Abstract
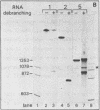
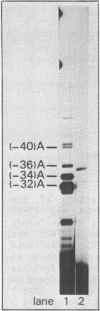
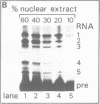
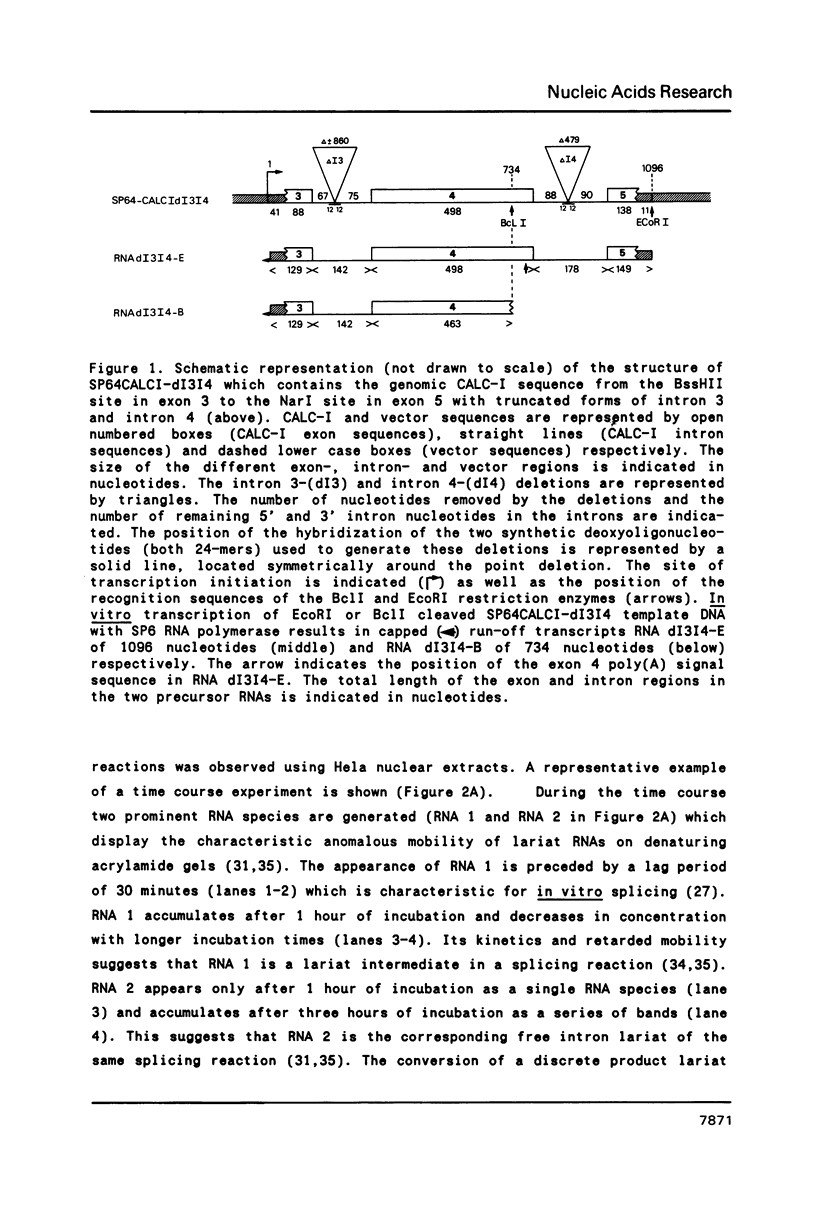
The Calcitonin/CGRP-I (CALC-I) gene is known to be expressed in a tissue specific fashion resulting in the production of Calcitonin mRNA in thyroid C-cells and CGRP-I mRNA in particular nerve cells. The alternative RNA processing reactions include splicing of exons 1, 2 and 3 to exon 4 and poly (A) addition at exon 4 (Calcitonin mRNA) or splicing of exons 1, 2 and 3 to exons 5 and 6 and poly (A) addition at exon 6 (CGRP-I mRNA). Using a model precursor RNA containing the exon 3 to exon 5 region of the human CALC-I gene we have investigated the Calcitonin- and CGRP-I mRNA-specific processing reactions in vitro, in nuclear extracts of Hela, PC12 and Ewing-1B cells, respectively. Extracts of PC12- and Ewing-1B cells were expected to perform CGRP mRNA-specific splicing, whereas Calcitonin mRNA specific processing was expected to occur in Hela cell extracts. Surprisingly, CGRP mRNA-specific splicing of exon 3 to exon 5 was the predominant reaction in all three extracts. Significant Calcitonin mRNA-specific splicing of exon 3 to exon 4 only took place upon elimination of the dominant downstream 3' splice site used in CGRP mRNA-specific splicing. This elimination occurs most definitively by cleavage at the Calcitonin mRNA specific poly (A) site at exon 4 which may then be the major regulatory mechanism for tissue-specific expression of the CALC-I gene.
Full text
PDF















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amara S. G., Evans R. M., Rosenfeld M. G. Calcitonin/calcitonin gene-related peptide transcription unit: tissue-specific expression involves selective use of alternative polyadenylation sites. Mol Cell Biol. 1984 Oct;4(10):2151–2160. doi: 10.1128/mcb.4.10.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amara S. G., Jonas V., Rosenfeld M. G., Ong E. S., Evans R. M. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature. 1982 Jul 15;298(5871):240–244. doi: 10.1038/298240a0. [DOI] [PubMed] [Google Scholar]
- Bovenberg R. A., van de Meerendonk W. P., Baas P. D., Steenbergh P. H., Lips C. J., Jansz H. S. Model for alternative RNA processing in human calcitonin gene expression. Nucleic Acids Res. 1986 Nov 25;14(22):8785–8803. doi: 10.1093/nar/14.22.8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabot B., Steitz J. A. Multiple interactions between the splicing substrate and small nuclear ribonucleoproteins in spliceosomes. Mol Cell Biol. 1987 Jan;7(1):281–293. doi: 10.1128/mcb.7.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R. K., Hall L., Edbrooke M. R., Allison J., MacIntyre I. Partial nucleotide sequence of human calcitonin precursor mRNA identifies flanking cryptic peptides. Nature. 1982 Jan 28;295(5847):345–347. doi: 10.1038/295345a0. [DOI] [PubMed] [Google Scholar]
- Crenshaw E. B., 3rd, Russo A. F., Swanson L. W., Rosenfeld M. G. Neuron-specific alternative RNA processing in transgenic mice expressing a metallothionein-calcitonin fusion gene. Cell. 1987 May 8;49(3):389–398. doi: 10.1016/0092-8674(87)90291-1. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edbrooke M. R., Parker D., McVey J. H., Riley J. H., Sorenson G. D., Pettengill O. S., Craig R. K. Expression of the human calcitonin/CGRP gene in lung and thyroid carcinoma. EMBO J. 1985 Mar;4(3):715–724. doi: 10.1002/j.1460-2075.1985.tb03688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frendewey D., Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985 Aug;42(1):355–367. doi: 10.1016/s0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- Gerke V., Steitz J. A. A protein associated with small nuclear ribonucleoprotein particles recognizes the 3' splice site of premessenger RNA. Cell. 1986 Dec 26;47(6):973–984. doi: 10.1016/0092-8674(86)90812-3. [DOI] [PubMed] [Google Scholar]
- Grabowski P. J., Padgett R. A., Sharp P. A. Messenger RNA splicing in vitro: an excised intervening sequence and a potential intermediate. Cell. 1984 Jun;37(2):415–427. doi: 10.1016/0092-8674(84)90372-6. [DOI] [PubMed] [Google Scholar]
- Grabowski P. J., Seiler S. R., Sharp P. A. A multicomponent complex is involved in the splicing of messenger RNA precursors. Cell. 1985 Aug;42(1):345–353. doi: 10.1016/s0092-8674(85)80130-6. [DOI] [PubMed] [Google Scholar]
- Hartmuth K., Barta A. Unusual branch point selection in processing of human growth hormone pre-mRNA. Mol Cell Biol. 1988 May;8(5):2011–2020. doi: 10.1128/mcb.8.5.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N., Roeder R. G. Transcription of human histone genes in extracts from synchronized HeLa cells. Proc Natl Acad Sci U S A. 1984 May;81(9):2713–2717. doi: 10.1073/pnas.81.9.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornig H., Aebi M., Weissmann C. Effect of mutations at the lariat branch acceptor site on beta-globin pre-mRNA splicing in vitro. Nature. 1986 Dec 11;324(6097):589–591. doi: 10.1038/324589a0. [DOI] [PubMed] [Google Scholar]
- Höppener J. W., Steenbergh P. H., Slebos R. J., Visser A., Lips C. J., Jansz H. S., Bechet J. M., Lenoir G. M., Born W., Haller-Brem S. Expression of the second calcitonin/calcitonin gene-related peptide gene in Ewing sarcoma cell lines. J Clin Endocrinol Metab. 1987 Apr;64(4):809–817. doi: 10.1210/jcem-64-4-809. [DOI] [PubMed] [Google Scholar]
- Jonas V., Lin C. R., Kawashima E., Semon D., Swanson L. W., Mermod J. J., Evans R. M., Rosenfeld M. G. Alternative RNA processing events in human calcitonin/calcitonin gene-related peptide gene expression. Proc Natl Acad Sci U S A. 1985 Apr;82(7):1994–1998. doi: 10.1073/pnas.82.7.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedes D. H., Steitz J. A. Accurate 5' splice-site selection in mouse kappa immunoglobulin light chain premessenger RNAs is not cell-type-specific. Proc Natl Acad Sci U S A. 1987 Nov;84(22):7928–7932. doi: 10.1073/pnas.84.22.7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller E. B., Noon W. A. Intron splicing: a conserved internal signal in introns of animal pre-mRNAs. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7417–7420. doi: 10.1073/pnas.81.23.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M. M., Padgett R. A., Sharp P. A. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984 Oct;38(3):731–736. doi: 10.1016/0092-8674(84)90268-x. [DOI] [PubMed] [Google Scholar]
- Krainer A. R., Maniatis T., Ruskin B., Green M. R. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984 Apr;36(4):993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- Krämer A. Analysis of RNase-A-resistant regions of adenovirus 2 major late precursor-mRNA in splicing extracts reveals an ordered interaction of nuclear components with the substrate RNA. J Mol Biol. 1987 Aug 5;196(3):559–573. doi: 10.1016/0022-2836(87)90032-5. [DOI] [PubMed] [Google Scholar]
- Langford C. J., Klinz F. J., Donath C., Gallwitz D. Point mutations identify the conserved, intron-contained TACTAAC box as an essential splicing signal sequence in yeast. Cell. 1984 Mar;36(3):645–653. doi: 10.1016/0092-8674(84)90344-1. [DOI] [PubMed] [Google Scholar]
- Le Moullec J. M., Jullienne A., Chenais J., Lasmoles F., Guliana J. M., Milhaud G., Moukhtar M. S. The complete sequence of human preprocalcitonin. FEBS Lett. 1984 Feb 13;167(1):93–97. doi: 10.1016/0014-5793(84)80839-x. [DOI] [PubMed] [Google Scholar]
- Leff S. E., Evans R. M., Rosenfeld M. G. Splice commitment dictates neuron-specific alternative RNA processing in calcitonin/CGRP gene expression. Cell. 1987 Feb 13;48(3):517–524. doi: 10.1016/0092-8674(87)90202-9. [DOI] [PubMed] [Google Scholar]
- Leff S. E., Rosenfeld M. G., Evans R. M. Complex transcriptional units: diversity in gene expression by alternative RNA processing. Annu Rev Biochem. 1986;55:1091–1117. doi: 10.1146/annurev.bi.55.070186.005303. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Reed R. The role of small nuclear ribonucleoprotein particles in pre-mRNA splicing. Nature. 1987 Feb 19;325(6106):673–678. doi: 10.1038/325673a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McLauchlan J., Gaffney D., Whitton J. L., Clements J. B. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3' termini. Nucleic Acids Res. 1985 Feb 25;13(4):1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris H. R., Panico M., Etienne T., Tippins J., Girgis S. I., MacIntyre I. Isolation and characterization of human calcitonin gene-related peptide. Nature. 1984 Apr 19;308(5961):746–748. doi: 10.1038/308746a0. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett R. A., Konarska M. M., Grabowski P. J., Hardy S. F., Sharp P. A. Lariat RNA's as intermediates and products in the splicing of messenger RNA precursors. Science. 1984 Aug 31;225(4665):898–903. doi: 10.1126/science.6206566. [DOI] [PubMed] [Google Scholar]
- Petermann J. B., Born W., Chang J. Y., Fischer J. A. Identification in the human central nervous system, pituitary, and thyroid of a novel calcitonin gene-related peptide, and partial amino acid sequence in the spinal cord. J Biol Chem. 1987 Jan 15;262(2):542–545. [PubMed] [Google Scholar]
- Peterson M. L., Perry R. P. Regulated production of mu m and mu s mRNA requires linkage of the poly(A) addition sites and is dependent on the length of the mu s-mu m intron. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8883–8887. doi: 10.1073/pnas.83.23.8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R., Maniatis T. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell. 1985 May;41(1):95–105. doi: 10.1016/0092-8674(85)90064-9. [DOI] [PubMed] [Google Scholar]
- Rosenfeld M. G., Mermod J. J., Amara S. G., Swanson L. W., Sawchenko P. E., Rivier J., Vale W. W., Evans R. M. Production of a novel neuropeptide encoded by the calcitonin gene via tissue-specific RNA processing. Nature. 1983 Jul 14;304(5922):129–135. doi: 10.1038/304129a0. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Green M. R. An RNA processing activity that debranches RNA lariats. Science. 1985 Jul 12;229(4709):135–140. doi: 10.1126/science.2990042. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Green M. R. Specific and stable intron-factor interactions are established early during in vitro pre-mRNA splicing. Cell. 1985 Nov;43(1):131–142. doi: 10.1016/0092-8674(85)90018-2. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Greene J. M., Green M. R. Cryptic branch point activation allows accurate in vitro splicing of human beta-globin intron mutants. Cell. 1985 Jul;41(3):833–844. doi: 10.1016/s0092-8674(85)80064-7. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Krainer A. R., Maniatis T., Green M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984 Aug;38(1):317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Zamore P. D., Green M. R. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988 Jan 29;52(2):207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- Sabate M. I., Stolarsky L. S., Polak J. M., Bloom S. R., Varndell I. M., Ghatei M. A., Evans R. M., Rosenfeld M. G. Regulation of neuroendocrine gene expression by alternative RNA processing. Colocalization of calcitonin and calcitonin gene-related peptide in thyroid C-cells. J Biol Chem. 1985 Mar 10;260(5):2589–2592. [PubMed] [Google Scholar]
- Schmitt P., Gattoni R., Keohavong P., Stévenin J. Alternative splicing of E1A transcripts of adenovirus requires appropriate ionic conditions in vitro. Cell. 1987 Jul 3;50(1):31–39. doi: 10.1016/0092-8674(87)90659-3. [DOI] [PubMed] [Google Scholar]
- Sharp P. A. Splicing of messenger RNA precursors. Science. 1987 Feb 13;235(4790):766–771. doi: 10.1126/science.3544217. [DOI] [PubMed] [Google Scholar]
- Solnick D. Alternative splicing caused by RNA secondary structure. Cell. 1985 Dec;43(3 Pt 2):667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]
- Steenbergh P. H., Höppener J. W., Zandberg J., Van de Ven W. J., Jansz H. S., Lips C. J. Calcitonin gene related peptide coding sequence is conserved in the human genome and is expressed in medullary thyroid carcinoma. J Clin Endocrinol Metab. 1984 Aug;59(2):358–360. doi: 10.1210/jcem-59-2-358. [DOI] [PubMed] [Google Scholar]
- Steenbergh P. H., Höppener J. W., Zandberg J., Visser A., Lips C. J., Jansz H. S. Structure and expression of the human calcitonin/CGRP genes. FEBS Lett. 1986 Dec 1;209(1):97–103. doi: 10.1016/0014-5793(86)81091-2. [DOI] [PubMed] [Google Scholar]
- Strauss M., Streuli C. H., Griffin B. E. Efficient oligodeoxyribonucleotide-directed deletion mutagenesis using pEMBL vectors: removal of early region introns from polyoma virus mutants. Gene. 1986;49(3):331–340. doi: 10.1016/0378-1119(86)90369-0. [DOI] [PubMed] [Google Scholar]
- Tazi J., Alibert C., Temsamani J., Reveillaud I., Cathala G., Brunel C., Jeanteur P. A protein that specifically recognizes the 3' splice site of mammalian pre-mRNA introns is associated with a small nuclear ribonucleoprotein. Cell. 1986 Dec 5;47(5):755–766. doi: 10.1016/0092-8674(86)90518-0. [DOI] [PubMed] [Google Scholar]
- Tschopp F. A., Henke H., Petermann J. B., Tobler P. H., Janzer R., Hökfelt T., Lundberg J. M., Cuello C., Fischer J. A. Calcitonin gene-related peptide and its binding sites in the human central nervous system and pituitary. Proc Natl Acad Sci U S A. 1985 Jan;82(1):248–252. doi: 10.1073/pnas.82.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]