Abstract
Voltage-gated Na+-permeable (Nav) channels form the basis for electrical excitability in animals. Nav channels evolved from Ca2+ channels and were present in the common ancestor of choanoflagellates and animals, although this channel was likely permeable to both Na+ and Ca2+. Thus, like many other neuronal channels and receptors, Nav channels predated neurons. Invertebrates possess two Nav channels (Nav1 and Nav2), whereas vertebrate Nav channels are of the Nav1 family. Approximately 500 Mya in early chordates Nav channels evolved a motif that allowed them to cluster at axon initial segments, 50 million years later with the evolution of myelin, Nav channels “capitalized” on this property and clustered at nodes of Ranvier. The enhancement of conduction velocity along with the evolution of jaws likely made early gnathostomes fierce predators and the dominant vertebrates in the ocean. Later in vertebrate evolution, the Nav channel gene family expanded in parallel in tetrapods and teleosts (∼9 to 10 genes in amniotes, 8 in teleosts). This expansion occurred during or after the late Devonian extinction, when teleosts and tetrapods each diversified in their respective habitats, and coincided with an increase in the number of telencephalic nuclei in both groups. The expansion of Nav channels may have allowed for more sophisticated neural computation and tailoring of Nav channel kinetics with potassium channel kinetics to enhance energy savings. Nav channels show adaptive sequence evolution for increasing diversity in communication signals (electric fish), in protection against lethal Nav channel toxins (snakes, newts, pufferfish, insects), and in specialized habitats (naked mole rats).
Keywords: gene duplication, nervous system
Multicellular animals evolved >650 million years ago (1). The nervous system and muscles evolved shortly thereafter. The phylogeny of basal metazoans is poorly resolved, likely because of the rapid radiation of these then-new life forms (2), so depending on the phylogeny one embraces, the nervous system evolved once, once with a loss in sponges, or twice independently in ctenophora and bilateria + cnidaria or bilateria and cnidaria + ctenophora (3, 4). However, in all animals with nervous systems, neurons generate action potentials (APs), release excitatory and inhibitory neurotransmitters, form circuits, receive sensory input, innervate muscle, and direct behavior.
The history of brain evolution and its key neural genes would fill volumes. I will use voltage-dependent Na+ (Nav, Na-permeable voltage-dependent = protein; scn, sodium channel = gene) channels as an exemplar to tell this story because all neuronal excitability depends on Nav channels, there is a good understanding of their function and regulation from biophysical, biochemical, and modeling studies, and there are fascinating examples of ecologically relevant adaptations. An additional rationale is that although many proteins, such as immunoglobins, sperm and egg receptors, olfactory receptors, opsins, and surface proteins of pathogens, are routinely studied in the field of molecular evolution, only recently have ion channels begun to receive greater attention (5–10); of these studies, the majority are on Nav channels.
Sodium Channel Genes Are Latecomers to the 6TM Family of Voltage-Dependent Ion Channels
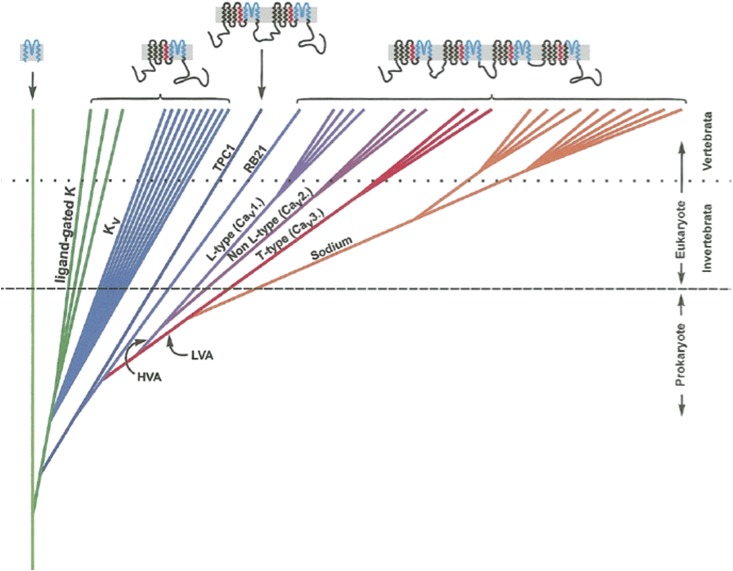
Voltage-gated ion channels are the basis of electrical excitability of all animals and many single-celled eukaryotes. Potassium leak and voltage-dependent K+ (Kv) channels appeared three billion years ago in bacteria and occur in all organisms (11) (Fig. 1). They establish resting potentials and repolarize membranes after excitatory events. Kv channels are the “founding members” of the family of ion-permeating channels whose basic structure is a protein of six transmembrane helices (6TM) that associate as tetramers to form a channel. At some point early in eukaryote evolution, the gene for a 6TM channel likely duplicated, giving rise to a protein with two domains. These proteins then dimerized to form a complete channel (12). Such a channel still exists in the two-pore channel family of Ca2+-permeable channels localized in endosomes and lysozomes (13). The gene for a two-domain channel likely duplicated to make a protein with four domains capable of forming a channel on its own (4x6TM). Eventually such a four-domain channel evolved (or retained) permeability to Ca2+, and these handily became involved in intracellular signaling. Other Ca2+-binding proteins and enzymes first appeared in single-celled eukaryotes (14). Additionally, there are single 6TM Na+-permeable channels in bacteria (15). Their relationship to eukaryotic Nav channels is unclear, and they will not be discussed in this review.
Fig. 1.
Schematic diagram of the evolutionary relationships among some key families in the ion channel superfamily. On the top of the figure is the structure of the channels moving from left to right showing a linear leak K+ channel that is composed of two membrane-spanning helices and a pore (blue), a 6TM channel with a single voltage sensor (red), and four domain 4x6TM channels with four voltage sensors. There is uncertainty about the origin of the 4x6TM family, which more likely evolved in eukaryotes than prokaryotes, as indicated in this figure. A more precise and detailed relationship among Cav and Nav channels in basal metazoans and their sister group, the choanoflagellates, is given in Fig. 3. Reprinted from Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology, 129/1, Peter A. V. Anderson, Robert M. Greenberg, Phylogeny of ion channels: Cues to structure and function, 12-17, Copyright (2001), with permission from Elsevier.
The three main types of Cav channels are L, N/P/Q/R, and T. Generally speaking, L type channels are found in muscle and neuronal dendrites, and N/P/Q/R are found in synaptic terminals and regulate transmitter release, whereas T types, which are sensitive to voltages close to resting potential, underlie spontaneous firing and pacemaking. These three subfamilies appear early in animals in a common ancestor of bilateria and cnidaria (16) (Fig. 2). Choanoflagellates, single-celled protists that are the sister group to metazoans, and sponges have a single Cav channel gene that is ancestral to the L and N/P/Q/R families. The origin of the T type channels is not clear.
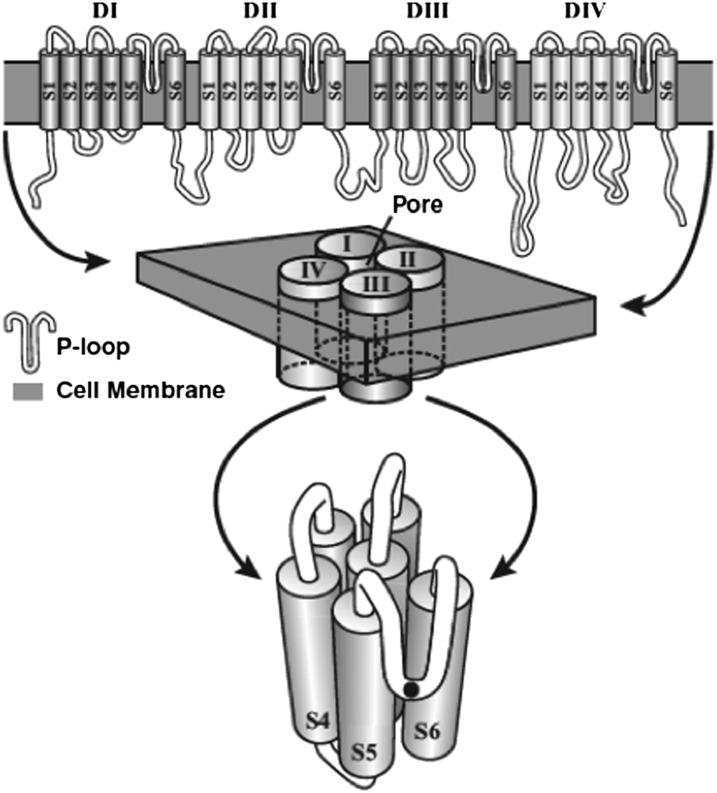
Fig. 2.
Hypothetical secondary structure of a Nav channel. Top: The Nav channel is composed of four repeating domains (I–IV), each of which has six membrane-spanning segments (S1–S6), and their connecting loops (in white). Middle: The four domains cluster around a pore. Bottom: The four P loops dip down into the membrane and line the outer mouth of the channel that is evident in an en face view of a single domain. The black dot represents the single amino acid at the deepest position of each of the four P loops that determines Na+ ion selectivity. From Liebeskind et al. (16).
Nav channels share the 4x6TM structure (Figs. 1 and 2) with Cav channels, and it has been suggested that Nav channels evolved from Cav channels (17). Analysis of putative Cav and Nav channel genes from fungi, choanoflagellates, and metazoans confirm this speculation and show that choanoflagellates have a channel that groups with recognized Nav channels with strong support (Fig. 3). The selectivity filter of 4x6TM channels depends on a single amino acid in each of the four domains that come together and face each other, presumably forming the deepest point in the pore. The selectivity filter of the choanoflagellate and other basal metazoans (DEEA) is midway between bona fide Cav (EEEE) and Nav1 (DEKA) channel pores and lives on in metazoans in a Nav channel found only in invertebrates (Nav2) (18) (Fig. 3). This pore sequence and studies of the invertebrate Nav2 suggest that the choanoflagellate Nav channel is likely permeable to both Ca2+ and Na+ and may not be a pure Na+-selective channel. This will be determined when the choanoflagellate Nav channel is expressed and studied in detail.
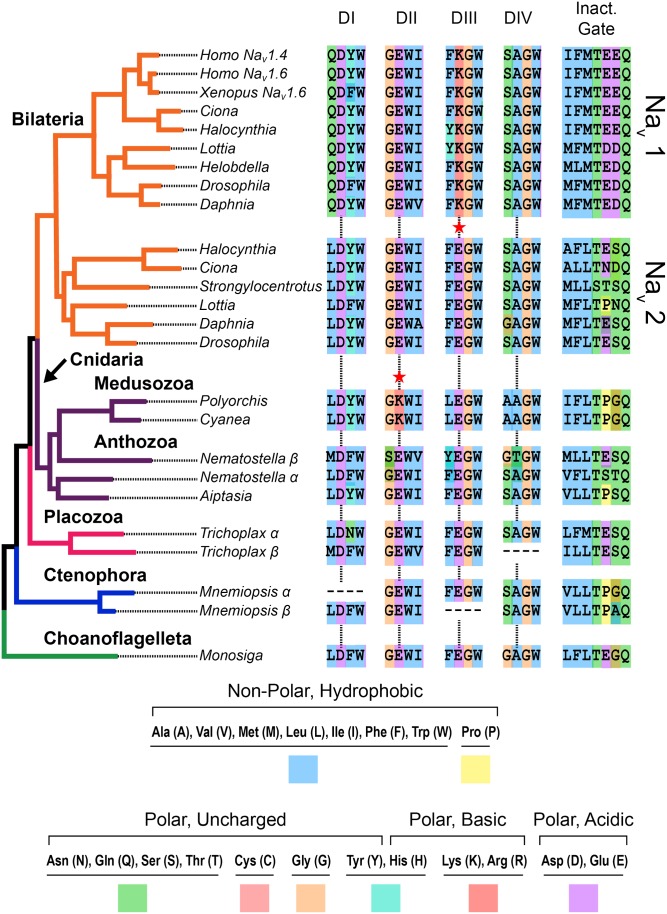
Fig. 3.
Maximum likelihood phylogeny of the voltage-gated sodium channel family. The common ancestor of choanoflagellates (represented by Monosiga in green) and animals had a Nav channel that was likely permeable to Ca2+ and Na+ (pore motif = DEEA). This motif is present in the Nav channels of anthozoan cnidaria (anemones, coral) and the Nav2 channel of invertebrates. The presence of a lysine (K) in the pore improves Na+ selectivity (indicated by red star). A lysine is found in the Nav1 channels of bilaterians (DEKA) and Nav channel of medusozoan cnidaria (jellyfish) (DKEA), both of which have more centralized nervous systems than anthozoans and are motile. Additionally, there is strong conservation of a hydrophobic (blue) triplet of amino acids in the “inactivation gate” region. From Liebeskind et al. (16).
The presence of a K in domain III of the pore, as in the bilaterian Nav1, increases Na+ selectivity substantially (Fig. 3). There is a K in domain II in the Nav channel pore of motile jellyfish (medusozoa) but not in sedentary anemones (anthozoa). The selectivity filter DKEA enhances Na+ selectivity less than DEKA but more than DEEA (19, 20). The nervous system of jellyfish has clusters of neurons approaching a real central nervous system, whereas that of anemones is more of a nerve net. Thus, enhanced Na+ selectivity occurred in parallel in medusozoan and bilaterian Nav channels along with increasing structural complexity of the nervous system (21).
There is little question as to the adaptive advantage conferred by Na+-selective channels in early animals. It was not only that, with the advent of multicellularity, they fulfilled the need in a newly evolved nervous system for rapid communication across distant parts of organisms, but that they did so by marshalling an ion that was abundant in the ocean and would minimally perturb intracellular Ca2+ levels and, therefore, intracellular signaling (17).
Beside the obvious change from Ca2+ to Na+ permeability, other changes occurred as well. The short intracellular loop between domains III and IV evolved function as the inactivation “ball” (22). In voltage-dependent K+ channels all four voltage sensors must be “engaged” for the channel to open. In the Na+ channel activation is accomplished by the three voltage sensors in domains I–III; the voltage sensor in domain IV initiates inactivation (23, 24). No Cav channel has been examined in such a way so we do not know whether they also have equivalently acting voltage sensors or whether the voltage sensor in domain IV had already evolved a novel function.
Evolution of Na+ Channel Clustering at the Axon Initial Segment and the Nodes of Ranvier
Myelination and saltatory conduction are key innovations of the vertebrate nervous system that markedly increase axonal conduction velocity [myelination evolved multiple times in some invertebrate lineages as well despite a widespread and persistent belief to the contrary (25, 26)]. Myelination is not present in agnathans but occurs in all gnathostomes, likely appearing first in a placoderm ancestor (27). Saltatory conduction depends on high densities of Nav channels at the nodes of Ranvier that inject sufficient current into the axon to depolarize the adjacent node to threshold. KCNQ-type K+ channels, which help to repolarize the AP, cluster at nodes as well, both channels tethered to ankyrin and thence to the cytoskeleton.
Remarkably, both Nav and KCNQ K+ channels evolved the same specific nine amino acid motif for ankyrin binding (28). This motif first appears in the Nav channels of ascidians and agnathans and, indeed, Nav channels cluster at axon initial segments (AIS) in the lamprey. In lampreys, and presumably nonvertebrate chordates, the high-density clustering of Nav channels adjacent to the soma ensures sufficient current injection into the high-resistance axon in the face of current shunting by the low-resistance soma (29). Shiverer mice, which have a mutation that prevents the formation of compact myelin, retain a high density of Nav channels (Nav1.6) at the AIS but not along the axon (30). This emphasizes the distinction between older non–myelin-dependent mechanisms for clustering Nav channels at the AIS and more recent myelin-dependent clustering of Nav channels at nodes. A surprising observation is that the AIS is mobile, moving toward the soma when a neuron’s firing rate is low and away from the some when it is high (31). This is likely different from the nodes of Ranvier, which are smaller and constrained by the myelin sheath. However, this remains to be investigated.
KCNQ channels only occur in gnathostomes. Once KCNQ channels appeared, all of the molecular components for construction of the nodes of Ranvier were in place. By this time the key genes for myelin components had also evolved (32, 33).
Making Up for Lost Time: Vertebrate Nav Channel Genes Duplicated Extensively in Teleosts and Tetrapods
Invertebrates have two Nav channel genes, Nav1 and Nav2, each in single copy. We have little information on the normal physiological role of Nav2 channels in invertebrates [knockouts in Drosophila are not lethal and produce only a mild phenotype (34, 35)]. It is interesting that both genes have been lost in nematodes (36), most of which are small and depend on passive transmission of electrical activity. The predominant Nav channel gene in invertebrates (para in Drosophila), and the only Nav channel gene in vertebrates, is Nav1. However, Nav1 has duplicated in vertebrates.
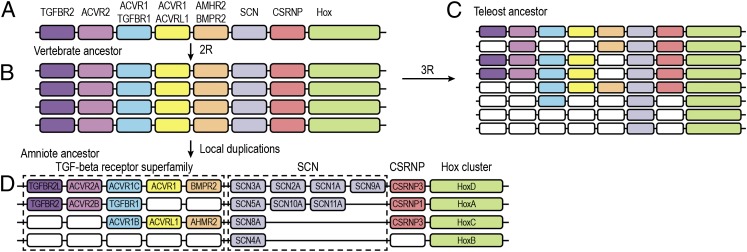
In a prescient insight in 1970, Susumu Ohno suggested that vertebrates underwent two rounds of whole-genome duplication (WGD) at their origin (2R hypothesis) and that a subsequent third WGD occurred in teleost fishes (3R) (37). Ohno believed that these ploidy events provided the raw genetic material from which emerged many of the defining features of vertebrates. Although originally controversial, his view has been empirically confirmed (38, 39). Nav1 channel genes show a perfect read-out of this history. A single Nav1 channel gene is present in tunicates, two in lampreys, four in elasmobranchs and in the common ancestor of teleosts and tetrapods (7, 8, 40, 41). As expected from a teleost-specific WGD, eight Nav channel genes are found in teleosts (Fig. 4).
Fig. 4.
The Nav channel gene family underwent an expansion in parallel in teleosts and tetrapods. (A) A schematic chromosome with Nav channel genes (SCN, sodium channel) surrounded by other genes. (B) This chromosome, along with all of the other ancestral chordate chromosomes, duplicated twice at the origin of vertebrates (2R). (C) There was an additional round of genome duplication in teleosts (3R) and (D) tandem duplications of Nav channel genes in ancestral tetrapods and amniotes. There is no indication of any loss of Nav channel genes despite losses of surrounding genes in both teleosts and tetrapods. Furthermore, although not shown here, no other ion channel gene family duplicated after the teleost and tetrapod divergence. Thus, there was likely to be strong selection for the preservation of Nav channel gene duplicates. Reprinted from Jenny Widmark, Görel Sundström, Daniel Ocampo Daza, Dan Larhammar, Differential evolution of voltage-gated sodium channels in tetrapods and teleost fishes, Molecular Biology and Evolution, 2011, by permission of Oxford University Press.
However, further gene duplication/retention occurred in tetrapods above and beyond that predicted by 2R. Two of the four Nav channel genes of our tetrapod ancestors underwent a series of tandem duplications in early amniotes, so that the stem reptilian ancestor of modern-day reptiles, birds, and mammals had nine Nav1 channel genes (8, 41). A final duplication occurred early in the mammalian lineage, giving us 10 Nav channel genes.
Was the retention of these duplicate genes in tetrapods adaptive? We can approach this by comparing the fates of Nav channel genes with other genes in tetrapods throughout 2R and beyond. In tetrapods, the genes surrounding the Nav channel genes that would have duplicated along with them in 2R show little or no evidence of further duplication and retention; indeed, some show a loss of one or more 2R duplicate (Fig. 4). This pattern of duplication and retention of Nav channel genes is statistically significantly different compared with that of the immediately surrounding genes (8). A similar analysis in teleosts shows that nearby genes, such as members of the TGF-β receptor superfamily, were also more likely to be lost than retained (41). Furthermore, an analysis of Cav, transient receptor potential, and various K+ channel subfamilies shows that there was no widespread duplication and retention of other ion channel genes in the tetrapod 6TM family since the teleost–tetrapod divergence (8). Thus, we infer that selection acted on the Nav channel duplicates independently in teleosts and tetrapods to preserve them. Future work detailing where Nav channels are expressed and how they behave in ray-finned fish, lungfish, and nonmammalian tetrapods will shed light on this question.
The addition of new Nav channels to the existing repertoire likely realized two benefits: enhanced computational ability and increased energetic efficiency. For example, Nav1.1 is expressed in fast-firing inhibitory cortical interneurons, and its properties allow these neurons to fire at sustained high rates (42). In pyramidal neurons Nav1.6 is found in the distal part of the AIS, whereas Nav1.2, which activates at voltages around 20 mV more positive than Nav1.6, is found more proximally. This will ensure that APs that are first generated in the most distal AIS propagate down the axon and these are followed by APs generated in the proximal AIS that back-propagate into the soma (43).
The extent to which Na+ channels inactivate before K+ currents commence influences energy consumption; optimally Nav channels should completely inactivate before the K+ channels open to minimize use of the ATP-dependent Na+/K+ pump (44–46). It is possible that variation in the properties of Nav channels allows more precision in matching their inactivation with Kv channels to save energy.
Adaptive Evolution of Nav Channels: Weakly Electric Fish
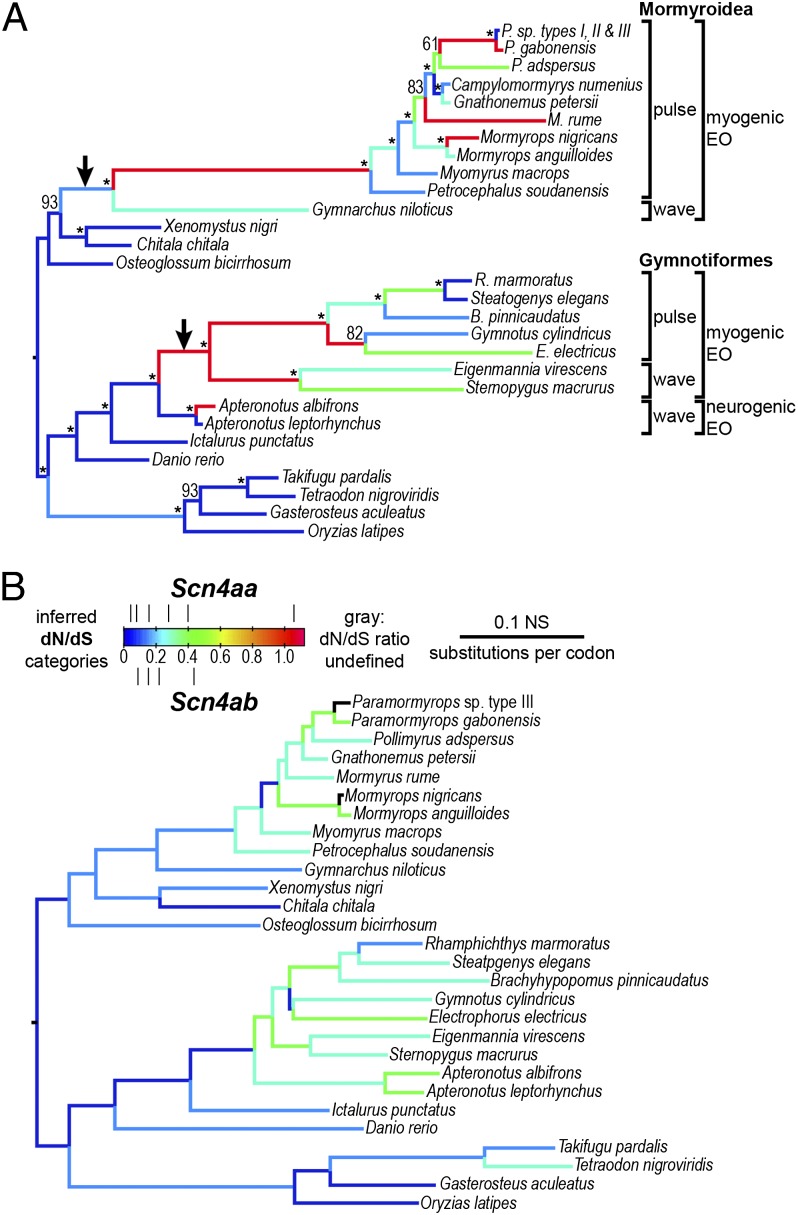
In most organisms ion channels cause behavior indirectly by triggering muscle movements. Weakly electric fish, however, emit electric signals directly into the water, and these are shaped by the biophysical properties of Nav and Kv channels in their electric organs. In nonteleost vertebrates the Nav channel Nav1.4 is expressed in muscle; because of the teleost-specific WGD, teleosts have two paralogs, Nav1.4a and Nav1.4b, in their muscles (5, 9) (Fig. 5). There must be strong selection for the retention of the expression of both paralogs in muscle because they are both expressed in muscles of most teleosts examined. In other words, the expression of both genes in fish muscle remains after 250 million years of teleost history. The only exceptions are two lineages of weakly electric fishes. These two groups—the South American gymnotiforms and African mormyriforms—evolved electric organs independently. In both lineages the gene for Nav1.4a (scn4aa) lost its expression in muscle and became compartmentalized in the electric organ. Nav1.4 in mammals is under strong purifying selection because mutations in the gene for this channel often cause muscle paralysis or myotonia. Freed from its constraints, Nav1.4a underwent a burst of evolutionary change at the origin of both groups of electric fishes, with numerous substitutions in key regions of the channel, many involved in inactivation (5, 9). The pace of evolutionary change quickened in similar regions of the channel in both groups; in some cases the same or neighboring amino acids changed in both groups. Although these substitutions have not yet been introduced into a channel and their effects tested, the implication is that these substitutions have facilitated the diversity of species-specific signals in these fish. An unanswered question is this: if nonelectric teleosts need two Nav channel paralogs, how do electric fish cope with only a single channel?
Fig. 5.
Nav1.4a is a fast-evolving Nav channel expressed in the electric organs of two independently derived lineages of weakly electric fish. Two paralogous genes, (A) scn4aa, which encodes Nav1.4a, and (B) scn4ab, which encodes Nav1.4ab, are expressed in the muscles of teleost fish. In the two lineages of weakly electric fishes, the mormyroidea and gymnotiformes, the gene for Nav1.4a (scn4aa) lost its expression in muscle and is only expressed in the electric organ. Nav1.4a underwent a burst of accelerated evolution at the origin of each lineage of electric fish. Nav1.4b, which is expressed in muscle and may also be expressed in the electric organ, evolved at a lower rate. The rate of nonsynonymous substitutions/nonsynonymous sites/rate of synonymous substitutions/synonymous site (dN/dS) in each gene is shown by a color scale in which cool colors represent low rates of sequence evolution and hot colors represent high rates. The arrows indicate where Nav1.4a gene expression was lost from muscle in both lineages. The production of either a highly regular wave type or an irregular pulse type of electric organ discharge is indicated in both groups. In both lineages of electric fishes, the electric organ develops from muscle (myogenic), except for one group (Apteronotidae) in which it is derived from the axons of motorneurons. From Arnegard et al. (2010) (5).
Muscles have diversified in other lineages of fishes. For example, rapidly contracting sound-producing muscles evolved independently in at least three lineages of fishes (47), and heater muscles that no longer contract but that engage in futile Ca2+ cycling to generate heat, in two lineages (48). It would be intriguing to know whether Nav channels show a similar pattern of compartmentalized expression and rapid evolutionary change in specialized muscles and muscle-derived organs of these lineages. Has the duplication of a muscle-expressing Nav channel gene facilitated the evolution of multiple novel muscle-derived structures in teleosts?
Adaptive Evolution of Nav Channels: Tetrodotoxin Resistance
The best-studied cases of adaptive evolution of Nav channel genes involve the evolution of resistance to the various neurotoxins that act on Nav channels. A number of animals use the neurotoxin tetrodotoxin (TTX), mainly for protection against predators (49) but in a few cases as a weapon to subdue prey (50). Animals associated with TTX span the animal kingdom. This is because TTX is likely produced by bacteria symbiotically associated with their hosts, or else, taken up from the food chain by animals that prey on TTX-accumulating organisms (51). In any event, unlike peptide toxins that are sequestered within a gland, TTX passes through cell membranes so that although it may be concentrated in certain tissues, all tissues are more or less exposed to it (52). Thus, those animals that sequester high concentrations of TTX have evolved mechanisms to protect themselves from its effects (53, 54). Because invertebrates possess only a single Nav channel gene, TTX resistance could occur easily enough with a single amino acid substitution. However, TTX resistance in vertebrates is more complex because vertebrates have multiple Nav channel genes. Evolution of TTX resistance in vertebrates offers an interesting case of parallel molecular evolution.
Pufferfishes, the most famous being the culinary delicacy Fugu of Japan, sequester TTX. This is a general trait of tetraodontiform fishes of which there are more than 120 species. Sequencing of Nav channel genes from Fugu and other pufferfishes shows that many of the same TTX-resistant amino acid substitutions have occurred multiple times in various Nav channels and lineages of pufferfishes (55–57). We still do not know how pufferfish were able to survive with only one or a few TTX-resistant Nav channels. The most likely scenario is that TTX-resistant mutations accumulated gradually in the Nav channel genes as fish were initially exposed to a light load of TTX. Gradually, as more channels gained resistance, they were able to carry a greater toxic load. This is suggested by the fact that certain substitutions were present in ancestral tetraodontids, with other substitutions appearing in different lineages of pufferfish and in different Nav channels (55).
Some of the most remarkable work in this field concerns the rich and extensively studied garter snake–newt system. Newts such as the California newt (Taricha torosa) sequester high levels of TTX for protection against predators. However, in some regions in the Pacific Northwest and northern California, the common garter snake (Thamnopis sirtalis) overlaps with some populations of the newt. Garter snakes that do not overlap with the newts are severely affected by ingesting newts and will vomit up the newt if they are lucky and die if they are not. However, populations of garter snakes sympatric with the newts are resistant to TTX and handily take newts. Variation in the extent of TTX resistance in different garter snakes populations suggests that each population has evolved resistance independently. Even more striking, TTX resistance has evolved multiple times in populations of other species of garter snakes that are also sympatric with Taricha in the Pacific Northwest and California, as well as other snake species sympatric with other newts or frogs that use TTX in South America and Asia (58, 59). Finally, sequencing and testing of expressed Nav channels (Nav1.4, a muscle-expressing Nav channel encoded by the scn4a gene) have highlighted that these channels show amino acid substitutions in the pore where TTX binds (10, 59). Not surprisingly, the Nav channels of the newts also have evolved TTX resistance to keep the newts from poisoning themselves (60).
However, this story is richer still. Newts lay their eggs in streams and ponds, and these eggs hatch into gill-bearing larvae. The larvae do not produce much TTX. Adults, however, do. The adults are carnivorous and may be cannibalistic. Larval newts that are “downwind” of adults will flee if they smell TTX wafting toward them in the water (61). Thus, TTX is used as a chemical signal [it is similarly used as an attractive pheromone in pufferfish, in which males can detect nanomolar levels of TTX that diffuse into the water from the TTX that females place in their eggs (62)]. It is not known yet what receptor detects the TTX in either newts or pufferfish. One possibility is that it is a Nav channel that has evolved to open, rather than close, upon TTX binding.
Newt eggs are protected from most vertebrate predators because of their high titer of TTX. Nevertheless, caddis fly (Limnephilus flavastellus) larvae have evolved TTX resistance and will eat newt eggs (63). It is not yet known whether this is due to a substitution in the pore of the Nav channel. Given that invertebrates have only a single Nav channel gene, this seems likely, and it will be interesting to see whether other invertebrate egg-predators are resistant to TTX.
Adaptive Evolution of Nav Channels: Proton Insensitivity
Naked mole rats (Heterocephalus glaber) live at high density in subterranean tunnels and seldom emerge into the light. They have evolved a number of adaptations for this life history, among them insensitivity to acid (64). The levels of CO2 that build up in their tunnels make carbonic acid; humans exposed to these levels of CO2 report stinging pain. However, naked mole rats show no pain-related behaviors and their C-fiber nociceptors are not activated by acid. Molecular and physiological examination of the naked mole rat’s acid-sensing (ASIC) and transient receptor V1 (TRPV1) channels, the channels in vertebrates that subserve acid sensitivity, showed no unusual behavior in these animals. Insofar as protons are also small monovalently positively charged molecules, these interact with and block Na+ channels. The Nav channel Nav1.7 sets the threshold for firing of C-fiber nociceptors. Naked mole rat Nav1.7, indeed, is extremely sensitive to proton block, ensuring that, at low pH, Nav1.7 will be blocked and the C-fiber nociceptors are not activated (65).
Adaptive Evolution of Nav Channels in Real Time: Insecticide Resistance
One unintended consequence of the liberal and worldwide use of dichlorodiphenyltrichloroethane, pyrethrin, and pyrethroid insecticides has been the rapid, massively parallel evolution of resistance to these pesticides in insects (66–69). Starting with their use in the 1940s, the first indications of resistance, so-called knockdown resistance because insects were no longer knocked down by normal concentrations of the insecticide, were evident in the early 1950s. These insecticides target the Nav1 channels of insects. They cross the cell membrane and lodge in a hydrophobic pocket in the inner mouth of the channel, where they are believed to prevent the inactivation gate (domain III–IV linker) from occluding the inner mouth of the channel. This allows Na+ ions to continue flowing into the cell, causing hyperexcitabiity. Amino acid substitutions have been discovered in a variety of insects at a number of sites in the inner mouth of the insect Nav channel (para in Drosophila) that either reduce pesticide binding or alter the channel properties to counteract the effects of insecticides. An example of the latter is a substitution that causes the channel to open at more positive potentials and to enhance the rate at which Nav channels enter closed state-inactivation. This minimizes the number of open channels counteracting the prolonged channel opening caused by insecticides.
The rapid evolution of Nav channels in insects exposed to insecticides is one of many warnings we have about the robust abilities of insect pests to overcome our best attempts to wipe them out.
Conclusions and Future Directions
Like many key components of the nervous system, Nav channels existed before neurons. It is likely that the Nav channels of choanoflagellates and early metazoans were permeable to both Na+ and Ca2+ and evolved enhanced selectivity to Na+ in parallel in early bilaterians and jellyfish. Although it is convenient to think that invertebrates possess only a single Nav1 channel gene, it is worth scouring the wealth of new genomes to determine whether there are any lineage-specific duplications, and if so, what this might mean. Further, we have little information on the Nav2 channels of invertebrates.
The parallel expansion of Nav channel genes in tetrapods and teleosts occurred along with an increase in the number of telencephalic nuclei in both groups. This was coincident with or just after the great Devonian extinction, during which teleosts began their domination of the aquatic and tetrapods of the terrestrial habitats. More types of Nav channels may allow for more sophisticated computational possibilities and energy savings. It will be intriguing to study the locations and types of Nav channels in lungfish, basal ray-fin fishes (e.g., bichirs, gars), a variety of tetrapods, and teleosts to know whether there is parallel evolution of different channel “types” in teleosts and tetrapods. For example, fast-firing inhibitory neurons in mammals express different Nav channels than more slowly firing pyramidal neurons. Do we see a similar functional partitioning of Nav channel types in teleosts? Are those groups with only four Nav channel genes (elasmobranchs, basal actinpoterygian fishes, basal sarcopterygian fishes) hampered in the complexity of their neural processing?
Finally, on a microevolutionary level, we see that Nav channels can be targets of adaptive changes for increasing diversity in signaling (electric fish), in the arms race against lethal naturally occurring or synthetic toxins (snakes, newts, pufferfish, insects), and in specialized habitats (naked mole rats). There are likely to be more examples of this, especially in animals with unique life histories, and we should keep an eye out for potentially interesting subjects.
Acknowledgments
I thank Francisco Ayala, John Avise, and Georg Striedter for organizing a stimulating Arthur M. Sackler Colloquium of the National Academy of Sciences and for their invitation to participate. Thank you also to Francisco Ayala for facilitating the dinner conversation with excellent wines from his vineyards. Much of the work from my laboratory discussed in this article was funded by National Institutes of Health Grant R01 NS025513.
Footnotes
The author declares no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “In the Light of Evolution VI: Brain and Behavior,” held January 19–21, 2012, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at www.nasonline.org/evolution_vi.
This article is a PNAS Direct Submission.
References
- 1.Love GD, et al. Fossil steroids record the appearance of Demospongiae during the Cryogenian period. Nature. 2009;457:718–721. doi: 10.1038/nature07673. [DOI] [PubMed] [Google Scholar]
- 2.Rokas A, Krüger D, Carroll SB. Animal evolution and the molecular signature of radiations compressed in time. Science. 2005;310:1933–1938. doi: 10.1126/science.1116759. [DOI] [PubMed] [Google Scholar]
- 3.Moroz LL. On the independent origins of complex brains and neurons. Brain Behav Evol. 2009;74:177–190. doi: 10.1159/000258665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schierwater B, et al. Concatenated analysis sheds light on early metazoan evolution and fuels a modern “urmetazoon” hypothesis. PLoS Biol. 2009;7:e20. doi: 10.1371/journal.pbio.1000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnegard ME, Zwickl DJ, Lu Y, Zakon HH. Old gene duplication facilitates origin and diversification of an innovative communication system—twice. Proc Natl Acad Sci USA. 2010;107:22172–22177. doi: 10.1073/pnas.1011803107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Z, et al. Parallel evolution of KCNQ4 in echolocating bats. PLoS ONE. 2011;6:e26618. doi: 10.1371/journal.pone.0026618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopreato GF, et al. Evolution and divergence of sodium channel genes in vertebrates. Proc Natl Acad Sci USA. 2001;98:7588–7592. doi: 10.1073/pnas.131171798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zakon HH, Jost MC, Lu Y. Expansion of voltage-dependent Na+ channel gene family in early tetrapods coincided with the emergence of terrestriality and increased brain complexity. Mol Biol Evol. 2011;28:1415–1424. doi: 10.1093/molbev/msq325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zakon HH, Lu Y, Zwickl DJ, Hillis DM. Sodium channel genes and the evolution of diversity in communication signals of electric fishes: Convergent molecular evolution. Proc Natl Acad Sci USA. 2006;103:3675–3680. doi: 10.1073/pnas.0600160103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geffeney SL, Fujimoto E, Brodie ED, 3rd, Brodie ED, Jr, Ruben PC. Evolutionary diversification of TTX-resistant sodium channels in a predator-prey interaction. Nature. 2005;434:759–763. doi: 10.1038/nature03444. [DOI] [PubMed] [Google Scholar]
- 11.Anderson PAV, Greenberg RM. Phylogeny of ion channels: Clues to structure and function. Comp Biochem Physiol B Biochem Mol Biol. 2001;129:17–28. doi: 10.1016/s1096-4959(01)00376-1. Available at http://www.journals.elsevier.com/comparative-biochemistry-and-physiology-part-b-biochemistry-and-molecular-biology/ [DOI] [PubMed] [Google Scholar]
- 12.Strong M, Chandy KG, Gutman GA. Molecular evolution of voltage-sensitive ion channel genes: on the origins of electrical excitability. Mol Biol Evol. 1993;10:221–242. doi: 10.1093/oxfordjournals.molbev.a039986. [DOI] [PubMed] [Google Scholar]
- 13.Galione A, et al. The acid test: The discovery of two-pore channels (TPCs) as NAADP-gated endolysosomal Ca(2+) release channels. Pflugers Arch. 2009;458:869–876. doi: 10.1007/s00424-009-0682-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai X. Unicellular Ca2+ signaling ‘toolkit’ at the origin of metazoa. Mol Biol Evol. 2008;25:1357–1361. doi: 10.1093/molbev/msn077. [DOI] [PubMed] [Google Scholar]
- 15.Koishi R, et al. A superfamily of voltage-gated sodium channels in bacteria. J Biol Chem. 2004;279:9532–9538. doi: 10.1074/jbc.M313100200. [DOI] [PubMed] [Google Scholar]
- 16.Liebeskind BJ, Hillis DM, Zakon HH. Evolution of sodium channels predates the origin of nervous systems in animals. Proc Natl Acad Sci USA. 2011;108:9154–9159. doi: 10.1073/pnas.1106363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hille B. Ion Channels of Excitable Membranes. Sunderland, MA: Sinauer Press; 2001. [Google Scholar]
- 18.Zhou W, Chung I, Liu Z, Goldin AL, Dong K. A voltage-gated calcium-selective channel encoded by a sodium channel-like gene. Neuron. 2004;42:101–112. doi: 10.1016/s0896-6273(04)00148-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lipkind GM, Fozzard HA. Voltage-gated Na channel selectivity: The role of the conserved domain III lysine residue. J Gen Physiol. 2008;131:523–529. doi: 10.1085/jgp.200809991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlief T, Schönherr R, Imoto K, Heinemann SH. Pore properties of rat brain II sodium channels mutated in the selectivity filter domain. Eur Biophys J. 1996;25:75–91. doi: 10.1007/s002490050020. [DOI] [PubMed] [Google Scholar]
- 21.Liebeskind BJ. Evolution of sodium channels and the new view of early nervous system evolution. Commun Integr Biol. 2011;4:679–683. doi: 10.4161/cib.17069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.West JW, et al. A cluster of hydrophobic amino acid residues required for fast Na(+)-channel inactivation. Proc Natl Acad Sci USA. 1992;89:10910–10914. doi: 10.1073/pnas.89.22.10910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chanda B, Asamoah OK, Bezanilla F. Coupling interactions between voltage sensors of the sodium channel as revealed by site-specific measurements. J Gen Physiol. 2004;123:217–230. doi: 10.1085/jgp.200308971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chanda B, Bezanilla F. Tracking voltage-dependent conformational changes in skeletal muscle sodium channel during activation. J Gen Physiol. 2002;120:629–645. doi: 10.1085/jgp.20028679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartline DK, Colman DR. Rapid conduction and the evolution of giant axons and myelinated fibers. Curr Biol. 2007;17:R29–R35. doi: 10.1016/j.cub.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 26.Wilson CH, Hartline DK. Novel organization and development of copepod myelin. ii. nonglial origin. J Comp Neurol. 2011;519:3281–3305. doi: 10.1002/cne.22699. [DOI] [PubMed] [Google Scholar]
- 27.Zalc B, Goujet D, Colman D. The origin of the myelination program in vertebrates. Curr Biol. 2008;18:R511–R512. doi: 10.1016/j.cub.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 28.Hill AS, et al. Ion channel clustering at the axon initial segment and node of Ranvier evolved sequentially in early chordates. PLoS Genet. 2008;4:e1000317. doi: 10.1371/journal.pgen.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kole MHP, et al. Action potential generation requires a high sodium channel density in the axon initial segment. Nat Neurosci. 2008;11:178–186. doi: 10.1038/nn2040. [DOI] [PubMed] [Google Scholar]
- 30.Boiko T, et al. Functional specialization of the axon initial segment by isoform-specific sodium channel targeting. J Neurosci. 2003;23:2306–2313. doi: 10.1523/JNEUROSCI.23-06-02306.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grubb MS, Burrone J. Activity-dependent relocation of the axon initial segment fine-tunes neuronal excitability. Nature. 2010;465:1070–1074. doi: 10.1038/nature09160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Richardson WD. The evolution of Olig genes and their roles in myelination. Neuron Glia Biol. 2008;4:129–135. doi: 10.1017/S1740925X09990251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schweigreiter R, Roots BI, Bandtlow CE, Gould RM. Understanding myelination through its evolution. Int Rev Neurobiol. 2006;73:219–273. doi: 10.1016/S0074-7742(06)73007-0. [DOI] [PubMed] [Google Scholar]
- 34.Kulkarni NH, Yamamoto AH, Robinson KO, Mackay TFC, Anholt RRH. The DSC1 channel, encoded by the smi60E locus, contributes to odor-guided behavior in Drosophila melanogaster. Genetics. 2002;161:1507–1516. doi: 10.1093/genetics/161.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stern M, Kreber R, Ganetzky B. Dosage effects of a Drosophila sodium channel gene on behavior and axonal excitability. Genetics. 1990;124:133–143. doi: 10.1093/genetics/124.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bargmann CI. Neurobiology of the Caenorhabditis elegans genome. Science. 1998;282:2028–2033. doi: 10.1126/science.282.5396.2028. [DOI] [PubMed] [Google Scholar]
- 37.Ohno S. Evolution by Gene Duplication. Berlin: Springer; 1970. [Google Scholar]
- 38.Jaillon O, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- 39.Meyer A, Schartl M. Gene and genome duplications in vertebrates: The one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr Opin Cell Biol. 1999;11:699–704. doi: 10.1016/s0955-0674(99)00039-3. [DOI] [PubMed] [Google Scholar]
- 40.Novak AE, et al. Gene duplications and evolution of vertebrate voltage-gated sodium channels. J Mol Evol. 2006;63:208–221. doi: 10.1007/s00239-005-0287-9. [DOI] [PubMed] [Google Scholar]
- 41.Widmark J, Sundström G, Ocampo Daza D, Larhammar D. Differential evolution of voltage-gated sodium channels in tetrapods and teleost fishes. Mol Biol Evol. 2011;28:859–871. doi: 10.1093/molbev/msq257. [DOI] [PubMed] [Google Scholar]
- 42.Ogiwara I, et al. Nav1.1 localizes to axons of parvalbumin-positive inhibitory interneurons: A circuit basis for epileptic seizures in mice carrying an Scn1a gene mutation. J Neurosci. 2007;27:5903–5914. doi: 10.1523/JNEUROSCI.5270-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu W, et al. Distinct contributions of Na(v)1.6 and Na(v)1.2 in action potential initiation and backpropagation. Nat Neurosci. 2009;12:996–1002. doi: 10.1038/nn.2359. [DOI] [PubMed] [Google Scholar]
- 44.Hasenstaub A, Otte S, Callaway E, Sejnowski TJ. Metabolic cost as a unifying principle governing neuronal biophysics. Proc Natl Acad Sci USA. 2010;107:12329–12334. doi: 10.1073/pnas.0914886107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt-Hieber C, Bischofberger J. Fast sodium channel gating supports localized and efficient axonal action potential initiation. J Neurosci. 2010;30:10233–10242. doi: 10.1523/JNEUROSCI.6335-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sengupta B, Stemmler M, Laughlin SB, Niven JE. Action potential energy efficiency varies among neuron types in vertebrates and invertebrates. PLOS Comput Biol. 2010;6:e1000840. doi: 10.1371/journal.pcbi.1000840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bass A, Ladich F. Vocal-acoustic communication: From behavior to neurons. In: Popper RF, Webb J, editors. Fish Bioacoustics—Springer Handbook of Auditory Research. New York: Springer Press; 2008. pp. 253–278. [Google Scholar]
- 48.Block BA, Finnerty JR, Stewart AF, Kidd J. Evolution of endothermy in fish: Mapping physiological traits on a molecular phylogeny. Science. 1993;260:210–214. doi: 10.1126/science.8469974. [DOI] [PubMed] [Google Scholar]
- 49.Gladstone W. The eggs and larvae of the sharpnose pufferfish Canthigaster valentini (Pisces: Tetraodontidae) are unpalatable to other reef fishes. Copeia. 1987;(1):227–230. [Google Scholar]
- 50.Ritson-Williams R, Yotsu-Yamashita M, Paul VJ. Ecological functions of tetrodotoxin in a deadly polyclad flatworm. Proc Natl Acad Sci USA. 2006;103:3176–3179. doi: 10.1073/pnas.0506093103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee M-J, et al. A tetrodotoxin-producing Vibrio strain, LM-1, from the puffer fish Fugu vermicularis radiatus. Appl Environ Microbiol. 2000;66:1698–1701. doi: 10.1128/aem.66.4.1698-1701.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams BL, Caldwell RL. Intra-organismal distribution of tetrodotoxin in two species of blue-ringed octopuses (Hapalochlaena fasciata and H. lunulata) Toxicon. 2009;54:345–353. doi: 10.1016/j.toxicon.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 53.Flachsenberger W, Kerr DIB. Lack of effect of tetrodotoxin and of an extract from the posterior salivary gland of the blue-ringed octopus following injection into the octopus and following application to its brachial nerve. Toxicon. 1985;23:997–999. doi: 10.1016/0041-0101(85)90393-9. [DOI] [PubMed] [Google Scholar]
- 54.Kidokoro Y, Grinnell A, Eaton D. Tetrodotoxin sensitivity of muscle action potentials in pufferfishes and related fishes. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1974;89:59–72. [Google Scholar]
- 55.Jost MC, et al. Toxin-resistant sodium channels: Parallel adaptive evolution across a complete gene family. Mol Biol Evol. 2008;25:1016–1024. doi: 10.1093/molbev/msn025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Venkatesh B, et al. Genetic basis of tetrodotoxin resistance in pufferfishes. Curr Biol. 2005;15:2069–2072. doi: 10.1016/j.cub.2005.10.068. [DOI] [PubMed] [Google Scholar]
- 57.Yotsu-Yamashita M, et al. Binding properties of (3)H-PbTx-3 and (3)H-saxitoxin to brain membranes and to skeletal muscle membranes of puffer fish Fugu pardalis and the primary structure of a voltage-gated Na(+) channel alpha-subunit (fMNa1) from skeletal muscle of F. pardalis. Biochem Biophys Res Commun. 2000;267:403–412. doi: 10.1006/bbrc.1999.1974. [DOI] [PubMed] [Google Scholar]
- 58.Feldman CR, Brodie ED, Jr, Brodie ED, 3rd, Pfrender ME. The evolutionary origins of beneficial alleles during the repeated adaptation of garter snakes to deadly prey. Proc Natl Acad Sci USA. 2009;106:13415–13420. doi: 10.1073/pnas.0901224106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feldman CR, Brodie ED, Jr, Brodie ED, 3rd, Pfrender ME. Constraint shapes convergence in tetrodotoxin-resistant sodium channels of snakes. Proc Natl Acad Sci USA. 2012;109:4556–4561. doi: 10.1073/pnas.1113468109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaneko Y, Matsumoto G, Hanyu Y. TTX resistivity of Na+ channel in newt retinal neuron. Biochem Biophys Res Commun. 1997;240:651–656. doi: 10.1006/bbrc.1997.7696. [DOI] [PubMed] [Google Scholar]
- 61.Zimmer RK, Ferrer RP. Neuroecology, chemical defense, and the keystone species concept. Biol Bull. 2007;213:208–225. doi: 10.2307/25066641. [DOI] [PubMed] [Google Scholar]
- 62.Matsumura K. Tetrodotoxin as a pheromone. Nature. 1995;378:563–564. doi: 10.1038/378563b0. [DOI] [PubMed] [Google Scholar]
- 63.Gall BG, Brodie ED. Survival and growth of the caddisfly Limnephilus flavastellus after predation on toxic eggs of the Rough-skinned Newt (Taricha granulosa) Can J Zool. 2011;89:483–489. [Google Scholar]
- 64.Park TJ, et al. Selective inflammatory pain insensitivity in the African naked mole-rat (Heterocephalus glaber) PLoS Biol. 2008;6:e13. doi: 10.1371/journal.pbio.0060013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith ESJ, et al. The molecular basis of acid insensitivity in the African naked mole-rat. Science. 2011;334:1557–1560. doi: 10.1126/science.1213760. [DOI] [PubMed] [Google Scholar]
- 66.Davies TG, Field LM, Usherwood PN, Williamson MS. DDT, pyrethrins, pyrethroids and insect sodium channels. IUBMB Life. 2007;59:151–162. doi: 10.1080/15216540701352042. [DOI] [PubMed] [Google Scholar]
- 67.Jones CM, et al. Footprints of positive selection associated with a mutation (N1575Y) in the voltage-gated sodium channel of Anopheles gambiae. Proc Natl Acad Sci USA. 2012;109:6614–6619. doi: 10.1073/pnas.1201475109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu Z, Valles SM, Dong K. Novel point mutations in the German cockroach para sodium channel gene are associated with knockdown resistance (kdr) to pyrethroid insecticides. Insect Biochem Mol Biol. 2000;30:991–997. doi: 10.1016/s0965-1748(00)00074-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Taylor MF, Heckel DG, Brown TM, Kreitman ME, Black B. Linkage of pyrethroid insecticide resistance to a sodium channel locus in the tobacco budworm. Insect Biochem Mol Biol. 1993;23:763–775. doi: 10.1016/0965-1748(93)90064-y. [DOI] [PubMed] [Google Scholar]