Abstract
Egr-1 is an inducible transcription factor that recognizes 9-bp target DNA sites via three zinc finger domains and activates genes in response to cellular stimuli such as synaptic signals and vascular stresses. Using spectroscopic and computational approaches, we have studied structural, dynamic, and kinetic aspects of the DNA-scanning process in which Egr-1 is nonspecifically bound to DNA and perpetually changes its location on DNA. Our NMR data indicate that Egr-1 undergoes highly dynamic domain motions when scanning DNA. In particular, the zinc finger 1 (ZF1) of Egr-1 in the nonspecific complex is mainly dissociated from DNA and undergoes collective motions on a nanosecond timescale, whereas zinc fingers 2 and 3 (ZF2 and ZF3, respectively) are bound to DNA. This was totally unexpected because the previous crystallographic studies of the specific complex indicated that all of Egr-1’s three zinc fingers are equally involved in binding to a target DNA site. Mutations that are expected to enhance ZF1’s interactions with DNA and with ZF2 were found to reduce ZF1’s domain motions in the nonspecific complex suggesting that these interactions dictate the dynamic behavior of ZF1. By experiment and computation, we have also investigated kinetics of Egr-1’s translocation between two nonspecific DNA duplexes. Our data on the wild type and mutant proteins suggest that the domain dynamics facilitate Egr-1’s intersegment transfer that involves transient bridging of two DNA sites. These results shed light on asymmetrical roles of the zinc finger domains for Egr-1 to scan DNA efficiently in the nucleus.
Keywords: NMR spectroscopy, target search process, interdomain dynamics, protein-DNA interactions, simulation
In cellular responses to various stimuli such as signals and stresses, gene regulation by transcription factors is of fundamental importance. Egr-1 (also known as Zif268) is an inducible transcription factor with crucial roles particularly in the brain and cardiovascular systems in mammals. In the brain, Egr-1 is induced by synaptic signals in an activity-dependent manner and activates genes for long-term memory formation and consolidation (1, 2). In the cardiovascular system, Egr-1 is a stress-inducible transcription factor that activates the genes for initiating defense responses against vascular stress and injury (3, 4). Given the short lifetime of induced Egr-1 (typically ∼2 h) (3), rapid gene activation by Egr-1 is important in these biological processes that require an immediate response to the stimuli.
The induced Egr-1 protein has to initiate its role by searching for its target DNA sites among billions of DNA base pairs in the nucleus. In the DNA scanning process, transcription factors need to discriminate their target sites from nonspecific sites based on relatively minor differences in DNA structure and sequence. Crystallographic studies demonstrated that Egr-1 recognizes its 9-bp target sequence, GCGTGGGCG, as a monomer via zinc finger domains 1, 2, and 3 (hereafter referred to as ZF1, ZF2, and ZF3) that contact 3 bp each (Fig. 1A) (5). Although the three zinc finger domains are comprised of only 87 residues combined, Egr-1’s interface with target DNA is as wide as 2,870 Å2. Although such extensive contact with DNA is favorable for high specificity in target recognition as well as for high stability of the specific complex, it is unfavorable for rapid scanning of DNA because the protein has to break a larger number of interactions, such as hydrogen bonds and ion pairs, whenever it moves from one nonspecific DNA site to another (6). This is problematic, particularly because the vast majority of Egr-1 molecules undergoing a target search should be bound to nonspecific DNA sites due to the extremely high density of DNA in the nucleus and Egr-1’s micromolar affinity to nonspecific DNA. This situation represents the “speed-stability” paradox (6, 7).
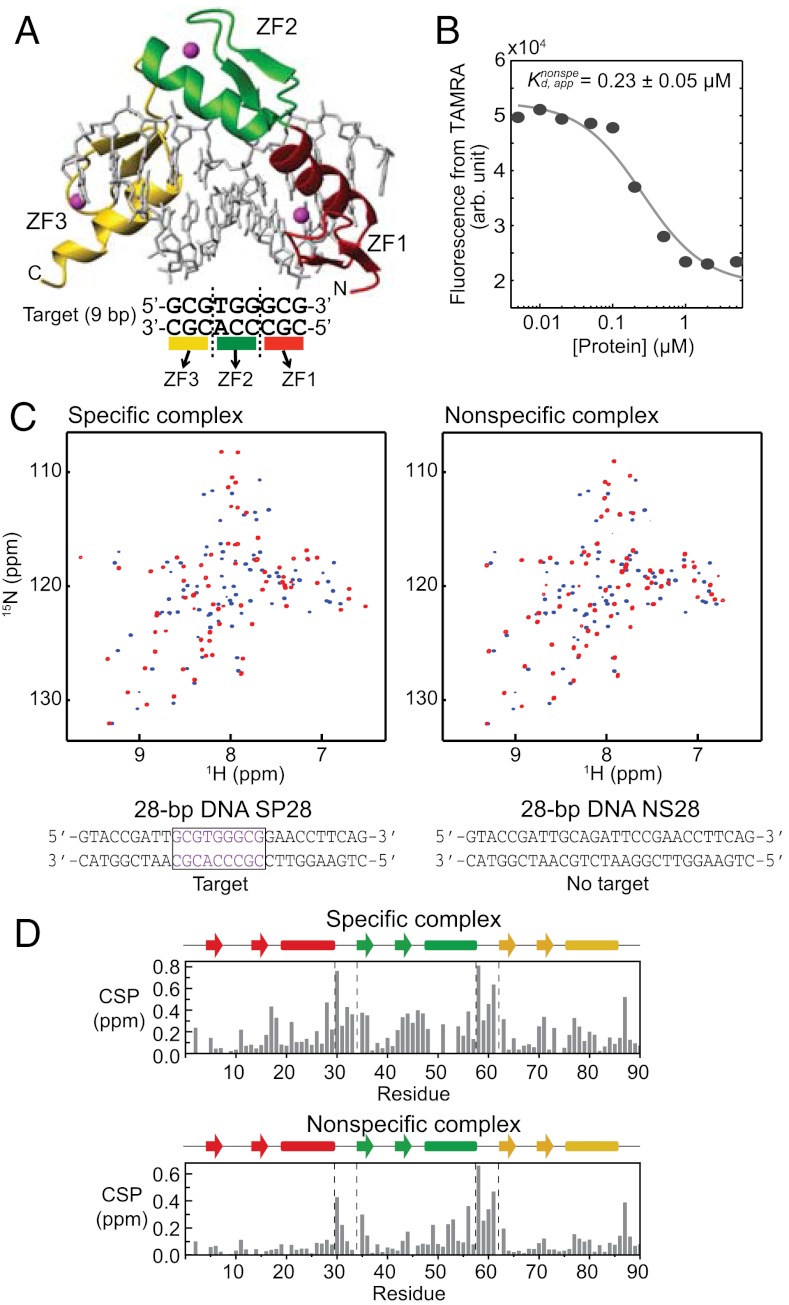
Fig. 1.
Specific and nonspecific DNA complexes of Egr-1. (A) Crystal structure (PDB 1AAY) of a complex of Egr-1 with a target DNA (5, 8). (B) Changes in fluorescence arising from TAMRA conjugated to NS28 upon Egr-1’s binding to nonspecific DNA. The fluorophore was conjugated to a 3′-terminus. The apparent dissociation constant for the nonspecific complex (Kd,app) was determined from three independent experimental datasets. (C) 1H-15N TROSY spectra recorded for 2H/15N-labeled Egr-1 proteins in the free state (blue) and the complexes (red) with the specific (SP28) or nonspecific (NS28) DNA duplexes. (D) Weighted-average chemical shift perturbation (CSP) as defined {(|ΔδH|2 + |0.2ΔδN|2)/2}1/2 (57), where ΔδH and ΔδN represent chemical shift differences of backbone amide 1H and 15N, respectively, between the free and DNA-bound states.
How does Egr-1 achieve high specificity in recognition and rapidity in scanning? Even though there have been many structural studies on zinc finger proteins bound to their target DNA sites or free in solution (e.g., refs. 5, 8–13), this question remains unanswered because very little is known about their binding to nonspecific DNA. In fact, there is a lack of knowledge on DNA-scanning by eukaryotic transcription factors. The vast majority of the previous literature on the experimental studies of target DNA search is for prokaryotic gene-regulatory proteins or restriction enzymes (7, 14, 15); however, because of the much higher DNA density and the presence of nucleosomes in eukaryotic nuclei, it is natural to consider that eukaryotic transcription factors may use a different strategy for their rapid target search. Our present work shows that it is indeed the case. Here, we apply spectroscopic and computational methods to the highly dynamic complex in which Egr-1 is nonspecifically bound to DNA and perpetually changes its location on DNA. This work provides structural, dynamic, and kinetic information on how Egr-1 efficiently scans DNA to find its target sites rapidly. Our results suggest that Egr-1’s asymmetrical domain dynamics in the DNA-scanning process can play an important role in resolving the speed-stability paradox.
Results
NMR of Specific and Nonspecific DNA Complexes.
In our studies, we used the Egr-1 DNA-binding domain comprised of three zinc fingers (Egr-1 residues 349–421; hereafter, referred to as “Egr-1” for simplicity’s sake) and two 28-bp DNA duplexes: SP28 and NS28 (Fig. 1). These two duplexes are identical except that SP28 contains a 9-bp target site (GCGTGGGCG) whereas the DNA duplex NS28 does not. The nonspecific 28-bp DNA NS28 does not contain any bp triplets recognized by individual domains. We chose these 28-bp DNA duplexes for our experiments for two reasons: (i) at this length, the negative charge of DNA governs molecular alignment promoted by Pf1 phage that makes interpretation of residual dipolar couplings (RDCs) easier (16, 17); and, (ii) the solubility and stability of nonspecific complexes with Egr-1 are lower for shorter, nonspecific DNA. Using fluorescence arising from tetramethylrhodamine (TAMRA) conjugated to the 3′-terminus of DNA NS28, we measured the apparent dissociation constant Kd to be 0.23 ± 0.05 μM at 40 mM KCl (Fig. 1B). This affinity for nonspecific DNA is ∼1,000-fold weaker than the affinity for target DNA under the same conditions (18). If an Egr-1 protein covers 11 bp, as observed in the crystal structure, the 28-bp nonspecific DNA can accommodate up to two protein molecules. To avoid complications from two proteins binding to the same DNA molecule, we studied the nonspecific complex using at least a two-fold molar excess of nonspecific DNA. Under these conditions, the population of the singly bound complexes should exceed 90%. We recorded 1H–15N transverse relaxation optimized spectroscopy (TROSY) spectra for nonspecific and specific complexes of 2H/15N-labeled Egr-1 with unlabeled 28-bp DNA. As shown in Fig. 1C, the NMR spectra of the complexes were substantially different from the corresponding TROSY spectrum for the protein in the free state. Although Egr-1 can potentially bind 18 different sites (28 - 11 + 1) on a nonspecific 28-bp DNA, the NMR spectra of nonspecific complexes showed only one set of resonances as was previously found for nonspecific DNA complexes with HoxD9 (16) and HMGB1 (19). This, along with additional data to be discussed indicates that translocation of the protein between different sites on DNA occurs in a fast exchange regime on NMR chemical shift timescale.
Using TROSY-based triple resonance experiments with deuterium decoupling (20), we assigned the 1H/13C/15N resonances of the protein backbone for the nonspecific and specific complexes as well as for the free protein. Fig. 1D shows chemical shift perturbation (CSP) upon DNA-binding for each Egr-1 amide group (Numbering of residues is according to previous literature) (5). CSP magnitudes were smaller for the nonspecific complex, though Kd data (e.g. Fig. 1B) together with molecular concentrations suggest that > 99% protein should be bound to DNA for both complex samples. Interestingly, ZF1’s CSP upon binding to the nonspecific DNA duplex was particularly small (only two residues exhibit a weighted-average CSP larger than 0.1 ppm); whereas, all ZF domains, including ZF1, exhibit substantial chemical shift perturbations upon binding to the specific DNA duplex (Fig. 1D). This implies that Egr-1’s binding mode for nonspecific DNA is quite different from that for specific DNA.
RDC Data Indicate ZF1’s Motions in Egr-1 Bound To Nonspecific DNA.
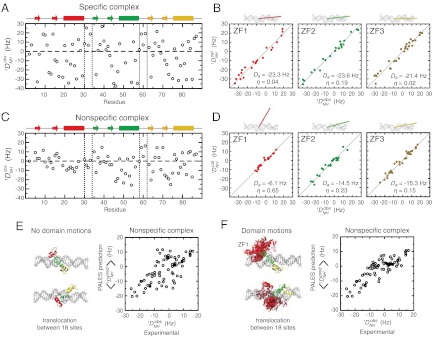
To obtain structural and dynamic information on Egr-1 bound to nonspecific DNA, we conducted a comparative study of the nonspecific and specific complexes by analyzing RDC 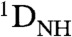 induced with 8 mg/mL Pf1 phage as an alignment medium (21). The RDC data for the specific complex (Fig. 2A) were consistent with the crystal structure of the specific complex of Egr-1, and the main principal axes were parallel to the DNA axis (Fig. 2B), which is reasonable because the tensor for the electrostatically driven molecular alignment should be governed by the negative charges on the 28-bp DNA (16, 17). The RDC data for the nonspecific complex (Fig. 2C) were consistent with the crystal structures of individual domains (Fig. 2D) indicating that the individual domain structures in the nonspecific complexes are maintained; however, the alignment tensor parameters for ZF1 were found to be substantially different from those for ZF2 and ZF3 (Fig. 2D). For example, whereas the main principal axes of alignment tensors for ZF2 and ZF3 in the nonspecific complex were almost parallel to the DNA axis with angles of 13° and 10°, respectively, the corresponding angle for ZF1 was as large as 57°. More importantly, the magnitude of the alignment tensor (Da) for ZF1 was much smaller than those for ZF2 and ZF3. These results are indicative of large-scale dynamics (22) and qualitatively suggest that ZF1 undergoes domain motions in the nonspecific complex. ZF2 and ZF3 in the nonspecific complex also seem to be more dynamic than in the specific complex because the experimental |Da| for the nonspecific complex is smaller than that for the specific complex (Fig. 2
B and D).
induced with 8 mg/mL Pf1 phage as an alignment medium (21). The RDC data for the specific complex (Fig. 2A) were consistent with the crystal structure of the specific complex of Egr-1, and the main principal axes were parallel to the DNA axis (Fig. 2B), which is reasonable because the tensor for the electrostatically driven molecular alignment should be governed by the negative charges on the 28-bp DNA (16, 17). The RDC data for the nonspecific complex (Fig. 2C) were consistent with the crystal structures of individual domains (Fig. 2D) indicating that the individual domain structures in the nonspecific complexes are maintained; however, the alignment tensor parameters for ZF1 were found to be substantially different from those for ZF2 and ZF3 (Fig. 2D). For example, whereas the main principal axes of alignment tensors for ZF2 and ZF3 in the nonspecific complex were almost parallel to the DNA axis with angles of 13° and 10°, respectively, the corresponding angle for ZF1 was as large as 57°. More importantly, the magnitude of the alignment tensor (Da) for ZF1 was much smaller than those for ZF2 and ZF3. These results are indicative of large-scale dynamics (22) and qualitatively suggest that ZF1 undergoes domain motions in the nonspecific complex. ZF2 and ZF3 in the nonspecific complex also seem to be more dynamic than in the specific complex because the experimental |Da| for the nonspecific complex is smaller than that for the specific complex (Fig. 2
B and D).
Fig. 2.
RDC data indicating ZF1’s substantial domain motions in the nonspecific DNA complex of Egr-1. (A, C) Experimental RDC 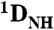 observed for Egr-1 in the specific (A) and nonspecific (C) complexes. (B, D) Fitting of RDC data to the 1.6 Å-resolution crystal structure of the specific complex (1AAY) for each domain. Correlations between calculated and observed RDCs are shown together with the obtained alignment tensor parameters (magnitude, Da and rhombicity, η). Orientations of the main principal axes determined by fitting are also shown along with DNA. (E, F) Correlations between experimental RDC data and those predicted by the program PALES (17, 23) based on the molecular shape and charge of the nonspecific complex. For the prediction, the nonspecific complex was represented by an ensemble of 18 states in which Egr-1 is bound to DNA at different locations (Details given in SI Text). For each state of the ensemble, the alignment tensor and RDCs were predicted with PALES. The correlation in E was obtained by assuming that individual domains are bound to each site in the nonspecific DNA in the same manner as is observed in the crystal structure of the specific complex. For F, the domain motions of ZF1 in the 18 states were taken into consideration with 50 different structures generated by high-temperature, rigid-body dynamics calculations (Details given in SI Text). Correlation coefficients for E and F were 0.66 and 0.86, respectively.
observed for Egr-1 in the specific (A) and nonspecific (C) complexes. (B, D) Fitting of RDC data to the 1.6 Å-resolution crystal structure of the specific complex (1AAY) for each domain. Correlations between calculated and observed RDCs are shown together with the obtained alignment tensor parameters (magnitude, Da and rhombicity, η). Orientations of the main principal axes determined by fitting are also shown along with DNA. (E, F) Correlations between experimental RDC data and those predicted by the program PALES (17, 23) based on the molecular shape and charge of the nonspecific complex. For the prediction, the nonspecific complex was represented by an ensemble of 18 states in which Egr-1 is bound to DNA at different locations (Details given in SI Text). For each state of the ensemble, the alignment tensor and RDCs were predicted with PALES. The correlation in E was obtained by assuming that individual domains are bound to each site in the nonspecific DNA in the same manner as is observed in the crystal structure of the specific complex. For F, the domain motions of ZF1 in the 18 states were taken into consideration with 50 different structures generated by high-temperature, rigid-body dynamics calculations (Details given in SI Text). Correlation coefficients for E and F were 0.66 and 0.86, respectively.
Using the program PALES that predicts alignment tensor as well as RDCs based on molecular shape and charge distribution (17, 23), we further examined the impact of ZF1’s domain motions on the RDC data for the nonspecific complex (details given in SI Text). For the specific complex, the RDCs predicted by PALES were in an excellent correlation with experimental data (Fig. S1A), which demonstrates the reliability of this approach. For PALES analysis, the nonspecific complex was represented by an ensemble of 18 states in which Egr-1 is bound to DNA at different locations (see Fig. S1 B and C). Under an assumption that the individual states of the nonspecific complex are similar to the specific complex, the structures that represent the nonspecific complex were generated from the crystal structure of the specific Egr-1/DNA complex and extension of B-form DNA (Details given in SI Methods and Fig. S1). For each structure in the ensemble, the alignment tensor and RDCs were predicted using PALES, and the ensemble averages were compared with the experimental RDC data for the nonspecific complex. When no domain motions were assumed for ZF1, the overall correlation between the predicted and observed RDC data was poor (Fig. 2E); however, the correlation was significantly improved when ZF1’s domain motions were taken into consideration with ensembles with randomized conformations of Linker 1 (Fig. 2F). These data suggest that ZF1 undergoes collective motions in the nonspecific complex, whereas ZF2 and ZF3 are bound to DNA with relative orientations as observed in the crystal structure of the specific complex.
ZF1’s Local Dissociation in the Nonspecific Complex.
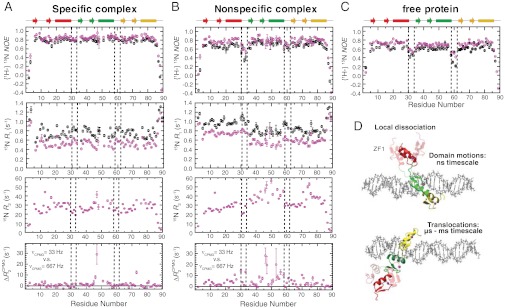
Unique dynamic properties of ZF1 in Egr-1 bound to nonspecific DNA were also evident from 15N relaxation studies of the nonspecific and specific complexes. Heteronuclear 1H–15N nuclear Overhauser effect (NOE), 15N longitudinal relaxation rates (R1), and 15N transverse relaxation rates (R2) for protein backbone amide groups in the specific and nonspecific Egr-1/DNA complexes are shown in Fig. 3
A and B, respectively. Comparison of heteronuclear NOE data for the specific and nonspecific complexes to those for the free protein (Fig. 3C) indicated that Linker 1 (between ZF1 and ZF2) in the nonspecific complex is as mobile as that in the free state, whereas Linker 2 in the nonspecific complex and both Linkers in the specific complex are immobilized upon complex formation. Furthermore, ZF1 in the nonspecific complex exhibited 15N R1 rates higher than those of ZF2 and ZF3 of the same complex (Fig. 3B), whereas 15N R1 rates are similar for all the three zinc finger domains in the specific complex (Fig. 3A). 15N R2 rates for many residues in the nonspecific complex were found to be substantially higher than those for the corresponding residues in the specific complex. CPMG relaxation dispersion data suggest that this is due to slow dynamics on a μs–ms timescale. The bottom graphs in Fig. 3
A and B show differences between apparent R2 rates measured at CPMG field strengths VCPMG of 33 Hz and 667 Hz [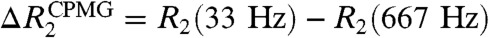 ] for the complexes. Corresponding data for the free Egr-1 protein are shown in Fig. S2A. From comparison of these CPMG data, it is clear that far more residues exhibit
] for the complexes. Corresponding data for the free Egr-1 protein are shown in Fig. S2A. From comparison of these CPMG data, it is clear that far more residues exhibit 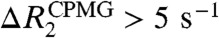 for the nonspecific complex.
for the nonspecific complex. 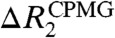 can be large in the presence of μs–ms dynamics involving multiple states with different 15N chemical shifts. Importantly, most residues with
can be large in the presence of μs–ms dynamics involving multiple states with different 15N chemical shifts. Importantly, most residues with 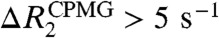 were found to be close to DNA for the nonspecific complex (Fig. S2B). It should be noted that 15N chemical shifts of such residues near DNA could significantly change when the Egr-1 protein transfers from one nonspecific site to another on DNA via inter- or intramolecular translocation. As shown below, intermolecular translocations of the Egr-1 protein between two nonspecific DNA duplexes were found to occur with an exchange rate constant of ∼1,000 s-1. The intramolecular translocation should be faster than the intermolecular translocation, as single molecule biophysical studies demonstrated that sliding of proteins on DNA occurs with a 1D diffusion coefficient in a range of 105–107 bp2 s-1 in general (7, 24). Considering these together, it is likely that the μs–ms dynamics detected for the nonspecific complex correspond to translocation of protein on DNA rather than to intrinsic conformational dynamics; however, it is difficult to obtain further information from CPMG relaxation dispersion data because popular analytical expressions for a two-state exchange (25, 26) are obviously inappropriate for the nonspecific complex that involves potentially 36 different states (18 binding sites and two opposite orientations for each).
were found to be close to DNA for the nonspecific complex (Fig. S2B). It should be noted that 15N chemical shifts of such residues near DNA could significantly change when the Egr-1 protein transfers from one nonspecific site to another on DNA via inter- or intramolecular translocation. As shown below, intermolecular translocations of the Egr-1 protein between two nonspecific DNA duplexes were found to occur with an exchange rate constant of ∼1,000 s-1. The intramolecular translocation should be faster than the intermolecular translocation, as single molecule biophysical studies demonstrated that sliding of proteins on DNA occurs with a 1D diffusion coefficient in a range of 105–107 bp2 s-1 in general (7, 24). Considering these together, it is likely that the μs–ms dynamics detected for the nonspecific complex correspond to translocation of protein on DNA rather than to intrinsic conformational dynamics; however, it is difficult to obtain further information from CPMG relaxation dispersion data because popular analytical expressions for a two-state exchange (25, 26) are obviously inappropriate for the nonspecific complex that involves potentially 36 different states (18 binding sites and two opposite orientations for each).
Fig. 3.
15N relaxation data indicating dynamics of Egr-1. (A, B) Backbone heteronuclear {1H-}15N NOE , 15N R1, and 15N R2 data for the specific (A) and nonspecific (B) complexes. Data collected at 1H-frequencies of 800 MHz and 600 MHz are shown in magenta and black, respectively. Differences between apparent 15N transverse relaxation rates at CPMG field strengths νCPMG = 33 Hz and νCPMG = 666 Hz [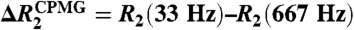 ] are also shown. (C) Heteronuclear {1H-}15N NOE data for the Egr-1 protein in the free state. (D) Domain motions and translocation of Egr-1 in the nonspecific complex as suggested by the current study.
] are also shown. (C) Heteronuclear {1H-}15N NOE data for the Egr-1 protein in the free state. (D) Domain motions and translocation of Egr-1 in the nonspecific complex as suggested by the current study.
To estimate the timescale of ZF1’s motions in the nonspecific complex, we used the extended model-free spectral density function (27, 28). First, from 15N relaxation data, we determined the rotational correlation time τr, rotational anisotropy D‖/D⊥, order parameters, and correlation times for internal motions of backbone NH bonds for the specific complex (see Fig. S3A). The same approach is inappropriate for the nonspecific complex because Egr-1’s translocation between different DNA sites substantially increases the R2 rates of many residues (Fig. 3B). Note that the presence of sliding on a μs timescale makes it difficult to suppress completely the exchange contribution Rex by CPMG. To estimate the timescale of ZF1’s domain motions we made two assumptions. The first assumption was that τr and D‖/D⊥ of the nonspecific complex were virtually the same as those of the specific complex because the inertia tensor of the complex should be governed by the long axis of the 28-bp DNA (∼95 Å) regardless of the position of the protein on the DNA. This assumption is supported by the fact that 15N R1 and heteronuclear NOE data for ZF2 and ZF3 are similar for the nonspecific and specific complexes. We also assumed that, internal motions of NH bonds within the coordinate frames of individual domains are identical for the nonspecific and specific complexes because the RDC data suggest that individual domain structures are the same for both complexes. Under these assumptions, we calculated the order parameters 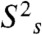 and the correlation time τs for domain motions of ZF1 with nonlinear least squares fitting to 15N R1 and NOE data measured for the nonspecific complex at 1H frequencies of 600 MHz and 800 MHz (details given in SI Text). The average correlation time for the domain motion was determined to be 2 ns (Fig. S3B). These data together with RDC data indicate that ZF1 is mainly dissociated from DNA and undergoes collective domain motions on a nanosecond timescale, while Egr-1 is bound to nonspecific DNA via ZF2 and ZF3.
and the correlation time τs for domain motions of ZF1 with nonlinear least squares fitting to 15N R1 and NOE data measured for the nonspecific complex at 1H frequencies of 600 MHz and 800 MHz (details given in SI Text). The average correlation time for the domain motion was determined to be 2 ns (Fig. S3B). These data together with RDC data indicate that ZF1 is mainly dissociated from DNA and undergoes collective domain motions on a nanosecond timescale, while Egr-1 is bound to nonspecific DNA via ZF2 and ZF3.
Rapid Intersegment Transfer of Egr-1 Between Nonspecific DNA.
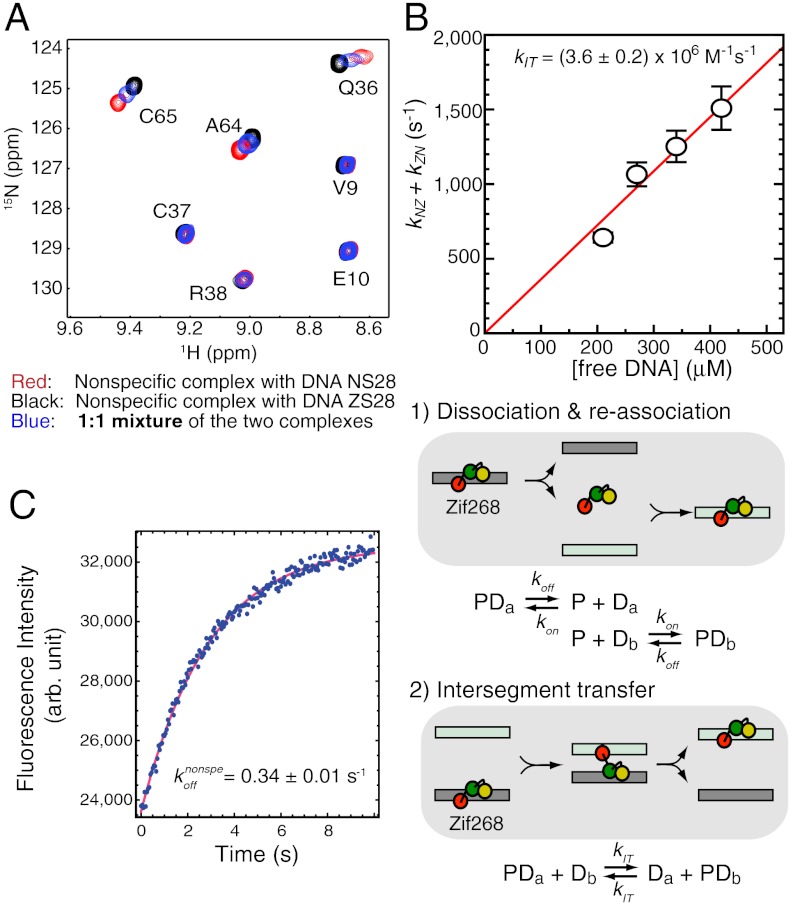
By using an NMR approach described previously (16), we have also analyzed the kinetics of Egr-1’s translocation between two nonspecific DNA duplexes. In this approach, translocation kinetics was analyzed with NMR line-shapes from the three samples: two nonspecific complexes with different 28-bp DNA duplexes and a 1∶1 mixture of these complexes. As shown in Fig. 4A, the translocation processes were found to occur in a fast exchange regime on the NMR chemical shift timescale. Due to additional exchange contributions arising from Egr-1’s translocation between the two DNA duplexes, the NMR line-shapes for the 1∶1 mixture are broader when the difference between the chemical shifts of the two complexes is large. By analyzing the NMR line-shapes for the individual complexes and the mixture, we determined the pseudo first-order rate constants for translocation between two nonspecific DNA duplexes at varying concentrations of DNA (Fig. 4B). Intermolecular translocation between two nonspecific DNA duplexes can occur via two possible mechanisms: (i) dissociation and reassociation and (ii) intersegment transfer (also known as direct transfer) (16, 29–31). Because the rate-limiting step in the former is dissociation when [DNA]≫Kd (note that this inequality leads to kon[DNA]≫koff), the overall rate of protein translocation via this mechanism should be virtually independent of the concentration of free DNA. Intersegment transfer, however, is a second-order reaction whose rate is proportional to the concentration of free DNA. Measurements of the apparent kinetic rate constants for intermolecular translocation at different DNA concentrations permit determination of the second-order rate constant kIT for intersegment transfer. By stopped-flow fluorescence experiment, we have also determined the rate constant koff for Egr-1’s dissociation from nonspecific DNA NS28 to be 0.34 ± 0.01 s-1 (Fig. 4C). From these kinetic data, we determined the second-order rate constant kIT for intersegment transfer between nonspecific DNA molecules to be (3.6 ± 0.2) × 106 M-1·s-1 (Fig. 4B). This second-order rate constant indicates that Egr-1’s intersegment transfer between nonspecific DNA molecules is extremely efficient. In fact, it is > 106-fold faster than intersegment transfer between target DNA sites (kIT,specific = 0.8 M-1·s-1), which was determined in our previous study (32). While the ratio of Kd (nonspecific complex)/Kd (specific complex) is ∼103 for Egr-1, the ratio of kIT(nonspecific)/kIT(specific) is ∼106 for intersegment transfer. This significant discrepancy suggests that weaker affinity alone cannot account for the observed efficient intersegment transfer between nonspecific DNA duplexes. The domain dynamics in the nonspecific complex seems to play an important role in intersegment transfer.
Fig. 4.
Translocation kinetics for Egr-1 bound to nonspecific DNA. (A) Overlay of TROSY spectra recorded for two nonspecific complexes of Egr-1 (with DNA NS28 or ZS28) and a 1∶1 mixture of the two complexes. ZS28 is a 28-bp DNA duplex with a sequence of 5′-GTACCGATAGACGTTTCCGAACCTTCAG-3′. Egr-1’s translocation between the two nonspecific DNA duplexes occurs in a fast exchange regime. (B) Sum of pseudo first-order rate constants for Egr-1’s translocation between two nonspecific DNA duplexes NS28 and ZS28 (kNZ + kZN). The rate constants were calculated by global fitting to NMR line-shape data for amide resonances of Q36, F44, I54, E60, C65, K71, and I84, as described previously (16). These residues exhibit a relatively large chemical shift difference (> 35 Hz) for the two nonspecific complexes. (C) Stopped-flow kinetics data for determination of the dissociation rate constant koff for the nonspecific complex of Egr-1 with NS28. Fluorescence arising from TAMRA conjugated to DNA was monitored immediately after the nonspecific complex with TAMRA-labeled DNA NS28 (40 nM) was mixed with unlabeled DNA NS28 (2,000 nM). The rate constant for intersegment transfer (kIT) was determined from the DNA-concentration dependence of the apparent rate constants for translocation, together with the koff constant.
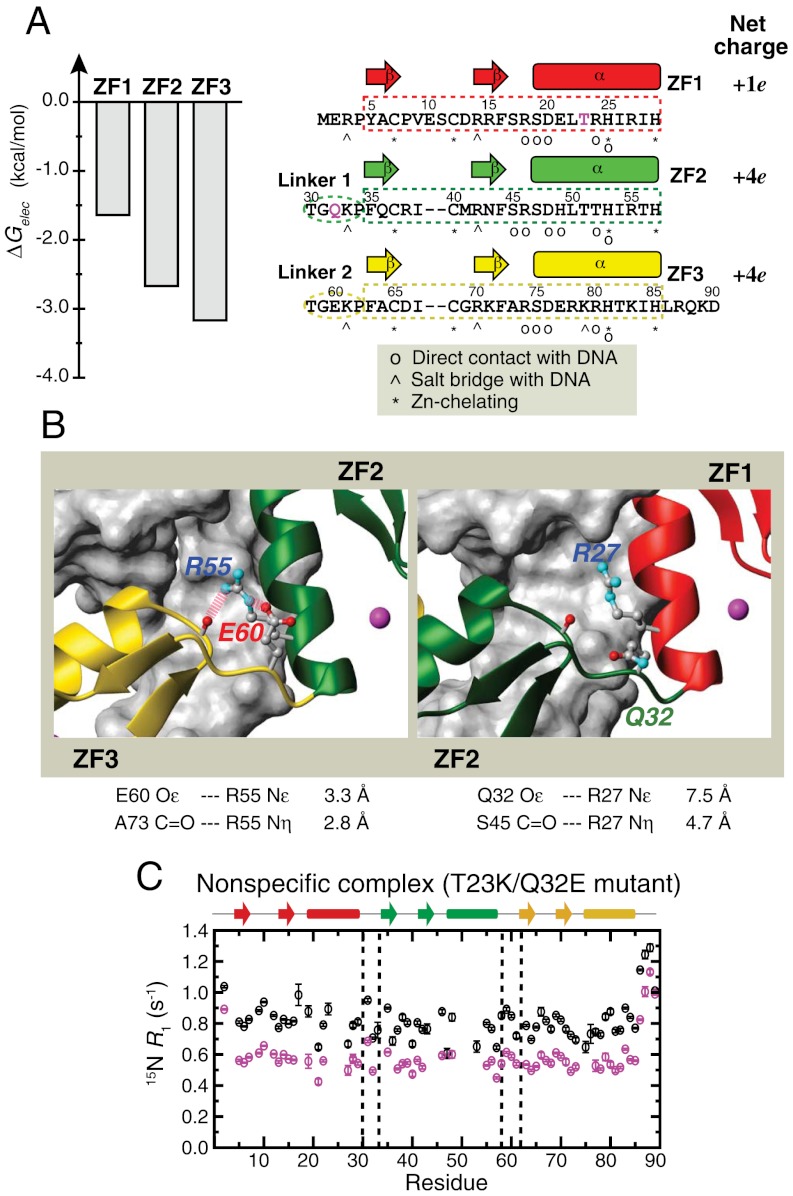
Determinants of ZF1’s Domain Motions in DNA-Scanning.
High degree of domain motions for ZF1 in the nonspecific complex can be ascribed to ZF1’s weaker interactions with DNA and with ZF2. Electrostatic binding free energy calculations (Fig. 5A) suggested that among the three zinc finger domains, ZF1 is the weakest DNA-binder with the lowest absolute value of electrostatic binding free energy (-1.6 kcal/mol as opposed to -3.2 kcal/mol for ZF3). Qualitatively, this is due to the smallest net charge of ZF1 (Fig. 5A). ZF1’s weak interdomain interaction with ZF2 can be another determinant (Fig. 5B). In all of 12 crystal structures available for wild type Egr-1 bound to its target DNA and for engineered Egr-1 proteins bound to their target DNAs (5, 8, 33–36), the interface between ZF2 and ZF3 involves two hydrogen bonds supported by a salt bridge between R55 (ZF2) and E60 (Linker 2); however, the corresponding interdomain interactions are absent between ZF1 and ZF2 presumably because there is no corresponding electrostatic stabilization between R27 (ZF1) and Q32 (Linker 1). To examine whether or not these features make ZF1 highly dynamic in Egr-1 bound to nonspecific DNA, we investigated the domain motions of the T23K/Q32E mutant protein. T23K mutation was intended to enhance ZF1’s electrostatic interaction with DNA by introducing an additional lysine/phosphate salt bridge found at the corresponding position in ZF3 (see Fig. 5A) whereas Q32E mutation was intended to enhance interdomain interaction between ZF1 and ZF2 by introducing a salt bridge corresponding to that found at the ZF2-ZF3 interface (see Fig. 5B). We measured backbone 15N R1 relaxation rates for the T23K/Q32E mutant protein bound to the 28-bp nonspecific DNA NS28 (Fig. 5C). Although ZF1 in the wild type nonspecific complex exhibited clearly elevated R1 rates (Fig. 3B), all zinc finger domains in the T23K/Q32E mutant nonspecific complex exhibited R1 rates in the same range as that for ZF2 and ZF3 in the wild type nonspecific complex. These results suggest that the T23K/Q32E double mutations indeed suppress ZF1’s domain motions in the nonspecific complex to a level comparable to those of ZF2 and ZF3.
Fig. 5.
Determinants of ZF1’s domain motions in Egr-1 bound to nonspecific DNA. (A) Electrostatic binding free energies for Egr-1’s three zinc finger domains calculated using the Adaptive Poisson-Boltzmann Solver (APBS) software package (58) for each domain in the crystal structure of the specific complex (PDB 1AAY). The energy calculation was carried out as described previously (39). The amino acid sequence and net charges of the three zinc-finger domains are also shown. Residues T23 and Q32 are highlighted in magenta. (B) Difference between interdomain interfaces in the crystal structure of the Egr-1/target DNA complex. Two hydrogen bonds and an ion pair stabilize interdomain interactions between ZF2 and ZF3; whereas, there is no corresponding stabilization between ZF1 and ZF2. (C) 15N R1 relaxation rates measured for the T23K/Q32E mutant protein bound to nonspecific DNA NS28. ZF1 of the T23K/Q32E mutant nonspecific complex does not exhibit elevated R1 rates, which is in a clear contrast to the R1 data for the wild type nonspecific complex (Fig. 3B). This suggests that the T23K/Q32E double mutation reduces ZF1’s motions in the nonspecific complex.
Significance of Domain Motions for Intersegment Transfer.
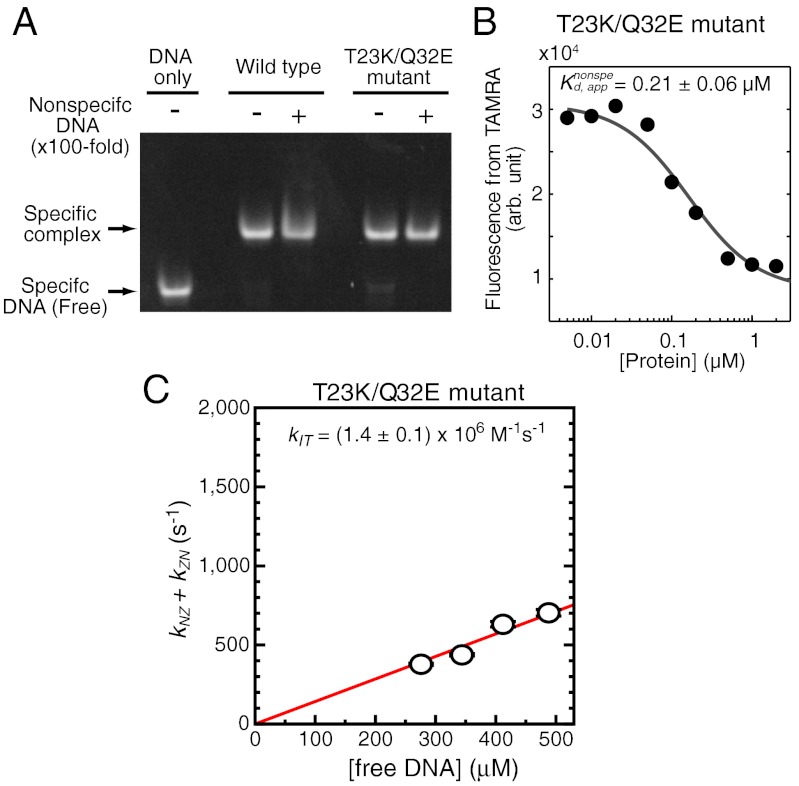
Because T23K/Q32E mutations cause the decrease of ZF1’s domain motions, this mutant protein is suitable for examining the role of ZF1’s domain motions in intersegment transfer. We confirmed that the T23K/Q32E mutant protein retains DNA-binding specificity and affinity to the target DNA (Fig. 6A). Interestingly, the nonspecific DNA-binding affinity of this mutant protein was found to be the same as that of the wild type protein (Fig. 6B), though one may expect a higher affinity for the mutant with more stable interactions between ZF1 and nonspecific DNA. This could be due to an entropy–enthalpy compensation arising from loss of ZF1’s freedom. Using the same NMR approach as that used for the wild type protein (Fig. 4B), we measured the kinetic rate constants for intermolecular translocation of the T23K/Q32E mutant protein between the two 28-bp nonspecific DNA duplexes at varying concentrations (Fig. 6C). From these data, we determined the second-order rate constant kIT for intersegment transfer to be (1.4 ± 0.1) × 106 M-1·s-1. Thus, T23K/Q32E mutations that reduce ZF1’s domain motions caused a 2.6-fold decrease of intersegment transfer efficiency. These results suggest that the domain dynamics is important for intersegment transfer of Egr-1.
Fig. 6.
Impact of T23K/Q32E mutations on DNA-binding specificity, affinity, and intersegment transfer. (A) EMSA competition assay using 100-nM TAMRA-labeled specific 28-bp DNA (the same sequence as SP28) and 200-nM protein in the absence and presence of 10,000-nM unlabeled 28-bp nonspecific DNA NS28. Protein and DNA were mixed in a buffer of 10 mM Tris•HCl (pH 7.5), 40-mM KCl, and 5% glycerol. Shown results indicate that the T23K/Q32E mutant retains affinity and specificity to the target DNA sequence. (B) Fluorescence-based affinity measurement for the interactions between the T23K/Q32E mutant protein and TAMRA-labeled nonspecific 28-bp DNA (the same sequence as NS28). The experimental conditions are the same as those for Fig. 1B. (D) Kinetics of intermolecular translocation of the T23K/Q32E mutant between nonspecific DNA duplexes. The experimental conditions are the same those for Fig. 4B. For the sake of comparison, the kinetic data are shown on the same scale as that for Fig. 4B. Intersegment transfer of the T23K/Q32E mutant protein is 2.6-fold slower than that of the wild type protein.
Egr-1’s Behavior on DNA in CGMD Simulations.
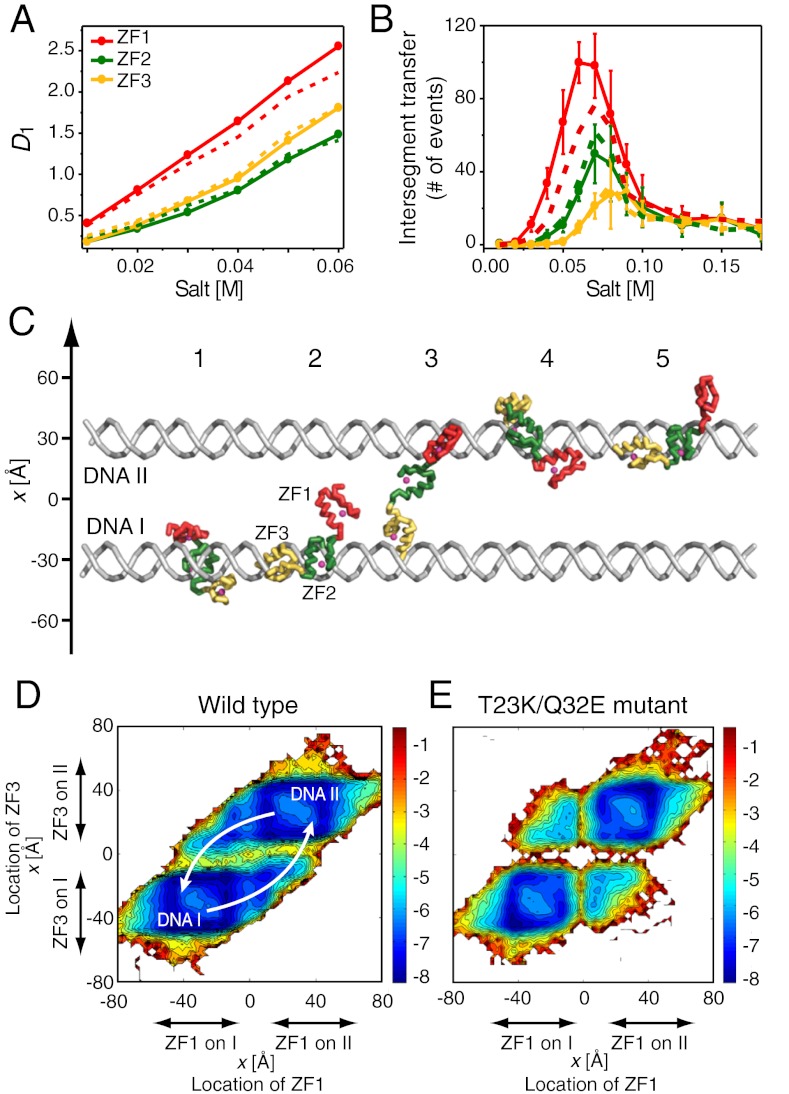
To gain more insight into the target DNA search by Egr-1, we have also studied behaviors of the wild type and T23K/Q32E mutant proteins on nonspecific DNA with coarse-grained molecular dynamics (CGMD) simulations as described by Levy and coworkers (37–41). Unique properties of ZF1 in the wild type Egr-1 protein were evident in the CGMD simulations as well. For example, 1D diffusion coefficients D1 for individual domains of Egr-1 bound to nonspecific DNA suggest that ZF1 is the most mobile of the three zinc finger domains (Fig. 7A). In simulations with two parallel 100-bp DNA duplexes separated by 60 Å, intersegment transfer of Egr-1 between the two DNA duplexes was observed. The intersegment transfer events occurred via an intermediate where an Egr-1 molecule transiently bridges two DNA molecules (Fig. 7C) that follows the “monkey-bar” mechanism (37, 40). For the wild type protein, the bridging events occurred most frequently by ZF1 capturing the other DNA as Fig. 7B and D indicate.
Fig. 7.
Results from the coarse-grained molecular dynamics (CGMD) simulations of Egr-1’s translocation on DNA. (A) 1D diffusion coefficients D1 calculated for individual domains as a function of salt concentration. Because the three domains are tethered, ZF1’s larger D1 indicates that this domain is more mobile than ZF2 and ZF3 in Egr-1 bound to nonspecific DNA. The values of D1 represent displacement of the domains on the DNA during sliding and hopping events as described (41). Continuous and dotted lines represent data for the wild type and T23K/Q32E mutant proteins, respectively. (B) The number of intersegment transfer events between two DNA duplexes (distance, 60 Å) by Egr-1 as a function of salt concentration during the simulation is shown. Transfers between the two DNA duplexes were counted independently for each of the tethered domains. (C) Snapshots of intersegment transfer in the CGMD simulation. Brachiation dynamics between two DNA molecules is promoted by splitting the protein into three regions that allow facilitated jumps with the “monkey-bar” mechanism. (D, E) Free energy landscape of intersegment transfer of Egr-1 from one DNA molecule to another. D and E show results for the wild type and T23K/Q32E mutant proteins, respectively. The free energy surface is projected along the locations of the center of mass of the recognition helices of ZF1 and ZF3 at a salt concentration of 0.06 M. The centers of the DNA molecules are placed at [30,0,z] and [-30,0,z]. The energy landscape data suggest that intersegment transfer of the wild type Egr-1 protein is led by the initial transfer of ZF1, which becomes less efficient by the T23K/Q32E mutation.
The computational results give an excellent explanation on why the T23K/Q32E mutations make Egr-1’s intersegment transfer slower as experimentally demonstrated (Figs. 4B and 6C). Our energy landscape data for the wild type and mutant proteins (Fig. 7 D and E, respectively) suggest that the energy barrier for intersegment transfer via ZF1 is significantly higher for the T23K/Q32E mutant protein making the process slower. Thus, the lower electrostatic affinity of ZF1 to nonspecific DNA increases the probability that it will lead the intersegment transfer of the wild type Egr-1 protein. This supports our previous computational findings: (i) Tethering domains with different affinities to DNA significantly facilitates intersegment transfer in which the domain with the higher affinity to DNA acts as an anchor and the domain with the lower affinity to DNA acts as an explorer (39) and (ii) the net positive charge of the explorer should be modest for optimal intersegment transfer (38). The current simulations show that, to a lesser extent, ZF3 can also locally dissociate from DNA in the nonspecific complex and lead to intersegment transfer. For the T23K/Q32E mutant protein, in which ZF1 is more constrained, efficiencies of intersegment transfers via ZF1 and ZF3 become more comparable in the simulations. These explain why the impact of the T23K/Q32E mutations on intersegment transfer is moderate rather than drastic. The asymmetric domain dynamics of the symmetric (repeat) protein may be governed not only by the overall net charges of individual domains but also by locations of positive and negative charges that affect the electrostatic potential. Studying additional mutants in the future may elucidate how the composition and distribution of charged residues in the binding interface may affect binding affinity, interdomain crosstalks, and consequently, intersegment transfer and DNA search kinetics.
Discussion
Mechanisms that allow transcription factors to find their target sites rapidly in a sea of nonspecific DNA have been the subject of considerable interest in molecular biophysics (7, 14, 15). Despite the long history of the field and a wealth of theoretical studies, very little has been experimentally known about structural details of target DNA search by transcription factors (16, 42, 43). Our present work demonstrates the asymmetrical roles of Egr-1’s three zinc fingers in the dynamic DNA-scanning process and provides important insights into how this mammalian transcription factor can achieve high specificity in binding and rapidity in DNA-scanning.
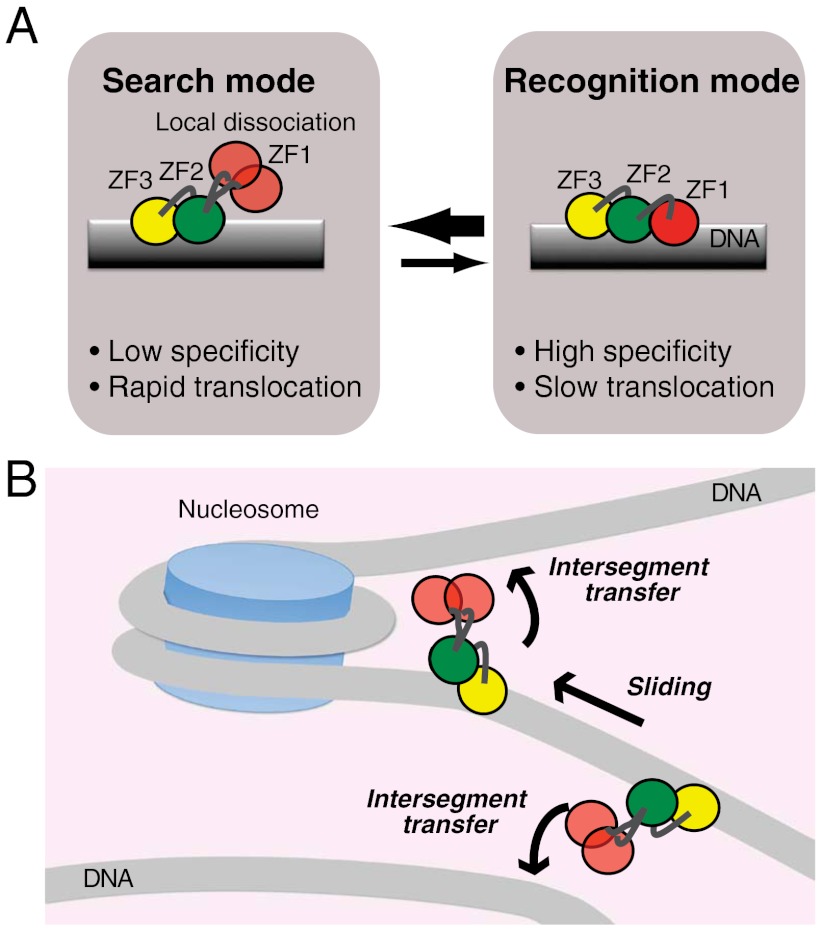
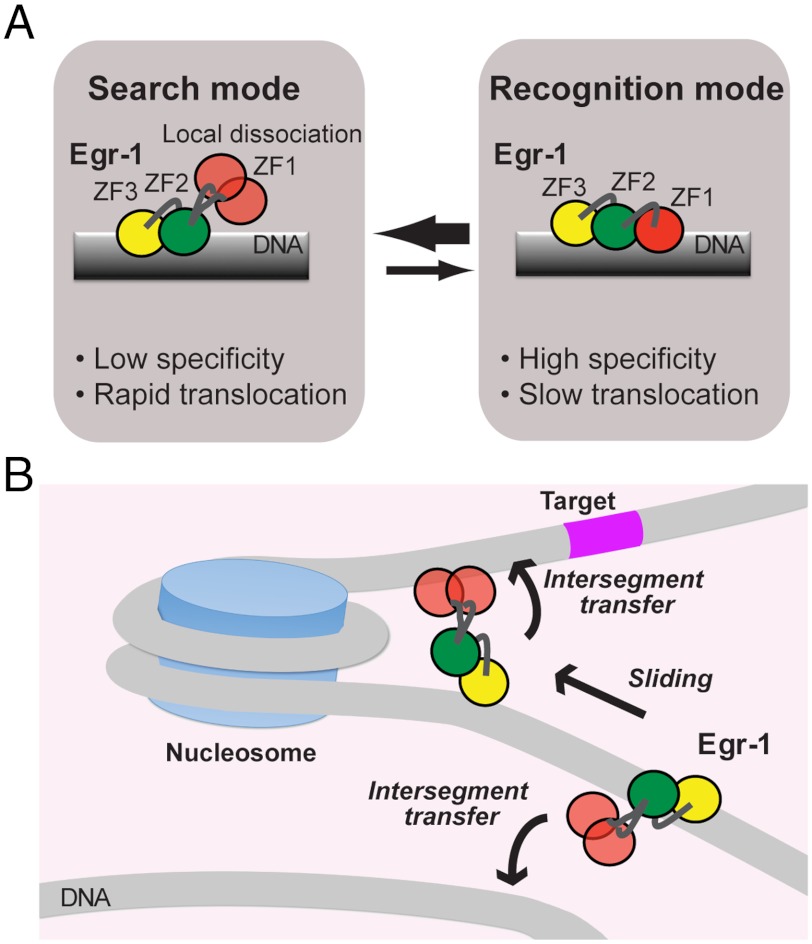
An important finding is that Egr-1’s binding modes for specific and nonspecific DNA duplexes are substantially different in structural and dynamic terms. Based on similar experimental observation for Escherichia coli lac repressor in a pioneering work by Kalodimos et al. (42), some groups theoretically pursued the conformational switch model as a mechanism that resolves the speed-stability paradox (6, 7, 44–46). Our finding for Egr-1 is remarkably consistent with this model, although the structures are completely different for lac repressor and Egr-1. In the conformational switch model, proteins on DNA undergo rapid transitions between two modes: the search mode and recognition mode. The search mode is suitable for proteins’ rapid translocation but unsuitable for high stability and specificity; whereas, the recognition mode is unsuitable for rapid translocation but suitable for high stability and specificity (6, 46). The state observed in the crystal structure of the specific complex with all three zinc finger domains bound to DNA corresponds to the recognition mode. The dynamic state of Egr-1 on nonspecific DNA with only ZF2 and ZF3 being bound corresponds to the search mode (Fig. 8A). Domain motions seem to permit the transitions between the search and recognition modes of Egr-1. This is clearly different from the lac repressor’s case in which the corresponding transitions occur via conformational change in terms of intersubunit orientations and secondary structure of the hinge regions (42). Because of ZF1’s domain motions on a nanosecond timescale, transitions between the two modes can occur more rapidly than translocation. This rapid conformational switch would allow Egr-1 to efficiently scan DNA and specifically locate its target sites.
Fig. 8.
Dynamic mechanism for rapid target search by Egr-1. (A) Dynamic transitions between search and recognition modes via domain dynamics that can allow Egr-1 to resolve the speed-stability paradox. Local dissociation of ZF1 in the search mode can accelerate translocation of Egr-1 on DNA by decreasing energy barriers for sliding and enhancing intersegment transfer. (B) Bypassing a nucleosome via intersegment transfer. Because the two DNA ends of a nucleosome are separated by only ∼60 Å, Egr-1 may bypass nucleosomes via intersegment transfer and carry out continuous scanning.
The domain motions of locally dissociated ZF1 in Egr-1 bound to nonspecific DNA can also promote intersegment transfer via a transient bridging of two DNA molecules in close proximity as observed in the CGMD simulations (Fig. 7 C and D). While ZF2 and ZF3 are bound to the original DNA site and act as anchors, ZF1 acts as an explorer that transiently searches for potential DNA binding sites. In fact, our experimental data (Fig. 4) indicate highly efficient intersegment transfer between nonspecific DNA duplexes. Due to the extremely high DNA density in the nucleus (∼100 mg/mL) (47), intersegment transfer can be kinetically predominant in translocation between distant DNA sites. Even if 90% of the DNA is wound up by nucleosomes, the concentration of naked 30-bp segments in the nucleus is as high as ∼0.5 mM. At this concentration, a pseudo first-order rate constant kIT[DNA] for intersegment transfer is calculated to be ∼1,800 s-1. Thus, Egr-1’s intersegment transfer between nonspecific DNA at physiological concentrations is far faster than translocation via the “dissociation and reassociation” mechanism for which the rate-limiting step is dissociation with a rate constant of 0.3 s-1. Furthermore, we should point out that intersegment transfer might also occur between two DNA segments separated by a nucleosome as depicted in Fig. 8B. Crystal structures show that the two DNA ends of a nucleosome particle are only ∼60 Å apart in 3D space (48). This distance is the same as that used for the CGMD simulations shown in Fig. 7C. So, nucleosomal DNA geometry may allow Egr-1 to bypass nucleosomes effectively via intersegment transfer and to carry out continuous scanning for an efficient target search.
In conclusion, our spectroscopic and computational investigations of the highly dynamic complex between Egr-1 and nonspecific DNA shed light on the asymmetrical roles of zinc finger domains in Egr-1’s scanning of DNA. Although the previous crystallographic studies suggested that all three zinc finger domains are involved in recognition of a target DNA site, our NMR data indicate that ZF1 is mainly dissociated from DNA while Egr-1 is scanning nonspecific DNA. ZF1’s domain motions on a nanosecond timescale in the nonspecific complex permit dynamic transitions between the search and recognition modes thereby allowing Egr-1 to achieve high specificity in recognition and rapidity in scanning. The mechanism for Egr-1’s transitions between the search and recognition modes is completely different from those for lac repressor (42) and p53 (49, 50). Egr-1’s search mode is made primarily via local dissociation of ZF1 from DNA whereas more positively charged and tightly packed ZF2 and ZF3 are bound to nonspecific DNA. This search mode of Egr-1 seems suitable for intersegment transfer that involves a transient bridging of two DNA sites. The new insights into DNA-scanning by Egr-1 can help understand how other zinc finger proteins (the most common class of transcription factors in human genome) (51) find their target DNA sites in the sea of nonspecific DNA sites. The present work also opens up a way to modulate rationally zinc finger proteins’ search kinetics without perturbing DNA-binding specificity and affinity. In particular, modulation of the domain net charge and flexibility may allow us to optimize the efficiency of DNA-scanning by zinc finger proteins. This aspect is important because the zinc finger nuclease technology, which is becoming popular (yet not always successful) for in vivo gene manipulation (52), may be improved by such rational optimization of the target search efficiency.
Methods
Preparation of Nonspecific and Specific Egr-1/DNA Complexes.
2H/15N- or 2H/13C/15N-labeled Egr-1 DNA-binding domain was expressed in E. coli cultured in D2O-based minimal media supplemented by 0.5 g/L ISOGRO (Sigma-Aldrich). The protein was purified as described previously (32). A plasmid for gene expression of the T23K/Q32E mutant protein was made with the Quick Change Lightening site-directed mutagenesis kit (Agilent). The mutant protein was prepared in the same way as that for the wild type protein. Individual DNA strands for NMR studies were chemically synthesized and purified by anion-exchange chromatography. After annealing, 28-bp DNA duplexes were purified via anion exchange chromatography to remove excess single-stranded DNA. To prepare complexes, the Egr-1 protein was mixed with 28-bp DNA at a high ionic strength (∼0.5 M NaCl), and the buffer was exchanged to reduce the ionic strength. NMR samples contained 0.8 mM complex, 10 mM d11-Tris•HCl (pH 7.5), 20 mM KCl, and 7% D2O. The TAMRA-labeled DNA NS28 (for Figs. 1B, 4C, and 6B) and the TAMRA-labeled DNA SP28 (for Fig. 6A) were prepared from chemically synthesized DNA strands purified by PAGE. The labeled duplexes were prepared by annealing and subsequent PAGE purification using a 4–20% gradient gel.
NMR Spectroscopy.
All NMR experiments for Egr-1/DNA complexes were performed at 35 °C. NMR experiments for Egr-1 in the free state were performed at 25 °C. The lower temperature for the free protein was necessary due to its lower stability. Backbone 1H/13C/15N resonances of the specific and nonspecific Egr-1/DNA complexes and the free protein were assigned by HNCO, HN(CA)CO, HNCA, HN(CO)CA, HNCACB, and HN(CO)CACB spectra (20). 15N relaxation rates R1 and heteronuclear NOE were measured at 1H-frequencies of 600 and 800 MHz as previously described (53). 15N CPMG R2 relaxation dispersion experiments were performed at a 1H-frequency of 800 MHz using the CW-CPMG scheme (54). Residual dipolar couplings 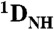 arising from partial molecular alignment due to 8-mg/mL Pf1 phage were measured for the specific and nonspecific complexes by using IPAP-HSQC spectra (55) recorded at a 1H frequency of 600 MHz. RDC data were analyzed using the PALES program (17, 23). Other details of the RDC analysis are given in the SI Text. Kinetic rate constants for translocation of Egr-1 between two 28-bp nonspecific DNA duplexes NS28 and ZS28 were determined from HSQC spectra recorded independently for two different nonspecific complexes and their 1∶1 mixture by using the NMR line-shape analysis as described previously (16).
arising from partial molecular alignment due to 8-mg/mL Pf1 phage were measured for the specific and nonspecific complexes by using IPAP-HSQC spectra (55) recorded at a 1H frequency of 600 MHz. RDC data were analyzed using the PALES program (17, 23). Other details of the RDC analysis are given in the SI Text. Kinetic rate constants for translocation of Egr-1 between two 28-bp nonspecific DNA duplexes NS28 and ZS28 were determined from HSQC spectra recorded independently for two different nonspecific complexes and their 1∶1 mixture by using the NMR line-shape analysis as described previously (16).
Fluorescence Spectroscopy.
Fluorescence experiments were performed at 20 °C with an ISS PC-1 spectrofluorometer. To determine the apparent dissociation constant Kd for NS28, fluorescence arising from 4 nM TAMRA-labeled DNA in a buffer of 10 mM Tris•HCl (pH 7.5) and 40 mM KCl was monitored as a function of protein concentration (0–5,000 nM). Stopped-flow experiments were conducted for measuring the dissociation rate constant koff for the nonspecific complex of Egr-1 with NS28. The time courses of fluorescence arising from TAMRA-labeled DNA were monitored immediately after mixing two solutions: (i) 40 nM TAMRA-labeled DNA and 400 nM protein and (ii) 2000 nM unlabeled DNA duplex NS28. The majority of TAMRA-labeled DNA duplex was bound to the protein in Solution 1 under this condition (see Fig. 1B). An Applied Photophysics Rx2000 stopped-flow device was used for rapid mixing. In the stopped-flow experiment, the excitation wavelength was 500 nm and the emissions were detected using a channel with a 560-nm-cutoff long-pass filter (Edmund Optics) without a monochromator for a higher sensitivity. The dissociation rate constant koff was determined via nonlinear least squares fitting to a monoexponential function (56).
Coarse-Grained Molecular Dynamics Simulations.
The dynamic nature of DNA scanning by Egr-1 was studied using a reduced model (37, 41) that allows sampling of long timescale processes such as sliding, hopping, 3D diffusion, and intersegment transfer. We modeled the DNA as having three beads per nucleotide representing the phosphate, sugar, and base. Each bead was located at the geometric center of the group it represents, and a negative point charge was assigned to beads representing the DNA phosphate groups. In the simulations, a 100 bp B-DNA molecule was used to study protein diffusion, and two 100-bp B-DNA separated by 60 Å molecules were used for investigation of intersegment transfer. The DNA remained in place and rigid throughout the simulations. The protein was represented by a single bead for each residue located at the Cα of that residue. Beads representing charged amino acids (Lys, Arg, Asp, and Glu) were charged in the model. Unlike the DNA, the protein remained flexible during the simulations and could undergo folding and unfolding events. Nonspecific protein–DNA interactions were modeled by electrostatic interactions between all charged residues of the protein and the phosphate beads of the DNA using the Debye–Huckel potential, which accounts for the ionic strength of a solute immersed in aqueous solution. The dynamics of each protein were studied at salt concentrations in the range of 0.01–0.2 M using a dielectric constant of 40 and a temperature at which the protein is completely folded. More details of the simulation can be found in previous publications from Levy’s group (37–41). Trajectories were analyzed to calculate the percentage of protein sliding and hopping, structural features during sliding, the number of intersegment transfer events, the linear diffusion coefficient (D1), and electrostatic free energies as described previously (39, 41).
Supplementary Material
ACKNOWLEDGMENTS.
We thank Dr. Markus Zweckstetter for providing the program PALES; Dr. Wlodek Bujalowski for advice in fluorescence studies; Dr. Tianzhi Wang for maintaining the NMR instruments; and Dr. Wayne Bolen for his leadership that rebuilt and enhanced SCSBMB’s infrastructure from devastation by a hurricane. Y.L. is grateful for the support by the Kimmelman Center Macromolecular Assemblies. Y.L. is the incumbent of the Lillian and George Lyttle Career Development Chair. This work was supported by Grant MCB-0920238 from the National Science Foundation (to J.I.), Grant 12BGIA8960032 from the American Heart Association (to J.I.), Grant 2010424 from the United States—Israel Binational Science Foundation (to Y.L. and J.I.), and funding from the Minerva Foundation with funding from the Federal German Ministry for Education and Research (to Y.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 10146 (volume 109, number 26).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1121500109/-/DCSupplemental.
References
- 1.Bozon B, Davis S, Laroche S. A requirement for the immediate early gene zif268 in reconsolidation of recognition memory after retrieval. Neuron. 2003;40:695–701. doi: 10.1016/s0896-6273(03)00674-3. [DOI] [PubMed] [Google Scholar]
- 2.Lee JL, Everitt BJ, Thomas KL. Independent cellular processes for hippocampal memory consolidation and reconsolidation. Science. 2004;304:839–843. doi: 10.1126/science.1095760. [DOI] [PubMed] [Google Scholar]
- 3.Khachigian LM, Lindner V, Williams AJ, Collins T. Egr-1-induced endothelial gene expression: A common theme in vascular injury. Science. 1996;271:1427–1431. doi: 10.1126/science.271.5254.1427. [DOI] [PubMed] [Google Scholar]
- 4.Yan SF, et al. Egr-1, a master switch coordinating upregulation of divergent gene families underlying ischemic stress. Nat Med. 2000;6:1355–1361. doi: 10.1038/82168. [DOI] [PubMed] [Google Scholar]
- 5.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: Crystal structure of a Zif268-DNA complex at 2.1 Å. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 6.Slutsky M, Mirny LA. Kinetics of protein–DNA interaction: Facilitated target location in sequence-dependent potential. Biophys J. 2004;87:4021–4035. doi: 10.1529/biophysj.104.050765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mirny L, et al. How a protein searches for its site on DNA: the mechanism of facilitated diffusion. J Phys A: Math Theor. 2009;42:434013–434035. [Google Scholar]
- 8.Elrod-Erickson M, Rould MA, Nekludova L, Pabo CO. Zif268 protein–DNA complex refined at 1.6 Å: A model system for understanding zinc finger–DNA interactions. Structure. 1996;4:1171–1180. doi: 10.1016/s0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]
- 9.Foster MP, et al. Domain packing and dynamics in the DNA complex of the N-terminal zinc fingers of TFIIIA. Nat Struct Biol. 1997;4:605–608. doi: 10.1038/nsb0897-605. [DOI] [PubMed] [Google Scholar]
- 10.Laity JH, Dyson HJ, Wright PE. Molecular basis for modulation of biological function by alternate splicing of the Wilms’ tumor suppressor protein. Proc Natl Acad Sci USA. 2000;97:11932–11935. doi: 10.1073/pnas.97.22.11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laity JH, Dyson HJ, Wright PE. DNA-induced alpha-helix capping in conserved linker sequences is a determinant of binding affinity in Cys2-His2 zinc fingers. J Mol Biol. 2000;295:719–727. doi: 10.1006/jmbi.1999.3406. [DOI] [PubMed] [Google Scholar]
- 12.Stoll R, et al. Structure of the Wilms tumor suppressor protein zinc finger domain bound to DNA. J Mol Biol. 2007;372:1227–1245. doi: 10.1016/j.jmb.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 13.Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 14.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Hippel PH, Berg OG. Facilitated target location in biological systems. J Biol Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 16.Iwahara J, Zweckstetter M, Clore GM. NMR structural and kinetic characterization of a homeodomain diffusing and hopping on nonspecific DNA. Proc Natl Acad Sci USA. 2006;103:15062–15067. doi: 10.1073/pnas.0605868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zweckstetter M, Hummer G, Bax A. Prediction of charge-induced molecular alignment of biomolecules dissolved in dilute liquid-crystalline phases. Biophys J. 2004;86:3444–3460. doi: 10.1529/biophysj.103.035790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamilton TB, Borel F, Romaniuk PJ. Comparison of the DNA binding characteristics of the related zinc finger proteins WT1 and EGR1. Biochemistry. 1998;37:2051–2058. doi: 10.1021/bi9717993. [DOI] [PubMed] [Google Scholar]
- 19.Iwahara J, Schwieters CD, Clore GM. Characterization of nonspecific protein–DNA interactions by 1H paramagnetic relaxation enhancement. J Am Chem Soc. 2004;126:12800–12808. doi: 10.1021/ja046246b. [DOI] [PubMed] [Google Scholar]
- 20.Pervushin K. Impact of transverse relaxation optimized spectroscopy (TROSY) on NMR as a technique in structural biology. Q Rev Biophys. 2000;33:161–197. doi: 10.1017/s0033583500003619. [DOI] [PubMed] [Google Scholar]
- 21.Hansen MR, Mueller L, Pardi A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat Struct Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- 22.Braddock DT, Cai M, Baber JL, Huang Y, Clore GM. Rapid identification of medium-to large-scale interdomain motion in modular proteins using dipolar couplings. J Am Chem Soc. 2001;123:8634–8635. doi: 10.1021/ja016234f. [DOI] [PubMed] [Google Scholar]
- 23.Zweckstetter M. NMR: Prediction of molecular alignment from structure using the PALES software. Nat Protoc. 2008;3:679–690. doi: 10.1038/nprot.2008.36. [DOI] [PubMed] [Google Scholar]
- 24.Gorman J, Greene EC. Visualizing one-dimensional diffusion of proteins along DNA. Nat Struct Mol Biol. 2008;15:768–774. doi: 10.1038/nsmb.1441. [DOI] [PubMed] [Google Scholar]
- 25.Loria JP, Rance M, Palmer AG. A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J Am Chem Soc. 1999;121:2331–2332. [Google Scholar]
- 26.Tollinger M, Skrynnikov NR, Mulder FAA, Forman-Kay JD, Kay LE. Slow dynamics in folded and unfolded states of an SH3 domain. J Am Chem Soc. 2001;123:11341–11352. doi: 10.1021/ja011300z. [DOI] [PubMed] [Google Scholar]
- 27.Baber JL, Szabo A, Tjandra N. Analysis of slow interdomain motion of macromolecules using NMR relaxation data. J Am Chem Soc. 2001;123:3953–3959. doi: 10.1021/ja0041876. [DOI] [PubMed] [Google Scholar]
- 28.Clore GM, et al. Deviations from the simple 2-parameter model-free approach to the interpretation of N-15 nuclear magnetic-relaxation of proteins. J Am Chem Soc. 1990;112:4989–4991. [Google Scholar]
- 29.Fried MG, Crothers DM. Kinetics and mechanism in the reaction of gene regulatory proteins with DNA. J Mol Biol. 1984;172:263–282. doi: 10.1016/s0022-2836(84)80026-1. [DOI] [PubMed] [Google Scholar]
- 30.Iwahara J, Clore GM. Direct observation of enhanced translocation of a homeodomain between DNA cognate sites by NMR exchange spectroscopy. J Am Chem Soc. 2006;128:404–405. doi: 10.1021/ja056786o. [DOI] [PubMed] [Google Scholar]
- 31.Lieberman BA, Nordeen SK. DNA intersegment transfer, how steroid receptors search for a target site. J Biol Chem. 1997;272:1061–1068. doi: 10.1074/jbc.272.2.1061. [DOI] [PubMed] [Google Scholar]
- 32.Takayama Y, Sahu D, Iwahara J. NMR studies of translocation of the Zif268 protein between its target DNA Sites. Biochemistry. 2010;49:7998–8005. doi: 10.1021/bi100962h. [DOI] [PubMed] [Google Scholar]
- 33.Elrod-Erickson M, Benson TE, Pabo CO. High-resolution structures of variant Zif268-DNA complexes: Implications for understanding zinc finger-DNA recognition. Structure. 1998;6:451–464. doi: 10.1016/s0969-2126(98)00047-1. [DOI] [PubMed] [Google Scholar]
- 34.Miller JC, Pabo CO. Rearrangement of side-chains in a Zif268 mutant highlights the complexities of zinc finger-DNA recognition. J Mol Biol. 2001;313:309–315. doi: 10.1006/jmbi.2001.4975. [DOI] [PubMed] [Google Scholar]
- 35.Peisach E, Pabo CO. Constraints for zinc finger linker design as inferred from X-ray crystal structure of tandem Zif268-DNA complexes. J Mol Biol. 2003;330:1–7. doi: 10.1016/s0022-2836(03)00572-2. [DOI] [PubMed] [Google Scholar]
- 36.Wolfe SA, Grant RA, Elrod-Erickson M, Pabo CO. Beyond the “recognition code”: Structures of two Cys2His2 zinc finger/TATA box complexes. Structure. 2001;9:717–723. doi: 10.1016/s0969-2126(01)00632-3. [DOI] [PubMed] [Google Scholar]
- 37.Vuzman D, Azia A, Levy Y. Searching DNA via a “Monkey Bar” mechanism: The significance of disordered tails. J Mol Biol. 2010;396:674–684. doi: 10.1016/j.jmb.2009.11.056. [DOI] [PubMed] [Google Scholar]
- 38.Vuzman D, Levy Y. DNA search efficiency is modulated by charge composition and distribution in the intrinsically disordered tail. Proc Natl Acad Sci USA. 2010;107:21004–21009. doi: 10.1073/pnas.1011775107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vuzman D, Polonsky M, Levy Y. Facilitated DNA search by multidomain transcription factors: cross talk via a flexible linker. Biophys J. 2010;99:1202–1211. doi: 10.1016/j.bpj.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vuzman D, Levy Y. Intrinsically disordered regions as affinity tuners in protein–DNA interactions. Mol BioSyst. 2012;8:47–57. doi: 10.1039/c1mb05273j. [DOI] [PubMed] [Google Scholar]
- 41.Givaty O, Levy Y. Protein sliding along DNA: Dynamics and structural characterization. J Mol Biol. 2009;385:1087–1097. doi: 10.1016/j.jmb.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 42.Kalodimos CG, et al. Structure and flexibility adaptation in nonspecific and specific protein–DNA complexes. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 43.Takayama Y, Clore GM. Intra-and intermolecular translocation of the bi-domain transcription factor Oct1 characterized by liquid crystal and paramagnetic NMR. Proc Natl Acad Sci USA. 2011;108:E169–176. doi: 10.1073/pnas.1100050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marcovitz A, Levy Y. Frustration in protein–DNA binding influences conformational switching and target search kinetics. Proc Natl Acad Sci USA. 2011;108:17957–17962. doi: 10.1073/pnas.1109594108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murugan R. Theory of site-specific DNA-protein interactions in the presence of conformational fluctuations of DNA binding domains. Biophys J. 2010;99:353–359. doi: 10.1016/j.bpj.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou HX. Rapid search for specific sites on DNA through conformational switch of nonspecifically bound proteins. Proc Natl Acad Sci USA. 2011;108:8651–8656. doi: 10.1073/pnas.1101555108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lewin B. Genes VII. Oxford: Oxford University Press; 2000. [Google Scholar]
- 48.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 49.Tafvizi A, Huang F, Fersht AR, Mirny LA, van Oijen AM. A single-molecule characterization of p53 search on DNA. Proc Natl Acad Sci USA. 2011;108:563–568. doi: 10.1073/pnas.1016020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khazanov N, Levy Y. Sliding of p53 along DNA can be modulated by its oligomeric state and by cross-talks between its constituent domains. J Mol Biol. 2011;408:335–55. doi: 10.1016/j.jmb.2011.01.059. [DOI] [PubMed] [Google Scholar]
- 51.Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 52.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 53.Palmer AG., 3rd NMR probes of molecular dynamics: Overview and comparison with other techniques. Annu Rev Biophys Biomol Struct. 2001;30:129–155. doi: 10.1146/annurev.biophys.30.1.129. [DOI] [PubMed] [Google Scholar]
- 54.Hansen DF, Vallurupalli P, Kay LE. An improved 15N relaxation dispersion experiment for the measurement of millisecond time-scale dynamics in proteins. J Phys Chem B. 2008;112:5898–5904. doi: 10.1021/jp074793o. [DOI] [PubMed] [Google Scholar]
- 55.Ottiger M, Delaglio F, Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J Magn Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 56.Perez-Howard GM, Weil PA, Beechem JM. Yeast TATA binding protein interaction with DNA: fluorescence determination of oligomeric state, equilibrium binding, on-rate, and dissociation kinetics. Biochemistry. 1995;34:8005–8017. doi: 10.1021/bi00025a006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grzesiek S, et al. Refined solution structure and backbone dynamics of HIV-1 Nef. Protein Sci. 1997;6:1248–1263. doi: 10.1002/pro.5560060613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baker NA, Sept D, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]