Abstract
The regulation of iron homeostasis is essential for most organisms, because iron is required for a variety of conserved biochemical processes, yet can be toxic at high concentrations. Upon experiencing iron starvation in vitro, the obligate intracellular human pathogen Chlamydia trachomatis exhibits elevated expression of a putative iron-transport system encoded by the ytg operon. The third component of the ytg operon, CT069 (YtgCR), encodes a protein with two distinct domains: a membrane-anchored metal ion permease and a diphtheria toxin repressor (DtxR)-like transcriptional repressor. In this report, we demonstrate that the C-terminal domain of CT069 (YtgR) serves as an iron-dependent autorepressor of the ytg operon. Moreover, the nascent full-length metal permease-transcriptional repressor protein was processed during the course of infection, and heterologously when expressed in Escherichia coli. The products produced by heterologous cleavage in E. coli were functional in the repression of a reporter gene downstream of a putative YtgR operator. We report a bona fide mechanism of iron-dependent regulation of transcription in Chlamydia. Moreover, the unusual membrane permease-DNA-binding polypeptide fusion configuration was found in several bacteria. Therefore, the DNA-binding capability and liberation of the YtgR domain from a membrane-anchored permease in C. trachomatis could represent a previously uncharacterized mechanism for prokaryotic regulation of iron-homeostasis.
Keywords: fusion-protein, ABC-3 permease, metalloregulation, autorepression
Iron is an essential micronutrient for nearly all organisms, with its redox capability being both beneficial and harmful to the organism, because of iron readily catalyzing the formation of toxic free radicals via the Haber–Weiss reaction (1). Thus, both eukaryotes and prokaryotes, alike, have evolved regulatory networks for the tight control of intracellular iron homeostasis.
In prokaryotes, these regulatory networks are usually controlled through metal-dependent DNA-binding proteins. Two of the best-characterized metal-dependent transcriptional regulators are ferric uptake regulator (Fur) in Escherichia coli and diphtheria toxin repressor (DtxR) in Corynebacterium diphtheriae. Although nonhomologous and structurally distinct, both families of transcriptional repressors function similarly, in that the presence of a cognate metal cofactor activates DNA-binding activity (2). Homodimers of the metal-activated proteins recognize specific cis-regulatory elements that are often located proximally to the promoter operating regions of the regulated gene, effectively blocking RNA polymerase initiation of transcription (3, 4).
Chlamydia trachomatis is an obligate, intracellular human pathogen that is responsible for the leading cause of bacterial sexually transmitted infection (5, 6) and infection-derived blindness (7) worldwide. The chlamydial requirement for iron is well established, because iron restriction forces Chlamydia into an alternative persistent growth mode distinguished by abnormal morphology (8–10) and altered transcriptional profile of hallmark genes (11, 12). However, how Chlamydia acquires iron is not understood, because it does not possess genes that encode siderophore biosynthetic enzymes or receptors for host iron-binding proteins (13). Therefore, significant attention has been focused recently on a putative divalent metal ATP-binding-cassette (ABC) permease system encoded by the ytgABCD operon (13). The operon encodes for a periplasmic metal substrate-binding protein (YtgA), a cytosolic ATPase (YtgB), and a membrane channel formed by two proteins (YtgC and YtgD) in complex (14). More recent evidence points to the Ytg system functioning in iron transport. First, the periplasmic substrate-binding protein, YtgA, exhibited a significant preference toward iron over zinc or manganese in vitro (15). Also, the transcript expression of ytgA was induced in response to iron starvation of C. trachomatis (16), indicating an iron-dependent regulation of the ytg operon.
This report focuses on the ct069 (originally annotated as ytgC), which is predicted to encode an unusual protein consisting of distinct domains with seemingly incongruent functions: the N terminus putatively encoding an integral membrane protein that forms the system permease complex with YtgD and the C terminus putatively encoding a DtxR-like DNA-binding transcriptional repressor, termed YtgR (17). Despite the apparent membrane-anchored localization of the nascent full-length CT069 protein, we hypothesized that the putative metal-dependent transcriptional repressor domain may have some function in the iron-dependent transcriptional regulation of ytgA, as reported previously (16). Recently, a recombinant and purified YtgR repressor was reported to bind DNA specifically in a Zn2+-dependent manner, occupying an operator sequence near the promoter of the ytgABCD operon in vitro (17). However, this reported Zn2+-dependent function is not consistent with reports of preferential iron binding of YtgA and the iron responsiveness of the ytg operon. Moreover, how YtgR sequestered at the inner membrane by the permease domain would function as the potential transcriptional repressor was not addressed.
Multiple bacteria have evolved regulatory mechanisms to liberate membrane-anchored transcription factors into the cytosol (e.g., refs. 18–20). Here, we report on a possible two-level activation of YtgR: liberation by proteolytic cleavage from membrane sequestration and iron-dependent activation of its DNA-binding and transcriptional repressor function.
Results
C-Terminal Domain of CT069 Is Analogous to the DtxR Family of Metal-Dependent Transcriptional Repressors.
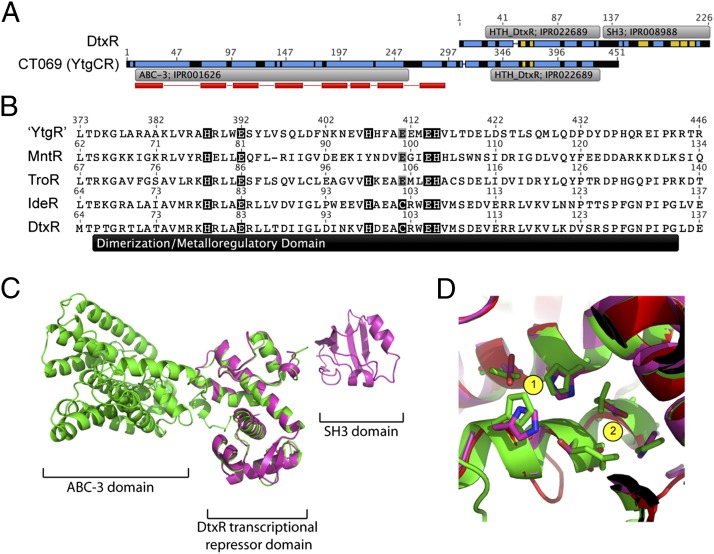
In addition to an N-terminal transmembrane permease domain [ABC-3; European Bioinformatics Institute (EBI) InterPro accession no. IPR001626], YtgC was also predicted to contain a C-terminal domain analogous to the DtxR family of metal-dependent DNA-binding repressors (HTH_DtxR; EBI InterPro accession no. IPR022689) (Fig. 1A). Therefore, to distinguish among the two domains and the nascent full-length protein, the gene product of ct069 will, henceforth, be referred to as YtgCR, with YtgC representing the permease domain and YtgR representing the DtxR-like transcriptional repressor domain. Because expression of ytgA was elevated upon iron starvation (16), we hypothesized that YtgR domain of YtgCR may be involved in the iron-dependent autoregulation of the ytg operon. In line with this hypothesis, the transmembrane hidden Markov model (TMHMM) algorithm (21, 22) predicted the YtgR domain to localize in the cytosolic side of the bacterial inner membrane. YtgR exhibited 24.5% pairwise protein sequence identity to DtxR, excluding the C-terminal Src-homology 3 (SH3) domain found in some DtxR family members. The aligned region of YtgR exhibited a predicted α-α-α-β-β-α-α-α secondary structure, similar to the DNA-binding and metalloregulatory domains of DtxR. In the DtxR holoenzyme, there are nine critical residues involved in metal cofactor stabilization, with His79, Glu83, His98, Glu170, and Gln173 coordinating site 1 and Met10, Cys102, Glu105, and His106 coordinating site 2 (23). Six of these residues are located within the dimerization/metalloregulatory domain. Fig. 1B depicts the dimerization domains of YtgR (C. trachomatis), MntR (Bacillus subtilis), TroR (Treponema pallidum), IdeR (Mycobacterium tuberculosis), and DtxR (C. diphtheriae), which were extracted from a global alignment with free end gaps. Compared with the DtxR dimerization domain, “YtgR” exhibited conservation at five of six sites involved in metal coordination, with a Cys→Glu change at site 411 that maintains stable coordinated binding of iron (24).
Fig. 1.
Bioinformatics analysis indicates that the ct069 (ytgCR) locus encodes a permease-repressor fusion protein. (A) Amino acid sequence alignment shows that CT069 (YtgCR) exhibits homology to DtxR in the C terminus. Recognized InterPro domains (EBI InterPro database) are annotated. Predicted membrane spanning regions are indicated by red bars below YtgCR sequence (TMHMM); the most probable arrangements of topology result in the C terminus of YtgCR localizing to the cytoplasmic side of the membrane. Secondary structures (predicted by Phyre or previously resolved) are indicated by blue (α-helix) and yellow (β-sheet). (B) Sequence alignment of the dimerization/metalloregulatory domain for the “YtgR” domain within Ct069 [C. trachomatis, MntR (B. subtilis), TroR (T. pallidum), IdeR (M. tuberculosis] and DtxR (C. diphtheriae). Residues involved in metal coordination in the DtxR holoenzyme (23) that are conserved are highlighted in black. Putative conservative substitutions are highlighted in gray. (C) Overlay of Phyre2 server modeled YtgCR (green) and DtxR (magenta) (PDB - 1g3w). (D) The divalent metal-binding sites of DtxR (magenta) are conserved in the YtgR domain (green). Site 1 residues are identical in both proteins, whereas Cys102 is replaced by Glu in the YtgR domain.
Moreover, the structure of the YtgCR was modeled using the Phyre2 Server (25), yielding a structure for the DNA-binding and dimerization domain that is remarkably similar to that of DtxR (Fig. 1C). Analysis of the divalent metal binding sites showed only slight structural repositioning in the residues involved in metal coordination (Fig. 1D). Thus, multiple lines of bioinformatics analysis provided support for the hypothesis that the YtgR domain, located at the C terminus of YtgCR, could be a functional divalent metal-dependent DNA-binding protein.
Unique Permease-Repressor Configuration Is Present in a Number of Bacteria.
To determine the prevalence of this permease-repressor protein configuration in the bacterial kingdom, all bacterial proteins containing the ABC-3 (EBI InterPro accession no. IPR001626) domain in the EBI InterPro database were analyzed for the presence of a C-terminal transcriptional repressor domain. Of the 6,560 total permease proteins, 295 (4.5%) contained C-terminal extensions of 50 aa or more beyond the defined ABC-3 domain. Of these extended permeases, 94 contained a recognized DNA-binding domain in the extension, with 65 of the 94 possessing the DtxR iron-dependent repressor domain (EBI InterPro accession no. IPR001367), with the remaining 29 possessing a general winged helix-turn-helix transcriptional repressor domain (EBI InterPro accession no. IPR011991). (See Table 1 for a summary and Dataset S1 for the full list.) The bacterial species possessing “extended” permeases belonged to eleven phyla, with the majority belonging to Chlamydiae (35) and Firmicutes (24). Of the permease-fusion proteins exhibiting the DtxR iron-dependent repressor domain, a high level of conservation was observed in the six putative metal-coordinating residues located within the dimerization region, with three of the six sites exhibiting identity (Fig. S1). Thus, the permease-repressor configuration is not unique to Chlamydiae and could represent an uncharacterized prokaryotic mechanism for iron-dependent transcriptional regulation.
Table 1.
Prevalence of genes encoding for putative permease-repressor fusion proteins
| Classification | No. of proteins |
| ABC-3-like domain (EBI InterPro accession no. IPR001626) | 6,560 |
| C-terminal extension of >50 amino acid residues | 295 (4.5%) |
| C-terminal extension with putative DNA-binding motifs | 94 (1.4%) |
| DtxR-like extension (EBI InterPro accession no. IPR001367) | 65 (1%) |
| Winged helix-turn-helix extension (EBI InterPro accession no. IPR011991) | 29 (0.4%) |
Recombinant YtgR Binds to an Operator Sequence Upstream of ytgA in an Iron-Dependent Manner.
Sequence analyses and structural modeling hinted at the functionality of the putative repressor domains within these fusion proteins. To ascertain whether these proteins could represent a mechanism for regulation of metal/iron homeostasis, the chlamydial representative YtgR was chosen for further analysis. We engineered an E. coli expression vector for the production of a polyhistidine (pHis) recombinant version of the final 147 aa (305–451) of YtgCR, spanning the repressor domain, but excluding the last predicted transmembrane region of the YtgC permease domain. The artificial start site for the recombinant YtgR was located within a Phyre2-predicted unstructured linker region (25). Recombinant YtgR was purified (Fig. S2) and filtered through a Chelex-100 resin (Bio-Rad) in an attempt to remove any chelatable (e.g., cofactor) metal ions, before use in in vitro DNA-binding assays.
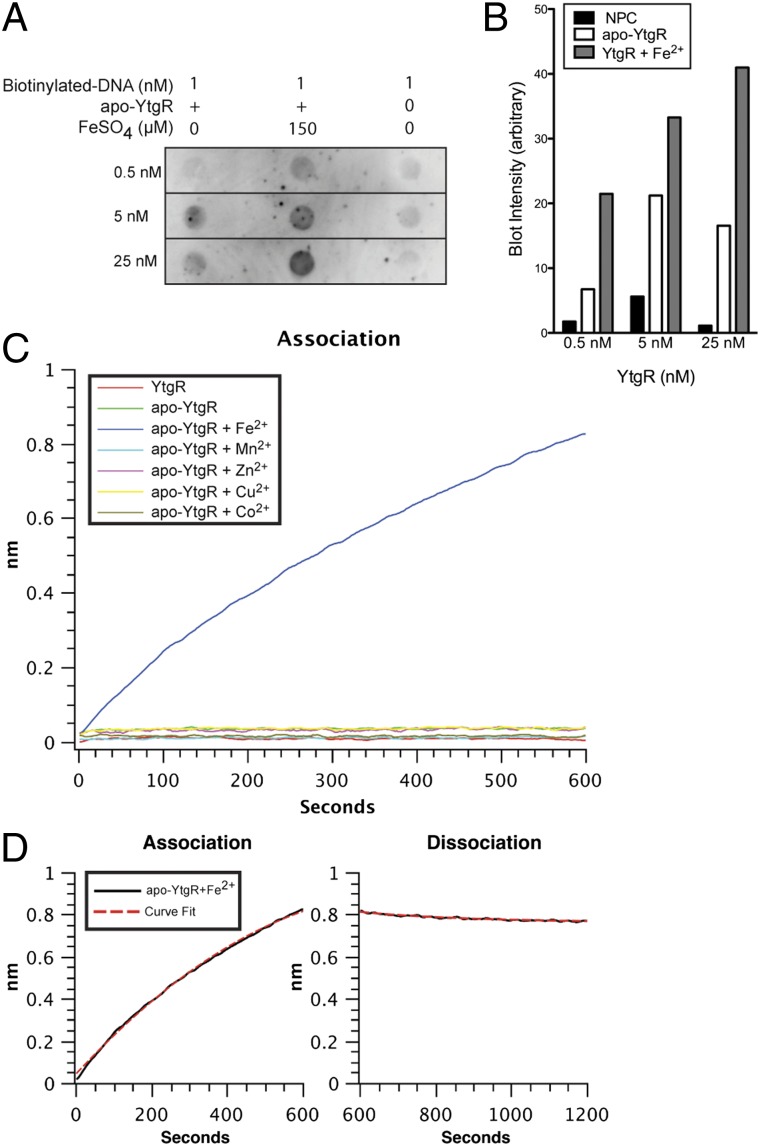
Initial biochemical characterization of the potential DNA-binding activity of YtgR used a dot-blot assay, in which the iron-dependent retention of a biotinylated dsDNA oligonucleotide on nitrocellulose (NC) membrane indicated the interaction of YtgR with the DNA probe. The probe chosen corresponded to the region upstream of the ytg operon. It was chosen with the assumption that the iron-responsive operon was autoregulated by YtgR, similar to the autoregulation of the T. pallidum tro operon by TroR (26). Fig. 2A shows the results of one representative experiment of three, with the corresponding quantification of blot intensities (Fig. 2B). Minimal interaction was observed between the apo-YtgR and the oligonucleotide compared with the no-protein control (NPC). However, upon supplementation with 150 μM ferrous iron before the addition of DNA, an increased retention of the oligonucleotide was observed for each concentration of YtgR tested. This suggested that iron-bound YtgR recognized a sequence within the region upstream of the ytgA coding sequence, and that the addition of ferrous iron was sufficient to activate the DNA-binding function of YtgR. Another divalent metal, Mn2+, failed to activate YtgR DNA-binding activity (Fig. S3), highlighting the specific requirement for iron of YtgR.
Fig. 2.
Recombinant version of the YtgR domain is a functional, iron-dependent DNA-binding polypeptide in vitro. (A) A labeled DNA molecule corresponding to the region upstream of ytgA was added to a solution containing various concentrations of apo-YtgR, in the presence or absence of iron. Blot represents one representative experiment of three. (B) Blot intensities were measured using ImageJ software, and the background level of an unused dot was subtracted from the measured values. Detection intensity increased within each apo-YtgR concentration upon supplementation of iron. (C) Real-time accumulation of biolayer was measured at the surface of an optical sensor that had been preloaded with a DNA oligonucleotide (corresponding to the region upstream of the ytgA coding sequence) and incubated in a solution containing purified YtgR or apo-YtgR in the presence of various divalent metals. Only the apo-YtgR plus Fe2+ reaction exhibited appreciable protein to DNA interaction. After allowing samples to associate with the sensor for 10 min, sensors were incubated in YtgR-free buffer and the dissociation was measured. (D) Rates of association (Kon) and dissociation (Koff) were determined by the “association-then-dissociation” function of the GraphPad Prism software platform for the apo-YtgR + Fe2+ binding reaction. A mean average KD of 3.45 × 10−8 M was measured over three independent experiments.
A more comprehensive analysis of the metal requirement of YtgR was conducted. To obtain quantitative DNA-binding kinetics for YtgR, we used a Bio-Layer Interferometry (BLI) assay using the Octet Platform (ForteBio) for measurement of DNA–protein interaction in real-time. This system quantitatively measures the accumulation of biomaterial on an optical sensor that has been preloaded with a specific bio-molecule. In our assay, streptavidin-coated optical sensors were saturated with a biotinylated version of the dot-blot DNA probe described above. Loaded sensors were then incubated in solutions containing YtgR supplemented with various divalent metals. By measuring changes in optical absorbance at the surface of the sensor, a direct measurement of biolayer accumulation (i.e., protein to DNA interaction) was observed in real time. In this assay, the purified, but non-Chelex-treated YtgR and apo-YtgR samples poorly associated with the oligonucleotide (Fig. 2C). Of the different divalent metals added to parallel samples, only the addition of iron led to appreciable DNA binding. Mn2+, Zn2+, Cu2+, and Co2+ all failed to activate DNA binding in apo-YtgR at the concentration tested (150 μM). Therefore, BLI analysis confirmed and extended the findings obtained from the dot-blot experiments.
The BLI analysis also enabled the determination of the dissociation constant for Fe2+-activated YtgR binding to the oligonucleotide by tracking the association and dissociation kinetics (Figs. 2D). The KD of YtgR binding to an oligonucleotide corresponding to the region upstream of ytgA was calculated as 3.45 × 10−8 M over three independent experiments. Together, the biochemical analysis of YtgR demonstrated its ability to bind DNA in a specific and iron-dependent manner, thus supporting the notion that YtgR functions as a DNA-binding protein, possibly as a transcriptional repressor in vivo.
YtgCR Is Processed During the Course of Chlamydial Infection.
Although in vitro binding experiments suggested that the YtgR domain within YtgCR was a functional iron-activated DNA-binding polypeptide, the ability of this polypeptide to affect chlamydial transcription efficiently while fused to the permease domain would be severely limited. Because multiple prokaryotes liberate membrane-sequestered transcription factors by proteolytic cleavage (e.g., refs. 18–20), we examined the possibility that YtgCR could undergo a processing event during infection to release YtgR from the YtgC permease domain. Cleavage during infection of cultured cells was monitored by Western blot with antisera raised against the full-length YtgCR protein (Fig. 3A). Immunoblots against infected cell lysates detected two distinct infection-specific proteins at 24 h postinfection, which migrated at ∼49 and 28 kDa, with the latter representing the cleavage product. Interestingly, only the 28-kDa fragment was detected in purified elementary bodies (EBs). The antisera generated against YtgCR recognized purified recombinant YtgR, confirming specific reactivity of the anti-YtgCR antisera (Fig. S4). Thus, it appeared that YtgR was generated during infection of cells in culture from the internal cleavage of YtgCR.
Fig. 3.
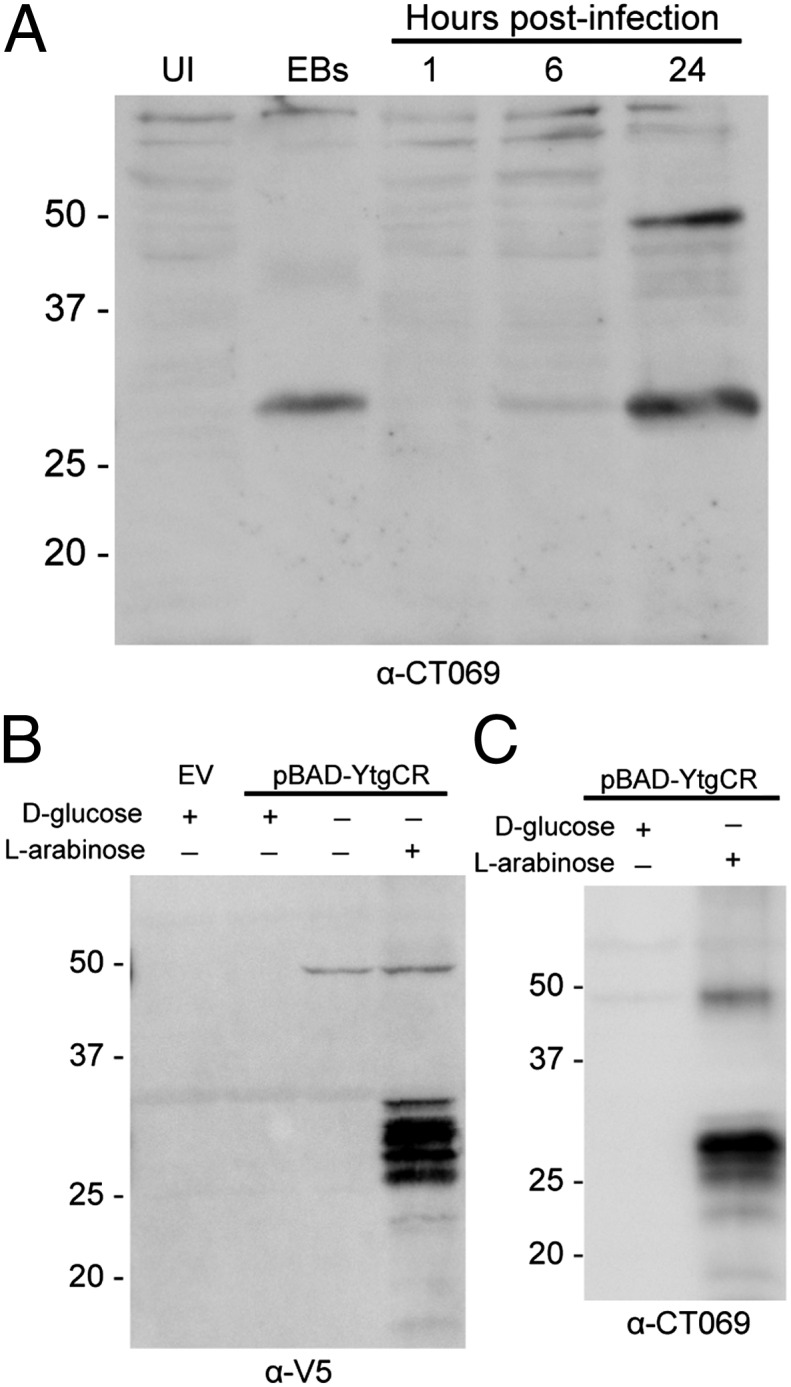
Nascent YtgCR is cleaved in vivo and heterologously in E. coli. (A) Two Chlamydia-specific proteins were detected using CT069 antisera in a Western blot of C. trachomatis-infected cell cultures at 24 h postinvasion, which presumably corresponded to the full-length YtgCR (∼49 kDa) and the putative YtgR domain (∼28 kDa). Only the putative YtgR domain was detected in purified EBs. (B) The YtgCR domain (49 kDa) was expressed with a C-terminal V5 epitope in E. coli, and found to be cleaved yielding a 28-kDa band that corresponded to YtgR. (C) Western blot of lysates from E. coli expressing YtgCR-V5 demonstrated a similar band pattern when probed with the CT069 antisera.
Surprisingly, when the full-length YtgCR fused to a V5 tag at the C terminus was expressed in E. coli, Western blot against the tag epitope also revealed the 28-kDa band in addition to the full-length 49 kDa species (Fig. 3B). However, instead of a single additional 28-kDa product (as observed in vivo), heterologous expression in E. coli produced a series of C-terminal cleavage products, which ranged from 26 to 32 kDa, likely because of imprecise cleavage arising from nonoptimal conditions (e.g., heterologous expression, absence of the YtgD binding partner, etc.). A Western blot of the same E. coli lysate using the YtgCR antisera also exhibited a band profile similar to the anti-V5 blot, indicating that the full-length and C-terminal cleavage products were detected by the anti-CT069 antisera (Fig. 3C). Combined with the observation that the antisera recognized a purified recombinant of YtgR, this result suggested that the 28-kDa band likely corresponds to the C-terminal YtgR fragment.
Heterologous Cleavage of CT069 in E. coli Produces Functional YtgR Polypeptides.
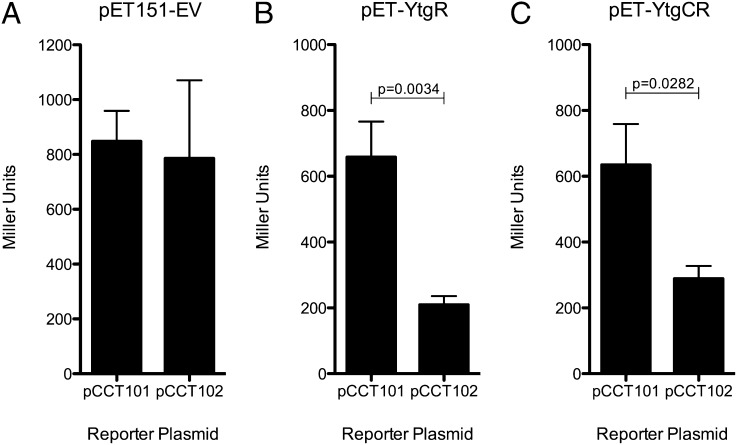
To determine whether the C-terminal cleavage products observed in E. coli functioned as a transcriptional regulator, a reporter gene assay system was implemented. In brief, a sequence containing the YtgR operator sequence from upstream of ytgA was inserted downstream of an arabinose inducible promoter (PBAD), but upstream of a lacZ reporter gene, giving rise to pCCT102. The pCCT102 vector and a control vector (pCCT101) that lacked the insert were cotransformed into BL21* E. coli with the expression vectors for YtgR or for the full-length YtgCR. Cotransformants were cultured in an iron-replete medium at a low optical density, and expression of β-galactosidase activity was measured.
To control for any alterations in LacZ expression conferred purely by the inserted sequence, the reporter plasmids were also cotransformed into BL21* E. coli containing an empty expression vector (pET151). In these cotransformants, similar levels of Miller Units (MU) were observed (Fig. 4A), indicating that the insertion of the sequence containing the cis-regulatory element upstream of the ytg operon did not have unwanted inhibitory or enhancing effects on the reporter gene expression independent of YtgR. For an additional control, pCCT102 was grown in the presence of d-glucose, which inhibits transcription from the PBAD promoter driving the lacZ reporter. The lack of β-galactosidase expression in the presence of glucose suggested that the transcription of the reporter gene was initiated from PBAD and not from the putative ytgA promoter present within the inserted sequence.
Fig. 4.
Recombinant expression of YtgCR results in repression of a reporter gene downstream of the putative YtgR operator. Two reporter plasmids, pCCT101 (PBAD:lacZ) and pCCT102 (PBAD:YtgR operator:lacZ), were cotransformed with an empty expression vector (A), a YtgR expression vector (B), or a YtgCR expression vector (C). The YtgR operator-containing sequence insert conferred repression in the YtgR and YtgCR expression vector cotransformants, but not the empty expression vector control. (n = 3; P values represent unpaired, one-tailed t test).
In contrast, when the reporter plasmids were cotransformed with the YtgR expression vector, the presence of the ytg promoter region in pCCT102 led to a significant decrease in β-galactosidase activity, compared with the no insert control vector (Fig. 4B; one-tailed, unpaired t test: P < 0.004). In conjunction with the dot blot and BLI data, these findings suggested that YtgR was able to bind to an operator site within the inserted sequence to repress expression of the lacZ reporter.
To determine whether the cleavage of YtgCR in E. coli produced a functional YtgR polypeptide, the reporter plasmids were cotransformed with full-length YtgCR expression vector and β-galactosidase activity was monitored. Enzyme activity was approximately twofold lower from pCCT102 than from pCCT101 cotransformants (one-tailed, unpaired t test: P < 0.03). This suggested that the heterologous cleavage of YtgCR in E. coli generated functional transcriptional repressor that recognized an operator sequence within the flanking insert engineered into pCCT102.
Discussion
In this study, we present a previously uncharacterized class of fusion proteins that consist of an N-terminal ABC-3 metal permease domain and a C-terminal DNA-binding domain, of which a majority are homologous to the DtxR family of metal-dependent transcriptional repressors. Examples of this family were identified in members of 11 different bacterial phyla, including our organism of interest, C. trachomatis. Our current study focuses on a chlamydial representative of this family, which we have termed YtgCR. Characterization of the DtxR-like domain of this protein (YtgR) revealed that it was capable of binding an operator sequence upstream of its own operon in an iron-dependent manner. Because the operon in which ytgCR resides is thought to encode an iron import system, our results demonstrate a bona fide molecular mechanism for the regulation of iron-homeostasis in Chlamydia. However, the likely targeting of the fusion protein to the inner membrane would require proteolytic processing to liberate the YtgR domain, so that it may function efficiently as an iron-dependent repressor. Indeed, the full-length YtgCR was cleaved during chlamydial infection and heterologous expression in E. coli, in both cases yielding a C-terminal fragment of ∼28 kDa in size. This cleavage event correlated with the transcriptional repression of the lacZ reporter gene under iron-replete conditions. This represents a previously uncharacterized mechanism for iron-dependent transcriptional regulation in Chlamydia and other bacteria.
Contrary to our findings, a recent report characterized YtgR DNA-binding activity as zinc-dependent. An explanation for this discrepancy could be the difference in DNA-binding assays or preparations of recombinant YtgR used. For example, residual metals present within the gel and buffering system used in an electromobility shift assays (EMSA) have been reported to provide false-positive activation of metal-dependent repressors (e.g. ref. 27). Akers et al. (17) did not include a prechelation step in their EMSA, which resulted in a small, but detectable amount of mobility shift observed in their “apo-YtgR” sample. In contrast, the BLI method used in this study utilized chelex-treated YtgR that exhibited minimal background binding. Another explanation could be the difference in the concentrations of metals used. Tao and Murphy reported that DtxR from C. diphtheriae required a 100-fold greater [Zn2+] compared with [Fe2+] (28), and it is possible that a similar difference in activation for YtgR may account for the inability of Zn2+ to activate DNA-binding in our hands. Without testing a higher [Zn2+], our results nonetheless demonstrate the preferential activation of YtgR by Fe2+. Unfortunately, Akers et al. did not make a similar comparison between Zn2+ and Fe2+. The apparent preference of YtgR for iron, instead of zinc is consistent with iron-related function of Ytg iron import system (15) and the iron responsiveness of ytgA expression (15, 16).
One of the long-standing problems in the iron biology in Chlamydia is the elucidation of the genomic response to iron fluctuation, which has been hindered by technical issues precluding the identification of primary response genes, and distinguishing them from secondary and tertiary responses involved in the onset of the persistent phenotype (16). The later responses are themselves important, but for the purpose of elucidating primary iron-dependent transcriptional regulation, they present complications in analysis. This complication is highlighted by the very restrictive, and somewhat arbitrary, classification of genes/open-reading frames into early/mid-/late timing of expression. Despite the availability of the transcriptional profile of Chlamydophila pneumoniae that have been starved for iron (29), the genetic nature of the primary response remains poorly characterized. With the identification of YtgR, which has homologs in all chlamydial species for which genome sequences are available, additional research avenues related to iron-dependent regulation and metabolism have been opened. It may now be possible to identify the genes potentially regulated by this repressor and address the extent to which the YtgR regulon determines the iron-dependent regulon of Chlamydia. By comparing the YtgR regulon to the total regulon induced by general iron starvation, we may find hints of additional iron-dependent gene regulation mechanisms.
Interestingly, nearly 5% of all ABC-3 metal permease proteins listed in the EBI InterPro database contained C-terminal extensions of greater than 50 aa in length. Because our search was limited to proteins with recognized DNA-binding domains, our estimate of the prevalence of permease-repressor fusion proteins may actually be conservative. Moreover, it is important to note that we began our analysis with all proteins containing the ABC-3 metal permease domain, and it is unknown whether other permease domains (ABC or other) may be found in the same configuration. For instance, the IrtAB system in Mycobacterium smegmatis sets a precedent for dual function metal–permease fusion proteins in bacteria (30). In this system, a ferric-chelate permease is formed by two transmembrane proteins (IrtA and IrtB) in complex. One of the permease proteins, IrtA, contains an additional amino-terminal ferric reductase domain. Therefore, upon translocation of the ferric-loaded siderophore into the bacterial cytoplasm, the amino-terminal IrtA domain is able to catalyze the reduction of the ferric iron into the ferrous form, which subsequently forces its release from the ferri-siderophore. However, in this example the fused ferric reductase domain is in a prime location for its cellular function, which is seemingly in contrast to the YtgCR permease-repressor configuration.
So, what might be the implications of the fusion configuration of YtgCR and other orthologs? In such a configuration, DNA binding would depend on both cleavage/liberation and metal binding. Furthermore, cleavage itself may be subjected to regulation, such as the requirement for prior cleavage of the preprotease form, akin to the Chlamydia-secreted protease-like activity factor (CPAF) (31), or for metal binding, as in metalloproteases. An added level of regulation of the repressor would provide a means of further fine-tuning gene regulation. Currently, the identity of the chlamydial protease responsible is unknown. At this time, we cannot speculate on the nature of the protease, except that it was present/active at any time point that exhibited YtgCR expression in intracellular Chlamydia and was ubiquitous/active in E. coli grown iron-replete conditions. However, we cannot discount a protease that is regulated in the context of a human infection where levels of iron are in constant flux.
Proteolytic cleavage generating YtgR in the E. coli heterologous system correlated with the transcriptional repression of the lacZ reporter gene under the control of an operator sequence vicinal to the ytg promoter, highlighting the importance of the protease in this regulatory system. Although it would be desirable to confirm that cleavage is required for efficient DNA-binding using an E. coli mutant that lacks the responsible protease, the protease must first be identified. There are precedents in bacteria for the proteolytic liberation of transcription factors from membrane sequestration (e.g. refs. 18--20), which may provide clues to the identity of the protease in question.
In this report, we have described the identification of a family of metal permease-transcriptional repressor fusion proteins. A representative protein of this family, YtgCR, was cleaved under native (C. trachomatis) and heterologous (E. coli) experimental systems. Evidence for the functionality of the YtgR repressor domain was presented, including an iron-dependent recognition of an operating sequence located upstream of its own operon, which putatively encodes for an iron import system. The involvement of proteolytic processing and iron binding in the activation of YtgR indicates a more complex two-step regulatory network for the maintenance of iron homeostasis in Chlamydia. This system is likely to exist in other bacteria as indicated by our discovery of similar permease-repressor fusion proteins in members of 11 bacterial phyla. Data presented here expands the bacterial repertoire of metal-dependent transcriptional regulation.
Methods
Bioinformatics Analysis.
All protein alignments, operon schematics, and transmembrane predictions (21, 22) were completed within the Geneious software platform (32). Secondary structures were predicted using the Phyre2 Server (25).
Plasmids.
A list of plasmids used and generated in this study is provided in Table S1, and a list of primers used is provided in Table S2. Specific methods for the generation of each plasmid are provided in SI Methods. All transformations were made into Top10 (Invitrogen) E. coli, and vectors were verified by sequencing (GATC).
Recombinant Protein Expression and Purification of YtgR.
Expression of recombinant YtgR was induced from the pET-YtgR plasmid in BL21* (DE3) E. coli. The recombinant was purified using a method for the purification of insoluble transcription factors, as described previously (26, 33). After purification, recombinant protein samples were incubated with Chelex-100 resin to remove any metal cofactors that may have been copurified with the protein. Complete descriptions of these methods are provided in SI Methods.
DNA-Binding Assays.
The dot-blot and Biolayer interferometry assays were performed as described in SI Methods. The biotinylated-oligonucleotide probes for these assays were generated by PCR using primers that had been custom synthesized with a 5′ Biotin molecule (Invitrogen). Binding kinetics were calculated with GraphPad Prism using the association then dissociation function.
Western Blots.
Western blotting was performed using standard Tris-glycine (TGS)/PAGE and transfer techniques. All blots show equal amounts loaded and are representative of at least three experiments showing the same cleavage pattern. The rabbit polyclonal anti-CT069 antisera were a kind gift of Guangming Zhong (University of Texas Health Sciences Center, San Antonio, TX). Complete descriptions of the induction conditions for Western blots of plasmid-based proteins are found in SI Methods.
Reporter Gene Expression Assay.
Reporter (pCCT101 and pCCT102) and expression (pET-EV, pET-YtgR, and pET-YtgCR) plasmids were sequentially transformed into BL21* (DE3) E. coli, and single colonies resistant to both carbenicillin and tetracycline were used for each individual experiment. Isolates were grown at low density in LB broth supplemented with exogenous iron. LacZ expression was measured via the Miller assay, as described (34). The experiment was performed three times independently, with each trial including the full complement of cotransformant samples. Interexperimental error was corrected by transforming the values of each trial, such that the mean of all samples within each batch were identical. The mean MU of each sample from the three batch corrected experiments is shown. Error bars represent 1 SD from the mean.
Supplementary Material
Acknowledgments
We thank Dr. Guangming Zhong for providing the CT069 antisera, Prof. Myra McClure for critical review of the manuscript, Dr. Robert Fagan for critical discussion, and Dr. Nadine Lossi for laboratory assistance. This work was funded by National Institutes of Health Grant AI065545 and MRC Grant G0900213.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201398109/-/DCSupplemental.
References
- 1.Kehrer JP. The Haber-Weiss reaction and mechanisms of toxicity. Toxicology. 2000;149:43–50. doi: 10.1016/s0300-483x(00)00231-6. [DOI] [PubMed] [Google Scholar]
- 2.Pennella MA, Giedroc DP. Structural determinants of metal selectivity in prokaryotic metal-responsive transcriptional regulators. Biometals. 2005;8:413–428. doi: 10.1007/s10534-005-3716-8. [DOI] [PubMed] [Google Scholar]
- 3.Schmitt MP, Twiddy EM, Holmes RK. Purification and characterization of the diphtheria toxin repressor. Proc Natl Acad Sci USA. 1992;89:7576–7580. doi: 10.1073/pnas.89.16.7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Lorenzo V, Wee S, Herrero M, Neilands JB. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J Bacteriol. 1987;169:2624–2630. doi: 10.1128/jb.169.6.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CDC . Sexually Transmitted Disease Surveillance. Atlanta, GA: Centers for Disease Control and Prevention; 2008. [Google Scholar]
- 6.CDC Chlamydia trachomatis Genital Infections–United States, 1995. Morb Mortal Wkly Rep. 1997;46:193–198. [PubMed] [Google Scholar]
- 7.Resnikoff S, et al. Global data on visual impairment in the year 2002. Bull World Health Organ. 2004;82:844–851. [PMC free article] [PubMed] [Google Scholar]
- 8.Beatty WL, Morrison RP, Byrne GI. Persistent chlamydiae: From cell culture to a paradigm for chlamydial pathogenesis. Microbiol Rev. 1994;58:686–699. doi: 10.1128/mr.58.4.686-699.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hogan RJ, Mathews SA, Mukhopadhyay S, Summersgill JT, Timms P. Chlamydial persistence: Beyond the biphasic paradigm. Infect Immun. 2004;72:1843–1855. doi: 10.1128/IAI.72.4.1843-1855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wyrick PB. Chlamydia trachomatis persistence in vitro: An overview. J Infect Dis. 2010;201(Suppl 2):S88–S95. doi: 10.1086/652394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ouellette SP, et al. Global transcriptional upregulation in the absence of increased translation in Chlamydia during IFNgamma-mediated host cell tryptophan starvation. Mol Microbiol. 2006;62:1387–1401. doi: 10.1111/j.1365-2958.2006.05465.x. [DOI] [PubMed] [Google Scholar]
- 12.Belland RJ, et al. Transcriptome analysis of chlamydial growth during IFN-gamma-mediated persistence and reactivation. Proc Natl Acad Sci USA. 2003;100:15971–15976. doi: 10.1073/pnas.2535394100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephens RS, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 14.Claverys JP. A new family of high-affinity ABC manganese and zinc permeases. Res Microbiol. 2001;152:231–243. doi: 10.1016/s0923-2508(01)01195-0. [DOI] [PubMed] [Google Scholar]
- 15.Miller JD, Sal MS, Schell M, Whittimore JD, Raulston JE. Chlamydia trachomatis YtgA is an iron-binding periplasmic protein induced by iron restriction. Microbiology. 2009;155:2884–2894. doi: 10.1099/mic.0.030247-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson CC, Carabeo RA. An optimal method of iron starvation of the obligate intracellular pathogen, Chlamydia trachomatis. Front Microbiol. 2011;2:20. doi: 10.3389/fmicb.2011.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akers JC, HoDac H, Lathrop RH, Tan M. Identification and functional analysis of CT069 as a novel transcriptional regulator in Chlamydia. J Bacteriol. 2011;193:6123–6131. doi: 10.1128/JB.05976-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alba BM, Leeds JA, Onufryk C, Lu CZ, Gross CA. DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma(E)-dependent extracytoplasmic stress response. Genes Dev. 2002;16:2156–2168. doi: 10.1101/gad.1008902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehara K, Ito K, Akiyama Y. YaeL (EcfE) activates the sigma(E) pathway of stress response through a site-2 cleavage of anti-sigma(E), RseA. Genes Dev. 2002;16:2147–2155. doi: 10.1101/gad.1002302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King-Lyons ND, Smith KF, Connell TD. Expression of hurP, a gene encoding a prospective site 2 protease, is essential for heme-dependent induction of bhuR in Bordetella bronchiseptica. J Bacteriol. 2007;189:6266–6275. doi: 10.1128/JB.00629-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 22.Möller S, Croning MD, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- 23.Pohl E, Holmes RK, Hol WG. Crystal structure of a cobalt-activated diphtheria toxin repressor-DNA complex reveals a metal-binding SH3-like domain. J Mol Biol. 1999;292:653–667. doi: 10.1006/jmbi.1999.3073. [DOI] [PubMed] [Google Scholar]
- 24.Hsin K, Sheng Y, Harding M, Taylor P, Walkinshaw M. MESPEUS: A database of the geometry of metal sites in proteinsitle. J Appl Cryst. 2008;41:963–968. [Google Scholar]
- 25.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 26.Posey JE, Hardham JM, Norris SJ, Gherardini FC. Characterization of a manganese-dependent regulatory protein, TroR, from Treponema pallidum. Proc Natl Acad Sci USA. 1999;96:10887–10892. doi: 10.1073/pnas.96.19.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mills SA, Marletta MA. Metal binding characteristics and role of iron oxidation in the ferric uptake regulator from Escherichia coli. Biochemistry. 2005;44:13553–13559. doi: 10.1021/bi0507579. [DOI] [PubMed] [Google Scholar]
- 28.Tao X, Murphy JR. Binding of the metalloregulatory protein DtxR to the diphtheria tox operator requires a divalent heavy metal ion and protects the palindromic sequence from DNase I digestion. J Biol Chem. 1992;267:21761–21764. [PubMed] [Google Scholar]
- 29.Mäurer AP, Mehlitz A, Mollenkopf HJ, Meyer TF. Gene expression profiles of Chlamydophila pneumoniae during the developmental cycle and iron depletion-mediated persistence. PLoS Pathog. 2007;3:e83. doi: 10.1371/journal.ppat.0030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryndak MB, Wang S, Smith I, Rodriguez GM. The Mycobacterium tuberculosis high-affinity iron importer, IrtA, contains an FAD-binding domain. J Bacteriol. 2010;192:861–869. doi: 10.1128/JB.00223-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen D, et al. Autoprocessing and self-activation of the secreted protease CPAF in Chlamydia-infected cells. Microb Pathog. 2010;49:164–173. doi: 10.1016/j.micpath.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drummond AJ, et al. 2010. Geneious v5.3. Available at http://www.geneious.com.
- 33.Nguyen LH, Jensen DB, Burgess RR. Overproduction and purification of sigma 32, the Escherichia coli heat shock transcription factor. Protein Expr Purif. 1993;4:425–433. doi: 10.1006/prep.1993.1056. [DOI] [PubMed] [Google Scholar]
- 34.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.