the fusion proteins. B. Analytical ultracentrifugation of (blue line) wildtype SOD1 and (red line) A4VSOD1 protein in PBS. Dimer peaks were seen in both fractions as expected for the native active form of the metalloenzyme. The A4VSOD1 fraction contained multimeric species most notably a relatively large peak at the tetramer position while the wildtype SOD1 fraction primarily contained dimer. C. AFM fluid scan and particle sizing (inset) reveal the presence of large oligomers in the A4VSOD1 protein fraction measuring 12.4 nm in diameter. D. AFM fluid scan and particle sizing (inset) show smaller dimer-sized structures present in the WTSOD1 fraction with diameters measuring 5.5 nm.
Atomic Force Microscopy
The AFM measurements were carried out with a multimode Nanoscope IIIa, (Veeco Instruments, Santa Barbara, CA) equipped with an E-type scanner. The AFM cantilevers used were V-shaped and made of silicon nitride (TR400, Olympus, Japan). All AFM scans were taken under PBS at room temperature.
Preparations of A4VSOD1 protein incorporated into lipid bilayer
1 mg of DOPC was dried against a glass tube in a vacuum chamber overnight. PBS was used to resuspend the lipid cake to a final concentration of 1 mg/mL. A stock A4VSOD1 protein solution was added and diluted to achieve a lipid to protein ratio of 20:1 (w/w) and vortexing was applied for 20 sec. The lipoprotein was then exposed to ice-bath sonication for 10 min. The proteoliposomes were adsorbed to freshly cleaved mica in 30 μl for 30 min enclosed in a humid chamber. Vesicle fusion events provided a large area supported lipid bilayer loaded with protein (Engel and Gaub, 2008). The sample was then rinsed 10 times with 60 ml of PBS to remove any unbound material. The sample was then loaded into the AFM for imaging at room temperature.
AFM Imaging
Samples were analyzed by AFM using both tapping and contact modes under liquid. The oxide sharpened silicon nitride cantilevers used have force constants of 0.08 N/m and 0.02 N/m for tapping and contact modes, respectively. The cantilevers were oscillated near 9 kHz at low amplitude for tapping mode, or, held to the minimum deflection set-point (just above probe pull-off) for contact mode. To etch one DOPC bilayer from the sample surface, contact mode was used to rub the surface briefly at a highly elevated contact force followed by re-scanning the same area using fluid tapping mode to reveal the square etch allowing determination of the presence of the supported bilayer from the depth of the etch.
Electrophysiology
Formation of planar lipid bilayer
To generate planar lipid bilayers, we employed the so-called “painting technique” which consists on applying a solution of lipids over a pore (Mueller et al., 1962). The following lipids 1-Palmitoyl-2-Oleoyl-sn-Glycero-3-[Phospho-rac-(1-glycerol)] (POPG) and 1-Palmitoyl-2-Oleoyl-sn-Glycero-3-Phosphoethanolamine (POPE) (Avanti Polar Lipids, Alabaster, AL) were dissolved either in hexane or heptane (Sigma, St. Louis, MO) at a 1:1 (w/w) ratio (for vertical bilayer) or in decane at a 3:1 (w/w) ratio (for horizontal bilayer). The final lipid concentration in solvents was 10 to 20 mg mL-1. We used vertical bilayer cups made of Delrin with a pore diameter of ~250 μm in a Delrin septum (Warner Instruments, Delrin perfusion cup volume 1 mL) and a home-made horizontal bilayer chamber made of Delrin with a ~250 μm pore diameter. The chamber's pores were pretreated with the lipid mixture and dried prior to the addition of the recording solution. Bilayer formation was obtained by swapping air bubbles over the chamber's pores from pipette tips dipped into the lipid mixture.
Current recordings: Vertical bilayer
Prior to painting the membrane, we verified that the electrode asymmetry was always less than 1 mV. Upon membrane formation we screened for membranes that were stable and held conductances of less than 7 pS up to voltages of ± 100 mV for a period of at least 10 min with capacitances higher than 100 pF (typically ~ 150 pF) (Capone et al., 2010). When the above three criteria were fulfilled, we added 9 -13.5 μl of SOD1 samples (1-2.5 μM final concentration of wild type SOD1 and A4VSOD1 dissolved in PBS) to the cis side and stirred for 1-2 min. We performed all recordings in “voltage clamp mode” using Ag/AgCl electrodes. We used a filter-cutoff frequency of 3 kHz, and a sampling frequency of 15 kHz for all bilayer recordings. For representation in figures, we filtered the current traces with a digital Gaussian low-pass filter with a cutoff frequency of 100 Hz. Our recording solution consisted on a non-buffered 150mM KCl solution.
Horizontal bilayer
For electrical connections, we used Ag/AgCl electrodes and 1% agar borosilicate bridges containing 1M KCl. Bilayer formation was controlled by a voltage-ramp protocol pulsing 1mV/ms from +100mV to -100mV. In these conditions, typical bilayers exhibited capacitances of ~ 0.3-0.5μF/cm2 (Mueller et al., 1962). Bilayers exhibiting stable capacitance over 5min were further treated with 1 - 5 μL of proteoliposomes prepared by reconstituting SOD1 proteins with POPE: POPG at 1:3 (w/w) at a protein to lipid ratio of 1:20 – 1:80 (w/w/). Our recording solution contained 100mM NaCl, 2mM KCl, 1.8mM CaCl2, 1mM MgCl2, 0.1mM EDTA and 10mM HEPES pH7.4. Data were acquired using a home-made low-pass filter set at 800 Hz and sampled at 20 kHz using in-house software. For figure representation, data were digitally filtered at 800 Hz.
Cell culture
Mouse neuroblastoma cell line (N2A) (American Type Culture Collection, Manassas, VA) was maintained in culture following ATCC recommendations. Cells were split 48 hrs before the experiment at 5000 cells/cm2 on glass bottomed petri dishes (MatTek, Ashland, MA) or chambered cover glass systems (Nunc, Rochester, NY).
Membrane Potential Measurement
Cell membrane potential changes were assessed with voltage sensitive fluorescent dye, DiBAC4(3) (Invitrogen, CA). Briefly, cells were washed 3 times in HBSS and loaded with the dye (100 nM) for 20 minutes at room temperature. Basal fluorescence (F0) was recorded for one minute before challenging with 1 μM SOD1 protein(s). Changes in fluorescence intensity specific to membrane potential were recorded at 5 sec intervals for 20 minutes. Relative change in mean fluorescence intensity at a given time ‘t’ (Ft), with respect to basal fluorescence was expressed as a percentage [(Ft/F0)*100] and plotted against time.
Cell Calcium Assay
Molecular Probes Fluo-4 NW calcium assay kit (Invitrogen) was used to monitor qualitative calcium changes within cells. Cells were loaded with the dye following the manufacturer's recommended protocol. Briefly, cells were washed free of serum proteins in the HBSS and 500 μl of dye mixed with 2.5 mM Probenecid was added to each assay well and incubated for 45 min at 37°C. For imaging, cells were transferred to the onstage incubator system (20/20 Technologies, USA) mounted on an inverted optical microscope (Olympus IX71) equipped with a FITC filter set (Semrock). Calcium ratio imaging was carried out with the RatioPlus module of IPLab (BD Biosciences) software. A random field was imaged every 5 seconds for 20 min or more. Typically, 6 frames were acquired first to establish the baseline distribution of Ca2+ within the cells. SOD1 protein (either wildtype or A4V) was added at a concentration of 1 μM to the buffer and imaging continued. Images were acquired with a 1024 × 1024 monochrome camera (Cascade II, Photometrics) and stored as X,Y,T stacks for analysis. A script was written in IPLab to interact with the RatioPlus module to plot the mean ratio of fluorescence change [(Ft/F0)*100] with time. Ft is the calcium specific fluorescence of the field at a given time ‘t’ and F0 is the calcium specific fluorescence of the same field at time t = 0. For visual representation, the intensity changes were color-coded using the ‘pseudocolor’ function of the program.
Statistical analysis
Comparisons between A4V mutant versus wild-type SOD1 treated samples were performed by a two-sample ‘t’ test for membrane depolarization and calcium uptake studies. Data are expressed as mean ± SE and P values less than 0.05 were considered statistically significant.
RESULTS
Mutant A4VSOD1 forms tetrameric oligomers
WTSOD1 and mutant A4VSOD1 proteins were purified to near homogeneity (Fig 1, top left). The oligomeric features of each of the two purified SOD1 protein fractions were analyzed under identical non-reducing, non-denaturing conditions using analytical ultracentrifugation. The A4VSOD1 fraction contained a relatively higher abundance of oligomeric species not found in the WTSOD1 fraction. The plot in Figure 1 (top right) shows a slightly greater amount of tetramer (37.6%) than dimer (31.7%) in the case of A4VSOD1 total protein, while there was a predominant dimer peak (68.3%) compared to tetramer (13.9%) for the WTSOD1. Consistent with these results, AFM images of the mutant A4VSOD1 proteins adsorbed on a mica substrate revealed a preponderance of large oligomeric structures averaging 12.4 nm in diameter (sd= 2.76 nm, n= 21) (Figure 1, bottom left), while AFM images of the WTSOD1 protein revealed much smaller globules, with diameters measuring 5.5 nm (sd= 1.56 nm, n= 42) (Figure 1, bottom right), likely representing dimers, the native enzymatic form.
Mutant A4VSOD1 forms pore-like structures in lipid bilayer
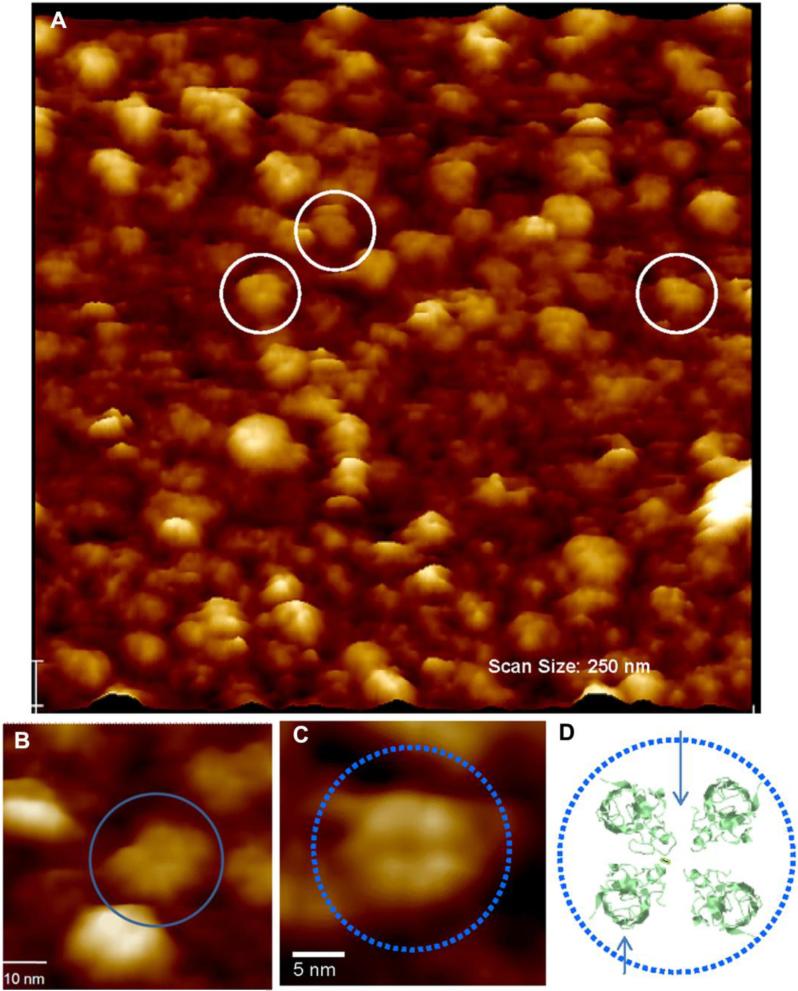
High resolution AFM images of mutant A4VSOD1 peptides reconstituted in bilayer show discrete pore-like structures (Figure 2) These structures have an outer diameter of ~12.2 nm (sd=1.6, n=32), inner diameter of ~ 3.0 nm (sd=0.5, n=31) and they protrude ~1.4 nm (sd=0.3, n=32) above the plane of the lipid bilayer. Using the same preparative and AFM methods, WTSOD1 showed evidence of interaction with the DOPC bilayer, coating the surface with a globular protein layer. However, we were unable to resolve any ultra-structural detail on the globules using AFM.
Fig. 2.
(A) Lower (250 nm × 250 nm) and (B, C) high resolution AFM scans taken in PBS buffer of mutant A4VSOD1 incorporated into a supported DOPC lipid bilayer. A4V forms 12.2 ±0.09 nm channel-like structures incorporated into the bilayer (white circles in A; blue circles in B,C). B. The channel-like structures are multimeric assemblies protruding 1.4 ±0.05 nm above the plane of the bilayer and exhibiting a central pore 3.0 ±0.05 nm in diameter. Scale bar, 5 nm. C. High resolution AFM topographic scan of an individual tetrameric A4VSOD1 ion pore incorporated into a fluid phase DOPC bilayer. Scale bar, 5 nm. D. Tetramer model (bottom right) is based on the crystal structure of A4VSOD1 found in a public database (PDB ID: 1UXM). The four SOD1 monomers were rotated so that the hydrophilic loops faced inward and the beta barrels out toward the hydrophobic core of the bilayer, in the center a small opening is formed.
Mutant A4VSOD1 pores have ion channel conductance
Mutant and wildtype SOD1 were tested for their ionic conductance across planar lipid bilayers. These measurements were carried out using two complementary approaches, (i) proteins added in a reconstituted form (i.e., in proteoliposomes) in horizontal bilayer (Figure 3a); and ii) proteins added in a free, soluble form in vertical bilayer systems (Figure 3b).
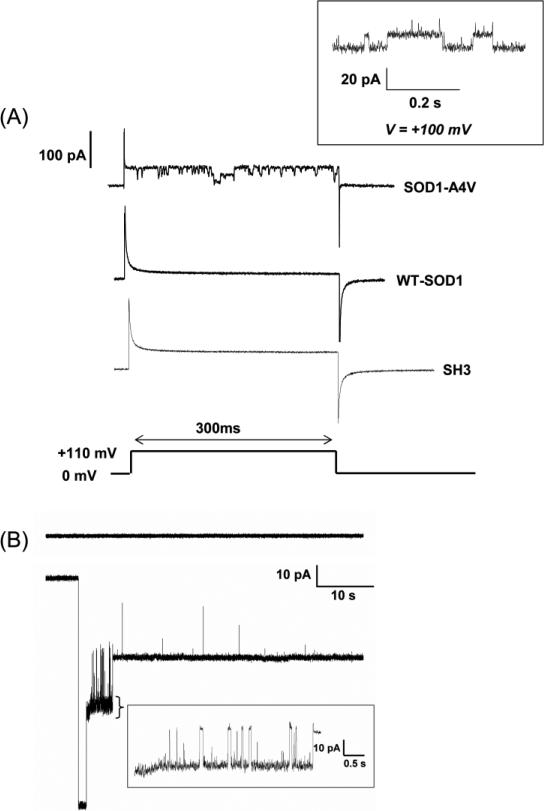
Fig. 3.
SOD1-A4V forms conducting channel-like structures in artificial membrane bilayers either by (A) proteoliposome or (B) direct A4V protein addition. A. A4V conducts when proteoliposmes are fused to the bilayer. The membrane was first held at 0 mV and then a 110 mV voltage pulse was applied for 300 ms. This step-protocol was used with mutant SOD1-A4V (upper trace), WT-SOD1 (middle trace) and the SH3 domain protein as negative control (bottom trace). Statistically significant channel openings were only observed with A4VSOD1. Inset: SOD1-A4V activity measured in POPG:POPE (1:3) bilayers held at constant voltage (+100mV). B. A4VSOD1 (1 μM) forms conducting channel-like structures also when added directly to one side of a POPG:POPE (1:1) bilayer (lower trace) and held at constant voltage. Time magnification (indicated by callout under the ‘spiky’ part of trace) shows an increased time resolution of A4VSOD1 bilayer activity. As a control, the upper trace in B shows a representative recording of wild type SOD1 under identical conditions. Both traces shown in panel B were at -80mV applied potential.
In experiments using the A4VSOD1 in proteoliposomes, we observed conductance events 17 times out of 33 trials (51%). When activity was observed, the mean time prior to the first event was 6.5 ±1.8 min. In most cases, the activity disappeared after about 10-15 minutes if no additional sample was added. As controls, we used samples made of pure liposomes or liposomes mixed with the SH3 domain from the oncogenic protein, Src. These control samples did not show any activity (out of 12 trials for the SH3 domain and 5 trials for empty liposomes) when tested under the same conditions. On the other hand, WTSOD1 showed no electrical activity in over 15 experiments.
Figure 3a shows a typical recording for the A4VSOD1 reconstituted in proteoliposomes. Pulses from 0 mV to +110 mV show channel-like activity for the mutant but not for the wildtype or the SH3 control. For the mutant, the ionic conductances were about 150 to 200 pS and clearly showed two well defined current levels. This may either indicate the presence of 2 distinct pores of similar conductance or a unique pore with two levels of conductance of approximately the same value. The inset at the top right corner of Figure 3a shows a similar experiment using the mutant but recorded in steady-state condition with a holding potential of +110 mV. Since proteoliposomes were added to the side that has the negative potential, these stepped-currents strongly indicate that A4VSOD1 can potentially create transmembrane conductances in resting neurons.
When mutant A4VSOD1 protein was directly added in a free and soluble form to the bilayer, channel-like activity was observed in 70% (5 out of 7) of cases and with an average appearance time of ~ 5min. This time is comparable to the lead time using horizontal bilayers (6.5±1.8 min), which indicates both proteoliposomes fusion and direct protein incorporation into the bilayer occur at similar rates. Figure 3b shows typical conductance events measured using the direct protein incorporation method. The results are nearly identical to the ones observed with proteoliposomes (Figure 3a). The mutant A4VSOD1 disrupts the integrity of the membrane and creates ionic conductances typical of ion-channel behavior (step-sized conductances and also flickering).
In contrast to the mutant, the wildtype when added directly to the bilayer showed no channel-like activity but the membranes became destabilized within an average of 21min. While we have not seen channel activity by the wildtype, our results so far indicate that the WTSOD1 alters bilayer stability, suggesting an interaction between the wildtype protein and the lipid bilayer.
Mutant A4VSOD1 induces membrane depolarization in neuronal cell line
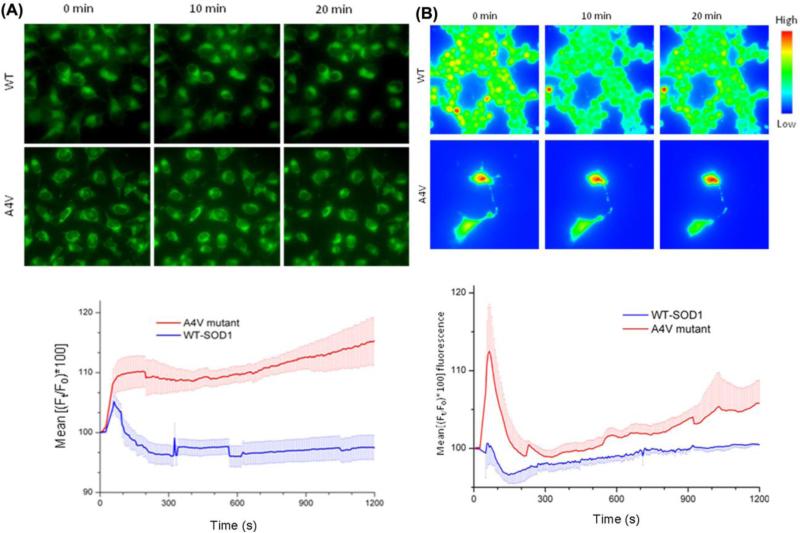
The effect of wildtype and mutant SOD1 on membrane potential was evaluated using a voltage sensitive dye DiBAC4(3) on mouse neuroblastoma cell line (N2A). DiBAC4(3) is a lipophilic, anionic dye that localizes on the positive potential side of the membrane. Upon depolarization, the dye enters the cell, binds to cytoplasmic proteins and exhibit concentration dependent increase in fluorescence intensity. As shown microscopically in Figure 4a, membrane depolarization could be observed in the cells at 10 minutes post-exposure to A4VSOD1 added directly to the culture medium. The lower panel in Figure 4b shows an approximately 15% increase in basal fluorescence due to mutant A4VSOD1 compared to WTSOD1.
Fig. 4.
A4V mutant causes membrane depolarization and an increase in intracellular calcium in N2A cells. A. Membrane potential specific fluorescence was recorded with DiBAC4(3) dye. Relative change in mean fluorescence [(Ft/F0)*100] intensity at a given time ‘t’, (Ft) with respect to basal fluorescence (F0) is expressed as a percentage. A4V mutant causes significant membrane depolarization at 10 min (p< 0.0001, n=8 experiments), at 20 min (P<0.002, n=7 experiments) compared to its wildtype (WT) counterpart. The error bars denote standard error (S.E.) of the mean fluorescence. B. Calcium specific fluorescence (Fluo-4) assay following treatment with 1 μM WT and A4V mutant. Mean values of all experiments plotted with S.E. of mean (n=4 for WT and n=6 for A4V). Significant calcium flux was only observed after 15 min (p<0.03) between the two groups.
Mutant A4VSOD1 alters intracellular calcium levels
Membrane depolarization could be due to addition of positive charges or loss of negative charges from the interior of the cell. Since ALS is one of the neurodegenerative diseases, we questioned whether it could be due to increased permeability to Ca2+ ions, which is tightly linked to cell stress and injury consistent with amyloid ion channel mediated toxicity (Jang et al., 2010a; Lin et al., 2001). To test this possibility, we assayed for intracellular Ca2+ changes following the addition of A4V mutant. The dye is an acetoxymethyl ester form of Fluo-4, which is a cell permeant, non-fluorescing dye. Upon cell entry, it is cleaved by esterase and become fluorescent and cell impermeant. However, the dye is slowly excreted by anionic transporters, which are inhibited by addition of probenecid to the dye mixture. Upon binding to calcium ions, its fluorescence increases proportionately. The results (Figure 4B) indicate a qualitative increase in intracellular Ca2+ for A4V mutant at the late stages of the experiment (after 15 min).
DISCUSSION
The major findings from the present study include: the presence of abnormal oligomeric stoichiometry of A4VSOD1 vs. WTSOD1 protein, specifically, tetrameric vs. dimeric, respectively; incorporation of the mutant A4VSOD1 protein into lipid membranes as discrete tetrameric pores; channel-like ionic conductance of these pore-like structures; and the cellular calcium uptake and cell membrane depolarization affected by the mutant SOD1.
WTSOD1 is normally a soluble dimeric metalloenzyme that dismutases superoxide in the host cell. However, under certain conditions (e.g., genetic mutation, oxidative stress, demetallation, monomerization, di-sulfide reduction etc.,), SOD1 could misfold and oligomerize differently as evident in the present results. Interestingly, as early as 1986, Getzoff and Tainer proposed SOD1 as a prototypical ion channel due to its structural and physicochemical properties (Getzoff, 1986). In separate unrelated studies, Elam and coworkers obtained x-ray diffraction patterns from crystalline water-filled helical filaments composed of misfolded mutated SOD1 protein (Elam et al., 2003) with striking similarity to an ion channel.
Interestingly, the SOD1 monomer possesses an 8-strand beta barrel. In pore-forming toxins, beta-barrels are known to form stable transmembrane domains even though their hydropathy plots do not strongly predict it (Jang et al., 2010b). SOD1 beta-barrels should also support stable transmembrane topology. However, two hydrophilic loops associated with electrostatic and zinc-binding domains protruding along one side of the beta-barrel would disfavor association with the bilayer's hydrophobic core. In the MD simulation model of the SOD1 ion channel (Fig. 2 bottom right), four SOD1 monomers are rotated within the bilayer plane such that the hydrophilic loops face inward forming the interior of a channel, while the beta barrels are positioned exteriorly. This tetrameric arrangement appears to be energetically stable, allowing acidic residues along the channel's interior to interact with ions, such as calcium. Importantly, copper and zinc atom deficiencies- a key feature of MTSOD1 permits unfolding of the hydrophilic loops (Galaleldeen et al., 2009; Hayward et al., 2002; Seetharaman et al., 2010). This would allow these loops to swing freely out of the bilayer plane and permit the passage of ions across it.
Considering A4VSOD1 and WTSOD1 differ by only one amino acid (A V), our electrophysiology results- which show highly repeatable channel-like activity for A4VSOD1 but not WTSOD1- cannot be explained satisfactorily without invoking a major conformational change for A4VSOD1. To form a pore, this conformational change must result in the formation of abnormal subunit aggregates larger than the native WTSOD1 dimer. It would appear this requirement for abnormal oligomers in the ability of A4VSOD1 to render lipid membrane permeable to ions is well supported by our ultracentrifugation (Figure 1) and AFM (Figure 2) results, which reveal the presence of tetrameric structures.
Specifically, we show that adding A4VSOD1 (free or reconstituted in liposomes) forms ion-permeable pores on lipid bilayers (Figure 3). The activity appears about 5 minutes after A4VSOD1 addition. The ability of A4VSOD1 to develop into pores provides a strong molecular basis to explain FALS. A similar “ion channel mechanism” by amyloid β is currently supported to explain Alzheimer's disease (Arispe et al., 1993a; Arispe et al., 1993b; Hirakura et al., 1999; Quist et al., 2005; Shirwany et al., 2007).
Challenge to cultured neuroblastoma cells by extracellular A4VSOD1 provides an in situ test bed for pore formation that might occur at the outer cell membrane. The voltage sensitive probe DiBAC4(3) reports a 15% change in fluorescence upon addition of A4VSOD1 compared to WTSOD1, suggesting a membrane depolarizing effect for A4VSOD1. The A4V mutant can bring about this change by either directly forming pores in the outer cell membrane and/or indirectly due to its interaction with other ion channels/transporters on the cell membrane. In support of direct pore formation in the neuroblastoma cells’ outer cell membrane, both the AFM and the electrophysiological data in this study suggest that A4VSOD1 is capable of forming functional pores in model lipid membranes by direct interaction.
Intracellular calcium levels are tightly regulated in cells whose imbalance is linked to cell death in several neurodegenerative diseases (Marambaud et al., 2009). Consistent with this, the cell calcium assay with Fluo-4 indicates a rise in intracellular Ca2+ at the late stages of the experiment (after 15 min). The A4V mutant could cause this disturbance in several ways: i) by forming calcium selective pores on the membrane directly, ii) by interacting with other ion channels/transporters that affect intracellular calcium, iii) by activating voltage sensitive calcium channels secondary to membrane depolarization, and iv) by any combination of the above. The slow kinetics of calcium rise suggests the third mechanism, i.e., primary membrane depolarization induced by mutant protein activates voltage regulated calcium channels in the cell membrane. However, we do not rule out the involvement of other possible mechanisms; detailed and quantitative studies are required to pinpoint the selectivity of these pores to any particular ionic species, and such a study is beyond the scope of this current work.
Relevant to the experiments in which neuroblastoma cells treated with A4VSOD1 is the recognition that MTSOD1 is secreted from the cell and therefore can be present extracellular as well as in the cytosol (Urushitani et al., 2008). The extracellular MTSOD1 has been reported to induce a microglial-mediated MN injury (Zhao et al., 2010). While localization of extracellular MTSOD1 to the outer cell membrane has not been reported, it may be that the incorporation of MTSOD1 as toxic channels in the MN's outer cell membrane makes these cells especially vulnerable to the microglial-induced injury. On the other hand, MTSOD1 has been localized to the outer mitochondrial membrane (Pasinelli et al., 2004; Vande Velde et al., 2008). Reportedly, this membrane association is so tight that the misfolded SOD1 behaves indistinguishably from that of an integral membrane protein. Furthermore, this finding, in particular, would seem to lend support to our observations of a membrane-associated structure and activity for A4VSOD1. Since the bulk of MTSOD1 is in the cytosol, we hypothesize that MTSOD1 may become incorporated as “toxic channels” primarily in mitochondrial membranes in MNs, causing the mitochondrial pathology that has been described – and ultimately leading to MN death.
Taken together, these data suggest that misfolded mutant A4VSOD1 forms pores that render lipid membranes permeable to ionic flow. Such disturbances to cellular membranes, continued and longstanding, could cause irreversible damage to the neuronal cells and cell death in ALS through a variety of mechanisms including: defective calcium buffering capacity (Alexianu et al., 1994; Beers et al., 2001), increased metabolic demands causing oxidative stress (Barber et al., 2006) and mitochondrial dysfunction (Beal, 2000; Borthwick et al., 1999) leading to cell death in ALS. Here, we propose that the wildtype SOD1 protein, normally active in healthy individuals as a soluble metalloenzyme, is transformed with respect to its structure/function by a mutation or environmental stress into a “toxic channel”. Furthermore these toxic channels in the lipid membranes of MNs (such as the outer cell and mitochondrial membranes) could create ionic imbalances (such as calcium overload) resulting in the neurotoxicity that characterizes ALS. The delineation of this new mechanism by which MTSOD1 carries out its toxic function identifies a common pathogenesis for a number of protein-misfolding diseases and provides a new target for ALS treatment.
Highlights.
Compelling new evidence for toxic channel mechanism of cell injury in ALS
A4V mutant was studied using complementary techniques
High resolution AFM revealed tetrameric pore structures in lipid bilayers
Heterogeneous single channel conductance was observed in lipid bilayers
Plasma membrane depolarization and cell calcium loading was observed in N2A cells
Acknowledgements
We thank Dr. Elena Solohama for her expertise and technical support regarding the analytical ultracentrifugation measurements.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alexianu ME, et al. The role of calcium-binding proteins in selective motoneuron vulnerability in amyotrophic lateral sclerosis. Ann Neurol. 1994;36:846–58. doi: 10.1002/ana.410360608. [DOI] [PubMed] [Google Scholar]
- Arimura K, et al. Isaacs’ syndrome as a potassium channelopathy of the nerve. Muscle Nerve. Suppl. 2002;11:S55–8. doi: 10.1002/mus.10148. [DOI] [PubMed] [Google Scholar]
- Arispe N, et al. Giant multilevel cation channels formed by Alzheimer disease amyloid beta-protein [A beta P-(1-40)] in bilayer membranes. Proc Natl Acad Sci U S A. 1993a;90:10573–7. doi: 10.1073/pnas.90.22.10573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arispe N, et al. Alzheimer disease amyloid beta protein forms calcium channels in bilayer membranes: blockade by tromethamine and aluminum. Proc Natl Acad Sci U S A. 1993b;90:567–71. doi: 10.1073/pnas.90.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber SC, et al. Oxidative stress in ALS: a mechanism of neurodegeneration and a therapeutic target. Biochim Biophys Acta. 2006;1762:1051–67. doi: 10.1016/j.bbadis.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Beal MF. Mitochondria and the pathogenesis of ALS. Brain. 2000;123(Pt 7):1291–2. doi: 10.1093/brain/123.7.1291. [DOI] [PubMed] [Google Scholar]
- Beers DR, et al. Parvalbumin overexpression alters immune-mediated increases in intracellular calcium, and delays disease onset in a transgenic model of familial amyotrophic lateral sclerosis. J Neurochem. 2001;79:499–509. doi: 10.1046/j.1471-4159.2001.00582.x. [DOI] [PubMed] [Google Scholar]
- Boillee S, et al. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Borthwick GM, et al. Mitochondrial enzyme activity in amyotrophic lateral sclerosis: implications for the role of mitochondria in neuronal cell death. Ann Neurol. 1999;46:787–90. doi: 10.1002/1531-8249(199911)46:5<787::aid-ana17>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Bruening W, et al. Up-regulation of protein chaperones preserves viability of cells expressing toxic Cu/Zn-superoxide dismutase mutants associated with amyotrophic lateral sclerosis. J Neurochem. 1999;72:693–9. doi: 10.1046/j.1471-4159.1999.0720693.x. [DOI] [PubMed] [Google Scholar]
- Bruijn LI, et al. Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science. 1998;281:1851–4. doi: 10.1126/science.281.5384.1851. [DOI] [PubMed] [Google Scholar]
- Bruijn LI, et al. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu Rev Neurosci. 2004;27:723–49. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- Camerino DC, et al. Ion channel pharmacology. Neurotherapeutics. 2007;4:184–98. doi: 10.1016/j.nurt.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Capone R, et al. Antimicrobial protegrin-1 forms ion channels: molecular dynamic simulation, atomic force microscopy, and electrical conductance studies. Biophys J. 2010;98:2644–52. doi: 10.1016/j.bpj.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Rothstein JD. From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat Rev Neurosci. 2001;2:806–19. doi: 10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- Elam JS, et al. Amyloid-like filaments and water-filled nanotubes formed by SOD1 mutant proteins linked to familial ALS. Nat Struct Biol. 2003;10:461–7. doi: 10.1038/nsb935. [DOI] [PubMed] [Google Scholar]
- Engel A, Gaub HE. Structure and mechanics of membrane proteins. Annu Rev Biochem. 2008;77:127–48. doi: 10.1146/annurev.biochem.77.062706.154450. [DOI] [PubMed] [Google Scholar]
- Ezzi SA, et al. Wild-type superoxide dismutase acquires binding and toxic properties of ALS-linked mutant forms through oxidation. J Neurochem. 2007;102:170–8. doi: 10.1111/j.1471-4159.2007.04531.x. [DOI] [PubMed] [Google Scholar]
- Galaleldeen A, et al. Structural and biophysical properties of metal-free pathogenic SOD1 mutants A4V and G93A. Archives of Biochemistry and Biophysics. 2009;492:40–47. doi: 10.1016/j.abb.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getzoff E. a. T. In: JA Superoxide dismutase as a model ion channel. Miller C, editor. Ion Channel Reconstitution Plenum Press; New York: 1986. pp. 57–74. [Google Scholar]
- Ghadge GD, et al. Truncated wild-type SOD1 and FALS-linked mutant SOD1 cause neural cell death in the chick embryo spinal cord. Neurobiol Dis. 2006;21:194–205. doi: 10.1016/j.nbd.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Grosskreutz J, et al. Role of mitochondria in kainate-induced fast Ca2+ transients in cultured spinal motor neurons. Cell Calcium. 2007;42:59–69. doi: 10.1016/j.ceca.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Gruzman A, et al. Common molecular signature in SOD1 for both sporadic and familial amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2007;104:12524–9. doi: 10.1073/pnas.0705044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney ME, et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–5. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- Hayward LJ, et al. Decreased metallation and activity in subsets of mutant superoxide dismutases associated with familial amyotrophic lateral sclerosis. J Biol Chem. 2002;277:15923–31. doi: 10.1074/jbc.M112087200. [DOI] [PubMed] [Google Scholar]
- Hirakura Y, et al. Alzheimer amyloid abeta1-42 channels: effects of solvent, pH, and Congo Red. J Neurosci Res. 1999;57:458–66. [PubMed] [Google Scholar]
- Ionov ID. Survey of ALS-associated factors potentially promoting Ca2+ overload of motor neurons. Amyotroph Lateral Scler. 2007;8:260–5. doi: 10.1080/17482960701523124. [DOI] [PubMed] [Google Scholar]
- Israelson A, et al. Misfolded mutant SOD1 directly inhibits VDAC1 conductance in a mouse model of inherited ALS. Neuron. 2010;67:575–87. doi: 10.1016/j.neuron.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H, et al. Truncated beta-amyloid peptide channels provide an alternative mechanism for Alzheimer's Disease and Down syndrome. Proc Natl Acad Sci U S A. 2010a;107:6538–43. doi: 10.1073/pnas.0914251107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H, et al. Structural convergence among diverse, toxic beta-sheet ion channels. J Phys Chem B. 2010b;114:9445–51. doi: 10.1021/jp104073k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JA, et al. Formation of high molecular weight complexes of mutant Cu, Zn-superoxide dismutase in a mouse model for familial amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2000;97:12571–6. doi: 10.1073/pnas.220417997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, et al. Oxidized/misfolded superoxide dismutase-1: the cause of all amyotrophic lateral sclerosis? Ann Neurol. 2007;62:553–9. doi: 10.1002/ana.21319. [DOI] [PubMed] [Google Scholar]
- Kuwabara S, Kanai K. [Altered axonal ion channel function in amyotrophic lateral sclerosis]. Brain Nerve. 2007;59:1109–15. [PubMed] [Google Scholar]
- Lin H, et al. Amyloid beta protein forms ion channels: implications for Alzheimer's disease pathophysiology. Faseb J. 2001;15:2433–44. doi: 10.1096/fj.01-0377com. [DOI] [PubMed] [Google Scholar]
- Marambaud P, et al. Calcium signaling in neurodegeneration. Mol Neurodegener. 2009;4:20. doi: 10.1186/1750-1326-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies FM, et al. Mitochondrial dysfunction in a cell culture model of familial amyotrophic lateral sclerosis. Brain. 2002;125:1522–33. doi: 10.1093/brain/awf167. [DOI] [PubMed] [Google Scholar]
- Mueller P, et al. Reconstitution of cell membrane structure in vitro and its transformation into an excitable system. Nature. 1962;194:979–80. doi: 10.1038/194979a0. [DOI] [PubMed] [Google Scholar]
- Pasinelli P, et al. Amyotrophic lateral sclerosis-associated SOD1 mutant proteins bind and aggregate with Bcl-2 in spinal cord mitochondria. Neuron. 2004;43:19–30. doi: 10.1016/j.neuron.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Pieri M, et al. Altered excitability of motor neurons in a transgenic mouse model of familial amyotrophic lateral sclerosis. Neurosci Lett. 2003;351:153–6. doi: 10.1016/j.neulet.2003.07.010. [DOI] [PubMed] [Google Scholar]
- Quist A, et al. Amyloid ion channels: a common structural link for protein-misfolding disease. Proc Natl Acad Sci U S A. 2005;102:10427–32. doi: 10.1073/pnas.0502066102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaume AG, et al. Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat Genet. 1996;13:43–7. doi: 10.1038/ng0596-43. [DOI] [PubMed] [Google Scholar]
- Rosen DR, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Sandelin E, et al. Amyotrophic lateral sclerosis-associated copper/zinc superoxide dismutase mutations preferentially reduce the repulsive charge of the proteins. J Biol Chem. 2007;282:21230–6. doi: 10.1074/jbc.M700765200. [DOI] [PubMed] [Google Scholar]
- Scholle MD, et al. In vivo biotinylated proteins as targets for phage-display selection experiments. Protein Expr Purif. 2004;37:243–52. doi: 10.1016/j.pep.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Seetharaman SV, et al. Disrupted zinc-binding sites in structures of pathogenic SOD1 variants D124V and H80R. Biochemistry. 2010;49:5714–25. doi: 10.1021/bi100314n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirwany NA, et al. The amyloid beta ion channel hypothesis of Alzheimer's disease. Neuropsychiatr Dis Treat. 2007;3:597–612. [PMC free article] [PubMed] [Google Scholar]
- Tiwari A, et al. Aberrantly increased hydrophobicity shared by mutants of Cu,Znsuperoxide dismutase in familial amyotrophic lateral sclerosis. J Biol Chem. 2005;280:29771–9. doi: 10.1074/jbc.M504039200. [DOI] [PubMed] [Google Scholar]
- Urushitani M, et al. The endoplasmic reticulum-Golgi pathway is a target for translocation and aggregation of mutant superoxide dismutase linked to ALS. Faseb Journal. 2008;22:2476–2487. doi: 10.1096/fj.07-092783. [DOI] [PubMed] [Google Scholar]
- Urushitani M, et al. N-methyl-D-aspartate receptor-mediated mitochondrial Ca(2+) overload in acute excitotoxic motor neuron death: a mechanism distinct from chronic neurotoxicity after Ca(2+) influx. J Neurosci Res. 2001;63:377–87. doi: 10.1002/1097-4547(20010301)63:5<377::AID-JNR1032>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Vande Velde C, et al. Selective association of misfolded ALS-linked mutant SOD1 with the cytoplasmic face of mitochondria. Proc Natl Acad Sci U S A. 2008;105:4022–7. doi: 10.1073/pnas.0712209105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, et al. Extracellular mutant SOD1 induces microglial-mediated motoneuron injury. Glia. 2010;58:231–243. doi: 10.1002/glia.20919. [DOI] [PMC free article] [PubMed] [Google Scholar]