Abstract
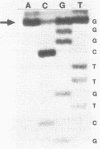
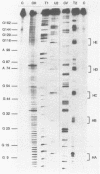
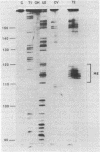
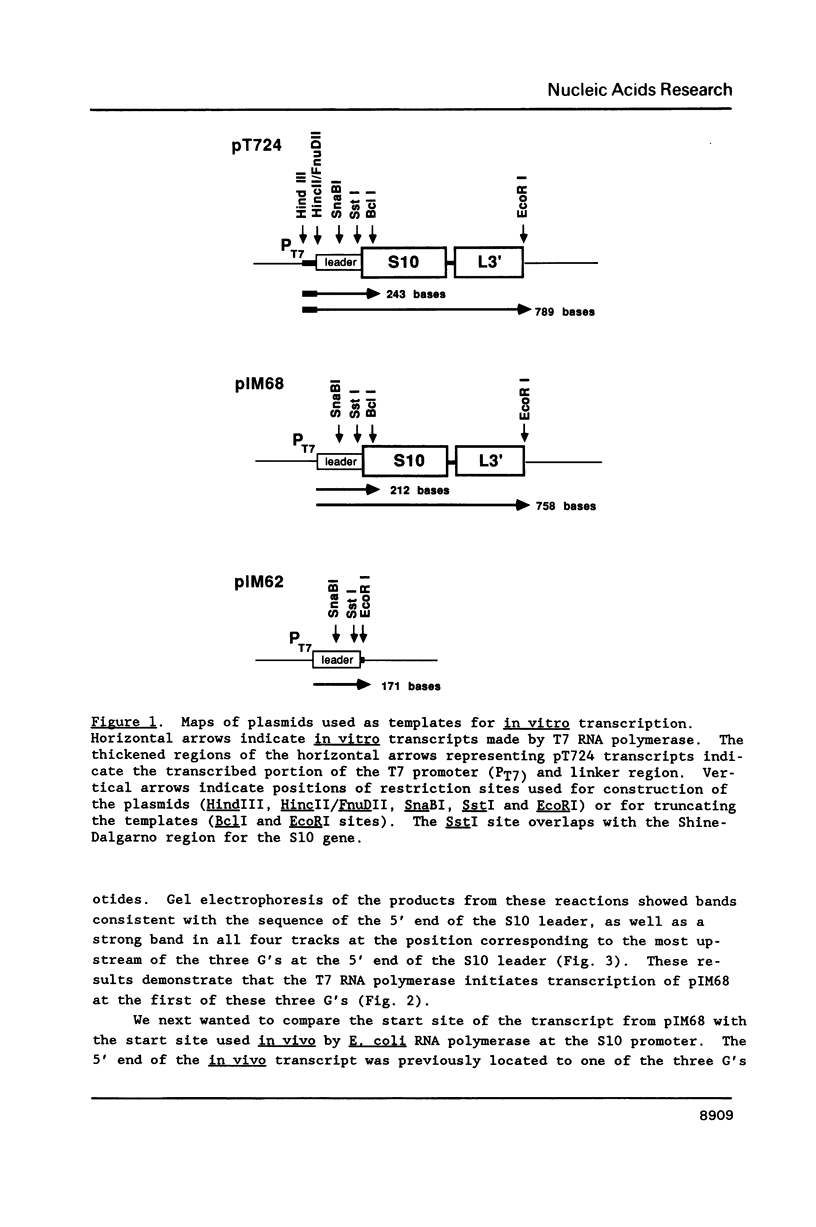
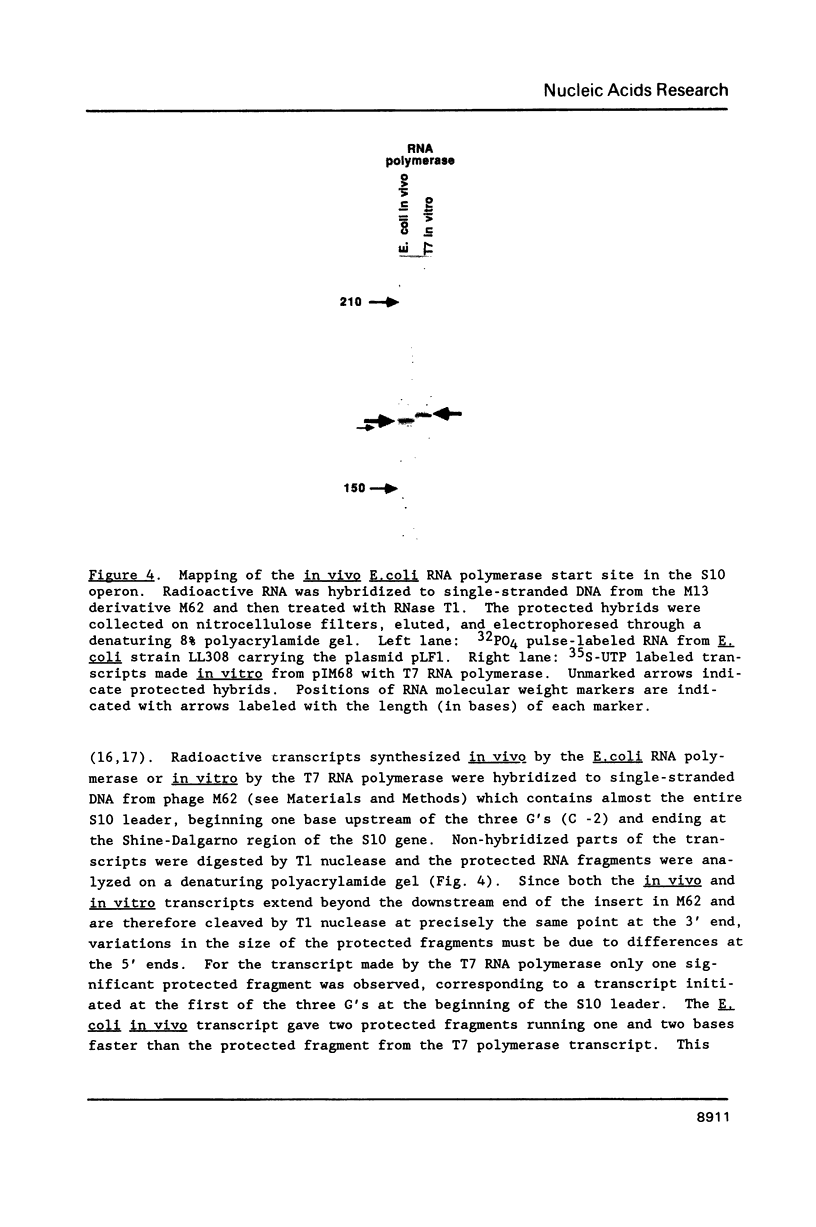
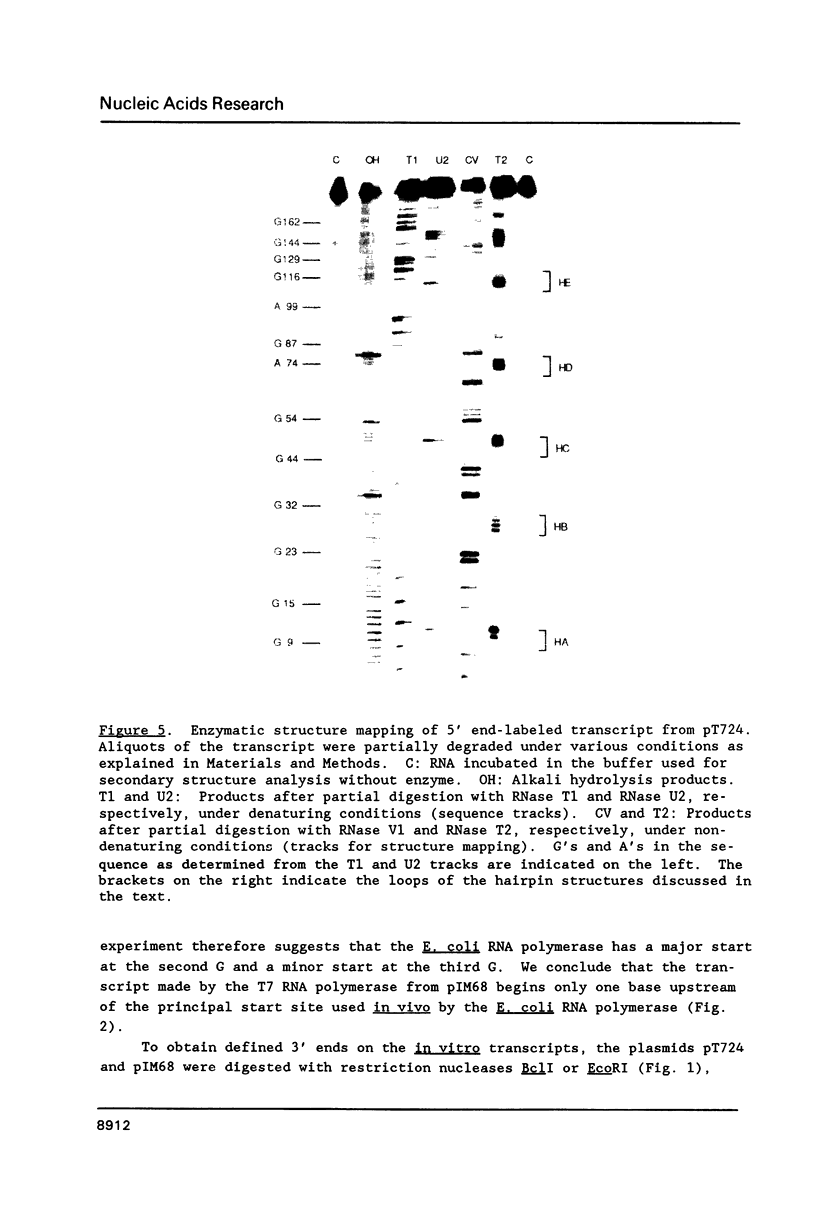
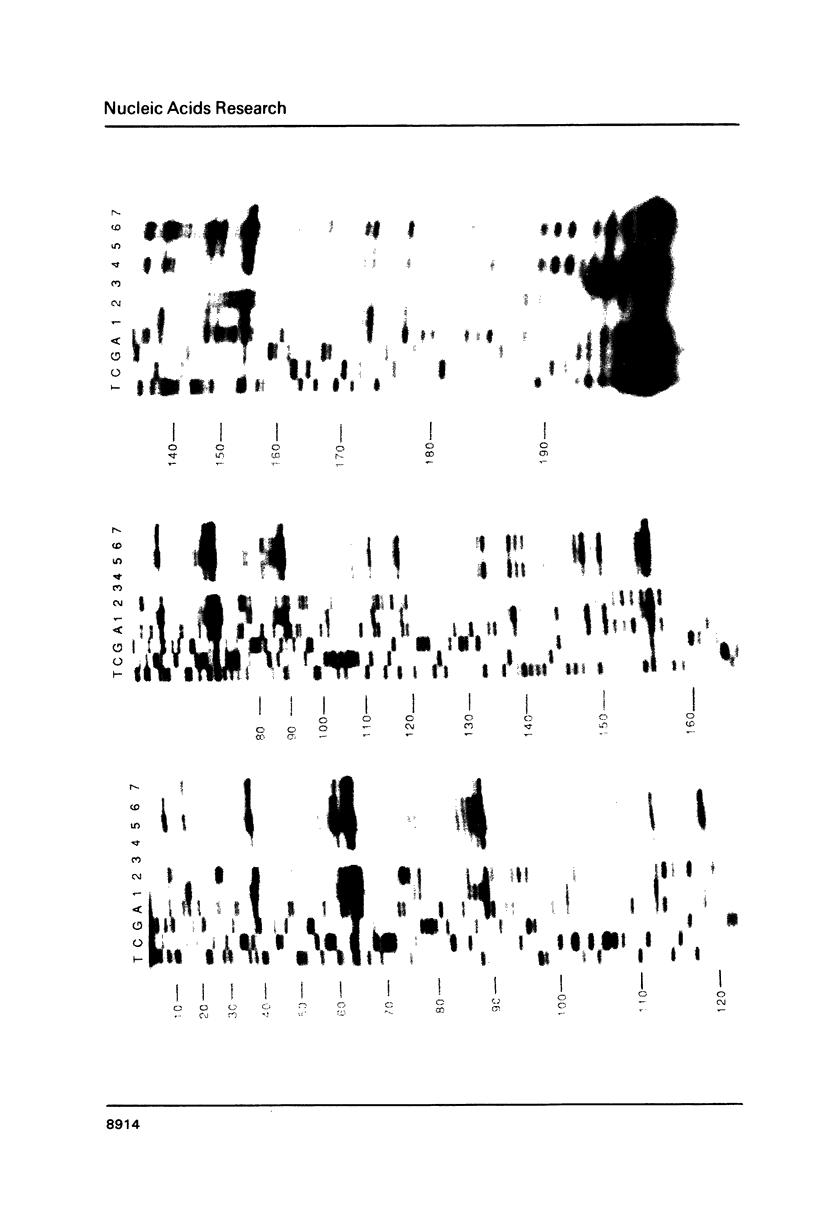
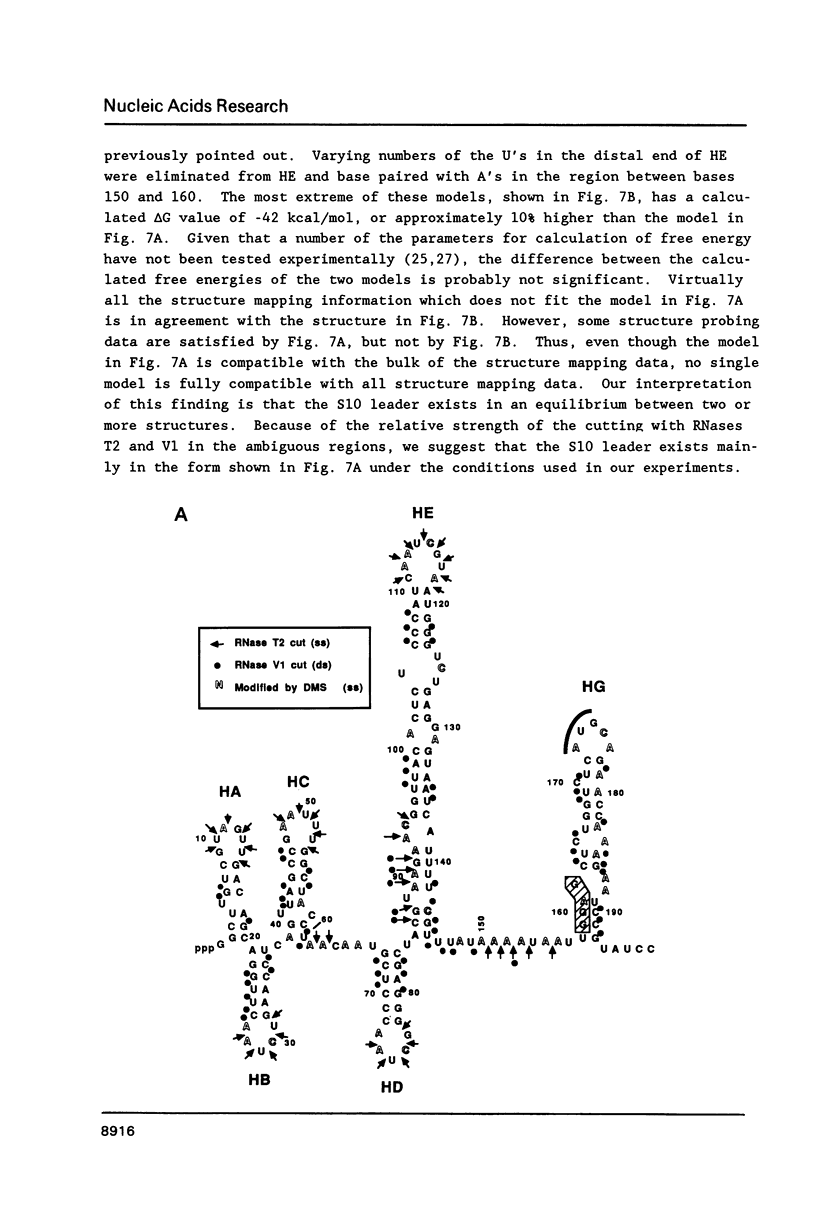
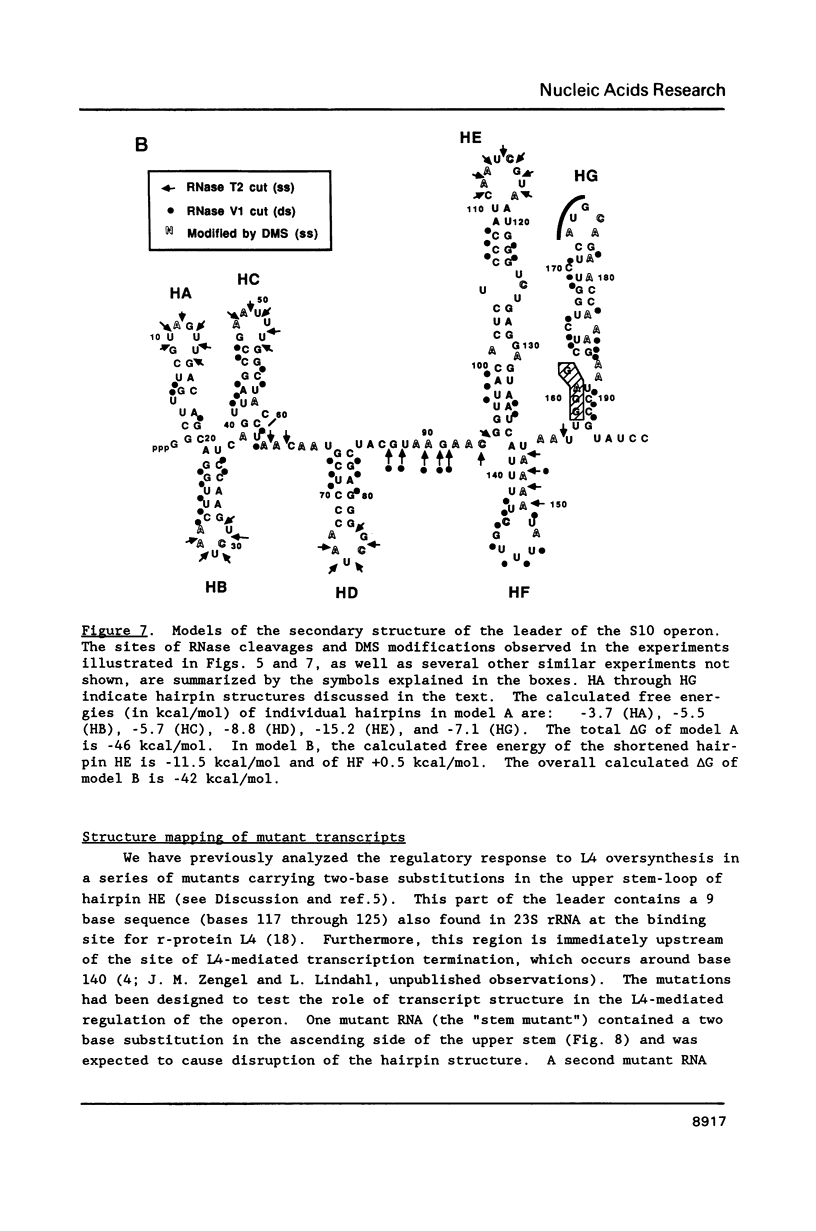
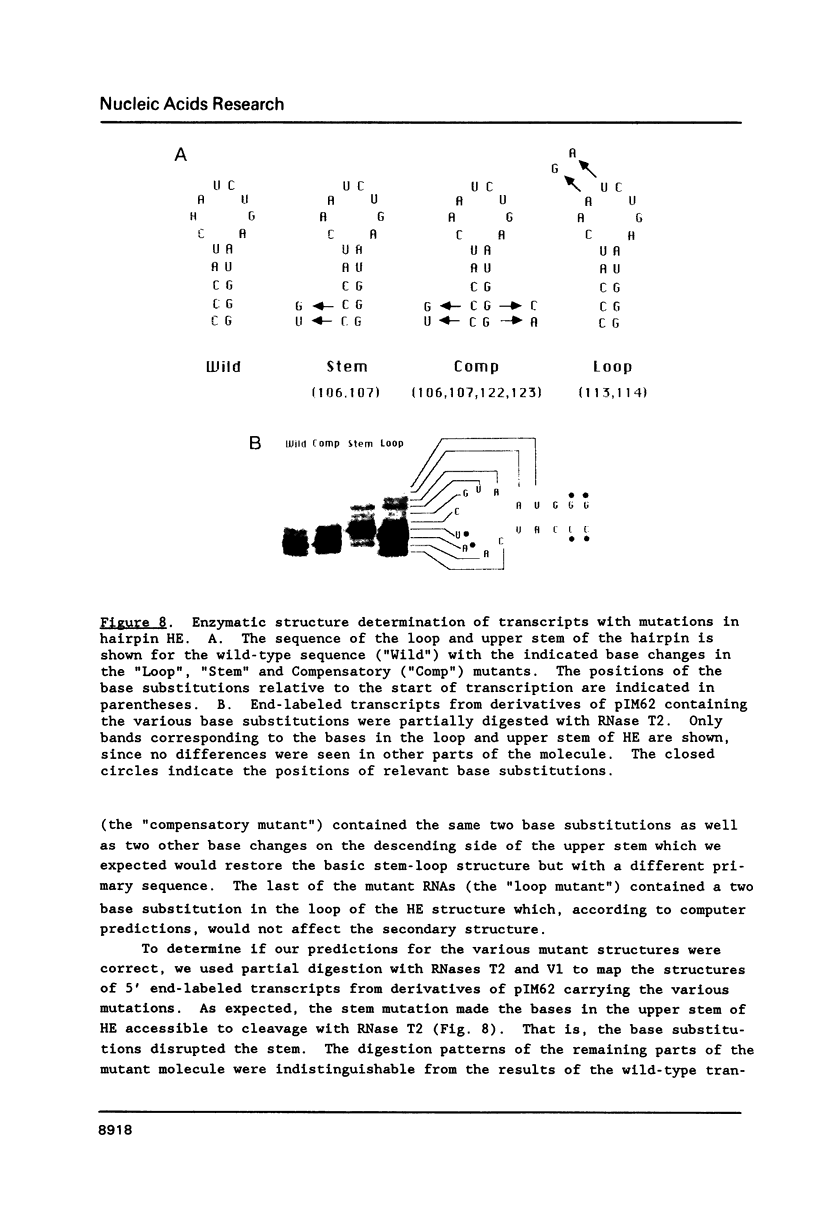
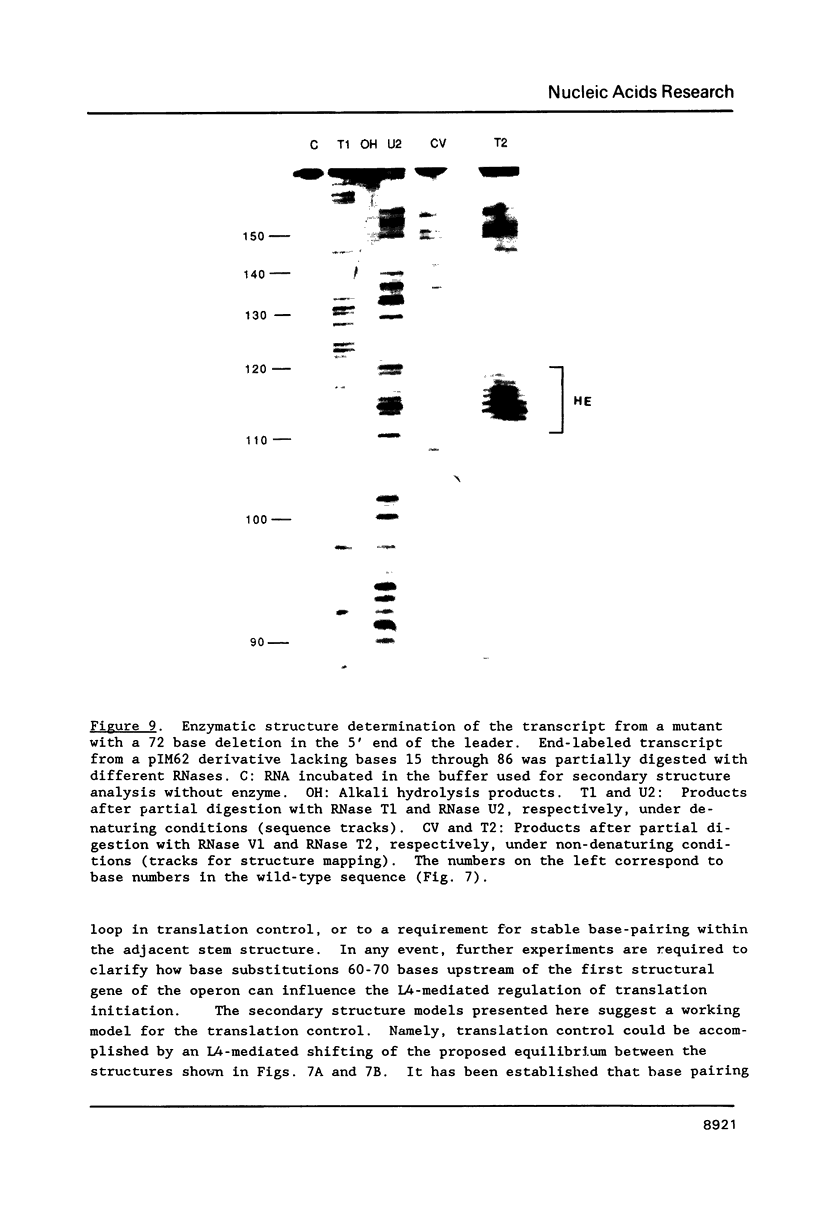
Genetic analysis of the autogenous control of the S10 ribosomal protein operon of Escherichia coli has suggested that the secondary or tertiary structure of the leader transcript is important for this regulation. We have therefore determined the secondary structure of the leader by enzyme digestion and chemical modification. Our results suggest that the 172 base leader exists in two forms, differing only immediately upstream of the Shine-Dalgarno sequence of the first gene. We discuss the possibility that the equilibrium between these alternate structures is important for the L4-mediated regulation of translation of the S10 operon. We have also determined the structure of several mutant transcripts. Correlation of these structures with the regulatory phenotypes suggest that a hairpin about 50 bases upstream of the first gene is essential for the control of translation of the operon. Finally, our results show that a two base substitution in an eight base loop destabilizes the attached stem.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BROOKES P., LAWLEY P. D. EFFECTS OF ALKYLATING AGENTS ON T2 AND T4 BACTERIOPHAGES. Biochem J. 1963 Oct;89:138–144. doi: 10.1042/bj0890138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B., Schmidt B. F., van Strien A., van Boom J., van Westrenen J., van Duin J. Lysis gene of bacteriophage MS2 is activated by translation termination at the overlapping coat gene. J Mol Biol. 1987 Jun 5;195(3):517–524. doi: 10.1016/0022-2836(87)90180-x. [DOI] [PubMed] [Google Scholar]
- Blumer K. J., Ivey M. R., Steege D. A. Translational control of phage f1 gene expression by differential activities of the gene V, VII, IX and VIII initiation sites. J Mol Biol. 1987 Oct 5;197(3):439–451. doi: 10.1016/0022-2836(87)90557-2. [DOI] [PubMed] [Google Scholar]
- Deckman I. C., Draper D. E. Specific interaction between ribosomal protein S4 and the alpha operon messenger RNA. Biochemistry. 1985 Dec 31;24(27):7860–7865. doi: 10.1021/bi00348a002. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman L. P., Zengel J. M., Archer R. H., Lindahl L. Autogenous control of the S10 ribosomal protein operon of Escherichia coli: genetic dissection of transcriptional and posttranscriptional regulation. Proc Natl Acad Sci U S A. 1987 Sep;84(18):6516–6520. doi: 10.1073/pnas.84.18.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman L. P., Zengel J. M., Lindahl L. Genetic dissection of stringent control and nutritional shift-up response of the Escherichia coli S10 ribosomal protein operon. J Mol Biol. 1985 Oct 20;185(4):701–712. doi: 10.1016/0022-2836(85)90055-5. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulle H., Hoppe E., Osswald M., Greuer B., Brimacombe R., Stöffler G. RNA-protein cross-linking in Escherichia coli 50S ribosomal subunits; determination of sites on 23S RNA that are cross-linked to proteins L2, L4, L24 and L27 by treatment with 2-iminothiolane. Nucleic Acids Res. 1988 Feb 11;16(3):815–832. doi: 10.1093/nar/16.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen U., Sharp P. A. Sequences controlling in vitro transcription of SV40 promoters. EMBO J. 1983;2(12):2293–2303. doi: 10.1002/j.1460-2075.1983.tb01737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Cech T. R. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc Natl Acad Sci U S A. 1985 Feb;82(3):648–652. doi: 10.1073/pnas.82.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen M., Christensen T., Dennis P. P., Fiil N. P. Autogenous control: ribosomal protein L10-L12 complex binds to the leader sequence of its mRNA. EMBO J. 1982;1(8):999–1004. doi: 10.1002/j.1460-2075.1982.tb01284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond A. I., Travers A. A. Genetically separable functional elements mediate the optimal expression and stringent regulation of a bacterial tRNA gene. Cell. 1985 Feb;40(2):319–326. doi: 10.1016/0092-8674(85)90146-1. [DOI] [PubMed] [Google Scholar]
- Lindahl L., Archer R., Zengel J. M. Transcription of the S10 ribosomal protein operon is regulated by an attenuator in the leader. Cell. 1983 May;33(1):241–248. doi: 10.1016/0092-8674(83)90353-7. [DOI] [PubMed] [Google Scholar]
- Lindahl L., Zengel J. M. Operon-specific regulation of ribosomal protein synthesis in Escherichia coli. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6542–6546. doi: 10.1073/pnas.76.12.6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maly P., Rinke J., Ulmer E., Zwieb C., Brimacombe R. Precise localization of the site of cross-linking between protein L4 and 23S ribonucleic acid induced by mild ultraviolet irradiation of Escherichia coli 50S ribosomal subunits. Biochemistry. 1980 Sep 2;19(18):4179–4188. doi: 10.1021/bi00559a007. [DOI] [PubMed] [Google Scholar]
- Neidhardt F. C., Bloch P. L., Smith D. F. Culture medium for enterobacteria. J Bacteriol. 1974 Sep;119(3):736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins P. O., Nomura M. Regulation of the S10 ribosomal protein operon in E. coli: nucleotide sequence at the start of the operon. Cell. 1981 Oct;26(2 Pt 2):205–211. doi: 10.1016/0092-8674(81)90303-2. [DOI] [PubMed] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- Stern S., Wilson R. C., Noller H. F. Localization of the binding site for protein S4 on 16 S ribosomal RNA by chemical and enzymatic probing and primer extension. J Mol Biol. 1986 Nov 5;192(1):101–110. doi: 10.1016/0022-2836(86)90467-5. [DOI] [PubMed] [Google Scholar]
- Tuerk C., Gauss P., Thermes C., Groebe D. R., Gayle M., Guild N., Stormo G., d'Aubenton-Carafa Y., Uhlenbeck O. C., Tinoco I., Jr CUUCGG hairpins: extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1364–1368. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilenko S. K., Ryte V. C. [Isolation of highly purified ribonuclease from cobra (Naja oxiana) venom]. Biokhimiia. 1975 May-Jun;40(3):578–583. [PubMed] [Google Scholar]
- Vournakis J. N., Celantano J., Finn M., Lockard R. E., Mitra T., Pavlakis G., Troutt A., van den Berg M., Wurst R. M. Sequence and structure analysis of end-labeled RNA with nucleases. Gene Amplif Anal. 1981;2:267–298. [PubMed] [Google Scholar]
- Wurst R. M., Vournakis J. N., Maxam A. M. Structure mapping of 5'-32P-labeled RNA with S1 nuclease. Biochemistry. 1978 Oct 17;17(21):4493–4499. doi: 10.1021/bi00614a021. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yates J. L., Nomura M. E. coli ribosomal protein L4 is a feedback regulatory protein. Cell. 1980 Sep;21(2):517–522. doi: 10.1016/0092-8674(80)90489-4. [DOI] [PubMed] [Google Scholar]
- Zengel J. M., Mueckl D., Lindahl L. Protein L4 of the E. coli ribosome regulates an eleven gene r protein operon. Cell. 1980 Sep;21(2):523–535. doi: 10.1016/0092-8674(80)90490-0. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]