Signaling by phosphatidylinositol 4-kinases (PI4Ks) in the Golgi apparatus controls lipid homeostasis and protein-sorting pathways. Signaling is shown to be terminated on the medial cisterna by a complex of a PI4K effector, Vps74, and Sac1, the major PtdIns4P phosphatase in the cell.
Abstract
In the Golgi apparatus, lipid homeostasis pathways are coordinated with the biogenesis of cargo transport vesicles by phosphatidylinositol 4-kinases (PI4Ks) that produce phosphatidylinositol 4-phosphate (PtdIns4P), a signaling molecule that is recognized by downstream effector proteins. Quantitative analysis of the intra-Golgi distribution of a PtdIns4P reporter protein confirms that PtdIns4P is enriched on the trans-Golgi cisterna, but surprisingly, Vps74 (the orthologue of human GOLPH3), a PI4K effector required to maintain residence of a subset of Golgi proteins, is distributed with the opposite polarity, being most abundant on cis and medial cisternae. Vps74 binds directly to the catalytic domain of Sac1 (KD = 3.8 μM), the major PtdIns4P phosphatase in the cell, and PtdIns4P is elevated on medial Golgi cisternae in cells lacking Vps74 or Sac1, suggesting that Vps74 is a sensor of PtdIns4P level on medial Golgi cisternae that directs Sac1-mediated dephosphosphorylation of this pool of PtdIns4P. Consistent with the established role of Sac1 in the regulation of sphingolipid biosynthesis, complex sphingolipid homeostasis is perturbed in vps74Δ cells. Mutant cells lacking complex sphingolipid biosynthetic enzymes fail to properly maintain residence of a medial Golgi enzyme, and cells lacking Vps74 depend critically on complex sphingolipid biosynthesis for growth. The results establish additive roles of Vps74-mediated and sphingolipid-dependent sorting of Golgi residents.
INTRODUCTION
Phosphatidylinositol 4-kinases (PI4Ks) regulate key aspects of the structure and function of the Golgi apparatus via effector proteins that bind phosphatidylinositol 4-phosphate (PtdIns4P) and coordinate lipid metabolism with vesicle-mediated trafficking pathways (Graham and Burd, 2011; Santiago-Tirado and Bretscher, 2011). PtdIns4P is most abundant on membranes of the trans-Golgi network (TGN; Godi et al., 2004; Cheong et al., 2010), in which PI4Ks reside at steady state (Strahl et al., 2005). It has been proposed that restriction of PtdIns4P to late Golgi compartments is enforced by dephosphorylation of PtdIns4P in early Golgi compartments by Sac1, an integral membrane phosphoinositide phosphatase (Cheong et al., 2010). In growing cells, Sac1 resides predominantly in the endoplasmic reticulum (ER), but it cycles through the Golgi (Whitters et al., 1993; Schorr et al., 2001; Rohde et al., 2003). Despite its restricted localization to organelles of the early secretory pathway, evidence in budding yeast (Saccharomyces cerevisiae) suggests that Sac1 accesses PtdIns4P (and other phosphoinositides) on multiple other organelles. The major pool of PtdIns4P controlled by Sac1 resides in the plasma membrane (PM; Foti et al., 2001; Roy and Levine, 2004), and it has been suggested that access to this pool is conferred in trans at specialized membrane sites at which the ER and PM are closely apposed (Stefan et al., 2011). A 70–amino acid flexible tether between the Sac1 phosphatase domain and its membrane-spanning segments may facilitate recruitment of the Sac1 catalytic domain from the ER to the PM, and possibly other organelles, via Sac1 receptors (Manford et al., 2010; Stefan et al., 2011).
Little is known about the specific functions of Sac1-mediated dephosphorylation of PtdIns4P. Regarding the Golgi, loss-of-function mutations in SAC1 rescue lethal mutations in SEC14, which encodes a sensor of lipid homeostasis that functions at the TGN–endosome interface (Cleves et al., 1989; Whitters et al., 1993). In addition, sac1 mutants fail to retain the cell wall–remodeling enzyme Chs3 in the Golgi apparatus, resulting in constitutive delivery of Chs3 to the PM (Schorr et al., 2001). It has been proposed that Sac1 regulates complex sphingolipid biosynthesis by modulating a Golgi pool of phosphatidylinositol, which provides the head group for complex sphingolipid synthesis via dephosphorylation of PtdIns4P (Brice et al., 2009). Initiation of sphingolipid synthesis by serine palmitoyl transferase (SPT) in the ER also appears be regulated by Sac1 (Breslow et al., 2010; Han et al., 2010). These observations suggest that Sac1-mediated regulation of PtdIns4P pools at multiple sites within the cell constitutes a signaling nexus that controls sphingolipid homeostasis and secretion.
We, and others, have recently described a conserved Golgi PI4K effector called Vps74 in yeast and GOLPH3 in human (Dippold et al., 2009; Wood et al., 2009). In yeast cells, loss of Vps74 function results in a failure to maintain residence of a subset of Golgi mannosyltransferases that localize predominantly to early (cis and medial) Golgi compartments, resulting in their degradation in the lysosome-like vacuole and severe glycosylation defects of secretory cargo (Schmitz et al., 2008; Tu et al., 2008; Wood et al., 2009). In this paper, we present data that reveal a key role for Vps74 in the regulation of Golgi PI4K signaling and lipid homeostasis.
RESULTS
Genetic interaction profiling of vps74Δ mutant
Although loss of Vps74 function results in severe defects in glycosylation of secretory cargo and Golgi organization (Corbacho et al., 2005, 2010; Schmitz et al., 2008; Tu et al., 2008; Wood et al., 2009), vps74Δ cells are viable, suggesting that other gene products buffer against the loss of Vps74 function. To identify such factors, we measured relative growth rates of ∼4700 double mutant strains containing a vps74Δ deletion allele and each nonessential gene deletion allele. Thirty-two double mutant combinations resulted in a “synthetic sick” growth defect indicative of a synergistic, aggravating genetic interaction (Figure 1A). Most of these genetic interactions resulted from the loss of gene products that function in the early secretory pathway, among them several key glycosylation enzymes and factors that function in transport to, through, and out of the Golgi. In addition, there was an enrichment of genes encoding enzymes of the sphingolipid biosynthetic pathway. A requirement for complex sphingolipid biosynthesis for the fitness of vps74Δ cells was confirmed by growth assays showing that these cells are hypersensitive to sublethal concentrations of aureobasidin A (Figure 1B), an inhibitor of the inositol phosphorylceramide (IPC) synthase Aur1, which resides in the medial Golgi compartment (Levine et al., 2000). (Aur1 is encoded by an essential gene, so it was not assayed in the genetic profiling.) In further support of these results, published large-scale genetic interaction and chemical genetic data sets report interactions between vps74Δ and other mutations (e.g., sac1Δ, ipt1Δ, ncr1Δ, tor2, orm2Δ) and small molecules affecting sphingolipid and sterol biosynthesis/homeostasis (Hillenmeyer et al., 2008; Costanzo et al., 2010; Koh et al., 2010). We also noted that a null allele of SAC1, which encodes an established regulator of sphingolipid synthesis (Brice et al., 2009; Breslow et al., 2010), scored as an aggravating mutation in our vps74Δ profiling; however, we filtered this mutant (vps74Δ sac1Δ) from our final data set, because the sac1Δ mutant itself grew poorly in our screening conditions. Moreover, in a sac1Δ profiling study, a vps74Δ allele scored as the fifth-most-correlated mutation with a sac1Δ allele, out of ∼1500 mutants affecting the early secretory pathway that were screened (Breslow et al., 2010), indicating that sac1 and vps74 mutants have highly related genetic interaction networks. Results presented in the next sections suggest that some functions of Vps74 and Sac1 are intimately linked.
FIGURE 1:
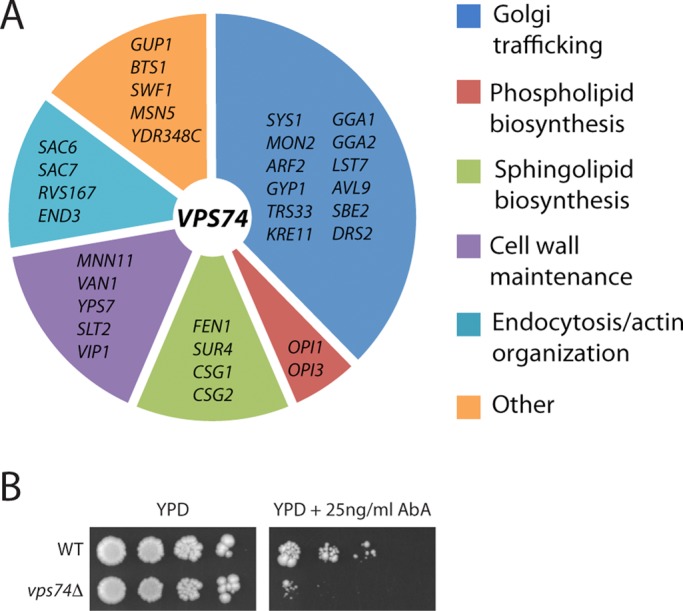
Genetic profiling of vps74Δ cells. (A) Gene deletions that aggravate cell growth when combined with the vps74Δ allele are listed and categorized by their functions. (B) vps74Δ cells are hypersensitive to aureobasidin A, an inhibitor of complex sphingolipid biosynthesis. Tenfold serial dilutions of wild-type and vps74Δ cells were spotted onto YPD medium containing 25 ng/ml aureobasidin A and grown for 4 d.
Vps74 and Sac1 phosphoinositide phosphatase form a complex
To gain insight into Vps74 function, we used affinity purification to identify proteins in a detergent yeast cell lysate that bind directly to Vps74. A yeast cell extract prepared with 1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) was incubated with glutathione S-transferase (GST)-Vps74 or GST-Snx3 (a peripheral membrane protein that localizes to endosomes) resins. After successive washes with buffers containing 50 and 150 mM NaCl, the resins were eluted with buffer containing 0.5 M NaCl and finally by denaturation with SDS–PAGE sample buffer. Protein bands that eluted only from the GST-Vps74 resin (unpublished data) were excised from the gel and subjected to mass spectrometry analysis. From the most prominent protein band in the 0.5 M NaCl eluate fraction, 25 unique peptides derived from Sac1, comprising ∼57% of the entire protein, were identified.
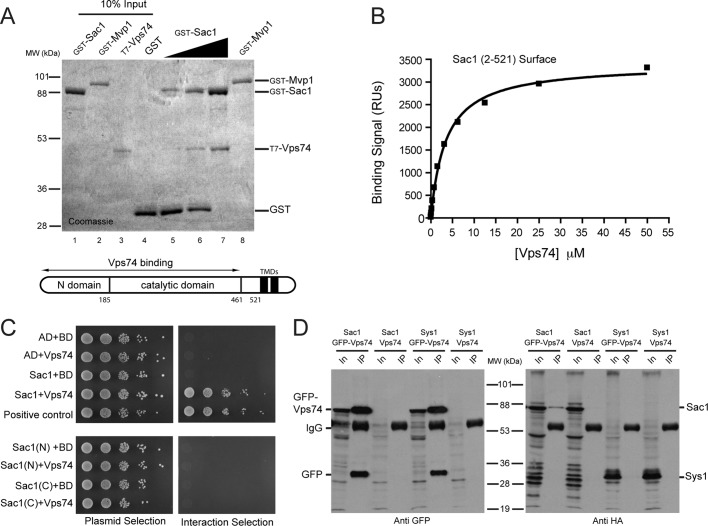
Sac1 is made up of a cytoplasmic amino terminal region (amino acids 1–521) containing the Sac1 homology phosphoinositide phosphatase domain, followed by ∼70 unstructured amino acids, two membrane-spanning segments, and a carboxy terminal cytoplasmic segment (Figure 2). Using purified fusion proteins produced in bacteria, we first determined that Vps74 binds to amino acids 1–521 of Sac1 (Figure 2A). By contrast, Vps74 did not bind GST or GST-Mvp1, a control phosphoinositide-binding protein that is approximately the same size as the GST-Sac1 fusion protein (Figure 2A). Surface plasmon resonance (SPR) experiments employing untagged recombinant proteins indicated that Vps74 binds Sac1 (amino acids 2–521) with a KD value of 3.8 ± 0.4 μM (Figure 2B). We also determined that the first 59 unstructured amino acids of Vps74 do not contribute to this interaction (KD value of Vps74Δ59 is 4.2 ± 0.7 μM; unpublished data). Sac1 and Vps74 also interacted in a yeast two-hybrid assay, and this assay was used to further elucidate the requirements for Sac1-Vps74 recognition (Figure 2C). The Sac1 homology region is composed of an N-terminal subdomain (amino acids 1–182) that is proposed to mediate interactions with other proteins, followed by the catalytic subdomain (amino acids 183–461) (Manford et al., 2010). In a two-hybrid assay, recognition of Sac1 by Vps74 required the entire intact Sac1 homology region (amino acids 1–461), as no interaction signal was observed with constructs expressing either of the Sac1 subdomains.
FIGURE 2:
Vps74 and Sac1 form a complex. (A) Purified Vps74 binds immobilized GST-Sac1 phosphatase domain. A series of binding reactions was set up in which the amount of immobilized GST-Sac1 (amino acids 2–521, comprising the entire N-terminal cytoplasmic portion) fusion protein was increased (lanes 5–7) while keeping the total amount of immobilized bait protein constant by adding a compensatory amount of GST. The position of T7- and His-tagged Vps74, confirmed by anti-T7 immunoblotting (unpublished data), is indicated. Note that Vps74 does not bind to GST-Mvp1, an endosomal protein that serves as a specificity control. A schematic of Sac1 structure is shown below the gel, with the amino acid positions of the domains indicated. (B) SPR analysis of Vps74 binding to Sac1 (amino acids 2–251). A series of samples of Vps74 at the indicated concentrations was passed over a CM5 chip to which Sac1 had been immobilized. The curve indicates the fit of a representative data set to a simple one-site binding equation. The KD value, determined from the fit to at least three independent data sets, is 3.8 ± 0.4 μM. (C) Yeast two-hybrid analysis of Vps74–Sac1 interaction. Cells containing plasmids that express the indicated proteins were spotted by limited dilution (1:10) onto nonselective plates (left) and onto interaction-selective plates (right) containing 10 mM 3-amino-triazole. The plates were photographed after 2 d incubation at 30°C. All Sac1-derived constructs were expressed as C-terminal fusions to the Gal4 activation domain. “Sac1” refers to amino acids 1–461 (encompassing the entire Sac1 homology region), “Sac1(N)” indicates the N-terminal subdomain (amino acids 1–185), and “Sac1(C)” indicates the catalytic subdomain (amino acids 186–421) of the Sac1 homology region. A schematic diagram of Sac1 summarizes the positions of the structural features of Sac1. The positive-control assay consisted of a fragment of Pan1 (amino acids 96–715) and Yap180 (amino acids 432–637) (Wendland and Emr, 1998). (D) Coimmunopurification of GFP-Vps74 and Sac1-HA. Cells expressing GFP-Vps74 and HA epitope-tagged Sac1 (Sac1-HA), or Sys1-HA as a control, were converted to spheroplasts and incubated with a bifunctional, cleavable cross-linking reagent. After the cells were detergent-solubilized, GFP-Vps74 was immunopurified with anti-GFP antiserum, and the bound material was probed with anti-HA antiserum. In the lanes marked “In,” 1% of the material used for the immunopurification is loaded. The positions of molecular-size standards (kilodaltons) are indicated.
We confirmed that Vps74 and Sac1 associate in vivo by using a cross-linking coimmunopurification assay (Figure 2D). Hemagglutinin (HA) epitope-tagged Sac1 coimmunopurified with green fluorescent protein (GFP)-tagged Vps74 in cross-linked detergent lysate prepared from cells expressing GFP-Vps74, but not from cells expressing untagged Vps74 (Figure 2D) or GFP (unpublished data). In further support of the specificity of Sac1-HA and GFP-Vps74 coimmunopurification, GFP-74 did not coimmunopurify with an HA epitope-tagged Golgi integral membrane control protein, Sys1 (Figure 2D). A small amount of Vps74 and Sac1 were observed to associate at steady state in this experiment, which is in line with the similarly small amount of Sac1 associated with Osh7, a proposed in vivo regulator of Sac1 activity, in similar experiments (Stefan et al., 2011).
Localization of Vps74-Sac1 complexes in vivo
We next sought to determine where Vps74 and Sac1 associate within the cell. ER and Golgi membranes can be effectively resolved by cell fractionation techniques; however, this was not a feasible approach, because Vps74 is essentially soluble after cell lysis (Tu et al., 2008; unpublished data). Therefore we turned to live-cell imaging and applied a yellow fluorescent protein (YFP)-based bimolecular fluorescence complementation (BiFC) assay, which requires association of query proteins fused to fragments of YFP to elicit fluorescence (Ghosh et al., 2000; Hu et al., 2002). In cells engineered to express an N-terminal portion of YFP fused to the N-terminus of Vps74 (N·YFP-Vps74), and a C-terminal portion of YFP fused to the N-terminus of Sac1 (C·YFP-Sac1), a fluorescence signal was observed (Figure 3A), further confirming that Vps74 and Sac1 associate in vivo. By contrast, no BiFC signal was observed in a control strain expressing C·YFP-Sac1 and appropriately tagged Snx3 (N·YFP-Snx3), an endosome-localized sorting nexin (unpublished data). The Vps74-Sac1 BiFC signal was observed on multiple compartments, with the brightest fluorescence localizing to punctate structures that resembled Golgi compartments, and a lower signal that appeared to emanate from ER membranes.
FIGURE 3:
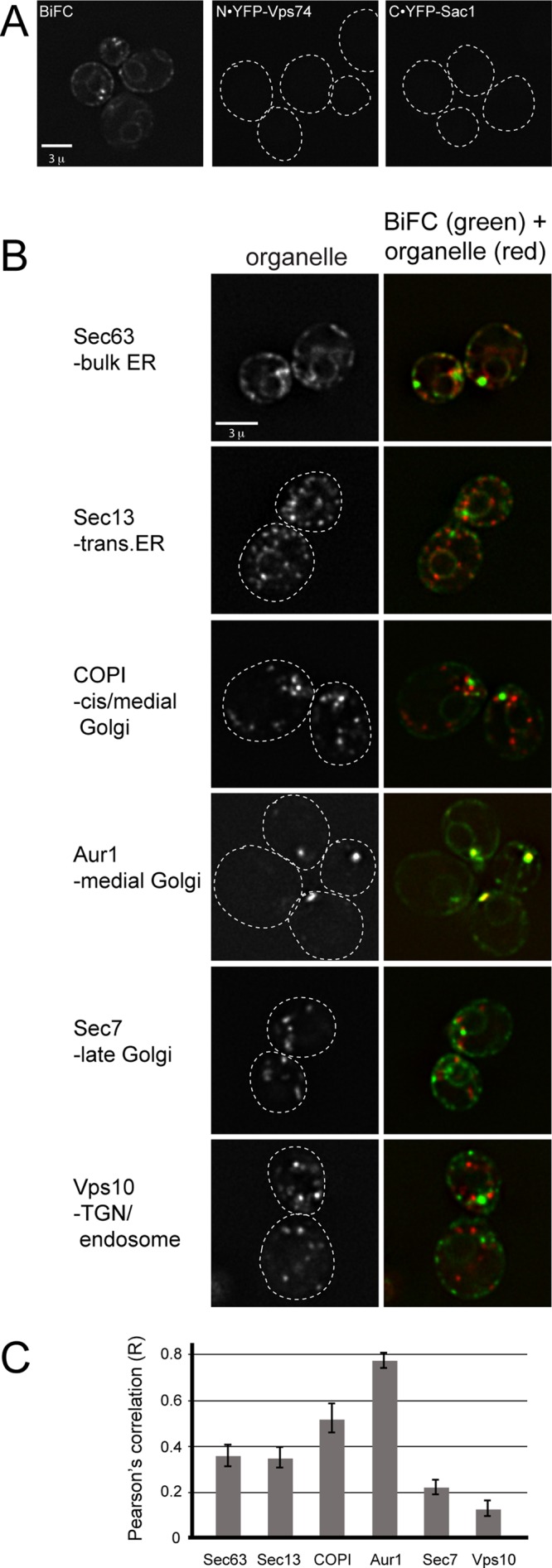
Visualization of Vps74-Sac1 complexes in vivo. (A) Identification of Vps74-Sac1 complexes in vivo by BiFC. Fluorescence micrographs of cells expressing the N-terminal portion of YFP fused to the N-terminus of Vps74 (N·YFP-Vps74), and the C-terminal portion of YFP fused to the N-terminus of Sac1 (C·YFP-Sac1), or both fusion proteins (BiFC) are shown. Fluorescence complementation is observed when both proteins are coexpressed. (B) Vps74-Sac1 BiFC colocalization with secretory organelles. The indicated resident proteins of the ER and Golgi were tagged on their C-termini with mKate in cells expressing N·YFP-Vps74and C·YFP-Sac1 and visualized by deconvolution microscopy. One optical focal plane from one Z-stack is shown as an example. (C) Pearson's correlation coefficients (r) were determined for the entire volumes (i.e., voxels) of at least 25 cells in at least three different fields. The mean r values (± SE) are plotted.
To more precisely define where the Vps74-Sac1 BiFC complex localizes, we determined Pearson's correlation coefficients between the Vps74-Sac1 BiFC signal and resident proteins of secretory organelles, from the ER to the TGN/endosome. Each of these strains expressed the N·YFP-Vps74 and C·YFP-Sac1 fusion proteins and one mKate-tagged marker protein expressed from its native locus (Figure 3, B and C). There was a striking correlation between Vps74-Sac1 BiFC and Aur1-mKate (r = 0.77), reported to be a resident of medial Golgi cisternae (Levine et al., 2000), and to a lesser extent with Cop1 (r = 0.52), a component of the coatamer I vesicle coat (COPI) that marks early (i.e., cis and medial) Golgi compartments. A substantially lower correlation was observed between Vps74-Sac1 BiFC and Sec7 (r = 0.22), a marker of late Golgi compartments, and Vps10 (r = 0.13), a protein that cycles between the TGN and endosomes. The Vps74-Sac1–BiFC complex also localized to ER membranes with Sec63 (r = 0.38); ER localization probably resulted from enhanced stability of the Sac1-Vps74 complex due to the BiFC tags, as we rarely observed ER localization of GFP-Vps74.
Elevated PtdIns4P on medial Golgi cisternae in vps74Δ cells
To test the possibility that Vps74 influences Sac1 function in vivo, we used metabolic [3H]inositol radiolabeling to assess PtdIns4P levels in wild-type and vps74Δ cells (Figure 4A). This analysis showed that the level of PtdIns4P is increased ∼1.5-fold in vps74Δ cells compared with wild-type cells (p < 0.002), whereas the level of PtdIns3P, an endosome-localized phosphoinositide, was unchanged. We considered the possibility that the increase in PtdIns4P resulted from reduced abundance of Sac1 in vps74Δ cells due to a failure to retrieve Sac1 from the Golgi; however, neither the gross localization nor steady-state abundance of Sac1 was affected in vps74Δ cells (Supplemental Figure S1). We also considered the possibility that Vps74 might directly influence Sac1 activity; however, we observed no effect of pure, recombinant Vps74 on PtdIns4P phosphatase activity of a soluble form of Sac1 in vitro (Figure S1). This may reflect a requirement for native (i.e., full-length, membrane-embedded) Sac1 and/or any of the other proteins reported to associate with Sac1.
FIGURE 4:
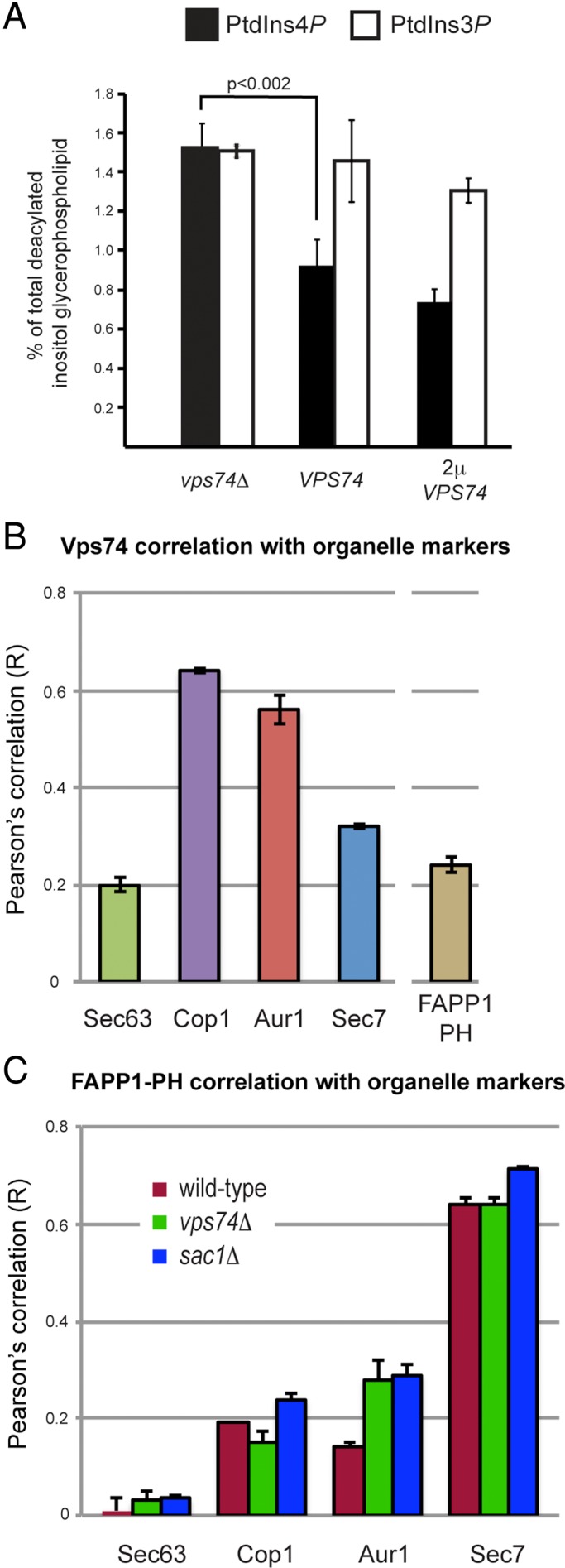
PtdIns4P is elevated in the Golgi of vps74Δ cells. (A) Elevated PtdIns4P in vps74Δ cells. Cells were labeled to steady state with [3H]myoinositol and the amounts of PtdIns4P and PtdIns3P (as a reference) were determined by high-performance liquid chromatography analysis of the corresponding deacylated glycero-phosphoinositol-phosphate derivatives. The analysis was conducted in a sec14-1 kes1Δ mutant background with the indicated VPS74 genotype to facilitate accurate determinations of changes in PtdIns4P levels. (B) Vps74 is enriched on cis and medial compartments of the Golgi. The distribution of GFP-Vps74 across ER and Golgi compartments was determined by calculating Pearson's correlation coefficients (r) for GFP-Vps74 and the indicated ER and Golgi residents expressed with a C-terminal fusion to mKate. A similar analysis was carried to compare colocalization of mCherry-Vps74 and GFP-FAPP1·PH. The mean r values (± SE) are plotted. (C) Distribution of the GFP-FAPP1·PH PtdIns4P probe across ER and Golgi compartments. GFP-FAPP1·PH was expressed from a low-copy CEN plasmid in strains expressing the mKate-tagged Sec63 (ER), Cop1, Aur1, or Sec7, and Pearson's correlation coefficients (r) were determined for voxels from deconcolved Z-series micrographs. The mean r values (± SE) are plotted.
We next sought to localize the sites of Vps74 function with respect to elevated PtdIns4P in vps74Δ cells. As a first step, the distribution of Vps74 across Golgi cisternae was determined by comparing Pearson's correlation coefficients between GFP-Vps74 and mKate-tagged markers of ER and early, medial, and late Golgi compartments (Figure 4B). In agreement with our previously published data (Schmitz et al., 2008), GFP-Vps74 correlated most closely with Cop1-mKate (r = 0.64) and Aur1-mKate (r = 0.56), and substantially less with the late Golgi resident, Sec7-mKate (r = 0.32). The intra-Golgi distribution of GFP-Vps74 was in close agreement with the high degree of correlation between the Vps74-Sac1 BiFC signal and early Golgi residents (i.e., Cop1-mKate and Aur1-mKate), indicating that the BiFC approach did not grossly alter the distribution of Vps74 within the Golgi.
Using a similar experimental strategy, we determined the distribution of PtdIns4P across Golgi compartments, which was inferred by determining correlation coefficients for Golgi-resident proteins and GFP-FAPP1·PH, an established in vivo reporter of the Golgi PtdIns4P pool (Godi et al., 2004; Figure 4C). This analysis confirmed that GFP-FAPP1·PH predominantly decorates late Golgi compartments identified by Sec7-mKate (Levine and Munro, 2002), with detectable, but substantially lower, amounts on early cisternae. In light of these results, and the requirement for PtdIns4P in recruiting Vps74 to Golgi membranes (Dippold et al., 2009; Wood et al., 2009), recognition of the cytoplasmic portions of early Golgi-resident proteins (e.g., Kre2; Schmitz et al., 2008; Tu et al., 2008) must make a substantial contribution for targeting to Golgi compartments enriched in these proteins.
We next determined the intra-Golgi distribution of GFP-FAPP1·PH in vps74Δ cells (Figure 4C). There was a twofold increase in the correlation between GFP-FAPP1·PH and Aur1-mKate, compared with wild-type cells, with essentially no changes in the correlations with residents of other Golgi cisternae. This increase in correlation was not likely to be due to a broader distribution of Aur1 within the Golgi of vps74Δ cells, as the correlations between Aur1-mKate and Sec7-GFP were essentially identical (r = 0.40 for wild-type cells vs. r = 0.45 for vps74Δ cells; Figure S2). Significantly, there was a <15% change in GFP-FAPP1·PH correlation with all other markers of other Golgi compartments in sac1Δ cells, indicating that the overall distribution of GFP-FAPP1·PH across Golgi compartments was not grossly perturbed in vps74Δ or sac1Δ cells compared with wild-type cells. These results indicate that both Vps74 and Sac1 function to maintain a low level of PtdIns4P on medial Golgi compartments.
Complex sphingolipid biosynthesis is required for sorting of Golgi residents
In the ER, Sac1 is associated with SPT, the enzyme that condenses serine and fatty acyl-CoA to initiate sphingolipid synthesis and is predicted to be a negative regulator of SPT activity on the basis of lipid profiling and genetic analysis (Breslow et al., 2010; Han et al., 2010). It has been postulated that Sac1 in the Golgi regulates complex sphingolipid biosynthesis by modulating the availability of phosphatidylinositol (which provides inositol phosphate) via dephosphorylation of PtdIns4P (Brice et al., 2009). Given the association of Vps74 and Sac1 in the Golgi, we addressed the possibility that complex sphingolipid homeostasis might be perturbed in vps74Δ cells (Figure 5). Metabolic inositol-labeling experiments showed a 68% decrease in mannosylinositol phosphorylceramide (MIPC) in vps74Δ cells compared with wild-type cells, but surprisingly, there were no changes in the levels of IPC and mannose-(inositol phosphate)2-ceramide (M(IP)2C). Strikingly, this reduction in MIPC was similar in magnitude to the reduction in MIPC observed in csg1Δ and csg2Δ cells, which lack one of the subunits of MIPC synthase. The perturbation to complex sphingolipid homeostasis in vps74Δ cells was not due to changes in the localization or abundances of IPC and MIPC synthases (Figure S2), indicating that, like Sac1, these enzymes do not rely on Vps74 for their proper localization.
FIGURE 5:
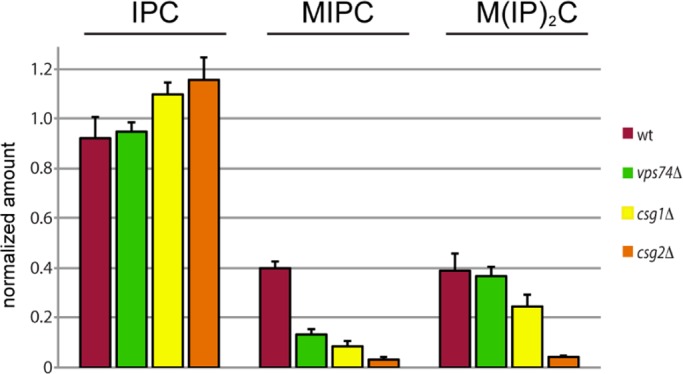
Complex sphingolipid analyses. The amount of each of the indicated sphingolipid species (normalized to phosphatidylinositol) in the indicated strains is plotted. The height of each bar represents the mean of three independent determinations.
Cells that do not make MIPC or M(IP)2C are viable, so the deficiency in MIPC observed in vps74Δ cells is unlikely to explain the synthetic phenotype of double mutants of vps74Δ and deletions of genes encoding complex sphingolipid enzymes. We therefore considered the possibility that the genetic dependency of vps74Δ cells on complex sphingolipid synthesis in the Golgi might reflect a contribution of sphingolipids to aspects of Golgi organization. The lipid composition of Golgi cisternae differed, with late compartments being enriched in sphingolipids and sterol, and lipid-based sorting mechanisms proposed on the basis of the physical properties of sphingolipid and cholesterol partitioning have been invoked to explain sorting of secretory cargo at the TGN (Simons and van Meer, 1988) and partitioning of Golgi residents against anterograde flow of secretory cargo (Bretscher and Munro, 1993). We therefore examined a GFP-tagged version of Kre2 (Kre2-GFP), a medial Golgi α-1,2-mannosyltransferase that depends on Vps74 to be retained in the Golgi (Schmitz et al., 2008; Tu et al., 2008; Wood et al., 2009). In each of the sphingolipid pathway mutants tested (Figure 6A), Kre2-GFP accumulated in the vacuole lumen, though the effect was not as severe as is observed in vps74Δ cells. Immunoblotting of myc epitope–tagged Kre2 (Kre2-myc) in cell extracts derived from each of these strains showed that the steady-state amount of Kre2-myc was reduced to ∼40–60% of the level in wild-type cells in each of these mutant strains (Figure 6B), even though Vps74 still localized to the Golgi in all of these cells. The data indicate that complex sphingolipid biosynthesis is required for efficient retention of Kre2, even in cells expressing functional Vps74.
FIGURE 6:
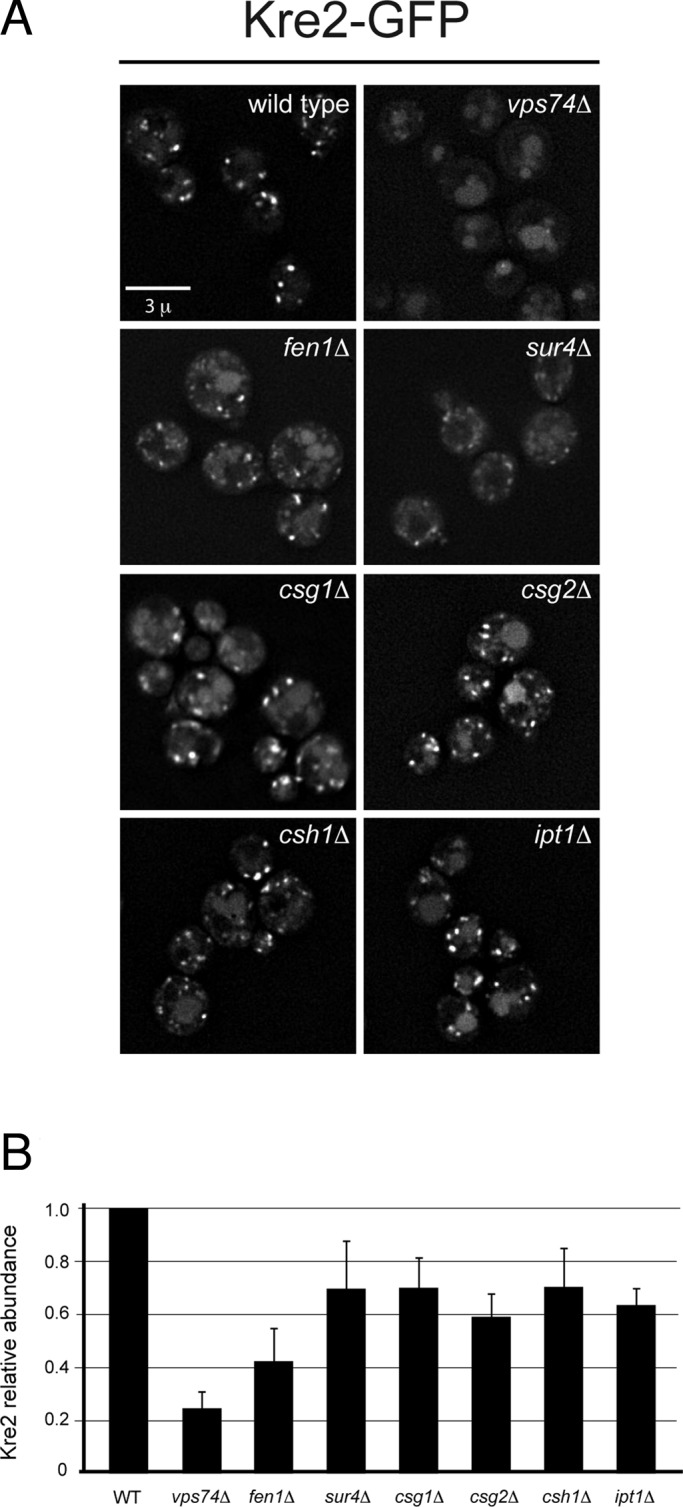
Disrupted localization of the medial Golgi mannosyltransferase, Kre2, in sphingolipid mutants. (A) Kre2-GFP was visualized in the indicated sphingolipid pathway mutants by deconvolution microscopy. One representative image from the Z-series is shown. For comparison, wild-type and the vps74Δ cells are shown. (B) Steady-state abundance of myc epitope–tagged Kre2 (Kre2-myc) in the indicated strains was determined by immunoblotting of detergent cell lysates. The means of three independent measurements are plotted, normalized to wild-type cells, with the SE of the measurements indicated.
DISCUSSION
Local control of PtdIns4P level by Vps74 and Sac1
The results presented here expand our understanding of Vps74, a PI4K effector that plays a key role in the retention of a subset of early Golgi-resident proteins, and Sac1, which spatially restricts PI4K signaling by dephosphorylating PtdIns4P. At the trans cisterna, Vps74 is proposed to sort early Golgi residents into retrograde-directed coatomer (COPI)-coated vesicles (Tu et al., 2008). Unless a mechanism exists that excludes PtdIns4P from these vesicles, they will deliver PtdIns4P to earlier cisternae, and thereby antagonize PtdIns4P restriction. In principle, the Vps74-Sac1 complex might act at to prevent incorporation of PtdIns4P into budding COPI vesicles by, for example, being copackaged with Golgi residents into the same vesicle. However, we feel that this is unlikely, because neither the gross localization nor the abundance of Sac1 was impacted in vps74Δ cells, indicating that retrograde sorting of Sac1 is independent of Vps74. The increase in PtdIns4P on the medial Golgi compartment in vps74Δ and sac1Δ cells, and the physical association of Vps74 and Sac1, instead suggest that Vps74 functions on medial Golgi cisternae as a PtdIns4P sensor that limits PI4K signaling by promoting Sac1-dependent turnover of PtdIns4P (Figure 7). Consistent with this, Vps74 and the Vps74-Sac1–BiFC complex were most abundant on the Aur1-marked (i.e., medial) cisternae. By restricting accumulation of PtdIns4P, Vps74 and Sac1 served to maintain a key functional difference between medial and trans-Golgi cisternae, thereby enforcing cisternal identity. RNA interference (RNAi)-mediated depletion of Sac1 in cultured human cells resulted in a qualitative increase in colocalization of FAPP1·PH and mannosidase II, a resident of the medial cisterna (Cheong et al., 2010), indicating that human Sac1 similarly prevents PtdIns4P accumulation on the medial cisterna. A second conclusion that follows from our PtdIns4P mapping data is that loss of Sac1 (or Vps74) does not result in wholesale distribution of PtdIns4P throughout the Golgi. This result is important because it demonstrates that the principal mode of PtdIns4P restriction within the Golgi is steady-state localization of Pik1 PI4K to the trans cisterna (Walch-Solimena and Novick, 1999; Strahl et al., 2005). Sac1 and Vps74 therefore function to maintain, rather than establish, PtdIns4P restriction. In addition, Sac1-mediated dephosphorylation of PtdIns4P will also ensure that a cytosolic pool of Vps74 is available for recruitment to the trans-Golgi to serve in sorting of medial residents (Figure 7).
FIGURE 7:
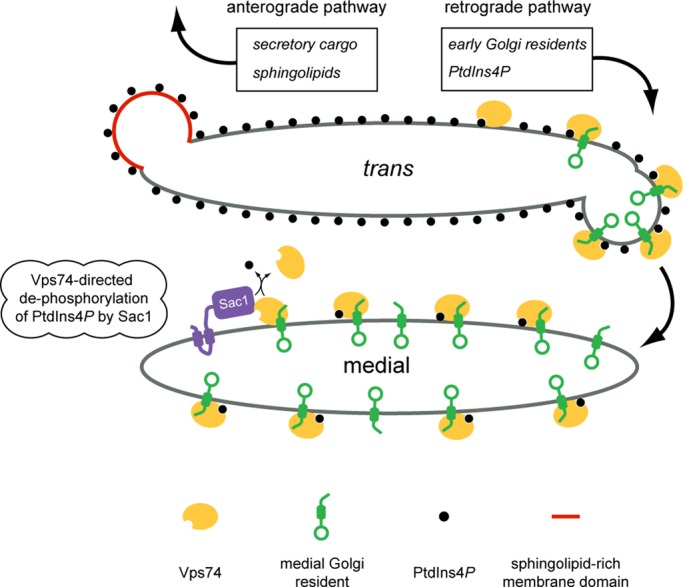
Model of Vps74-mediated protein sorting and PtdIns4P signaling in the Golgi apparatus. The medial and trans compartments of the Golgi apparatus are depicted with PtdIns4P (black circles) most abundant on the trans cisterna. Vps74 (yellow oval) recognizes PtdIns4P and early Golgi-resident mannosyltransferases (green). Vps74 is depicted to sort these enzymes into COPI-coated retrograde vesicles (COPI coat is not depicted). Also in the trans cisterna, complex sphingolipids are preferentially packaged with secretory cargo into anterograde-directed transport vesicles (red vesicle). We propose that the efficiency of Vps74-mediated sorting of Golgi residents into the retrograde pathway increases as a consequence of excluding Golgi residents from sphingolipid-rich domains. As a consequence of copackaging PtdIns4P with Golgi residents into retrograde vesicles, PtdIns4P is delivered to the medial cisterna. Sac1 (purple) cycles between the ER and Golgi apparatus and the data presented herein indicate that Sac1 and Vps74 function in the Golgi to maintain a low amount of PtdIns4P on the medial cisterna; we propose that this is mediated by the Vps74-Sac1 complex.
Vps74 and Sac1 are most likely to encounter each other as Sac1 cycles through the Golgi (i.e., an interaction in cis), but it is also possible that Vps74 recruits the Sac1 catalytic domain to Golgi membranes, while Sac1 resides in the ER (i.e., interaction in trans). In mammalian cells, sites of close apposition of ER and TGN are thought to be sites at which ceramide, a precursor to sphingomyelin, and cholesterol are transferred to the TGN under tight regulation by PtdIns4P signaling (Graham and Burd, 2011). Intriguingly, we have noted that the punctate Vps74-Sac1–BiFC compartments seem to be juxtaposed to an element of the ER, although the significance of this is not yet apparent, as there are no reports in the literature of such contacts in yeast cells. If they do exist in yeast cells, it may be that they are short-lived but stabilized by the BiFC approach used here.
Given the genetic links between vps74Δ and sphingolipid metabolism, and the codependence of sphingolipid and sterol metabolism in yeast cells (Guan et al., 2009), the possibility that the Vps74-Sac1 complex regulates sphingolipid and sterol trafficking at the Golgi via PtdIns4P signaling warrants further study.
A key question regards the relationship between the increase in Golgi PtdIns4P and perturbed sphingolipid level (i.e., the reduction in MIPC) in vps74Δ cells. At present, it is unclear whether the increase of PtdIns4P causes, or is a homeostatic response to, perturbed sphingolipid level, and whether Vps74 directly regulates sphingolipid metabolism. Mutations that ablate particular Vps74 functions while leaving others intact will be key for distinguishing these possibilities. A further curious aspect of vps74Δ cells is that, to our knowledge, this is the first description of a mutant with a deficit of MIPC but no significant changes in IPC and M(IP)2C; it is unclear how, based on the known activities of sphingolipid enzymes, the level of MIPC could be controlled independent of the levels of other complex sphingolipids. Intriguingly, MIPC has been implicated as a signaling molecule in a pathway that senses PM function (Roelants et al., 2010), and VPS74 has been identified as a multi-copy suppressor of a loss-of-function mutation (ypk1-1 ykr2Δ) in this pathway (Roelants et al., 2002). Together these studies suggest that lipid- and protein-processing reactions in the Golgi are modulated via a signaling pathway that originates at the PM and regulates Golgi function, possibly via Vps74.
Sphingolipid biosynthesis and Golgi organization
Our finding that retention of a Golgi resident (Kre2) is perturbed in complex sphingolipid mutants is relevant for understanding the mechanisms that establish and maintain the distributions of Golgi residents. Complex sphingolipid biosynthesis in yeast is initiated by Aur1, which resides in the medial compartment at steady state (Levine et al., 2000). Downstream processing of IPC to MIPC and M(IP)2C occurs in medial/late Golgi compartments, positioning complex sphingolipid biosynthetic reactions at the compartments in which Vps74-dependent Golgi residents reside and from which they are retrieved. Lipid-based sorting mechanisms proposed on the basis of the physical properties of sphingolipid and cholesterol domains have been invoked to explain sorting of secretory cargo at the TGN (Simons and van Meer, 1988) and partitioning of Golgi residents against anterograde flow of secretory cargo (Bretscher and Munro, 1993). Perturbations to Golgi membrane structure that arise as a result of genetic disruption of complex sphingolipid synthesis should reduce the extent to which Golgi residents partition laterally from anterograde secretory cargo (Figure 7), explaining the genetic dependence of vps74Δ and complex sphingolipid biosynthesis mutants. In further support of this, in large-scale genetic interaction profiling studies, a loss-of-function AUR1-damp allele has been highly correlated with mutations in genes encoding components of the COPI coat and the conserved oligomeric Golgi (COG) retrograde-trafficking complex (Costanzo et al., 2010). With this in mind, it is also notable that the COPI vesicle coat, which is thought to mediate retrograde trafficking of Golgi residents, preferentially associates with lipid-disordered domains of synthetic vesicles (Manneville et al., 2008). Our study elucidates an interface between lipid-based and coat protein–based sorting mechanisms, with PI4K signaling at its nexus, that coordinately maintains Golgi organization.
MATERIALS AND METHODS
Yeast methods
All yeast strains were constructed in the BY4742 background (MATα his3-1, leu2-0, met15-0, and ura3-0) unless indicated otherwise. Integration of DNA-encoding fluorescent and epitope tags at native chromosomal loci was done by recombination of gene-targeted, PCR-generated DNAs. Bi-molecular fluorescence complementation strains were constructed using PCR-generated DNAs by the method of Sung and Huh (2007). For microscopy and protein-interaction experiments, strains were grown in standard synthetic complete medium lacking nutrients required to maintain selection for auxotrophic markers (Sherman et al., 1979).
Synthetic genetic array screening
The procedure used was based on a previously published method (Tong et al., 2001). The query strain, CBY191 (vps74Δ:NatMX, constructed in Y7092), was crossed to a deletion mutant array of viable single-gene knockouts using a Singer RoToR robot (Singer Instrument, Somerset, UK) and vps74Δ-DMA double mutants were selected. Three independent screens were conducted, yielding a composite data set of six replicate double mutant colonies. An aggravating genetic interaction was scored if the sizes of at least four replicate colonies were <60% of the size the smallest single mutant colony.
Biochemical methods
For identification of Vps74-binding proteins, ∼200 μg GST-Vps74 (amino acids 1–345) or, as a control, ∼200 μg GST-Snx3 (amino acids 1–162), were immobilized on Glutathione Sepharose 4B resin (Amersham) and incubated with a yeast cell extract (strain: TVY614 MATα, ura3–52, his3-Δ200, trp1-Δ 901, lys2–801, suc2-Δ 9, leu2–3, 11, pep4Δ::LEU2, prb1Δ::HISG, prc1Δ::HIS3). The cell extract was prepared in lysis buffer (LB; 40 mM Tris, pH 7.0, 100 mM KCl, 10 mM MgCl2, 1X Complete Mini protease inhibitor cocktail [Roche, Indianapolis, IN] + 2 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride [AEBSF; Calbiochem, San Diego, CA]) by two passes through a French press. CHAPS was added to 1% (wt/vol), and the lysate was rocked at 4°C for 10 min. After clarification by centrifugation (10,000 × g for 30 min at 4°C), the resultant supernatant was incubated with 2 ml of glutathione (GSH) resin and the GSH-cleared lysate was divided into two 20-ml (14-mg/ml) portions; one aliquot was incubated with GST-Vps74 bound to GSH resin, and the other aliquot was incubated with GST-Snx3 resin. The resins were incubated for 2 h at 4°C with rocking, and were then collected by centrifugation (5 min at 2000 × g). After the supernatants were removed, the resins were washed once each with wash buffer 1 (20 mM Tris, pH 7.0, 50 mM KCl) and wash buffer 2 (50 mM Tris, pH 7.5, 150 mM KCl, 2.5 mM MgCl2, 2.5 mM CaCl2, 0.1 CHAPS). Bound proteins were eluted by four incubations with 0.5 ml elution buffer (1 M NaCl, 5 mM EDTA, 0.5% CHAPS) and concentrated by precipitation with trichloroacetic acid. Eluted proteins were resolved by SDS–PAGE, the gel was stained with Coomassie blue, and protein bands that were unique to the GST-Vps74 eluate were excised and identified by mass spectrometry at the Proteomics and Systems Core facility of the Abramson Cancer Center of the University of Pennsylvania.
Binding of purified His- and T7-tagged Vps74 to GST-Sac1 (amino acids 2–521) was done as follows: 10 μg of GST fusion protein was immobilized on Glutathione Sepharose 4B (Amersham) for 1 h at 4°C in 0.3 ml of binding buffer (1 mM KH2PO4, 10 mM Na2HPO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4, 5 mM MgCl2, 0.1% Triton X-100). To show concentration-dependent binding, the ratio of GST fusion protein to GST was varied to keep the total amount of immobilized protein constant. The resins were collected by centrifugation and washed with binding buffer, 5 μM of purified T7-tagged Vps74 was added, and the volume of the reaction was adjusted to 250 μl with binding buffer. The binding reaction was incubated at 4°C for 1 h, and then the resins were washed three times with 1 ml binding buffer. Bound proteins were eluted with 2X SDS sample buffer and analyzed by 10% SDS–PAGE.
A published protocol was used for the cross-linking immunopurification experiments (Strochlic et al., 2007). The strains used expressed epitope-tagged Sac1 or Sys1 from their native loci and carried a deletion of the VPS74 locus (vps74Δ), and GFP-Vps74 was expressed in these strains from a low-copy yeast centromere (CEN) vector.
Surface plasmon resonance-binding studies
Vps74 and Vps74ΔN59 were expressed and purified as previously described (Wood et al., 2009), except the lysis buffer was substituted with 50 mM Tris, 300 mM NaCl, 10% glycerol (pH 8.0) supplemented with 1μM phenylmethylsulfonyl fluoride. Sac1 2–251 was expressed from a modified pET-28 vector to produce protein with an N-terminal MAT/Smt3p (SUMO-like) tag. Proteins were expressed in Rosetta2(DE3)pLysS Escherichia coli by isopropyl β-d-1-thiogalactopyranoside induction at 37°C. Cells were lysed by sonication in the above lysis buffer. Sac1 2–251 was purified using nickel-nitrilotriacetic acid resin (Qiagen, Valencia, CA). The Mat/Smt3p tag was removed by addition of Smt3p-specific Ulp1 protease during overnight dialysis at 4°C against 10 mM HEPES, 150 mM NaCl, 5 mM 2-mercaptoethanol (pH 7.0). Ulp1-digested Sac1 2–251 was further purified by anion exchange (Source Q; GE Healthcare, Waukesha, WI) and size exclusion chromatography (Superose 12; GE Healthcare).
SPR experiments were performed on a Biacore 3000 instrument at 25°C in 25 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.005% Tween-20 (pH 7.5). The hydrogel matrix of a Biacore CM5 biosensor chip was activated with N-ethyl-N′-(dimethylaminopropyl)-carbodiimide hydrochloride and N-hydroxysuccinimide. Sac1 2–251 (100 μg/ml in 10 mM sodium acetate, pH 5.0) was flowed over this activated surface at a rate of 10 μl/min for 5 min. The remaining reactive sites were blocked with 1 M ethanolamine-HCl (pH 8.5). The signal contributed by immobilized Sac1 2–251 ranged from 9000 to 11,000 RU. Binding response values are reported after reference (mock-coupled surface binding) subtraction. Data were analyzed using Prism, version4 (GraphPad, La Jolla, CA).
Yeast two-hybrid analysis
The full-length Vps74 sequence was cloned into plasmid pGBT9, the first 1383 base pairs of the Sac1 gene (amino acids 1–461) were cloned into plasmid pGAD424, and the sequences were confirmed. To assay interaction, we transformed plasmids into strain HF7c, cell cultures were grown overnight with the density adjusted to OD600 = 1.0, and then 10-fold dilutions were spotted onto plasmid-selection plates (-leucine, -tryptophan) and interaction-selection plates (-leucine, -tryptophan, -histidine) containing 10 mM 3-amino-1,2,4-triazole (Sigma-Aldrich, St. Louis, MO). Plates were incubated at 30°C for 2 d and then photographed.
Fluorescence microscopy and image analysis
Cells were grown in selective media to log phase (OD600 = 0.6) and were mounted on a microscope slide in growth medium. Three-dimensional image stacks were collected at 0.5-μ Z-increments on a DeltaVision workstation (Applied Precision) based on an inverted microscope (IX-70; Olympus) using a 100×, 1.4 NA oil-immersion lens. Images were captured at 24°C with a 12-bit charge-coupled device camera (CoolSnap HQ; Photometrics, Tucson, AZ) and deconvolved using the iterative-constrained algorithm and the measured point spread function. The images were processed for presentation using the National Institute of Health's (NIH) ImageJ (http://rsb.info.nih.goc/ij). For colocalization analyses, Volocity imaging software (Perkin Elmer-Cetus, Waltham, MA) was used to analyze a minimum of 25 individual cells in at least three different fields and the mean of Pearson's correlation coefficients (r) were determined.
Lipid analyses
Metabolic [3H]inositol labeling, recovery and deacylation of phosphoinositides, and resolution of the glycero-phosphoinositol derivatives by anion-exchange chromatography was done according to a published method (Rivas et al., 1999; Schaaf et al., 2008). The genotype of the parental strain used for measurements of phosphoinositide levels is sec14-1 kes1Δ, which allows for accurate changes in PtdIns4P abundance due to elevated basal PtdIns4P. The VPS74 locus was deleted by gene replacement, or overexpressed from a multi-copy 2 μ VPS74 vector, in this strain.
Sphingolipid analyses were done by metabolic [3H]inositol labeling and thin layer chromotography according to a published method (Gaynor et al., 1999). Densitometry of autoradiographs using ImageJ was used to quantify the changes in the levels of IPC, MIPC, and M(IP)2C normalized to PtdIns.
Supplementary Material
Acknowledgments
We thank Todd Graham, David Katzmann, Mickey Marks, Mark Lemmon, Margaret Chou, and members of our laboratories for helpful discussions. We are grateful to Aaron Gitler for use of genetic profiling instrumentation and advice with data analysis, Andrea Stout for advice regarding image acquisition, and Scott Young (Perkin Elmer-Cetus) for advice regarding image analysis. This work is supported by grants from the American Heart Association (C.S.W. and K.M.F.), the National Science Council of Taiwan (C.-S.H.), and the NIH (C.G.B., K.M.F., and V.B.).
Abbreviations used:
- AEBSF
4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride
- BiFC
bimolecular fluorescence complementation
- CEN
yeast centromere
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- COPI
coatomer
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- GSH
glutathione
- GST
glutathione S-transferase
- IPC
inositol phosphorylceramide
- LB
lysis buffer
- MIPC
mannosylinositol phosphorylceramide
- M(IP)2C
mannose-(inositol phosphate)2-ceramide
- PI4K
phosphatidylinositol 4-kinase
- PM
plasma membrane
- PtdIns4P
phosphatidylinositol 4-phosphate
- RNAi
RNA interference
- SPR
surface plasmon resonance
- SPT
serine palmitoyl transferase
- TGN
trans-Golgi network
- YFP
yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-01-0077) on May 2, 2012.
REFERENCES
- Breslow DK, Collins SR, Bodenmiller B, Aebersold R, Simons K, Shevchenko A, Ejsing CS, Weissman JS. Orm family proteins mediate sphingolipid homeostasis. Nature. 2010;463:1048–1053. doi: 10.1038/nature08787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher MS, Munro S. Cholesterol and the Golgi apparatus. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- Brice SE, Alford CW, Cowart LA. Modulation of sphingolipid metabolism by the phosphatidylinositol-4-phosphate phosphatase Sac1p through regulation of phosphatidylinositol in Saccharomyces cerevisiae. J Biol Chem. 2009;284:7588–7596. doi: 10.1074/jbc.M808325200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong FY, Sharma V, Blagoveshchenskaya A, Oorschot VM, Brankatschk B, Klumperman J, Freeze HH, Mayinger P. Spatial regulation of Golgi phosphatidylinositol-4-phosphate is required for enzyme localization and glycosylation fidelity. Traffic. 2010;11:1180–1190. doi: 10.1111/j.1600-0854.2010.01092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleves AE, Novick PJ, Bankaitis VA. Mutations in the SAC1 gene suppress defects in yeast Golgi and yeast actin function. J Cell Biol. 1989;109:2939–2950. doi: 10.1083/jcb.109.6.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbacho I, Olivero I, Hernandez LM. A genome-wide screen for Saccharomyces cerevisiae nonessential genes involved in mannosyl phosphate transfer to mannoprotein-linked oligosaccharides. Fungal Genet Biol. 2005;42:773–790. doi: 10.1016/j.fgb.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Corbacho I, Olivero I, Hernandez LM. Identification of the MNN3 gene of Saccharomyces cerevisiae. Glycobiology. 2010;20:1336–1340. doi: 10.1093/glycob/cwq114. [DOI] [PubMed] [Google Scholar]
- Costanzo M, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dippold HC, et al. GOLPH3 bridges phosphatidylinositol-4-phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell. 2009;139:337–351. doi: 10.1016/j.cell.2009.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti M, Audhya A, Emr SD. Sac1 lipid phosphatase and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol Biol Cell. 2001;12:2396–2411. doi: 10.1091/mbc.12.8.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor EC, Mondesert G, Grimme SJ, Reed SI, Orlean P, Emr SD. MCD4 encodes a conserved endoplasmic reticulum membrane protein essential for glycosylphosphatidylinositol anchor synthesis in yeast. Mol Biol Cell. 1999;10:627–648. doi: 10.1091/mbc.10.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh I, Hamilton AD, Regan L. Antiparallel leucine zipper-directed protein reassembly: application to the green fluorescent protein. J Am Chem Soc. 2000;122:5658–5659. [Google Scholar]
- Godi A, et al. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- Graham TR, Burd CG. Coordination of Golgi functions by phosphatidylinositol 4-kinases. Trends Cell Biol. 2011;21:113–121. doi: 10.1016/j.tcb.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan XL, et al. Functional interactions between sphingolipids and sterols in biological membranes regulating cell physiology. Mol Biol Cell. 2009;20:2083–2095. doi: 10.1091/mbc.E08-11-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Lone MA, Schneiter R, Chang A. Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc Natl Acad Sci USA. 2010;107:5851–5856. doi: 10.1073/pnas.0911617107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer ME, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- Koh JL, Ding H, Costanzo M, Baryshnikova A, Toufighi K, Bader GD, Myers CL, Andrews BJ, Boone C. DRYGIN: a database of quantitative genetic interaction networks in yeast. Nucleic Acids Res. 2010;38:D502–D507. doi: 10.1093/nar/gkp820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine TP, Munro S. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr Biol. 2002;12:695–704. doi: 10.1016/s0960-9822(02)00779-0. [DOI] [PubMed] [Google Scholar]
- Levine TP, Wiggins CA, Munro S. Inositol phosphorylceramide synthase is located in the Golgi apparatus of Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:2267–2281. doi: 10.1091/mbc.11.7.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manford A, Xia T, Saxena AK, Stefan C, Hu F, Emr SD, Mao Y. Crystal structure of the yeast Sac1: implications for its phosphoinositide phosphatase function. EMBO J. 2010;29:1489–1498. doi: 10.1038/emboj.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manneville JB, Casella JF, Ambroggio E, Gounon P, Bertherat J, Bassereau P, Cartaud J, Antonny B, Goud B. COPI coat assembly occurs on liquid-disordered domains and the associated membrane deformations are limited by membrane tension. Proc Natl Acad Sci USA. 2008;105:16946–16951. doi: 10.1073/pnas.0807102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas MP, Kearns BG, Xie Z, Guo S, Sekar MC, Hosaka K, Kagiwada S, York JD, Bankaitis VA. Pleiotropic alterations in lipid metabolism in yeast sac1 mutants: relationship to “bypass Sec14p” and inositol auxotrophy. Mol Biol Cell. 1999;10:2235–2250. doi: 10.1091/mbc.10.7.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants FM, Baltz AG, Trott AE, Fereres S, Thorner J. A protein kinase network regulates the function of aminophospholipid flippases. Proc Natl Acad Sci USA. 2010;107:34–39. doi: 10.1073/pnas.0912497106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants FM, Torrance PD, Bezman N, Thorner J. Pkh1 and Pkh2 differentially phosphorylate and activate Ypk1 and Ykr2 and define protein kinase modules required for maintenance of cell wall integrity. Mol Biol Cell. 2002;13:3005–3028. doi: 10.1091/mbc.E02-04-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde HM, Cheong FY, Konrad G, Paiha K, Mayinger P, Boehmelt G. The human phosphatidylinositol phosphatase SAC1 interacts with the coatomer I complex. J Biol Chem. 2003;278:52689–52699. doi: 10.1074/jbc.M307983200. [DOI] [PubMed] [Google Scholar]
- Roy A, Levine TP. Multiple pools of phosphatidylinositol 4-phosphate detected using the pleckstrin homology domain of Osh2p. J Biol Chem. 2004;279:44683–44689. doi: 10.1074/jbc.M401583200. [DOI] [PubMed] [Google Scholar]
- Santiago-Tirado FH, Bretscher A. Membrane-trafficking sorting hubs: cooperation between PI4P and small GTPases at the trans-Golgi network. Trends Cell Biol. 2011;21:515–525. doi: 10.1016/j.tcb.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf G, et al. Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily. Mol Cell. 2008;29:191–206. doi: 10.1016/j.molcel.2007.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz KR, Liu J, Li S, Setty TG, Wood CS, Burd CG, Ferguson KM. Golgi localization of glycosyltransferases requires a Vps74p oligomer. Dev Cell. 2008;14:523–534. doi: 10.1016/j.devcel.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorr M, Then A, Tahirovic S, Hug N, Mayinger P. The phosphoinositide phosphatase Sac1p controls trafficking of the yeast Chs3p chitin synthase. Curr Biol. 2001;11:1421–1426. doi: 10.1016/s0960-9822(01)00449-3. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Lawrence LW. Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1979. [Google Scholar]
- Simons K, Van Meer G. Lipid sorting in epithelial cells. Biochemistry. 1988;27:6197–6202. doi: 10.1021/bi00417a001. [DOI] [PubMed] [Google Scholar]
- Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 2011;144:389–401. doi: 10.1016/j.cell.2010.12.034. [DOI] [PubMed] [Google Scholar]
- Strahl T, Hama H, Dewald DB, Thorner J. Yeast phosphatidylinositol 4-kinase, Pik1, has essential roles at the Golgi and in the nucleus. J Cell Biol. 2005;171:967–979. doi: 10.1083/jcb.200504104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strochlic TI, Setty TG, Sitaram A, Burd CG. Grd19/Snx3p functions as a cargo-specific adapter for retromer-dependent endocytic recycling. J Cell Biol. 2007;177:115–125. doi: 10.1083/jcb.200609161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung MK, Huh WK. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast. 2007;24:767–775. doi: 10.1002/yea.1504. [DOI] [PubMed] [Google Scholar]
- Tong AH, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Tu L, Tai WC, Chen L, Banfield DK. Signal-mediated dynamic retention of glycosyltransferases in the Golgi. Science. 2008;321:404–407. doi: 10.1126/science.1159411. [DOI] [PubMed] [Google Scholar]
- Walch-Solimena C, Novick P. The yeast phosphatidylinositol-4-OH kinase pik1 regulates secretion at the Golgi. Nat Cell Biol. 1999;1:523–525. doi: 10.1038/70319. [DOI] [PubMed] [Google Scholar]
- Wendland B, Emr SD. Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J Cell Biol. 1998;141:71–84. doi: 10.1083/jcb.141.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitters EA, Cleves AE, McGee TP, Skinner HB, Bankaitis VA. SAC1p is an integral membrane protein that influences the cellular requirement for phospholipid transfer protein function and inositol in yeast. J Cell Biol. 1993;122:79–94. doi: 10.1083/jcb.122.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CS, Schmitz KR, Bessman NJ, Setty TG, Ferguson KM, Burd CG. PtdIns4P recognition by Vps74/GOLPH3 links PtdIns 4-kinase signaling to retrograde Golgi trafficking. J Cell Biol. 2009;187:967–975. doi: 10.1083/jcb.200909063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.