Abstract
Fe-S clusters are critical prosthetic groups for proteins involved in various critical biological processes. Before being transferred to recipient apo-proteins, Fe-S clusters are assembled on the highly conserved scaffold protein Isu, the abundance of which is regulated posttranslationally on disruption of the cluster biogenesis system. Here we report that Isu is degraded by the Lon-type AAA+ ATPase protease of the mitochondrial matrix, Pim1. Nfs1, the cysteine desulfurase responsible for providing sulfur for cluster formation, is required for the increased Isu stability occurring after disruption of cluster formation on or transfer from Isu. Physical interaction between the Isu and Nfs1 proteins, not the enzymatic activity of Nfs1, is the important factor in increased stability. Analysis of several conditions revealed that high Isu levels can be advantageous or disadvantageous, depending on the physiological condition. During the stationary phase, elevated Isu levels were advantageous, resulting in prolonged chronological lifespan. On the other hand, under iron-limiting conditions, high Isu levels were deleterious. Compared with cells expressing normal levels of Isu, such cells grew poorly and exhibited reduced activity of the heme-containing enzyme ferric reductase. Our results suggest that modulation of the degradation of Isu by the Pim1 protease is a regulatory mechanism serving to rapidly help balance the cell’s need for critical iron-requiring processes under changing environmental conditions.
Keywords: proteolysis, Hsp70, aging, Saccharomyces cerevisiae
Iron, a critical element for virtually all living organisms, has a number of essential cellular roles. For example, heme, a critical iron-containing prosthetic group whose first and last biosynthetic steps occur in mitochondria, serves important roles in oxygen sensing and electron transfer (1). Most pertinent to the present work, iron-sulfur (Fe-S) clusters are a versatile prosthetic group found on proteins that play crucial roles in electron transport, catalytic reactions, environmental sensing, and regulation (1, 2). Given that iron enhances the production of intracellular reactive oxygen species that damage cellular macromolecules, it is not surprising that the level of cellular iron is tightly regulated. For example, under iron-limited conditions, the Aft transcription factors of the budding yeast Saccharomyces cerevisiae activate the so-called “iron regulon,” increasing expression of proteins important for cellular iron metabolism, including high-affinity iron transporters of the plasma and mitochondrial membranes (3).
A single Fe-S cluster biogenesis system closely related to the iron-sulfur cluster (ISC) assembly system of bacteria exists in the mitochondria of eukaryotes (2, 4). In both eukaryotes and prokaryotes, Fe-S clusters are not built de novo on proteins, but rather are first assembled on a scaffold protein and then transferred to recipient proteins. In S. cerevisiae, the scaffold Isu is encoded by two very closely related genes, ISU1 and ISU2, of which ISU1 is the more highly expressed (5). In the assembly process, a cysteine desulfurase, Nfs1, provides sulfur to Isu via the conserved cysteine located in the active site (6, 7). The ferredoxin Yah1 and ferredoxin reductase Arh1 supply the required electrons (8). Yfh1, the yeast frataxin homolog, has been proposed to serve as an iron donor and/or a regulator of Nfs1 function (9–11). Isu, Nfs1, and Yfh1 can bind with one another to form a so-called “Fe-S cluster assembly” complex (11–13). Transfer of the assembled clusters to recipient proteins requires a specialized Hsp70 chaperone system composed of the Hsp70 Ssq1 and the J-protein cochaperone Jac1, as well as the nucleotide release factor Mge1 (14, 15). The physical and functional interactions between Isu (a small 14-kDa protein) and the components critical for cluster assembly and transfer are just beginning to be understood. The conserved tripeptide PVK (residues 134–136 in Isu) is known to be critical for interaction with Ssq1. Alterations of single residues in this motif disrupt the interaction and result in cell inviability (16). The interface between Isu and Nfs1 is complex. Structural analysis of crystals of a complex formed between the Escherichia coli proteins IscU and IscS (orthologs of Isu and Nfs1, respectively) revealed a large interaction surface (17).
Along with generating Fe-S clusters per se, the mitochondrial ISC system is central to cellular iron homeostasis. Activity of the Aft transcription factors is negatively regulated by an as-yet unidentified signal generated by the ISC system and exported to the cytosol (18). Consistent with this idea, mutations that affect the functions of components of the ISC system, such as Ssq1 and Jac1 of the specialized Hsp70 cluster transfer system, result in Aft activation. Pertinent to this report, Isu levels are 10- to 15-fold higher in jac1 and ssq1 mutants than in WT cells. However, this up-regulation is due not to Aft activity, but rather to an increase in the stability of ISU protein (5). To better understand the mechanism behind the control of Isu levels, as well as the biological relevance of this control, we assessed the requirements for stabilization of Isu and the cellular consequence of higher Isu levels. We found that up-regulation of Isu, a substrate of the Pim1 protease, can be advantageous or disadvantageous depending on the physiological context, providing a biological rationale for posttranslational regulation of Isu.
Results
Isu Is a Substrate of the Pim1 Protease.
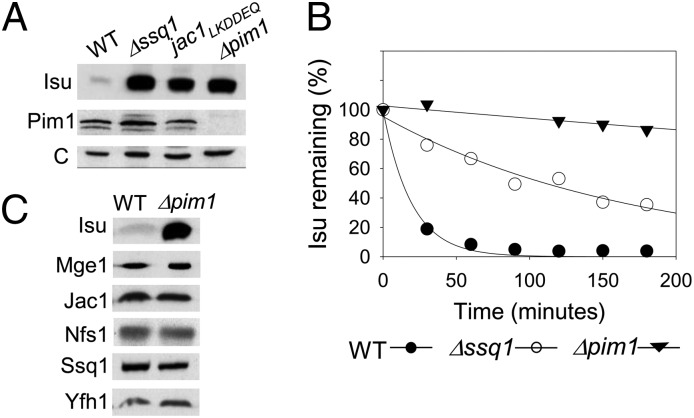
To better understand the posttranslational regulation of Isu, we sought to identify the protease responsible for its degradation. We first tested a strain lacking Pim1, the abundant ATP-dependent Lon-type protease of the mitochondria matrix (19). As determined by immunoblot analysis, the Isu level was ∼20-fold higher in lysates from Δpim1 than from WT cells, comparable to the increased Isu levels in lysates of SSQ1 and JAC1 mutants known to have defects in Fe-S cluster biogenesis (5) (Fig. 1A).
Fig. 1.
Degradation of Isu by Pim1. (A) Cell lysates of indicated strains were subjected to immunoblot analysis using Isu, Pim1, and, as a control, Mge1-specific antibodies (c). (B) Cells were pulse-labeled with [35S] amino acids. Lysates prepared from aliquots removed at indicated times after initiation of the chase were subjected to IP with Isu-specific antibodies. The amount of labeled Isu at time 0 was set at 100%. (C) Mitochondrial lysates from WT and Δpim1 cells were analyzed by immunoblotting using antibodies specific for the indicated proteins.
To confirm that the high Isu levels were due to slow degradation, we performed pulse-chase analysis. After a 2-min incubation with [35S] amino acids, an excess of nonradioactive amino acids was added. The decrease in radiolabeled Isu was monitored by immunoprecipitation (IP) using Isu-specific antibodies (Fig. 1B). As expected (5), Isu was rapidly degraded in WT cells, with only 20% remaining at the 30-min time point. Over the course of the experiment, Isu was so stable in Δpim1 cells that a half-life could not be determined; radiolabeled Isu did not drop below 90% of initial levels during the 200-min chase. In comparison, in Δssq1 cells, Isu had a half-life of ∼110 min. Jac1, Ssq1, Mge1, Nfs1, and Yfh1 levels were not significantly different in Δpim1 and WT cells (Fig. 1C), indicating that Isu is a specific target within the Fe-S cluster biogenesis system of the Pim1 protease.
To test whether Pim1 activity per se is altered in cells defective in Fe-S cluster biogenesis, we assessed the level of a known Pim1 substrate, the ribosomal protein Mrs20 (20). Although Mrp20 levels were increased in Δpim1 cells, as expected, they were not increased in Δssq1 cells (Fig. S1). Thus, our data do not indicate that altered Pim1 activity is the cause of Isu stabilization, suggesting that a change within the Fe-S cluster biogenesis system itself is responsible.
Stabilization of Isu1 Variants.
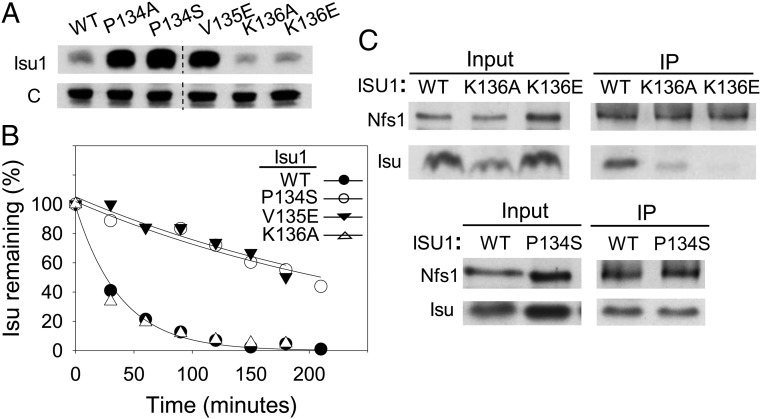
Given that the absence of Ssq1 results in stabilization of Isu, we next tested Isu1 variants with alterations of the well-defined PVK Ssq1 binding site. Because Hsp70 binding is essential, we constructed a strain expressing WT Isu fused to Myc epitopes. The Isu1–Myc fusion supported viability, but migrated more slowly during electrophoresis (Fig. S2), allowing assessment of the abundance and stability of Isu variants. We tested the following mutants encoding a single alteration: P134A, P134S, V135E, K136A, and K136E. Those with alterations at residues P134 and V135 were present at 5- to 20-fold higher levels than WT protein (Fig. 2A). The level of K136 variants did not differ significantly from that of WT Isu.
Fig. 2.
Effect of amino acid alterations on Isu1’s stability and association with Nfs1. (A) Lysates of cells expressing WT Isu1 or variants with indicated alterations were subjected to immunoblot analysis using antibodies specific for Isu and, as a control, Mge1 (c). Dotted line indicates position of a lane, which is not relevant to this report, removed from the image. (B) Cells were pulse-labeled with [35S] amino acids. Lysates prepared from aliquots removed at indicated times after initiation of the chase were subjected to IP with Isu-specific antibodies. The amount of labeled Isu at time 0 was set at 100%. (C) Mitochondrial lysates of cells expressing WT Isu1 or an indicated variant were subjected to IP using Nfs1-specific antibodies. Precipitated Nfs1 and Isu1 (IP) were detected by immunoblot analysis using specific antibodies, with 4% mitochondrial lysate used as a loading control (Input). Δisu2 ISU1-myc background was used to eliminate comigration of WT Isu1 with variants. In C, As discussed in the text, Δpim1 was included to ensure comparable protein levels.
To determine the basis of the difference, we compared the protein degradation rates of three variants with that of WT Isu using pulse-chase analysis (Fig. 2B). Degradation of Isu1K136A was indistinguishable from that of WT Isu1; however, the half-lives of Isu1P134S and Isu1V135E exceeded 200 min, consistent with their higher steady-state levels.
In light of previous results suggesting that the increase in Isu levels in the absence of Ssq1 requires the cysteine desulfurase Nfs1 (5), we compared the interaction of Isu1 variants with Nfs1 in mitochondrial lysates. Initially such comparisons proved difficult, because Nfs1 levels are significantly higher than Isu levels and thus more Nfs1–Isu complex is formed when Isu is present at higher levels (Fig. S3). Therefore, we took advantage of the stabilization of Isu in the Δpim1 background to assess the interaction between Nfs1 and Isu1. We performed coimmunoprecipitation (co-IP) experiments using antibodies specific for Nfs1, followed by detection of Isu by immunoblot analysis. The amounts of Isu1P134S and WT Isu coimmunoprecipitated with Nfs1 were not significantly different (Fig. 2C); however, the efficiency of co-IP of Isu1K136A and Isu1K136E were only ∼30% and 5%, respectively, of that seen for WT Isu1. These results suggest that residue K136 is important for interaction with Nfs1, as well as Ssq1. Consistent with this idea, structural analysis indicates that the analogous lysine residue of the E. coli ortholog K103IscU is in close proximity to IscS in the IscS–IscU complex (17).
Increased Isu1 Stability in the Presence of Nfs1 Variant Lacking Cysteine Desulfurase Activity.
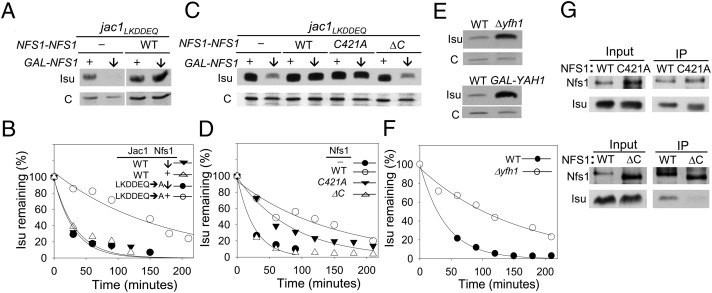
We next directly tested the idea that Nfs1 interaction is necessary for Isu stabilization. The rationale behind our approach was to engineer a strain background in which (i) Isu was stabilized due to a mutation in the JAC1 gene and (ii) NFS1 was under a regulatable promoter, allowing us to control WT Nfs1 concentration and thus test whether Nfs1 variants, which on their own could not support cell viability, affected Isu stability. Specifically, our strain background had the jac1LKDDEQ mutation (5) (Fig. 1A) and the chromosomal copy of WT NFS1 under the control of the GAL1 promoter, allowing depletion of the essential NFS1 protein on a shift from a galactose-based to a glucose-based medium. As a test of the system, we used two GAL-NFS1 jac1LKDDEQ strains, one with a plasmid harboring a NFS1 gene under the control of its endogenous promoter (NFS1-NFS1) and thus competent to express Nfs1 after the shift to a glucose-based medium, and a second with only a control vector, in which case the Nfs1 levels dropped dramatically after glucose addition. We observed a decrease in Isu levels after Nfs1 depletion (Fig. 3A).
Fig. 3.
Effect of Nfs1 on Isu up-regulation. (A) GAL-NFS1 jac1LKDDEQ cells harboring WT NFS1 under control of NFS1 promoter (WT) or empty control vector (−) were shifted from galactose-based to glucose-based medium to repress expression of NFS1 from the GAL1 promoter (GAL-NFS1). Cells were harvested either before (+) or 28 h after glucose addition (downward arrow). Cell extracts were prepared and analyzed by immunoblotting with antibodies specific for Isu and, as a control, Mge1 (c). (B) Indicated cells from A along with analogous controls with a WT rather than a jac1LKDDEQ background were pulse-labeled after depletion of Nfs1. Samples were removed at indicated times after initiation of chase, and the amount of radiolabeled Isu was monitored by IP using Isu-specific antibody. (C) Cell extracts from GAL-NFS1 jac1LKDDEQ cells described in A, harboring the control vector (−) or the following NFS1 genes under control of the NFS1 promoter: WT, C421A, and Δ476–497 (ΔC). Relative Isu and Mge1 (c) levels were determined by immunoblot analysis. Cells were harvested either before (+) or 28 h after addition of glucose (downward arrow) to repress expression from the GAL promoter. (D) Cells in C were pulse-labeled at 28 h after the shift to glucose-based medium and subjected to pulse-chase analysis. (E) Cell lysates from indicated strains were subjected to immunoblot analysis with Isu-specific antibodies or, as a loading control, Mge1-specific antibodies (c). To deplete Yah1, GAL-YAH1 cells were harvested at 28 h after the shift to a glucose-based medium. (F) WT and Δyfh1 cells were pulse-labeled and subjected to pulse-chase analysis. (G) GAL-NFS1 Δpim1 cells harboring a plasmid containing either WT NFS1 or indicated mutants under control of its endogenous promoter were harvested at 28 h after the addition of glucose. Mitochondrial extracts were prepared and subjected to IP using Nfs1-specific antibodies. Precipitated Nfs1 and Isu proteins (IP) were detected by immunoblot analysis using specific antibodies, with 4% mitochondrial lysate used as a loading control (Input).
We performed pulse-chase analysis with both strains after the addition of glucose (Fig. 3B). Isu had a half-life of 125 min in jac1LKDDEQ cells in which Nfs1 levels were maintained. Isu was less stable in cells depleted of Nfs1, with the amount of radiolabeled Isu dropping below 40% of initial levels by 25 min after initiation of the chase. As a control, we also assessed the stability of Isu in JAC1 cells and found rapid degradation of Isu regardless of whether or not Nfs1 was depleted. These results indicate that the longer Isu half-life observed in the jac1LKDDEQ background requires Nfs1.
Next, to ask whether the enzymatic activity of Nfs1 is required for Isu stabilization, we altered the codon for the essential and conserved active site cysteine, residue 421, such that it encoded alanine. Purified Nfs1C421A had a background desulfurase activity level of 1.3 ± 0.04 nmol sulfide/min/mg Nfs1, compared with 45.7 ± 4 nmol sulfide/min/mg Nfs1 for purified WT Nfs1. A plasmid harboring nfs1C421A was introduced into jac1LKDDEQ GAL-NFS1 cells. After the addition of glucose, the Nfs1C421A-expressing cells had nearly the same high levels of Isu found in cells expressing WT Nfs1 (Fig. 3C).
We next evaluated the stability of Isu in jac1LKDDEQ GAL-NFS1 cells expressing Nfs1C421A. Isu was more stable in cells expressing Nfs1C421A than in cells carrying the empty control vector and thus depleted of WT Nfs1 (Fig. 3D). For example, at 30 min into the chase, only 20% of the pulse-labeled Isu remained in Nfs1-depleted cells, compared with more than 75% in the cells expressing Nfs1C421A. The half-life of Isu was 60 min in Nfs1C421A-expressing cells, compared with 100 min in cells expressing WT Nfs1.
The longer half-life of Isu in Nfs1C421A-expressing cells compared with cells depleted of Nfs1 suggested to us that the presence of a cluster in Isu was not necessary for stabilization. Thus, we assessed the half-life of Isu in strains lacking Yfh1, the putative iron donor and Nfs1 regulator, whose absence causes a severe defect in cluster formation (11). The steady-state levels of Isu were approximately 10-fold higher in Δyfh1 than in WT cells (Fig. 3E). Moreover, the half-life of Isu in Δyfh1 of ∼100 min was approximately fourfold longer than that seen in WT cells (Fig. 3F). Isu levels were 10-fold higher in cells depleted of Yah1, the essential electron donor for Fe-S cluster formation, than in WT cells. Together, these results indicate that Fe-S cluster formation is not required for an increase in Isu stability.
No Increase in Isu1 Stability in the Presence of Nfs1 Variant with Defective Interaction with Isu.
Given that the catalytic activity of Nfs1 is not essential for increased stability of Isu, we next asked whether a physical interaction between the two proteins was important. We designed a NFS1 C-terminal truncation mutant, deleting codons 476–497, because this conserved segment of the orthologous IscS protein of E. coli is known to be important for interaction with the scaffold IscU (17, 21). Nfs1Δ476–497 (henceforth designated Nfs1ΔC) had similar cysteine desulfurase activity to WT Nfs1 (42.5 ± 4 vs. 45.7 ± 4 nmol sulfide/min/mg Nfs1). However, less Isu was coimmunoprecipitated from mitochondrial lysates with Nfs1ΔC compared with WT Nfs1 (Fig. 3G), whereas the amount of Nfs1C421A was only slightly less than with WT Nfs1. The slight decrease in Nfs1C421A might be related to the close proximity of the active site cysteine to the Isu interaction surface (17).
To test the effect of Nfs1ΔC on Isu stability in vivo, we again used the GAL-NFS1 system in the jac1LKDDEQ genetic background. On repression of WT Nfs1 synthesis, the Isu level dropped to that seen in WT cells (Fig. 3C), suggesting instability of Isu. Pulse-chase analysis confirmed this interpretation, showing similarly rapid decreases in the level of radiolabeled Isu in Nfs1ΔC-expressing cells and cells depleted of Nfs1 (Fig. 3D). We conclude that Nfs1ΔC, which is defective in interaction with Isu, is not capable of substituting for WT Nfs1 to increase the stability of Isu.
High Isu Levels Compromise Growth Under Iron-Depleted Conditions.
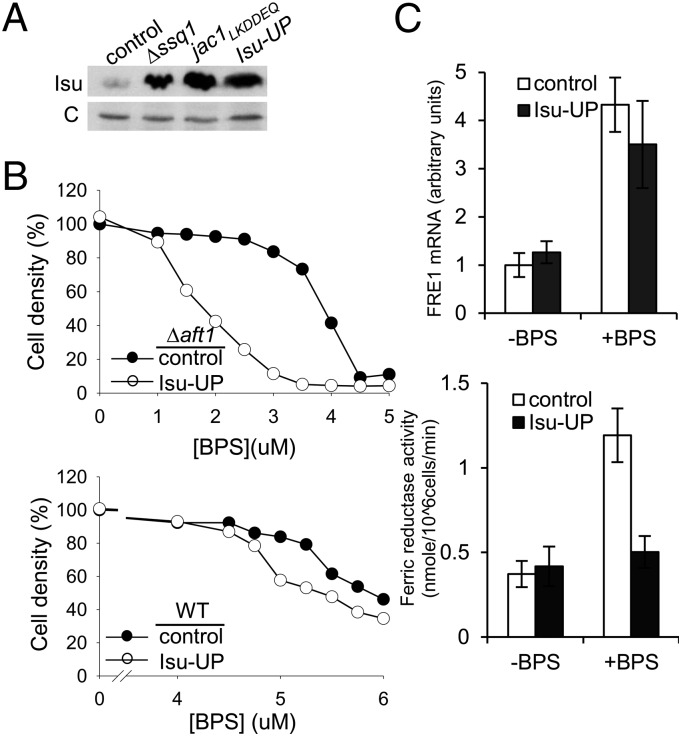
The dramatic increase in stability of Isu when Fe-S cluster biogenesis is compromised raises the question of the biological importance of cells maintaining the “normal” low levels of Isu. Specifically, we speculated that levels might be carefully regulated to help balance the needs of other iron-requiring processes. To test this idea, we constructed a strain (Isu-UP) expressing Isu at levels similar to those seen in cells defective in cluster biogenesis (Fig. 4A), making use of the strong tetR promoter system. We also included an aft1 deletion to diminish the transcriptional response to iron deprivation, thereby sensitizing the strain background. We carried out a growth assay to compare the cell density attained in liquid cultures with different concentrations of the iron chelator bathophenanthroline disulfonate (BPS) (Fig. 4B, Upper). In the absence of BPS, control and Isu-UP cells grew to the same density during the course of the experiment. At concentrations above 4 μM BPS, no growth of either strain was observed; however, at concentrations between 1 and 4 μM, Isu-UP cells were more sensitive to BPS. For example, in media containing 3 μM BPS, Isu-UP cells grew to only one-tenth of the density of cells with normal Isu levels. We also tested Isu-UP cells in an AFT1 background. Both AFT1 strains were more resistant to BPS than Δaft1 mutants, with no growth difference observed up to a concentration of 4.5 μM BPS. A reproducible growth difference between WT and Isu-UP cells was observed at higher concentrations, but was much less pronounced than in Δaft1 cells (Fig. 4B, Lower).
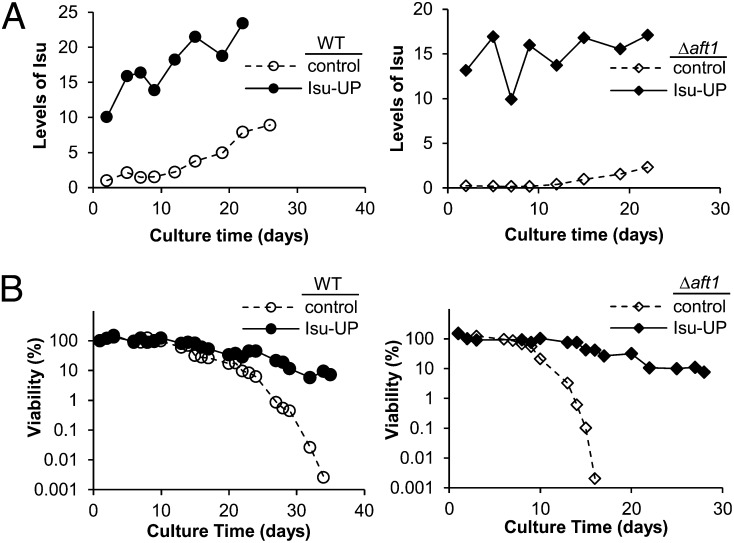
Fig. 4.
Effect of overexpression of Isu under iron-depleted conditions. (A) Cell extracts prepared from Δssq1, jac1LKDDEQ, and WT cells harboring empty vector (control) or plasmid with ISU1 under the control of the tetR promoter plasmid (Isu-UP) were subjected to immunoblot analysis with Isu- and, as a control, Mge1-specific antibodies (c). (B and C) Cells harboring either control vector or Isu-UP plasmid were compared using two genetic backgrounds, WT and Δaft1. Δaft1 cells were of the genetic background: Δaft1Δisu2 ISU1*. ISU1* denotes mutations in the Aft binding consensus site in the promoter of ISU1 (5). (B) Cells were inoculated into minimal media containing the indicated concentrations of BPS. The density of the culture with no BPS was monitored. When an OD600 of 1.0 was reached, OD600 measurements were also obtained from the cultures containing BPS and plotted as a percentage of the density reached by the control culture. (Upper) Δaft1. (Lower) WT background. (C) Exponentially growing Δaft1 cells were diluted to an OD600 of 0.1 in minimal media in the presence or absence of BPS. After 4 h, aliquots were removed. FRE1 mRNA levels were determined by quantitative PCR (Upper), and ferric reductase activity was measured (Lower). Error bars represent SDs from three or more independent experiments.
To more directly test the idea that high Isu levels disrupt other iron-dependent processes, we assessed the activity of the heme-dependent enzyme, ferric reductase (Fre1) (22). We measured both Fre1 mRNA levels and enzyme activity at 4 h after the addition of BPS and approximately threefold higher mRNA levels in both the control and Isu-UP cells (Fig. 4C, Upper), an expected response to iron limitation given that FRE1 is a target gene of the Aft transcription factor (23). However, although Fre1 enzyme activity in controls cells increased threefold in that 4-h span, the activity in Isu-UP cells did not change significantly (Fig. 4C, Lower). These results are consistent with the idea that high Isu levels can be deleterious to cells under iron-limiting conditions and may limit iron availability for other iron-requiring processes, such as maturation of heme-containing proteins.
High Isu Levels Prolong Chronological Lifespan.
The foregoing results indicate that Isu levels exceeding those normally present during exponential growth can be deleterious. We wanted to determine whether high Isu levels are advantageous under certain conditions. We noted during our studies that Isu-UP cultures appeared to retain a higher proportion of viable cells on prolonged storage; thus, we carried out chronological lifespan experiments to compare cell longevity. Exponentially growing liquid cultures of control and Isu-UP cells were grown to stationary phase, and incubation continued over a 35-d period. Cells were periodically removed from each culture. Aliquots were used to prepare extracts for determination of Isu levels or diluted before plating to determine the number of cells able to form colonies. As before, strains with AFT1 intact and AFT1 deleted were tested. Immunoblot analysis revealed 10- to 15-fold higher Isu1 levels in Isu-UP cells of both backgrounds at the beginning of the experiment, with levels remaining the same or rising slightly over 20 d. Isu levels also rose in control cells. In AFT1 cells, Isu levels were sixfold to eightfold higher at 3 wk after initiation of the experiment (Fig. 5A).
Fig. 5.
Effect of Isu overexpression on chronological lifespan. WT or Δaft1 cells harboring the control or Isu-UP plasmid, as described in Fig. 4, were inoculated and cultured in minimal media at 30 °C. (A) From day 1, cell extracts were prepared and relative levels of Isu and, as a control, Mge1 were determined. Data are plotted as the relative intensity of the Isu/Mge signal as arbitrary units. The Mge1 signal varied little over the time course of the experiment. Background: Left, WT; Right, Δaft1. (B) Cell viability was monitored by measuring colony-forming units. Viability of the cells at day 1 was set as 100%. Experiments were repeated a minimum of three times, with similar results obtained.
Control AFT1 cells lost viability slowly (Fig. 5B); at day 12, ∼80% of cells remained viable, with viability dropping slowly, such that by day 34 less than 0.01% were viable. AFT1 Isu-UP cells maintained viability even longer, with 10% of cells viable at day 34. Even greater differences were observed in the aft1Δ background. Less than 0.01% of the control Δaft1 cells were viable at 16 d, but 10% of Δaft1 Isu-UP cells were viable at 30 d. These findings indicate a longer chronological lifespan in cells expressing high levels of Isu than in cells with normal Isu expression, regardless of the presence of Aft1.
Discussion
A picture emerges from the work presented here in which the stability of the Fe-S cluster scaffold is regulated posttranslationally due to changes in its rate of degradation by the Lon-type protease of the mitochondrial matrix. Our results also strongly support the idea that a physical interaction between Nfs1 and Isu is a prerequisite for increased stability of Isu. Because the exact mechanism(s) by which Pim1, or indeed other Lon-type proteases, recognize substrates is not known, what makes Isu susceptible or resistant to degradation remains unclear. Yeast Pim1, like Lon-type proteases in bacteria and human mitochondria, is able to recognize both unfolded (24) and folded (25) segments as “degrons,” depending on the substrate protein. Adding to the challenge in understanding Isu degradation, structural studies of bacterial IscS and IscU, the orthologs of Nfs1 and Isu, respectively, found that their interaction stabilized the structure of IscU in one case and destabilized it in another (17, 26–28). Detailed in vitro analysis of Isu degradation by Pim1 are needed to identify the Isu degron recognized by Pim1.
Regardless of the exact mode of recognition by Pim1, the most straightforward model of Isu stabilization is that binding of Nfs1 is sufficient to protect the unknown recognition site from Pim1. In one sense, such a model has appeal, because only known factors are needed. However, such a mechanism requires that Nfs1 be able to protect Isu regardless of whether cluster formation or transfer is compromised, as demonstrated by the finding that mutations in every gene encoding a component of the mitochondrial Fe-S cluster biogenesis system tested other than NFS1 resulted in increased Isu stability. It can be envisioned that an intricate interplay of Isu-interacting proteins could have evolved to achieve this stabilization, given sufficient selective pressure favoring this regulatory mechanism. The involvement of other factors is possible as well. In this regard, it is intriguing that the mitochondrial ISC system produces an as-yet unidentified signal that inhibits activation of Aft transcription factors and whose export from mitochondria requires the ABC inner membrane transporter Atm1 (18). The idea that this elusive signal, whose absence appears to indicate a defect in cluster biogenesis (and in the native environment, likely a paucity of iron), may have both intramitochondrial and extramitochondrial affects, is appealing. Identification of this signaling molecule, as well as a better understanding of Isu1’s interaction with Nfs1 (including its interplay with other Isu-interacting proteins, such as Yfh1, Jac1 and Ssq1), is needed to better understand this regulatory mechanism.
The results presented here underscore the idea that the activity of the Fe-S biogenesis system can have strikingly beneficial or detrimental effects depending on the physiological context and/or environmental conditions. The finding that increasing the capacity to generate Fe-S clusters is deleterious under conditions of iron limitation without regard to other cellular needs for iron is perhaps not surprising. It is widely accepted that cells fine-tune a wide variety of cellular pathways when iron becomes limiting, with global changes in regulation in mRNA transcription and degradation (29–31) occurring when iron metabolism is perturbed. These mechanisms both increase iron availability and modulate metabolism, sparing only the most critical iron-using metabolic pathways. On the other hand, we found high Isu levels to be greatly beneficial during stationary phase, dramatically prolonging longevity. Although the mechanism behind this increased lifespan is not known, it is plausible that the increase in Isu molecules loaded with Fe-S clusters serves to repair clusters in proteins required for emergence from stationary phase, a time during which little protein synthesis occurs (32). Interestingly, defects in Fe-S cluster biogenesis have been implicated as a primary cause of the genome instability that occurs during replicative aging (33).
In summary, our results suggest that modulating the stability of Isu provides a built-in mechanism to rapidly alter Isu levels, and thus generate Fe-S clusters. Given that studies in the laboratory are generally poor mimics of the diversity and severity of environmental situations faced by organisms in the wild, it is likely that the level of Isu is regulated posttranslationally under a variety of extreme conditions.
Materials and Methods
Yeast Genetics.
All strains used were of the W303 genetic background. The Δssq strain contained Δaft1 to partially suppress the growth defect caused by iron accumulation (18). ISU1 under the control of the tetR promoter (18) is referred to as Isu-UP throughout. jac1LKDDEQ (34), Gal-Nfs1, and Δaft1 Δisu2 ISU1* (5) strains were described previously. The coding region of PIM1 was replaced with the KanMX4 cassette, generating Δpim1. ISU1 (16) and NFS1 mutants were generated in pRS314-ISU1 and pRS316-NFS1 using the Stratagene QuikChange protocol. Rich glucose-based (YPD) or minimal media, as described previously (5, 35) were used. Glucose was replaced by galactose to derepress GAL promoters. ISU1-Myc has three Myc tags at the C terminus of Isu1.
Protein Stability and Interaction.
Protein stability was determined essentially as described previously (18). First, 10 OD600 units of exponentially growing cells were concentrated, labeled with [35S]Express protein labeling mix (PerkinElmer) for 2 min, and diluted into minimal media containing 0.2 mg/mL of methionine and cysteine. Then the amount of labeled Isu in collected samples was determined by IP with antibody specific for Isu and detection of radioactivity by PhosphoImager (GE Healthcare) after separation by SDS/PAGE. Data were fit to a single two-parameter exponential decay (y = ae−bx, where y is the percent of radiolabeled Isu and x is minutes) using Sigmaplot 11.0 (Systat Software).
Whole-cell and mitochondrial lysates were prepared by bead beating (5). Co-IP was performed as described previously (36). In brief, anti-Nfs1 antibodies were crosslinked to protein A-Sepharose beads (Sigma Aldrich) and incubated with lysates for 1 h at 4 °C. After three washings, bound proteins were eluted with 0.1 M glycine, separated by SDS/PAGE, and detected by immunoblotting using anti-Nfs1 or anti-Isu antiserum.
Other Procedures.
For longevity tests, cells were inoculated to an OD600 of 0.05 in synthetic minimal medium (day 0) and cultured at 30 °C with continuous shaking. From day 1, when the cells reached stationary phase, samples were collected and levels of Isu determined by immunoblot analysis. Cell viability was monitored by plating diluted cultures onto YPD and counting the number of colonies formed.
To determine FRE1 mRNA levels and ferric reductase activity, total RNA and cell lysates were prepared from exponentially growing cells cultured in minimal media in the presence or absence of 200 μM BPS for 4 h. Reverse-transcription and quantitative real-time PCR were carried out according to the manufacturer’s instructions (Applied Biosystems). Ferric reductase activity was measured as described previously (22).
Supplementary Material
Acknowledgments
We thank Carolyn Suzuki for Pim1 and Mrp20 antibodies, Roland Lill for GAL-NFS1, and Brenda Schilke, June Pais, and Peter Kuhn for helpful comments. This work was supported by National Institutes of Health Grant GM27870 (to E.A.C.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206945109/-/DCSupplemental.
References
- 1.Levi S, Rovida E. The role of iron in mitochondrial function. Biochim Biophys Acta. 2009;1790:629–636. doi: 10.1016/j.bbagen.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 2.Lill R. Function and biogenesis of iron–sulphur proteins. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- 3.Yamaguchi-Iwai Y, Dancis A, Klausner RD. AFT1: A mediator of iron- regulated transcriptional control in Saccharomyces cerevisiae. EMBO J. 1995;14:1231–1239. doi: 10.1002/j.1460-2075.1995.tb07106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fontecave M, Ollagnier-de-Choudens S. Iron-sulfur cluster biosynthesis in bacteria: Mechanisms of cluster assembly and transfer. Arch Biochem Biophys. 2008;474:226–237. doi: 10.1016/j.abb.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 5.Andrew AJ, Song JY, Schilke B, Craig EA. Posttranslational regulation of the scaffold for Fe-S cluster biogenesis, Isu. Mol Biol Cell. 2008;19:5259–5266. doi: 10.1091/mbc.E08-06-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith AD, et al. Sulfur transfer from IscS to IscU: The first step in iron-sulfur cluster biosynthesis. J Am Chem Soc. 2001;123:11103–11104. doi: 10.1021/ja016757n. [DOI] [PubMed] [Google Scholar]
- 7.Zheng L, White RH, Cash VL, Dean DR. Mechanism for the desulfurization of L-cysteine catalyzed by the nifS gene product. Biochemistry. 1994;33:4714–4720. doi: 10.1021/bi00181a031. [DOI] [PubMed] [Google Scholar]
- 8.Alves R, Herrero E, Sorribas A. Predictive reconstruction of the mitochondrial iron-sulfur cluster assembly metabolism, I: The role of the protein pair ferredoxin–ferredoxin reductase (Yah1-Arh1) Proteins. 2004;56:354–366. doi: 10.1002/prot.20110. [DOI] [PubMed] [Google Scholar]
- 9.Lange H, Kaut A, Kispal G, Lill R. A mitochondrial ferredoxin is essential for biogenesis of cellular iron-sulfur proteins. Proc Natl Acad Sci USA. 2000;97:1050–1055. doi: 10.1073/pnas.97.3.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai CL, Barondeau DP. Human frataxin is an allosteric switch that activates the Fe-S cluster biosynthetic complex. Biochemistry. 2010;49:9132–9139. doi: 10.1021/bi1013062. [DOI] [PubMed] [Google Scholar]
- 11.Gerber J, Mühlenhoff U, Lill R. An interaction between frataxin and Isu1/Nfs1 that is crucial for Fe/S cluster synthesis on Isu1. EMBO Rep. 2003;4:906–911. doi: 10.1038/sj.embor.embor918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prischi F, et al. Structural bases for the interaction of frataxin with the central components of iron–sulphur cluster assembly. Nat Commun. 2010;1:95. doi: 10.1038/ncomms1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmucker S, et al. Mammalian frataxin: An essential function for cellular viability through an interaction with a preformed ISCU/NFS1/ISD11 iron-sulfur assembly complex. PLoS ONE. 2011;6:e16199. doi: 10.1371/journal.pone.0016199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vickery LE, Cupp-Vickery JR. Molecular chaperones HscA/Ssq1 and HscB/Jac1 and their roles in iron-sulfur protein maturation. Crit Rev Biochem Mol Biol. 2007;42:95–111. doi: 10.1080/10409230701322298. [DOI] [PubMed] [Google Scholar]
- 15.Craig EA, Marszalek J. A specialized mitochondrial molecular chaperone system: A role in formation of Fe/S centers. Cell Mol Life Sci. 2002;59:1658–1665. doi: 10.1007/PL00012493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dutkiewicz R, et al. Sequence-specific interaction between mitochondrial Fe-S scaffold protein Isu and Hsp70 Ssq1 is essential for their in vivo function. J Biol Chem. 2004;279:29167–29174. doi: 10.1074/jbc.M402947200. [DOI] [PubMed] [Google Scholar]
- 17.Shi R, et al. Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein–protein interactions. PLoS Biol. 2010;8:e1000354. doi: 10.1371/journal.pbio.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rutherford JC, et al. Activation of the iron regulon by the yeast Aft1/Aft2 transcription factors depends on mitochondrial, but not cytosolic, iron-sulfur protein biogenesis. J Biol Chem. 2005;280:10135–10140. doi: 10.1074/jbc.M413731200. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki CK, Suda K, Wang N, Schatz G. Requirement for the yeast gene LON in intramitochondrial proteolysis and maintenance of respiration. Science. 1994;264:273–276. doi: 10.1126/science.8146662. [DOI] [PubMed] [Google Scholar]
- 20.Bayot A, et al. Identification of novel oxidized protein substrates and physiological partners of the mitochondrial ATP-dependent Lon-like protease Pim1. J Biol Chem. 2010;285:11445–11457. doi: 10.1074/jbc.M109.065425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Urbina HD, Silberg JJ, Hoff KG, Vickery LE. Transfer of sulfur from IscS to IscU during Fe/S cluster assembly. J Biol Chem. 2001;276:44521–44526. doi: 10.1074/jbc.M106907200. [DOI] [PubMed] [Google Scholar]
- 22.Dancis A, Klausner RD, Hinnebusch AG, Barriocanal JG. Genetic evidence that ferric reductase is required for iron uptake in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2294–2301. doi: 10.1128/mcb.10.5.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Philpott CC, Protchenko O. Response to iron deprivation in Saccharomyces cerevisiae. Eukaryot Cell. 2008;7:20–27. doi: 10.1128/EC.00354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Janowsky B, et al. Structural properties of substrate proteins determine their proteolysis by the mitochondrial AAA+ protease Pim1. Biol Chem. 2005;386:1307–1317. doi: 10.1515/BC.2005.149. [DOI] [PubMed] [Google Scholar]
- 25.Ondrovicová G, et al. Cleavage site selection within a folded substrate by the ATP-dependent lon protease. J Biol Chem. 2005;280:25103–25110. doi: 10.1074/jbc.M502796200. [DOI] [PubMed] [Google Scholar]
- 26.Kim JH, et al. Structure and dynamics of the iron-sulfur cluster assembly scaffold protein IscU and its interaction with the cochaperone HscB. Biochemistry. 2009;48:6062–6071. doi: 10.1021/bi9002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramelot TA, et al. Solution NMR structure of the iron-sulfur cluster assembly protein U (IscU) with zinc bound at the active site. J Mol Biol. 2004;344:567–583. doi: 10.1016/j.jmb.2004.08.038. [DOI] [PubMed] [Google Scholar]
- 28.Bertini I, Cowan JA, Del Bianco C, Luchinat C, Mansy SS. Thermotoga maritima IscU: Structural characterization and dynamics of a new class of metallochaperone. J Mol Biol. 2003;331:907–924. doi: 10.1016/s0022-2836(03)00768-x. [DOI] [PubMed] [Google Scholar]
- 29.Hausmann A, Samans B, Lill R, Mühlenhoff U. Cellular and mitochondrial remodeling upon defects in iron-sulfur protein biogenesis. J Biol Chem. 2008;283:8318–8330. doi: 10.1074/jbc.M705570200. [DOI] [PubMed] [Google Scholar]
- 30.Shakoury-Elizeh M, et al. Metabolic response to iron deficiency in Saccharomyces cerevisiae. J Biol Chem. 2010;285:14823–14833. doi: 10.1074/jbc.M109.091710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puig S, Askeland E, Thiele DJ. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell. 2005;120:99–110. doi: 10.1016/j.cell.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 32.Herman PK. Stationary phase in yeast. Curr Opin Microbiol. 2002;5:602–607. doi: 10.1016/s1369-5274(02)00377-6. [DOI] [PubMed] [Google Scholar]
- 33.Veatch JR, McMurray MA, Nelson ZW, Gottschling DE. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell. 2009;137:1247–1258. doi: 10.1016/j.cell.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andrew AJ, Dutkiewicz R, Knieszner H, Craig EA, Marszalek J. Characterization of the interaction between the J-protein Jac1p and the scaffold for Fe-S cluster biogenesis, Isu1p. J Biol Chem. 2006;281:14580–14587. doi: 10.1074/jbc.M600842200. [DOI] [PubMed] [Google Scholar]
- 35.Sherman F, et al. Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1986. [Google Scholar]
- 36.Liu Q, D’Silva P, Walter W, Marszalek J, Craig EA. Regulated cycling of mitochondrial Hsp70 at the protein import channel. Science. 2003;300:139–141. doi: 10.1126/science.1083379. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.