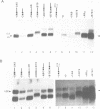
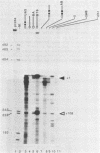
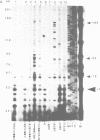
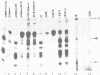Abstract
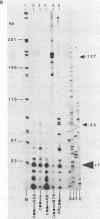
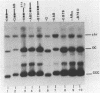
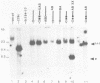
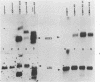
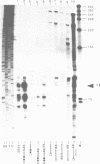
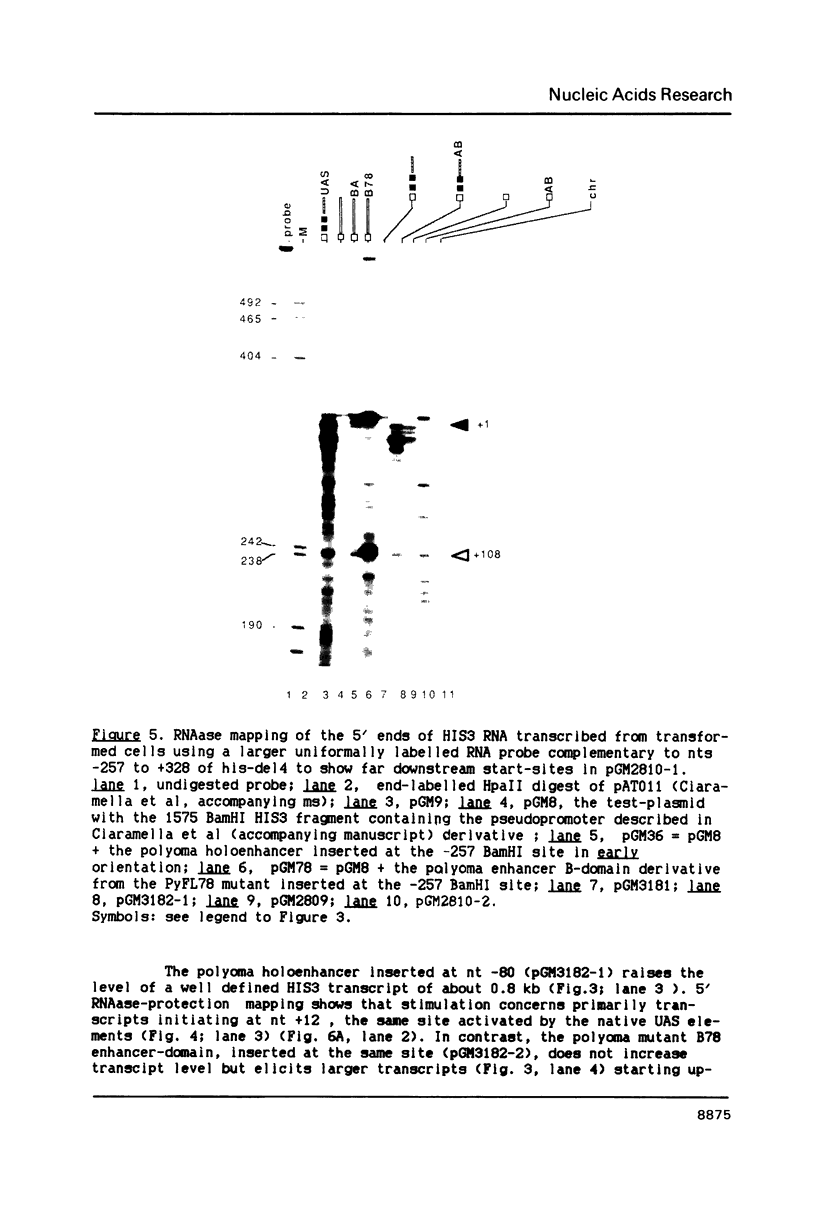
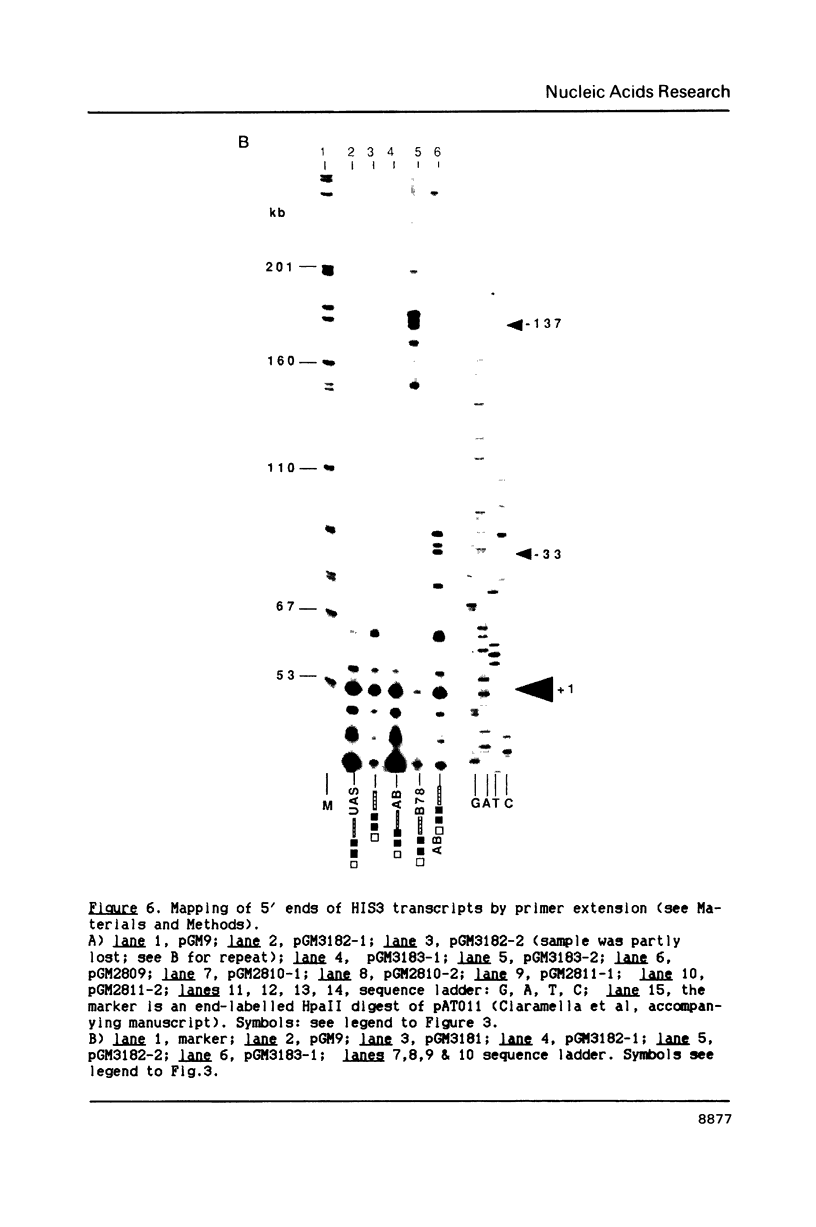
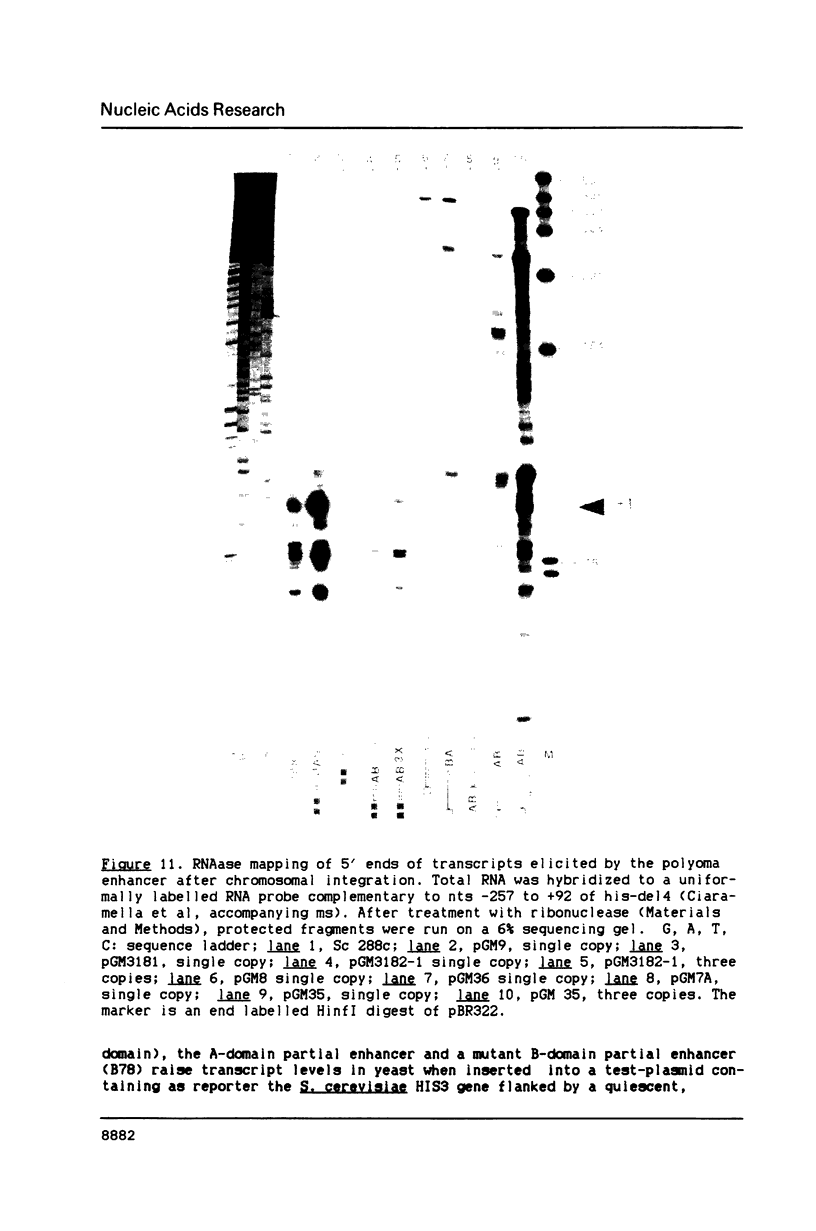
In this paper, to further analyze the function of the polyoma enhancer in Saccharomyces cerevisiae, we use as reporter-genes derivatives of the yeast HIS3 gene flanked by two types of partially deleted promoters: in one, UAS elements are removed by deletion of sequences upstream of nt -80 (pGM3181) in the second both TATA boxes and UAS elements are removed by deletion of sequences upstream of nt -35 (pGM2809). These constructs have been studied both as free plasmids and after integration at the TRP1 chromosomal locus. We find that in general the polyoma holoenhancer (A + B domains) elicits transcription from the physiological HIS3 RNA start sites when the native TATA boxes are present. In contrast, an altered enhancer B-domain from polyoma mutant Py-B78, although active when inserted downstream of the test-gene or when coupled to a pseudopromoter (Ciaramella et al, accompanying manuscript), does not work properly in concert with the native yeast TATA boxes. We describe experiments that suggest an important role for the foreign enhancer in RNA start-site selection in yeast.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Angel P., Allegretto E. A., Okino S. T., Hattori K., Boyle W. J., Hunter T., Karin M. Oncogene jun encodes a sequence-specific trans-activator similar to AP-1. Nature. 1988 Mar 10;332(6160):166–171. doi: 10.1038/332166a0. [DOI] [PubMed] [Google Scholar]
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Campbell B. A., Villarreal L. P. Functional analysis of the individual enhancer core sequences of polyomavirus: cell-specific uncoupling of DNA replication from transcription. Mol Cell Biol. 1988 May;8(5):1993–2004. doi: 10.1128/mcb.8.5.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghini S., Herbomel P., Jouanneau J., Saragosti S., Katinka M., Bourachot B., de Crombrugghe B., Yaniv M. Structure and function of the promoter-enhancer region of polyoma and SV40. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):935–944. doi: 10.1101/sqb.1983.047.01.107. [DOI] [PubMed] [Google Scholar]
- Chen W., Tabor S., Struhl K. Distinguishing between mechanisms of eukaryotic transcriptional activation with bacteriophage T7 RNA polymerase. Cell. 1987 Sep 25;50(7):1047–1055. doi: 10.1016/0092-8674(87)90171-1. [DOI] [PubMed] [Google Scholar]
- Guarente L., Hoar E. Upstream activation sites of the CYC1 gene of Saccharomyces cerevisiae are active when inverted but not when placed downstream of the "TATA box". Proc Natl Acad Sci U S A. 1984 Dec;81(24):7860–7864. doi: 10.1073/pnas.81.24.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. UASs and enhancers: common mechanism of transcriptional activation in yeast and mammals. Cell. 1988 Feb 12;52(3):303–305. doi: 10.1016/s0092-8674(88)80020-5. [DOI] [PubMed] [Google Scholar]
- Harshman K. D., Moye-Rowley W. S., Parker C. S. Transcriptional activation by the SV40 AP-1 recognition element in yeast is mediated by a factor similar to AP-1 that is distinct from GCN4. Cell. 1988 Apr 22;53(2):321–330. doi: 10.1016/0092-8674(88)90393-5. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Jones R. H., Moreno S., Nurse P., Jones N. C. Expression of the SV40 promoter in fission yeast: identification and characterization of an AP-1-like factor. Cell. 1988 May 20;53(4):659–667. doi: 10.1016/0092-8674(88)90581-8. [DOI] [PubMed] [Google Scholar]
- Kakidani H., Ptashne M. GAL4 activates gene expression in mammalian cells. Cell. 1988 Jan 29;52(2):161–167. doi: 10.1016/0092-8674(88)90504-1. [DOI] [PubMed] [Google Scholar]
- Lech K., Anderson K., Brent R. DNA-bound Fos proteins activate transcription in yeast. Cell. 1988 Jan 29;52(2):179–184. doi: 10.1016/0092-8674(88)90506-5. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987 Mar 13;48(5):847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982 Jul 23;217(4557):316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Nagawa F., Fink G. R. The relationship between the "TATA" sequence and transcription initiation sites at the HIS4 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8557–8561. doi: 10.1073/pnas.82.24.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Constitutive and inducible Saccharomyces cerevisiae promoters: evidence for two distinct molecular mechanisms. Mol Cell Biol. 1986 Nov;6(11):3847–3853. doi: 10.1128/mcb.6.11.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Genetic properties and chromatin structure of the yeast gal regulatory element: an enhancer-like sequence. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7865–7869. doi: 10.1073/pnas.81.24.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Struhl K. The DNA-binding domains of the jun oncoprotein and the yeast GCN4 transcriptional activator protein are functionally homologous. Cell. 1987 Sep 11;50(6):841–846. doi: 10.1016/0092-8674(87)90511-3. [DOI] [PubMed] [Google Scholar]
- Struhl K. The yeast his3 promoter contains at least two distinct elements. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7385–7389. doi: 10.1073/pnas.79.23.7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster N., Jin J. R., Green S., Hollis M., Chambon P. The yeast UASG is a transcriptional enhancer in human HeLa cells in the presence of the GAL4 trans-activator. Cell. 1988 Jan 29;52(2):169–178. doi: 10.1016/0092-8674(88)90505-3. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Cheng P. F., Conrad K. Expression of transfected DNA depends on DNA topology. Cell. 1986 Jul 4;46(1):115–122. doi: 10.1016/0092-8674(86)90865-2. [DOI] [PubMed] [Google Scholar]