It is somewhat ironic that an important finding concerned with cellulose biosynthesis in higher plants (1) appeared just before the paper in this issue of the Proceedings by Smant et al. (2) describing genes from a plant parasitic nematode that encode proteins that degrade cellulose. Cellulose is the most abundant polymer on the planet and is an essential component of the plant cell wall. Although the cell wall or extracellular matrix, as it is sometimes called, is complex (3), crystalline cellulose is the structural linchpin. Given this situation, it is not surprising that hungry pathogens commonly attack components of the plant cell wall, including cellulose.
Genetic evidence has established that cellulases that depolymerize cellulose often are required for high virulence (the quantitative degree of pathogenicity) by microbial pathogens (e.g., refs. 4 and 5). These entrepreneurs damage the plant cell wall to facilitate their movement through plant tissue and to obtain nutrients from the digested cell wall components or the contents of unclothed plant protoplasts. Curiosities abound, including questions such as: Where and when did cell wall degrading genes originate? How were such genes recruited by pathogens? What do plants do about this mode of attack? How can human activity intervene and prevent such plant diseases without secondary downsides? Answers are not abundant, but we are improving our understanding of plant pathogenesis and what to do about it so that humans reap more of the harvest.
First, pathogens are turning out to be copycats. For example, microbial pathogens of plants and animals share clusters of highly homologous genes involved in pathogenicity and virulence. These have come to be called “pathogenicity islands” (6), and there is compelling reason to think that an entire “island” containing dozens of genes can move from one organism to another, thus illuminating one of the questions posed above. Second, there is clear commonality in the way virulence mechanisms are delivered by pathogens to plant and animal cells. These secretion systems, encoded by particular gene clusters, deliver virulence proteins either to the surface of host cells or, in some cases, into the host cells. Some of the virulence proteins also are recognized by plants to initiate active defense mechanisms (7). Although certain of the virulence proteins delivered may also be conserved, the current view is that virulence mechanisms targeted to plant and animal cells differ considerably and indeed may be specific for one or a few host species or even only a single genotype. Cellulases are a good case in point because they are widely made by pathogens that attack plants but not those that attack animals. Animals are often pathogens or pests on plants, however, and one animal group with considerable economic impact is nematodes (8). As Smant et al. (2) have now shown, plant pathogenic nematodes appear to have borrowed cellulase genes from microbes to enhance their pathogenic success.
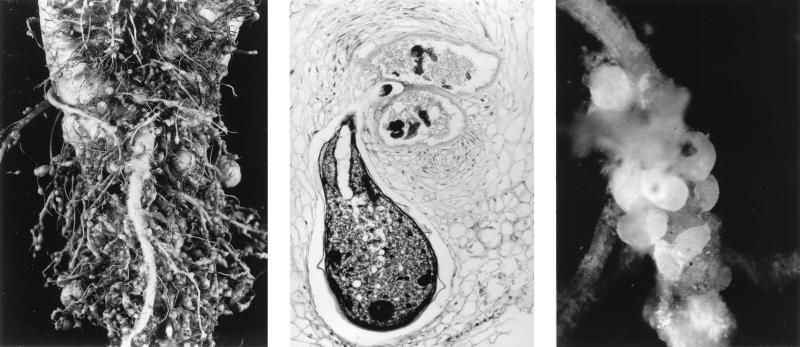
These nematodes begin life as larvae that are free-living in the soil, but to complete the life cycle they must find a plant host and establish an adequate nutritional relationship. Once the fledgling nematode locates a plant root, the question arises of “how to rob the bank.” Some nematodes do this by randomly puncturing plant cells with a lancet device called a “stylet,” and sucking out the cell contents for lunch. The style of these less discriminating “hit and run” artists is contrasted by other nematodes that establish specialized feeding sites on plant roots for their growth and reproduction. The latter nematodes trick plant cells into abnormal enlargement and division, eventually forming “giant cells” or “syncytia,” which may be enclosed in massive galls (Fig. 1) (9). These structures are made at the expense of the remainder of the plant, which becomes stunted. The nematode females establish residence among the modified plant cells and use their stylets to feed on the plant cell contents without killing them, thus ensuring tomorrow’s meal. As a consequence of their feast, the initially small, tubular nematode females grow rapidly, eventually becoming large and spherical and filled with thousands of eggs (Fig. 1). This is a clever and rather insidious modus operandi for subverting plant development to benefit the pathogen. It has been suspected that the production of cellulases may be required for initial infection by the nematodes when they migrate intracellularly from point of root entry to the feeding site, or possibly in the abnormal development of the enlarged plant cells.
Figure 1.
Nematode infections on plant roots. (Left) Root-knot galls caused by a member of the nematode genus Meloidogyne on plant roots. (Center) Microscopic view of a root-knot nematode (Meloidogyne sp.) female feeding on plant giant cells. Members of this group cause large galls as well as malformed and stunted roots and above-ground plant parts (photo courtesy of E. Bernard, University of Tennessee, Knoxville, TN). (Right) Cyst nematode (Heterodera avenae) females on oat roots. Note the swollen bodies. This nematode is closely related to those studied by Smant et al. (2). The females mature into egg-filled cysts and greatly reduce plant growth, development, and agricultural yield (photo courtesy of R. Cook, Institute of Grassland and Environmental Research, Aberystwyth, U.K.).
In a tour de force, the authors (10) previously isolated an mAb specific to protein in esophageal glands that presumably is also present in nematode stylet secretions. Smant et al. (2) used this antibody to painstakingly isolate enough target proteins from the tiny nematodes to obtain partial sequence data. This information was in turn employed to obtain nematode cDNA clones by reverse genetics. Two different cellulase genes were cloned in this way from each of two nematode species and, upon expression in Escherichia coli, their protein products were shown in fact to exhibit cellulase activity. Somewhat surprisingly, all four proteins looked very much like microbial rather than eukaryote cellulases despite conclusive evidence that the cloned genes in fact did originate from the nematodes. This result returns us to the queries at the outset on where such genes come from and how they move between organisms. The bet is that the nematodes borrowed the cellulase genes from microorganisms at some point in their evolution. It is quite possible that microbes in the digestive system of nonparasitic nematodes may have been the source of cellulase and other genes that converted the nematodes to a parasitic habit.
Although standard molecular genetic tricks are technically difficult with plant parasitic nematodes, the next step will be to transform nematodes with antisense constructs of the cellulase genes to reduce their expression. If the transgenic animals are reduced in virulence, the observation would strongly support the importance of the cellulases in virulence. The success in identifying and cloning the nematode cellulase genes naturally also has accelerated efforts to identify other components of nematode esophageal exudates and their roles in pathogenicity.
The paper by Smant et al. (2) graphically illustrates that the current “Golden Age of Biology” is extending into areas previously deemed difficult if not downright impenetrable! Plant pathogenic nematodes have quirks that make them difficult for application of standard molecular genetic and cell biology approaches to understanding their secrets, but as Smant et al. (2) have shown, such nematodes nonetheless are joining the celebration. This trend can be expected to increase. Why? First, plant pathogenic nematodes have a close cousin, Caenorhabditis elegans, a darling of developmental biologists that will have its complete genome sequenced very soon. That information will stimulate investigation not only of Caenorhabditis, but also its plant pathogenic cousins. Second, fruitful approaches are being taken to deduce plant genes that are stimulated by nematode infection (11, 12). For pathogens such as these that induce pronounced changes in plant development, study of the plant genes involved will tell us a great deal about basic plant biology as well as nematode biology. Finally, the kind of relentless yet clever assault exemplified by the paper of Smant et al. (2) bodes well for increased understanding of nematode pathogens of plants.
Footnotes
A commentary on this article begins on page 4906.
References
- 1.Arioli T, Peng L, Betzner A S, Burn J, Wittke W, Herth W, Camilleri C, Hofte H, Lazinski J, Birch R, et al. Science. 1998;279:717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- 2.Smant G, Stokkermans J P W G, Yan Y, de Boer J M, Baum T J, Wang X, Hussey R S, Gommers F J, Henrissat B, Davis E L, et al. Proc Natl Acad Sci USA. 1998;95:4906–4911. doi: 10.1073/pnas.95.9.4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carpita N, McCann M, Griffing L R. Plant Cell. 1996;8:1451–1463. doi: 10.1105/tpc.8.9.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts D P, Denny T P, Schell M A. J Bacteriol. 1988;170:1445–1451. doi: 10.1128/jb.170.4.1445-1451.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barras F, van Gijsegem F, Chatterjee A K. Annu Rev Phytopathol. 1994;32:201–234. [Google Scholar]
- 6.Groisman E A, Ochman H. Cell. 1996;87:791–794. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 7.Alfano J R, Collmer A. J Bacteriol. 1997;179:5655–5662. doi: 10.1128/jb.179.18.5655-5662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barker K R, Hussey R S, Krusberg L R, Bird G W, Dunn R A, Ferris H, Ferris V R, Freckman D W, Gabriel C J, Grewal P S, et al. J Nematol. 1994;26:127–137. [PMC free article] [PubMed] [Google Scholar]
- 9.Endo B Y. In: Electron Microscopy of Plant Pathogens. Mendgen K, Lesemann D E, editors. Berlin: Springer; 1991. pp. 291–305. [Google Scholar]
- 10.de Boer J M, Smant G, Goverse A, Davis E L, Overmars H A, Pomp H, van Gent-Pelzer M, Zilverentant J F, Stokkermans J P W G, Hussey R S, et al. Mol Plant–Microbe Interact. 1996;9:39–46. doi: 10.1094/mpmi-9-0039. [DOI] [PubMed] [Google Scholar]
- 11.Bird D M. J Parasitol. 1996;82:881–888. [PubMed] [Google Scholar]
- 12.Barthels N, van der Lee F M, Klap J, Goddijn O J M, Karimi M, Puzio P, Grundler F M W, Ohl S A, Lindsey K, Robertson L, et al. Plant Cell. 1997;9:2119–2134. doi: 10.1105/tpc.9.12.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]