Abstract
The lagging-strand DNA polymerase requires an oligoribonucleotide, synthesized by DNA primase, to initiate the synthesis of an Okazaki fragment. In the replication system of bacteriophage T7 both DNA primase and DNA helicase activities are contained within a single protein, the bifunctional gene 4 protein (gp4). Intermolecular interactions between gp4 and T7 DNA polymerase are crucial for the stabilization of the oligoribonucleotide, its transfer to the polymerase, and its extension by DNA polymerase. We have identified conditions necessary to assemble the T7 priming complex and characterized its biophysical properties using fluorescence anisotropy. In order to reveal molecular interactions that occur during delivery of the oligoribonucleotide to DNA polymerase, we have used four genetically altered gp4 to demonstrate that both the RNA polymerase and the zinc-finger domains of DNA primase are involved in the stabilization of the priming complex and in sequence recognition in the DNA template. We find that the helicase domain of gp4 contributes to the stability of the complex by binding to the ssDNA template. The C-terminal tail of gp4 is not required for complex formation.
Keywords: DNA replication, RNA primer, replisome
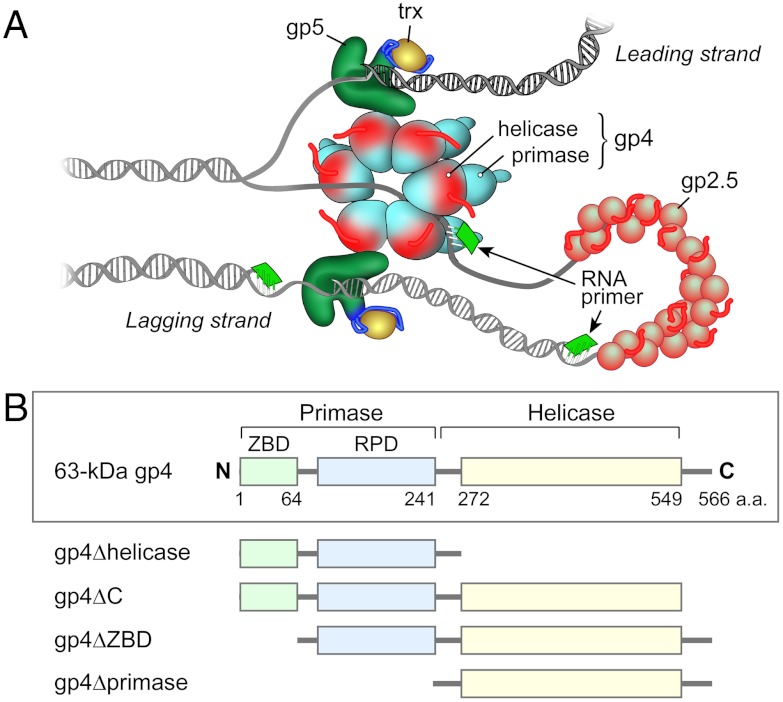
The relative simplicity of the replication system of bacteriophage T7 offers advantages for the study of molecular interactions that occur during DNA replication. All the fundamental activities that take place at a replication fork can be reconstituted with four proteins (Fig. 1A): the gene 5 DNA polymerase (gp5), gene 4 DNA primase-helicase (gp4), gene 2.5 ssDNA binding protein (gp2.5), and the processivity factor, Escherichia coli thioredoxin (trx) (1). Physical interactions between these proteins and DNA coordinate processive synthesis of the leading and lagging strands with a replication loop on the lagging strand containing the nascent Okazaki fragment (1).
Fig. 1.
The replisome of bacteriophage T7 and truncation variants of gp4. (A) The replisome of bacteriophage T7 contains DNA polymerase (gp5) and its processivity factor, Escherichia coli thioredoxin (trx), DNA primase-helicase (gp4), and ssDNA-binding protein (gp2.5). Gp4 unwinds dsDNA and generates two ssDNA templates for the leading and lagging-strand gp5/trx. Gp2.5 coats the lagging-strand ssDNA. The primase domain of gp4 catalyzes the synthesis of oligoribonucleotides that function as primers for the initiation of each Okazaki fragment. (B) Gp4 consists of two major domains connected by a flexible linker. The C-terminal half of gp4 contains the helicase domain and the N-terminal half the primase domain. The primase domain contains two subdomains: the N-terminal ZBD and the RPD that are also connected by a flexible linker. The carboxy-terminal 17 amino acids of the helicase domain are unstructured. The positions of residues important in the construction of the various forms of g4 are indicated. In order to assess the roles of individual domains of gp4 in stabilization of the priming complex, we constructed four truncated proteins: gp4∆helicase (residues 1-272) lacks the helicase domain but contains the flexible linker connecting the RPD with the helicase domain; gp4ΔC (residues 1-549) lacks the C-terminal 17 amino-acids; gp4ΔZBD (residues 64-566) lacks the N-terminal ZBD but includes the flexible linker that connects the ZBD and RPD; gp4Δprimase (residues 241-566) lacks both the N-terminal ZBD and RPD but contains the flexible linker that connects the RPD and the helicase domain.
Gp5 forms a 1∶1 complex (gp5/trx) with its processivity factor trx that is encoded by the host bacterium (2). The crystal structure of gp5/trx bound to DNA revealed that trx binds to a 76 amino acid loop [thioredoxin binding domain (TBD)] located between helices H and H1 in the “thumb” region of gp5 (3). Gp5 alone is a nonprocessive DNA polymerase, dissociating from the DNA every 15 to 50 nucleotides (4). Trx bound to the TBD resides over the duplex portion of the primer/template in the DNA binding cleft (3, 5) and increases the processivity to approximately 800 nucleotides per binding event (6).
Gp4 provides both helicase and primase activities with the helicase domain located within the C-terminal half of the protein and the primase domain in the N-terminal half (Fig. 1B) (7). Crystal structures of the helicase domain (8–10) and the primase domain (11) have been determined. The helicase domain of gp4 uses the energy derived from the hydrolysis of dTTP to translocate on ssDNA in a 5′ to 3′ direction relative to the strand to which it is bound and to unwind duplex DNA that it encounters (12, 13). The DNA primase domain is composed of two sub-domains. The zinc-binding domain (ZBD), located at the N terminus of the primase, is connected to the C-terminal RNA polymerase domain (RPD) where oligoribonucleotides are synthesized (7). The helicase domain, ZBD, and RPD are connected by flexible linkers.
The current model for lagging-strand synthesis in the T7 replication system involves several events. First, the ZBD recognizes the trinucleotide sequence, 5′-GTC-3′ in the lagging strand (14). At this trinucleotide recognition site T7 primase catalyzes the synthesis of the diribonucleotide 5′-pppAC-3′ and further extends it to the functional tetraribonucleotides 5′-pppACCC-3′, 5′-pppACCA-3′, and 5′-pppACAC-3′ provided cognate sequences are available in the template. Finally, the tetraribonucleotide is transferred to T7 DNA polymerase, where it serves as a primer to initiate the synthesis of Okazaki fragments (14). Interestingly, the interaction of the ZBD with the RPD can occur in cis or trans within the functional gp4 hexamer (15). In the trans mode the ZBD on one subunit of the hexamer contacts the RPD on an adjacent subunit to catalyze the synthesis of the oligonucleotide. The trans mode has been postulated to explain the halting of helicase movement observed during oligoribonucleotide synthesis of primers (16).
Earlier studies suggested that the ZBD alone plays a major role in sequence recognition and in the delivery of the oligoribonucleotide to DNA polymerase (11). However, the results obtained were dependent of relatively high concentrations of the ZBD. In addition, gp4 lacking the ZBD is also capable of primer delivery, necessitating the involvement of other elements (17). Here, we present conditions under which the bacteriophage T7 priming complex can be assembled in vitro. We use this reconstituted system to dissect protein–protein and protein–DNA interactions that occur during delivery of the primer by gp4 to gp5/trx. We find that both the RPD and ZBD of gp4 are involved in primer delivery.
Results
Genetically Altered Variants of gp4.
In the current study we have used wild-type 63-kDa gp4 as well as four altered proteins: gp4ΔZBD, gp4ΔC, gp4Δprimase, and gp4Δhelicase (Fig. 1B). The gp4ΔZBD lacks the N-terminal zinc-finger motif and thus cannot recognize primase recognition sites in DNA. The ZBD is connected to the RPD by a flexible linker, and that linker is present in gp4ΔZBD. Gp4ΔZBD is similar to the 56-kDa gp4 found in phage-infected cells, where it arises as a result of an internal initiation codon and ribosome-binding site within the full length 63-kDa gp4 (18). The 36.5-kDa gp4Δprimase is a more extensive deletion of the N terminus, resulting in removal of the entire primase domain. Gp4Δprimase has the same dTTPase activity as the wild-type 63-kDa gp4, but the unwinding activity is several hundred-fold lower (19). Gp4ΔC lacks the C-terminal 17 amino acids, a motif that is essential for interaction of gp4 with gp5/trx for loading onto a replication fork (20) and during the process of polymerase exchange at the replication fork (21). The wild-type 63-kDa gp4, gp4ΔZBD, gp4ΔC, and gp4Δprimase display similar oligomerization patterns (Fig. S1), and 63-kDa gp4, gp4ΔZBD, and gp4ΔC have nearly identical ssDNA-binding affinities (Fig. S2). Gp4Δprimase binds the ssDNA approximately threefold less tightly than does 63-kDa gp4 (Fig. S2). Gp4Δhelicase lacks the entire helicase domain but retains the flexible linker that connects the RPD with the helicase domain in the wild-type gp4. Gp4Δhelicase forms dimers in solution, it is devoid of dTTPase and unwinding activities, and catalyzes template-directed synthesis of tetraribonucleotides at a rate similar to the rate measured for 63-kDa gp4 (22).
Formation of a Lagging-Strand Priming Complex.
We define the lagging-strand priming complex as a complex containing the following components: gp5/trx, gp4, and an oligonucleotide hybridized to a ssDNA template. These proteins, bound to a primer/template, interact with each other such that the complex adopts a priming mode conformation. The multiple interactions that occur must stabilize the oligoribonucleotide and place it into the DNA binding crevice of gp5/trx for extension by gp5. The gp5 used in these studies is a genetically altered protein with two amino acid substitutions (D5A, E7A) in the active site of the 3′–5′ exonuclease domain. The altered protein has a normal DNA polymerase activity, but its exonuclease activity is reduced by a factor of 106 (23). The elimination of exonuclease activity prevents hydrolysis of the oligoribonucleotide by gp5/trx.
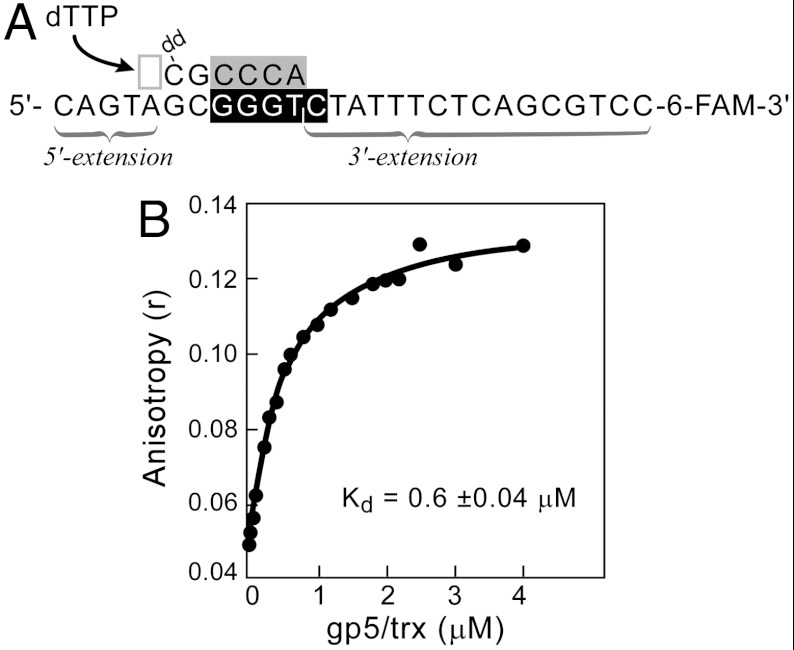
The primer/template consists of a 6-nt oligoribonucleotide annealed to a 27-nucleotide 3′-fluorescein-labeled template (Fig. 2A). The oligoribonucleotide (5′-rArCrCrCdGddC-3′) contains the sequence 5′-rArCrCrC-3′ that would be synthesized by DNA primase at the recognition sequence 5′-GGGTC-3′ to which it is annealed. The primer has an additional two nucleotides at the 3′ terminus with dideoxcytosine at the 3′ end. The resulting primer/template has a 5-nucleotide 5′ extension and a 16-nucleotide 3′ extension of the template (Fig. 2A). The addition of two deoxyribonucleotides at the 3′ end of the oligoribonucleotide increases the stability of the complex between gp5/trx and gp4 (24). The 3′-terminal ddC lacking a 3′-hydroxyl group prevents primer extension by gp5/trx and essentially locks gp5/trx in a polymerizing mode when the next incoming dNTP is present (3). dTTP is included in the reaction as it is the next incoming nucleotide that base-pairs with the template. dTTP is also the preferred nucleoside triphosphate for the helicase binding to DNA and for unwinding activity (25).
Fig. 2.
Formation of a lagging-strand priming complex. (A) The primer/template is constructed by annealing a 6-nucleotide primer with a 27-nucleotide 3′-fluorescein-labeled template such that a 5-nucleotide 5′-extension and a 16-nucleotide 3′ extension are formed. The primer contains the tetraribonucleotide sequence (gray box) complementary to that found in the primase recognition site (black box) and has an additional two deoxyribonucleotides (black letters). The primer contains a dideoxycytidine (ddC) at the 3′ terminus. The next incoming nucleotide (deoxythymidine 5′- triphosphate; dTTP) to base-pair with dA in the template is indicated by an arrow. (B) Reaction mixtures contained different concentrations of gp5/trx, 0.8 μM gp4, 5 mM dTTP, 0.1 μM template (shown in A), and 0.5 μM primer (shown in A). The components were incubated for 30 min at 25 °C in 20 mM TRIS pH 7.5, 200 mM NaCl, 2 mM MgCl2, and 5 mM DTT. After incubation anisotropy was measured using excitation and emission wavelengths of 495 nm and 518 nm, respectively. The apparent affinity (Kd) for gp5/trx binding to the complex is 0.6 ± 0.04 μM.
The formation of a complex between gp5/trx, gp4, and the primer/template can be observed and its stability determined by fluorescence anisotropy. Addition of gp5/trx and 63-kDa gp4 to the 6-carboxyfluorescein (6-FAM)-labeled primer/template in the presence of dTTP results in a dramatic increase in the anisotropy as the concentration of gp5/trx is increased (Fig. 2B). The apparent binding constant for gp5/trx is 0.6 ± 0.04 μM, and this value reflects the formation of a complex and its stability.
Characterization of Formation and Stoichiometry of the Lagging-Strand Priming Complex.
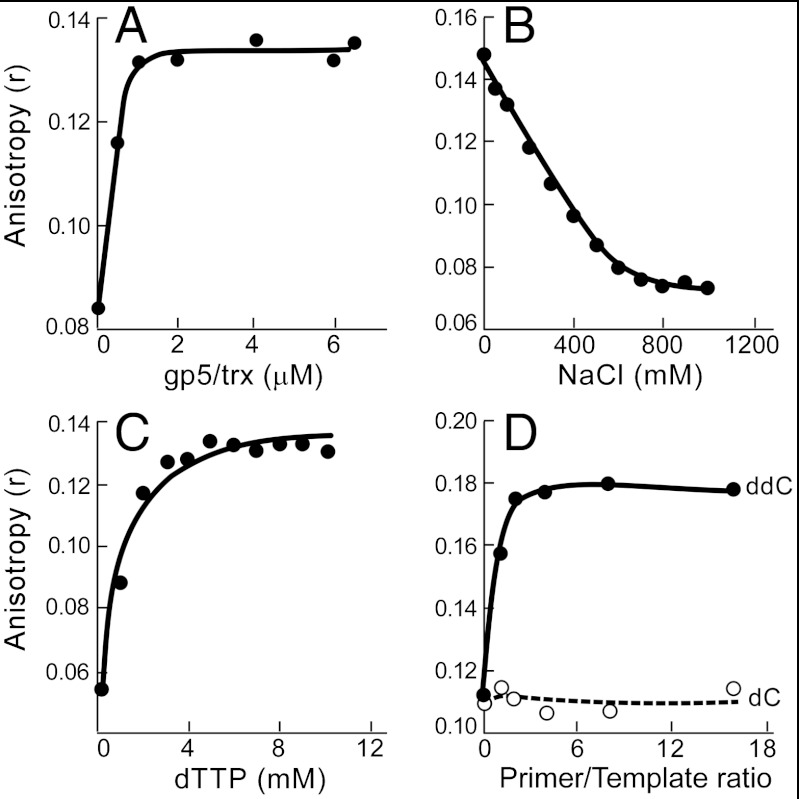
We have used fluorescence anisotropy measurements to identify the components and conditions necessary for the assembly of the priming complex demonstrated above. The stoichiometry of binding between gp5/trx and the 63-kDa gp4 in the presence of the primer/template shown in Fig. 3A was first measured. Increasing amounts of gp5/trx were added to a reaction mixtures containing 8 μM 63-kDa gp4 (monomeric concentration), 1 μM primer/template, and 5 mM dTTP. In the presence of dTTP the 63-kDa gp4 binds to ssDNA as a hexamer. Because 8 μM gp4 is sufficient to saturate l μM ssDNA we estimate the concentration of the gp4/primer/template complex in the reaction mixture to be 1 μM. Upon addition of gp5/trx the anisotropy increases until it reaches a plateau at approximately 1 μM gp5/trx indicating that the stoichiometry of binding between gp5/trx and the template is 1∶1.
Fig. 3.
Characterization of formation and stoichiometry of the lagging-strand priming complex. (A) Stoichiometry of binding between gp5/trx and gp4 in the priming complex. Reaction mixtures contained 8 μM 63-kDa gp4, 1 μM template (5′-CAG TAG CGG GTC TAT TTC TCA GCG TCG-6-FAM-3′), 5 μM primer (5′-rArCrCrCdGddC-3′), and different concentrations of gp5/trx. Reactions were incubated for 30 min, and anisotropy was measured. (B) Effect of NaCl on the stability of the priming complex. Reactions consisted of 0.5 μM gp5/trx, 1.5 μM 63-kDa gp4, 0.1 μM template (as above), 0.5 μM primer (as above) and were incubated for 30 min at 25 °C in 20 mM TRIS pH 7.5, 5 mM dTTP, 2 mM MgCl2, and the indicated concentrations of NaCl. (C) Stability of the priming complex is dependent on dTTP. Reaction mixtures contained 0.5 μM gp5/trx, 1.5 μM 63-kDa gp4, 0.1 μM template (as above), 0.5 μM primer (as above), and the indicated concentrations of dTTP. Samples were incubated for 30 min, and then the anisotropy was measured. (D) The presence of the chain-terminating ddC on the primer is necessary for formation of a stable complex. Reaction mixtures consisted of 1.5 μM 63-kDa gp4, 0.5 μM gp5/trx, 5 mM dTTP, 0.1 μM template (as above), and different concentrations of the primer containing the chain-terminating dideoxycytidine at the 3′ end (ddC; 5′-rArCrCrCdGddC-3′) or deoxycytidine at the 3′ end (dC; 5′-rArCrCrCdGdC-3′). The graph shows anisotropy values as a function of a primer to template ratio.
The priming complex is sensitive to salt concentration. In the experiment shown in Fig. 3B increasing concentrations of NaCl were added to reaction mixtures containing the components necessary for formation of the priming complex. At 500 mM NaCl only 15% complex formation is observed. In subsequent experiments, 200 mM NaCl is present because this value corresponds to approximately the concentration of NaCl in the bacterial cell and is the optimal concentration for DNA synthesis using purified T7 replication proteins.
As described above, in these experiments we have used dTTP and a dideoxynucleotide-terminated primer to stabilize the complex. In addition, gp4 requires dTTP for the optimal formation of hexamers and for any movement on the primer/template. The importance of dTTP is shown in Fig. 3C, where increasing concentrations of dTTP are added to the priming complex. Complex formation reaches a maximum at approximately 5 mM dTTP as measured by the increase in anisotropy. In subsequent experiments the concentration of dTTP is 5 mM.
Specific Engagement of Gp5/trx in the Priming Complex.
We examined the effect of primer concentration on the stability of the complex as described in Fig. 3D. In this experiment, increasing amounts of the primer containing the chain-terminating ddC (5′-rArCrCrCdGddC-3′) were added to the reaction mixture containing the remaining components of the priming complex. After the 30 min incubation, we measured anisotropy of the labeled template (Fig. 3D). If no primer is present (the first data point), no complex is formed. The maximal stability of the complex is observed when 0.2 μM primer is present in the reaction mixture. There is no significant change with the addition of increasing concentrations of primer. The slight excess of primer required may reflect instability of the primer/template duplex at the temperature of 25 °C.
To assess if gp5/trx engages the primer in the complex we substituted a primer containing dC at the 3′ terminus rather than ddC (5′-rArCrCrCdGdC-3′). If gp5/trx makes specific contacts with the primer lacking the chain-terminating ddC it should extend the primer and then dissociate from the primer/template. The low anisotropy values that we measured in this experiment suggest that gp5/trx makes specific contacts with the primer that enable it to extend the primer (Fig. 3D).
The Role of the ZBD, RPD, and the C-Terminal Tail of Gp4 in Stabilization of the Priming Complex.
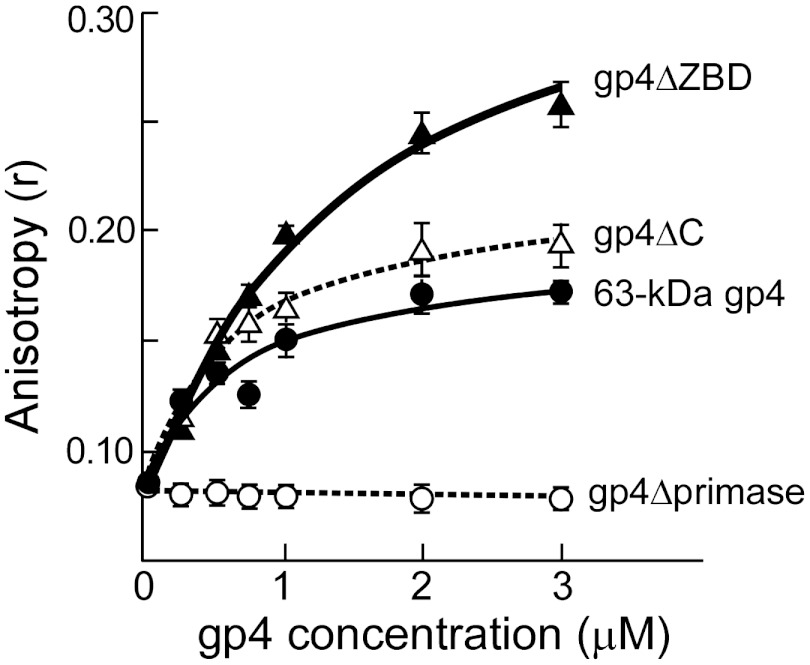
There are multiple modes of interaction between gp5/trx and gp4 in the T7 replisome (1). For example, the acidic C-terminal tail of gp4 interacts with a patch containing basic residues located near the O helix of gp5 to enable loading of the two proteins onto a replication fork (20). The C-terminal tail also binds to two basic loops in the TBD of gp5 to transiently capture gp5/trx if it dissociated from the primer/template during leading strand synthesis (21). To investigate the role of these electrostatic interactions in the priming complex we assessed binding of gp4ΔC, lacking the 17 C-terminal amino acids and compared it to the binding of 63-kDa gp4. Reaction mixtures contained 2 μM gp5/trx, 0.1 μM template, 0.5 μM primer, 5 mM dTTP, and different concentrations of gp4ΔC or 63-kDa gp4. Anisotropy readings obtained after 30 min incubation were plotted as a function of protein concentration in Fig. 4. The apparent affinities for gp4ΔC and 63-kDa gp4 binding to the priming complex were calculated using an algorithm for the single-site saturation. The Kd for gp4ΔC binding to the priming complex is 0.6 ± 0.02 μM, a value slightly lower than that measured for the wild-type gp4 (0.75 ± 0.05 μM). The above data suggest that interactions between gp5/trx and gp4 in the priming complex are not mediated through the C-terminal tail of gp4.
Fig. 4.
Molecular interactions between gp5/trx, gp4, and DNA in the priming complex. Anisotropy as a function of increasing concentrations of 63 kDa gp4 (filled circles), gp4∆ZBD (filled triangles), gp4ΔC (open triangles), gp4∆primase (open circles). The indicated gp4 were added to a reaction mixture containing 2 μM gp5/trx, 5 mM dTTP, 0.1 μM template (5′-CAG TAG CGG GTC TAT TTC TCA GCG TCG-6-FAM-3′), and 0.5 μM primer (5′-rArCrCrCdGddC-3′). After 30 min of incubation the anisotropy was measured. The error bars represent standard deviations of data points obtained in three independent experiments.
In order to assess the role of ZBD in the priming complex we examined complex formation using gp4ΔZBD lacking the N-terminal ZBD of the primase domain (Fig. 4). The apparent affinity for gp4ΔZBD binding to the priming complex is 1.5 ± 0.45 μM, a value two fold higher than that measured for 63-kDa gp4 (Kd of 0.75 ± 0.05 μM). This result suggests that the ZBD contributes to the formation of the complex, but its presence is not essential.
Finally, as a control we examined the ability of gp4 lacking the entire primase domain, gp4Δprimase, to form a priming complex. Not surprisingly, gp4Δprimase does not form the priming complex (Fig. 4).
Involvement of the Helicase Domain of Gp4 in Formation of the Priming Complex.
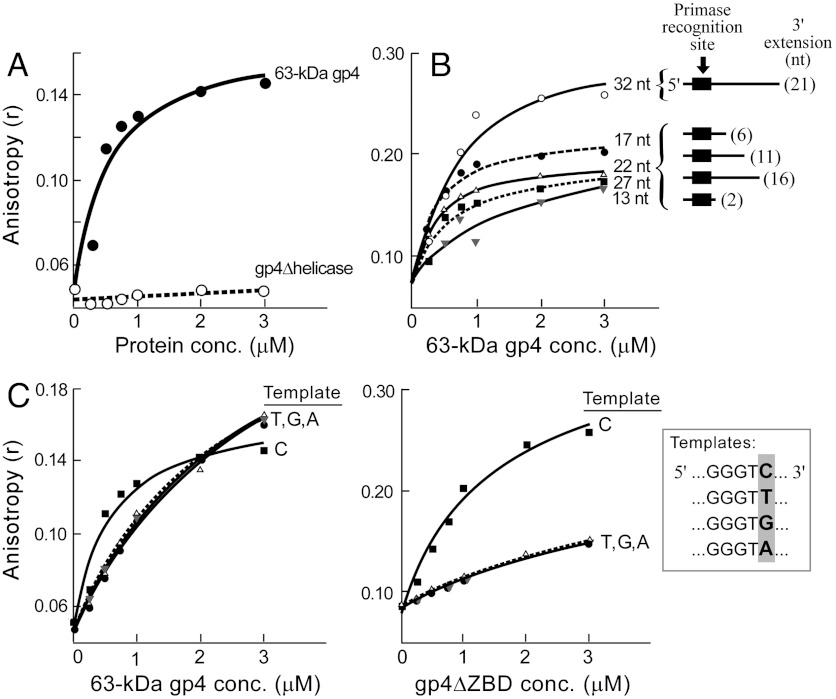
Having examined roles of the RPD, ZBD, and the C-terminal tail of gp4 in formation of the priming complex, we examined the role of the helicase domain of gp4. We assessed complex formation using gp4Δhelicase lacking the entire helicase domain. Reaction mixtures contained 2 μM gp5/trx, 0.1 μM template, 0.5 μM primer, 5 mM dTTP, and increasing concentrations of gp4Δhelicase. Following 30 min incubation anisotropy was measured, and the readings were plotted as a function of protein concentration in Fig. 5A. Gp4Δhelicase does not form the priming complex as indicated by the low anisotropy values. In one scenario, the helicase domain can contribute to the complex formation by binding to ssDNA, hence stabilizing the complex. In order to test this hypothesis we assessed complex formation using five different 3′-fluorescein-labeled templates (see Fig. 5B, Inset). These templates contain a different number of nucleotides (13, 17, 22, 27, or 32 nucleotides). After annealing with a 6-nucleotide primer (5′-rArCrCrCdGddC-3′), the 5-nucleotide 5′-extension is formed in all templates, but the 3′ extensions vary in length (2-, 6-, 11-, 16-, or 21-nucleotide 3′-extensions). The 3′ extension of the ssDNA template has to be sufficiently long for the helicase domain to bind. Reaction mixtures containing 2 μM gp5/trx, 0.5 μM primer, 5 mM dTTP, increasing concentrations of 63-kDa gp4, and 0.1 μM of each different templates were incubated for 30 min, and anisotropy readings were plotted as a function of the concentration of 63-kDa gp4 (Fig. 5B). The apparent affinities for the 63-kDa gp4 binding to the priming complexes containing templates with 17, 22, and 27 nucleotides are 0.2 ± 0.01 μM, 0.4 ± 0.05 μM, and 0.6 ± 0.2 μM, respectively. The data indicate that the optimal size of the 3′-extension for helicase binding ranges from 6 to 16 nucleotides. The 13-nucleotide template containing 2-nucleotide 3′ extension is not sufficiently long for helicase binding as reflected by Kd = 1.6 ± 1.1 μM. The template containing 32 nucleotides with the 21-nucleotide 3′-extension is too long, as reflected by high anisotropy readings that likely represent binding of multiple helicase rings to the 3′ extension. In summary, the results suggest that the helicase domain of gp4 contributes to the formation and stabilization of the priming complex, most likely by increasing the binding affinity of the complex to ssDNA.
Fig. 5.
The role of the helicase domain of gp4, and the “cryptic” C in the stabilization of the priming complex. (A) The helicase domain of gp4 stabilizes the priming complex. Reaction mixtures contained the indicated concentrations of 63 kDa gp4 (filled circles) or gp4∆helicase (open circles), 2 μM gp5/trx, 0.1 μM template (5′-CAG TAG CGG GTC TAT TTC TCA GCG TCG-6-FAM-3′), and 0.5 μM primer (5′-rArCrCrCdGddC-3′). After 30 min incubation at 25 °C anisotropy was measured, and the anisotropy values are plotted as a function of protein concentrations. (B) Priming complex binding to different templates. Five templates (see Inset) with varying 3′extensions: 21 nt (open circles), 16 nt (filled squares), 11 nt (open triangles), 6 nt (filled circles), and 2 nt (gray triangles) were incubated in a reaction mixture containing 0.5 μM primer (shown above), 2 μM gp5/trx, and increasing concentrations of 63 kDa gp4. Anisotropy was measured after 30 min incubation. (C) Stability of the priming complex using DNA templates (see Inset) in which the cryptic cytosine (template C) was substituted with thymine, guanine or adenine (templates: T, G, or A, respectively). Each template (0.1 μM) was incubated in a reaction mixture containing 0.5 μM primer, 2 μM gp5/trx, and increasing concentrations of 63 kDa gp4 or gp4∆ZBD. Anisotropy readings are shown as a function of increasing concentrations of 63 kDa gp4 or gp4∆ZBD.
The Role of the “Cryptic” Cytosine in Stabilization of the Priming Complex.
In the current model of lagging-strand synthesis the ZBD recognizes the trinucleotide sequence (5′-GTC-3′) within a DNA template. The cytosine in the template is “cryptic” in that it is essential for recognition but is not copied into the primer. We have examined the importance of the cryptic cytosine in the formation of a priming complex. In the experiment presented in Fig. 5C the ability of DNA templates in which the cytosine is replaced by either adenine, thymine, or guanine (see Inset) to support complex formation is examined. Reaction mixtures consisted of increasing concentrations of 63-kDa gp4, 2 μM gp5/trx, 0.5 μM primer, 5 mM dTTP, and 0.1 μM one of the four DNA templates. Following the 30 min incubation fluorescence anisotropy was measured, and the readings were plotted as a function of the increasing protein concentration. Substitution of the “cryptic” C with T, G, or A results in a decrease in the stability of the priming complex. Due to the weak 63-kDa gp4 binding to the complex only the lower bounds for the Kd could be determined. The wild-type gp4 binds to the priming complex containing templates T, G, and A with Kd > 2 μM. In comparison, the Kd for 63-kDa gp4 binding to the priming complex with the template C is 0.75 ± 0.05 μM. We conclude that the cryptic cytosine in the template is a significant determinant of stability of the priming complex.
Because the RPD is involved in stabilization of the priming complex (Fig. 4), we assessed the role of the RPD in recognition of the cryptic cytosine. Reaction mixtures containing increasing concentrations of gp4ΔZBD, 2 μM gp5/trx, 0.5 μM primer, 5 mM dTTP, and 0.1 μM one of the four DNA templates (C, T, G, or A) were incubated for 30 min, and fluorescence anisotropy was measured (Fig. 5C). gp4ΔZBD binds to the priming complex containing templates T, G, and A with Kd > 3 μM.
In comparison, the Kd for gp4ΔZBD binding to the priming complex with the template C is 1.5 ± 0.45 μM. Taken together the data show that both the ZBD and the RPD are important in the recognition of the cryptic cytosine and thus stabilization of the priming complex.
Discussion
DNA primases play an essential role in DNA replication. These enzymes catalyze the synthesis of oligoribonucleotides at specific recognition sites on the lagging DNA strand. DNA primases also stabilize the short oligoribonucleotides on the DNA and promote their utilization by DNA polymerases as primers to initiate the synthesis of Okazaki fragments (26). During replication in Escherichia coli, the β clamp and ssDNA binding protein mediate the transfer of a DNA template with a bound primer from the DnaG primase to DNA polymerase III (27). In contrast, the primase of gp4 directly transfers newly synthesized primers to gp5/trx and enables primer extension by an intimate physical interaction with gp5/trx (26).
In the present study we have identified conditions necessary for the formation of a stable priming complex between gp4, gp5/trx, and a primer/template. The priming complex consists of multiple interactions of the proteins with one another and with the primer/template, interactions that are required not only for sequence recognition and oligoribonucleotide synthesis but also for the delivery of the primer by gp4 to gp5/trx. The complex is formed only when the 3′ end of the primer contains the chain-terminating ddC and the next incoming nucleoside 5′ triphosphate is present. This requirement in itself provides strong evidence for a specific engagement of gp5/trx with the 3′ terminus of the primer within the complex. The presence of ddC prevents gp5/trx from extension of the primer and consequently dissociation from the priming complex. When the chain-terminating ddC is substituted with dC the primer is extended and the complex is unstable. However, formation of the stable complex also requires the next incoming dNTP. This dNTP base-pairs with the next nucleotide in the template and locks gp5/trx into a polymerization mode because the 3′ ddC on the primer has no 3′ hydroxyl group to carry out a nucleophilic attack on the α-phosphate of the dNTP. In the present study the template sequence was such that the next incoming nucleotide is dTTP. dTTP is the preferred nucleotide for oligomerization of gp4 and its binding to DNA. The presence of the helicase domain of gp4 is required for priming complex formation, most likely to increase the stability of the complex by binding to the 3′ extension of the template strand. This extension must be between 6 and 16 nucleotides long for optimal binding. Anisotropy experiments suggest that the priming complex is formed in 1∶6 stoichiometry with one gp5/trx per gp4 hexamer. Gp4 can bind to gp5/trx via two different electrostatic interactions involving contact of the acidic C-terminal tail of gp4 with either of two basic regions in gp5. We find that the C-terminal tail of gp4 does not play a role in stabilization of the priming complex as evidenced by nearly identical binding affinities for gp4ΔC (Kd of 0.6 μM) and 63-kDa gp4 (Kd of 0.75 μM) within the complex.
Earlier studies suggested that the ZBD plays a major role in primer delivery (11, 14). However, gp4 lacking the ZBD is also capable of primer delivery, necessitating the involvement of other elements (17). In the current study, we demonstrate that gp4 lacking the N-terminal ZBD (gp4ΔZBD) forms a complex with gp5/trx and primer/template. However, the complex is two fold less stable than one formed by wild-type 63-kDa gp4 (Kd of 1.5 ± 0.45 and 0.75 ± 0.05 μM, respectively). A genetically altered gp4 harboring a deletion of the entire primase domain (gp4Δprimase) is unable to form the priming complex, confirming that the RPD is crucial in stabilization of the complex. The ssDNA binding affinity of gp4Δprimase is approximately threefold lower than that of wild-type 63kDa gp4 (Kd of 370 ± 48 vs. 143 ± 42 nM, respectively). The decreased binding to ssDNA contributes to the low anisotropy signal measured with gp4Δprimase. The earlier studies suggested that the ZBD is exclusively responsible for binding to the cryptic cytosine in DNA template (11, 14). However, our results show that RPD is also involved in recognition of the cryptic cytosine.
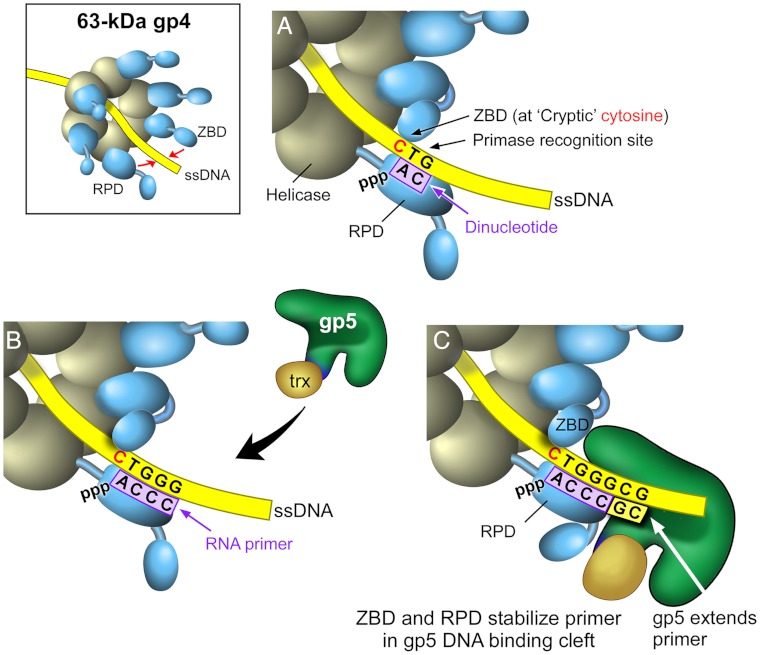
The model shown in Fig. 6 depicts our current understanding of the arrangement of gp4, gp5/trx, and primer/template in the priming complex. The hexameric gp4 encircles the lagging DNA strand and translocates in 5′ to 3′ direction relative to the strand to which it is bound to simultaneously unwind the duplex DNA that it encounters. The RPD of one subunit within the hexamer and the ZBD of an adjacent subunit (trans mode of primer synthesis) position ATP and CTP at the primase recognition site. The catalytic site within the RPD catalyzes the formation of the dinucleotide 5′ pppAC-3′ (Fig. 6A). The dinucleotide is subsequently extended to the tetraribonucleotide provided the correct sequence is present in the template (Fig. 6B). The cryptic cytosine is not copied into the primer. Primers can also be synthesized in a cis mode where the RPD and the ZBD of the same subunit physically interact. However, the trans interaction of the ZBD of one subunit with the RPD of an adjacent subunit described here likely triggers conformational changes that halt the sequential hydrolysis of dTTP by gp4 and consequently temporarily halts movement of the replication fork (15). This pause allows for the relatively slow process of primer synthesis and the multiple events that lead to the initiation of a new Okazaki fragment (16). Without such a pause, leading-strand synthesis would outpace lagging-strand synthesis. Once the tetranucleotide is synthesized it must be transferred into the DNA binding crevice of gp5 such that the 3′ terminus is situated in the active site (Fig. 6C). A loop of 4 amino acids (residues 401–404), not found in other members of the DNA polymerase I family, are located within the DNA-binding groove. Gp5 lacking this unique loop is defective in the use of tetranucleotides as primers to initiate DNA synthesis (28). Precisely how the tetranucleotide is stabilized and transferred to the DNA binding crevice is not known. Oligonucleotides less than 21 nt in length are ineffective primers for gp5/trx, indicating that the primase must interact with gp5/trx to secure the primer in the DNA binding cleft (3, 5). The current study shows that both the ZBD and RPD are involved in stabilization of the priming complex and that both domains recognize the cryptic cytosine.
Fig. 6.
The arrangement of gp4, gp5/trx, and the primer/template in the priming complex. (A) The RPD of one subunit within the hexamer and the ZBD of an adjacent subunit position ATP and CTP at the primase recognition site, and the catalytic site within the RPD catalyzes the formation of the dinucleotide pppAC. (B) The RPD synthesizes a tetraribonucleotide, and gp5/trx is recruited to the primer/template. (C) The contacts of the ZBD and RPD with gp5/trx stabilize the tetraribonucleotide in the DNA binding crevice. Both RPD and ZBD make contacts with the cryptic cytosine. Gp5/trx extends the tetraribonucleotide.
Materials and Methods
Fluorescence anisotropy measurements were conducted in order to determine formation and stability of the priming complex. Experiments were carried out at 25 °C in the following buffer: 20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 2 mM MgCl2 and 5 mM DTT. Each reaction contained 100 nM 3′-fluorescein-labeled DNA template, the indicated concentrations of dTTP, and the indicated concentrations of the primer (5′-rArCrCrCdGdC-3′ or 5′-rArCrCrCdGddC-3′). The HPLC purified oligonucleotides were annealed at 25 °C. The data were collected with a Photon Technology International C-60 spectrofluorimeter by using excitation and emission wavelengths of 495 nm and 518 nm, respectively. A correction for the G factor of the instrument was applied. Experimental data points were fitted using an algorithm for the single-site saturation in the software KaleidaGraph.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Seung-Joo Lee for providing gp4∆helicase, and Steve Moskowitz for assistance in figure preparation. This work was supported by a National Institutes of Health research grant (GM-54397) to C.C.R.
Footnotes
The authors declare no conflict of interest .
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207033109/-/DCSupplemental.
References
- 1.Hamdan SM, Richardson CC. Motors, switches, and contacts in the replisome. Annu Rev Biochem. 2009;78:205–243. doi: 10.1146/annurev.biochem.78.072407.103248. [DOI] [PubMed] [Google Scholar]
- 2.Huber HE, Tabor S, Richardson CC. Escherichia coli thioredoxin stabilizes complexes of bacteriophage T7 DNA polymerase and primed templates. J Biol Chem. 1987;262:16224–16232. [PubMed] [Google Scholar]
- 3.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 4.Tabor S, Huber HE, Richardson CC. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J Biol Chem. 1987;262:16212–16223. [PubMed] [Google Scholar]
- 5.Akabayov B, et al. Conformational dynamics of bacteriophage T7 DNA polymerase and its processivity factor, Escherichia coli thioredoxin. Proc Natl Acad Sci USA. 2010;107:15033–15038. doi: 10.1073/pnas.1010141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JB, et al. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439:621–624. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein JA, Richardson CC. A 7-kDa region of the bacteriophage T7 gene 4 protein is required for primase but not for helicase activity. Proc Natl Acad Sci USA. 1988;85:396–400. doi: 10.1073/pnas.85.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sawaya MR, Guo S, Tabor S, Richardson CC, Ellenberger T. Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7. Cell. 1999;99:167–177. doi: 10.1016/s0092-8674(00)81648-7. [DOI] [PubMed] [Google Scholar]
- 9.Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 10.Toth EA, Li Y, Sawaya MR, Cheng Y, Ellenberger T. The crystal structure of the bifunctional primase-helicase of bacteriophage T7. Mol Cell. 2003;12:1113–1123. doi: 10.1016/s1097-2765(03)00442-8. [DOI] [PubMed] [Google Scholar]
- 11.Kato M, Ito T, Wagner G, Richardson CC, Ellenberger T. Modular architecture of the bacteriophage T7 primase couples RNA primer synthesis to DNA synthesis. Mol Cell. 2003;11:1349–1360. doi: 10.1016/s1097-2765(03)00195-3. [DOI] [PubMed] [Google Scholar]
- 12.Matson SW, Tabor S, Richardson CC. The gene 4 protein of bacteriophage T7. Characterization of helicase activity. J Biol Chem. 1983;258:14017–14024. [PubMed] [Google Scholar]
- 13.Satapathy AK, Kulczyk AW, Ghosh S, Richardson CC. A critical residue for coupling dTTP hydrolysis with DNA unwinding by the helicase of bacteriophage T7. J Biol Chem. 2011;286:34468–34478. doi: 10.1074/jbc.M111.283796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato M, Ito T, Wagner G, Ellenberger T. A molecular handoff between bacteriophage T7 DNA primase and T7 DNA polymerase initiates DNA synthesis. J Biol Chem. 2004;279:30554–30562. doi: 10.1074/jbc.M403485200. [DOI] [PubMed] [Google Scholar]
- 15.Qimron U, Lee SJ, Hamdan SM, Richardson CC. Primer initiation and extension by T7 DNA primase. EMBO J. 2006;25:2199–2208. doi: 10.1038/sj.emboj.7601112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamdan SM, Loparo JJ, Takahashi M, Richardson CC, van Oijen AM. Dynamics of DNA replication loops reveal temporal control of lagging-strand synthesis. Nature. 2009;457:336–339. doi: 10.1038/nature07512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu B, Lee SJ, Richardson CC. Direct role for the RNA polymerase domain of T7 primase in primer delivery. Proc Natl Acad Sci USA. 2010;107:9099–9104. doi: 10.1073/pnas.1004220107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernstein JA, Richardson CC. Purification of the 56-kDa component of the bacteriophage T7 primase/helicase and characterization of its nucleoside 5'-triphosphatase activity. J Biol Chem. 1988;263:14891–14899. [PubMed] [Google Scholar]
- 19.Guo S, Tabor S, Richardson CC. The linker region between the helicase and primase domains of the bacteriophage T7 gene 4 protein is critical for hexamer formation. J Biol Chem. 1999;274:30303–30309. doi: 10.1074/jbc.274.42.30303. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, et al. Helicase-DNA polymerase interaction is critical to initiate leading-strand DNA synthesis. Proc Natl Acad Sci USA. 2011;108:9372–9377. doi: 10.1073/pnas.1106678108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loparo JJ, Kulczyk AW, Richardson CC, van Oijen AM. Simultaneous single-molecule measurements of phage T7 replisome composition and function reveal the mechanism of polymerase exchange. Proc Natl Acad Sci USA. 2011;108:3584–3589. doi: 10.1073/pnas.1018824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frick DN, Baradaran K, Richardson CC. An N-terminal fragment of the gene 4 helicase/primase of bacteriophage T7 retains primase activity in the absence of helicase activity. Proc Natl Acad Sci USA. 1998;95:7957–7962. doi: 10.1073/pnas.95.14.7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tabor S, Richardson CC. Selective inactivation of the exonuclease activity of bacteriophage T7 DNA polymerase by in vitro mutagenesis. J Biol Chem. 1989;264:6447–6458. [PubMed] [Google Scholar]
- 24.Kato M, et al. A complex of the bacteriophage T7 primase-helicase and DNA polymerase directs primer utilization. J Biol Chem. 2001;276:21809–21820. doi: 10.1074/jbc.M101470200. [DOI] [PubMed] [Google Scholar]
- 25.Matson SW, Richardson CC. DNA-dependent nucleoside 5'-triphosphatase activity of the gene 4 protein of bacteriophage T7. J Biol Chem. 1983;258:14009–14016. [PubMed] [Google Scholar]
- 26.Frick DN, Richardson CC. DNA primases. Annu Rev Biochem. 2001;70:39–80. doi: 10.1146/annurev.biochem.70.1.39. [DOI] [PubMed] [Google Scholar]
- 27.Yuzhakov A, Kelman Z, O’Donnell M. Trading places on DNA—A three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell. 1999;96:153–163. doi: 10.1016/s0092-8674(00)80968-x. [DOI] [PubMed] [Google Scholar]
- 28.Chowdhury K, Tabor S, Richardson CC. A unique loop in the DNA-binding crevice of bacteriophage T7 DNA polymerase influences primer utilization. Proc Natl Acad Sci USA. 2000;97:12469–12474. doi: 10.1073/pnas.230448397. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.