Apoptotic cells display various “eat me” signals that can be recognized through bridging molecules that cross-link the dying cells to phagocytes. This work illustrates the first full-length structure of such a bridging molecule, TTR-52. The study elucidates the binding of these bridging molecules with the apoptotic cell signals and phagocyte receptors, providing valuable new insight into the process of apoptotic cell recognition.
Keywords: apoptotic cell engulfment, bridging molecule, crystal structure, TTR-52
Abstract
During apoptosis, apoptotic cells are removed by professional phagocytes or neighboring engulfing cells either directly through phagocytic receptors or indirectly through bridging molecules that cross-link dying cells to phagocytes. However, how bridging molecules recognize “eat me” signals and phagocytic receptors to mediate engulfment remains unclear. Here, we report the structural and functional studies of Caenorhabditis elegans TTR-52, a recently identified bridging molecule that cross-links surface-exposed phosphatidylserine (PtdSer) on apoptotic cells to the CED-1 receptor on phagocytes. Crystal structure studies show that TTR-52 has an open β-barrel-like structure with some similarities to the PKCα-C2 domain. TTR-52 is proposed to bind PtdSer via an “ion-mediating” PtdSer-binding mode. Intensive functional studies show that CED-1 binds TTR-52 through its N-terminal EMI domain and that the hydrophobic region of the TTR-52 C terminus is involved in this interaction. In addition, unlike other PtdSer-binding domains, TTR-52 forms dimers, and its dimerization is important for its function in vivo. Our results reveal the first full-length structure of a bridging molecule and the mechanism underlying bridging molecule-mediated apoptotic cell recognition.
Phagocytosis and the removal of apoptotic cells are important events in tissue remodeling, suppression of inflammation, and regulation of immune responses. Defects in these processes cause autoimmune diseases and developmental retardation (Henson et al. 2001; Savill et al. 2002). During apoptosis, apoptotic cells are marked by “eat me” signals such as phosphatidylserine (PtdSer), which are exposed on the surface of apoptotic cells and recognized by phagocytes either directly through phagocyte receptors or indirectly through bridging molecules that cross-link apoptotic cells to phagocytes (Savill and Fadok 2000). The recognition of “eat me” signals by phagocytes triggers signaling cascades, leading to the internalization and degradation of apoptotic cells by phagocytes. Proteins such as hBAI1 (Park et al. 2007), Stabilin2 (Park et al. 2008), and T-cell immunoglobulin and mucin domain 4 (TIM4) (Kobayashi et al. 2007) act as phagocytic receptors, while milk fat globule EGF factor 8 (MFG-E8) (Hanayama et al. 2002) and growth arrest-specific 6 (Gas6) (Nakano et al. 1997) are extracellular bridging molecules in mammals that cross-link PtdSer signals on apoptotic cells with receptors on macrophages. The use of phagocyte receptors and bridging molecules in apoptotic cell recognition is evolutionarily conserved, although the detailed mechanisms of these two approaches are different.
In Caenorhabditis elegans, phagocytosis of apoptotic cells is controlled by two partially redundant signaling pathways. In one pathway, CED-2, CED-5, and CED-12 mediate the activation of the small GTPase CED-10, leading to the cytoskeletal reorganization needed for cell corpse engulfment (Reddien and Horvitz 2000; Wang et al. 2003). In the other pathway, CED-1, CED-6, CED-7, and TTR-52 are involved in recognizing and transducing certain “eat me” signals (Liu and Hengartner 1998; Wu and Horvitz 1998; Zhou et al. 2001). CED-1 is a phagocyte receptor with a single transmembrane domain and a large N-terminal extracellular region responsible for apoptotic cell recognition. The extracellular region of CED-1 contains a predicted EMI domain with an unknown structure at its extreme N terminus followed by 16 atypical EGF-like motifs (Zhou et al. 2001; Callebaut et al. 2003). The cytoplasmic region of CED-1 contains two motifs characterized by NPXY and YXXL, which are important for signal transduction (Su et al. 2002).
PtdSer is an evolutionarily conserved “eat me” signal exposed on the surface of apoptotic cells. The recognition of PtdSer by phagocytic receptor TIM4, the C2 domain of the bridging molecule MFG-E8 (MGF-E8-C2 domain), or intracellular PtdSer-binding proteins that are not involved in apoptotic cell recognition, such as Annexin V, the C2 domains of synaptotagmins, or PKCα, has been extensively studied by structural biology approaches (Sutton et al. 1995; Swairjo et al. 1995; Santiago et al. 2007; Shao et al. 2008; Guerrero-Valero et al. 2009). The C2 domains of synaptotagmins and PKCα fold into a β-sandwich structure containing eight β strands. One characterized feature of this domain is the successive β-bulge folds of some of these strands (Sutton et al. 1995). Annexin V is a helix bundle protein with channel-forming membrane characteristics. The PtdSer-binding domains of TIM4 have immunoglobulin-like features, while the C2 domain at the MFG-E8 N terminus has a β-sandwich structure that has some significant differences from that of PKCα-C2. With the exception of MFG-E8, all of these proteins have been reported to have a common “ion-mediating” mechanism such as calcium-bridging for PtdSer binding, although they fold into different structures. Although numerous extracellular bridging molecules were identified, how they cross-link apoptotic cells to phagocytes remains unclear, largely due to the lack of a complete structure of full-length bridging molecules.
TTR-52 is a secreted protein identified recently in C. elegans that recognizes surface-exposed PtdSer and binds to the extracellular domain of CED-1 (Wang et al. 2010). It therefore serves as a bridging molecule that mediates recognition of apoptotic cells by cross-linking the “eat me” signal PtdSer with the CED-1 receptor. Based on sequence analysis, TTR-52 has been predicted to have a transthyretin-like fold structure (Wang et al. 2010). Transthyretin family proteins are known to transport lipids such as retinol and thyroxine (Schreiber 2002; Prapunpoj and Leelawatwattana 2009). Nevertheless, no transthyretin family protein has yet been found to associate with PtdSer. If TTR-52, a secreted bridging molecule, indeed has a transthyretin-like fold structure, then how TTR-52 recognizes PtdSer and recruits CED-1 to clear apoptotic cells is not clear. To address these questions, we determined the crystal structure of a mutated full-length TTR-52. Our structural and functional study revealed functional mechanisms of the bridging molecule TTR-52 in the engulfment process.
Results
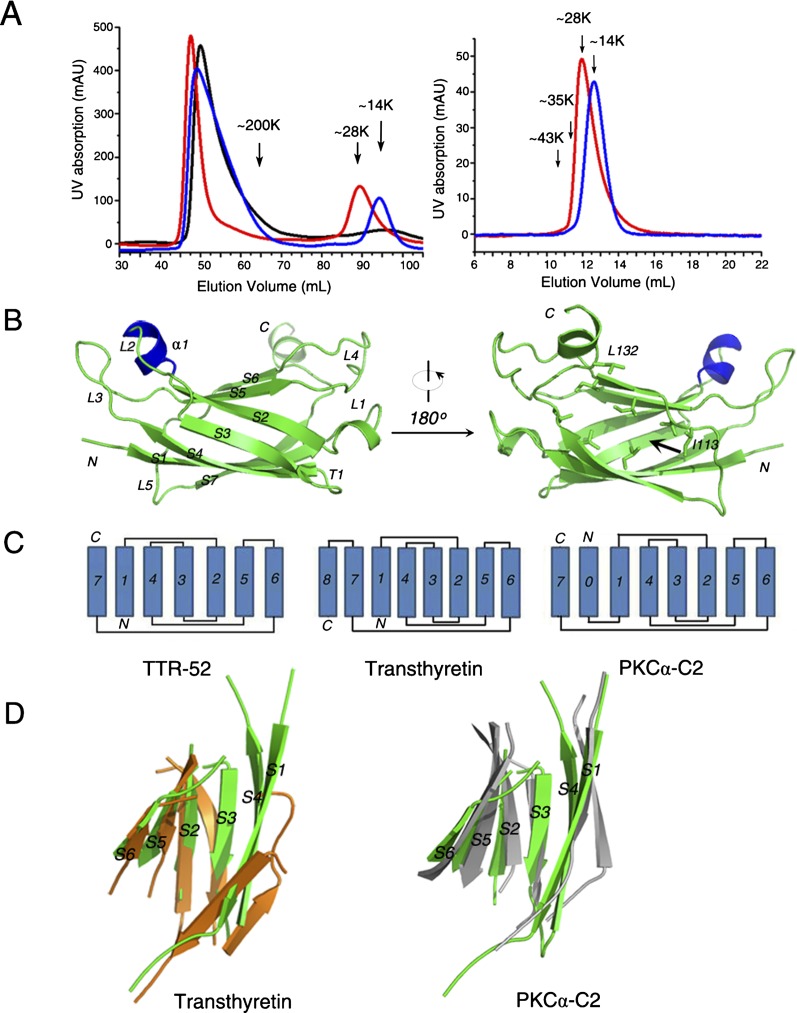
Full-length TTR-52 is composed of 135 amino acids, the first 20 residues being a signal peptide. To begin with, our work was hampered because aggregation of the wild-type protein purified from Escherichia coli prevented its crystallization (Fig. 1A). We evaluated the baculovirus expression system and the 293T mammalian cell expression system for production of large amounts of TTR-52, but both systems failed due to low expression of TTR-52. To overcome this problem, a series of mutant proteins were analyzed using alanine-block scanning mutagenesis. Although dozens of mutant proteins were tested, only one in which residues 50–55 (50EDSLPL55) were replaced by six successive alanines (termed TTR-52 [M5]) produced some proteins that were found to be stable dimers by gel filtration chromatography (Fig. 1A). Well-diffracted crystals were obtained using this protein.
Figure 1.
Structure of TTR-52 (M5). (A, left panel) Gel filtration chromatography results for wild-type TTR-52 (black line), TTR-52 (M5) (red line), and TTR-52 (M5)-R63D/W65Q/Q73A (blue line) purified from E. coli on a Superdex200 column. All three proteins gave large peaks of high molecular weight that represent protein aggregates in the left panel. When loaded on a Superdex200 column (GE Healthcare), TTR-52 (M5) gave a peak at ∼28 kDa, while TTR-52 (M5)-R63D/W65Q/Q73A gave a peak at ∼14 kDa. The oligomerization state of TTR-52 (M5) and TTR-52 (M5)-R63D/W65Q/Q73A are stable in solution. (Right panel) Nonaggregated samples of TTR-52 (M5) and TTR-52 (M5)-R63D/W65Q/Q73A separated on a Superdex200 column also showed constant 28-kDa and 14-kDa molecular weights, respectively, when loaded on a Superdex75 10/300 GL column. (B) Overview of the TTR-52 (M5) structure. The TTR-52 (M5) structure is shown as a cartoon diagram, colored in green. The artificial α helix produced by mutations of six consecutive residues to alanines in TTR-52 is colored in blue. Secondary structural elements are labeled in the left panel. The N-terminal region includes α1, L2, L3, and L5. The C-terminal region includes L1, L4, and T1. In the right panel, the C-terminal gap of the β barrel is indicated by a black arrow, and residues on the hydrophobic surface are shown as stick models. Residues I113 and L132 are labeled. (C) Topology of TTR-52, transthyretin, and the PKCα-C2 domain. The first β strand in PKCα-C2 was termed 0, rather than 1, to facilitate comparisons with TTR-52. (D) Superposition of strands 1, 4, 3, 2, 5, and 6 of TTR-52 (M5) (colored in green) with those corresponding strands of the transthyretin monomer (Protein Data Bank [PDB]: 3NEE, colored in orange) and PKCα-C2 (PDB 1DSY, colored in gray).
The 2.01 Å crystal structure of TTR-52 (M5) was determined using the single isomorphous replacement with anomalous scattering (SIRAS) method (Fig. 1; Supplemental Table S1). A summary of the crystallographic data and refinement statistics for TTR-52 (M5) is shown in Supplemental Table S1. The structure was refined to a final R value of 17.7% and a free R value of 23.8%. A clear and continuous electron density map indicates that the structure was properly defined (Supplemental Fig. S1).
Overall structure of the TTR-52 monomer
The TTR-52 (M5) monomer consists of seven β strands (S1–S7) that are connected by five loops (L1–L5) and one β turn (T1) (Fig. 1B). These β strands fold into an open β-barrel-like structure, with the sixth and seventh strands forming edges around the opening. Loops linking these β strands are located at the two sides of the barrel. Loop 2 (L2, between S2 and S3), loop 3 (L3, between S4 and S5), and loop 5 (L5, between S6 and S7) are located in the N-terminal region of the barrel, and loop 1 (L1, between S1 and S2), loop 4 (L4, between S5 and S6), and turn 1 (T1, between S3 and S4) are located in the C-terminal region. The six consecutive alanine residues introduced by mutagenesis form a short α helix (α1) in L2. We propose that helix α1 is an artifact resulting from mutation of those residues to alanine and that the corresponding region in wild-type TTR-52 would be a loop, according to the secondary structure prediction by PsiPred (Bryson et al. 2005). We further assume that this six-alanine peptide does not change the overall folding of TTR-52, since it is located on the flexible loop that is far away from the β strands. In order to perform structural and functional analysis of the wild-type protein, the structure of wild-type TTR-52, in which the six alanines were mutated back to their original residues, was built using Modeller (Eswar et al. 2006) based on the structure of TTR-52 (M5). No additional changes were introduced to other parts of the structure, except that helix α1 was modeled as a short loop. Overall, the structure exhibits extensive negatively charged and hydrophobic surfaces (Fig. 2B).
Figure 2.
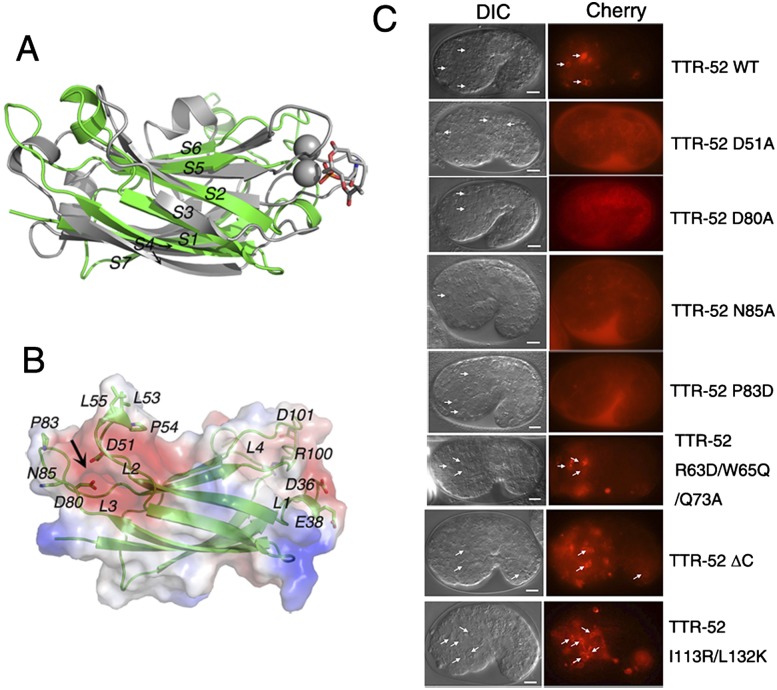
The PtdSer-binding site of TTR-52. (A) Superposition of TTR-52 (M5) with the first through seventh strands of the PKCα-C2 domain. The cartoon diagram of TTR-52 (M5) is colored in green, and the PKCα-C2 domain is in gray. The calcium ions in the PKCα-C2 domain are shown as spheres. The labels for strands 1–7 in both TTR-52 (M5) and the PKCα-C2 domain correspond to those in Figure 1C. (B) Surface electrostatic potential of TTR-52 is shown. (Red) Negatively charged residues; (blue) positively charged residues; (white) hydrophobic residues. The model of TTR-52 was generated by Modeller based on TTR-52 (M5). The residues on L1 and L4, and L2 and L3, believed to be involved in PtdSer binding, are shown as stick models. The black arrow indicates the cation-mediated PtdSer-binding site in the negatively charged N-terminal region. (C) Labeling of dying cells by wild-type and mutant TTR-52 in C. elegans embryos. Wild-type TTR-52::mCherry forms bright rings around dying cells (arrows), but point mutations D51A, D80A, N85A, and P83D abolished TTR-52 labeling of dying cells. Mutations at R63D/W65Q/Q73A on the dimer interface and mutations at the C terminus, ΔC, and I113R/L132K did not alter corpse labeling by TTR-52. Bars, 5 μm.
Structural comparison of TTR-52 (M5) with transthyretin
Based on sequence homology, it was predicted that TTR-52 may have a transthyretin protein fold (Wang et al. 2010). Structural alignment using DALI (Dietmann et al. 2001) showed that these two protein structures have an overall root mean square deviation (R.M.S.D.) of ∼3.0 Å. We found three major differences between these two structures. First, there is a hydrophobic region on the C terminus of TTR-52 that might serve as a functional surface (Fig. 1B) that is not present in transthyretin. Second, transthyretin has a β-sandwich structure in which strands 4, 1, 7, and 8 and strands 3, 2, 5, and 6 form two layers of the twisted β sheets (Hornberg et al. 2000). However, topological analysis showed that TTR-52 has only seven β strands, corresponding to the N-terminal seven strands in transthyretin (Fig. 1C). In TTR-52, strand 3 connects the second and fourth strands by backbone hydrogen bonds (Supplemental Fig. S1). Compared with the third and fourth strands in transthyretin, the corresponding strands in TTR-52 are located closer to the center of the structure and to each other, making the TTR-52 structure like an open β barrel (Fig. 1D). Third, intramolecular interactions are also different. Transthyretin monomers assemble into stable dimers via hydrogen bonds between the sixth and eighth strands (Hornberg et al. 2000). Two dimers then interact further to form a tetramer through hydrophobic interactions (Connelly et al. 2010). In contrast, gel filtration analysis showed that recombinant wild-type TTR-52 purified from 293T cells exists as dimers (Supplemental Fig. S2). In our structure, while TTR-52 (M5) is also shown to exist as a dimer, its monomer–monomer interaction mode is different from that of transthyretin. The monomers in the dimer of TTR-52 interact back to back, mainly through the side chains on the third and fourth strands (Fig. 6A, below).
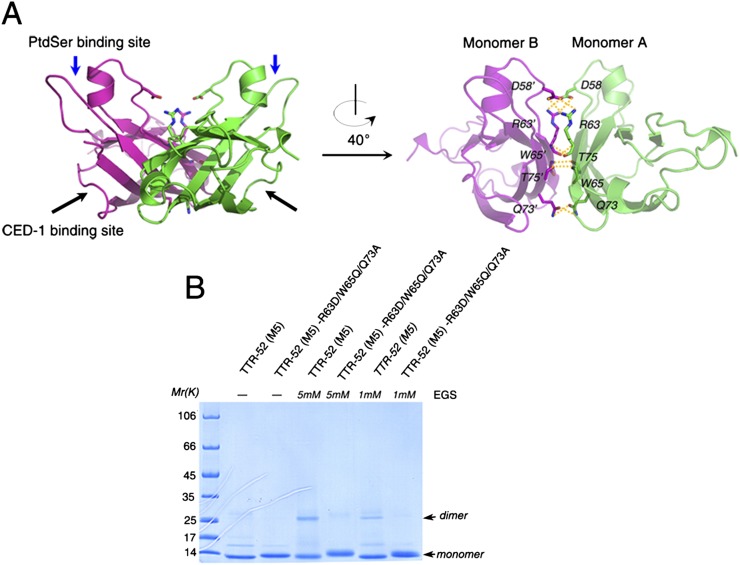
Figure 6.
Dimer interface of TTR-52 (M5). (A) The interface of the TTR-52 (M5) dimer shown as a cartoon. The proposed PtdSer- and CED-1-binding sites are indicated as blue and black arrows, respectively, in the left panel. The key residues on the surface (D58, R63, W65, Q73, and T75) are shown as stick models in the right panel. The interactions between D58/R63′, R63/R63′, W65/T75′, and Q73/Q73′ are labeled (residues from monomer B are labeled with a prime). (B) Cross-linking of TTR-52 (M5) and TTR-52 (M5)-R63D/W65Q/Q73A. Cross-linker EGS was added to recombinant proteins TTR-52 (M5) and TTR-52 (M5)-R63D/W65Q/Q73A purified from E. coli. TTR-52 (M5), but not TTR-52 (M5)-R63D/W65Q/Q73A, was cross-linked in a concentration-dependent manner and appeared as a dimer-sized band on a 15% SDS-PAGE gel.
Mechanism of TTR-52 binding to PtdSer
When DALI searching was used to identify proteins with a structure similar to TTR-52, none of them were PtdSer-binding proteins. When compared with the reported structures of PtdSer-binding proteins (Sutton et al. 1995; Swairjo et al. 1995; Gaboriaud et al. 2003; Shao et al. 2008), we found that TTR-52 has a structure similar to the PKCα-C2 domain, which is known for its intracellular PtdSer-binding ability, although sequence identity of the PKCα-C2 domain and TTR-52 is <20% (Supplemental Fig. S3). We compared the structure of TTR-52 with that of the PKCα-C2 domain (Guerrero-Valero et al. 2009). The PKCα-C2 domain has eight strands and folds into a β-sandwich structure. The seven-stranded TTR-52 has a topology similar to the C-terminal seven strands (strands 1–7) of the PKCα-C2 domain (for convenience, we refer to the first N-terminal strand of PKCα-C2 as S0) (Fig. 1C). The successive β-bulge folds that characterize C2 domain proteins do not exist in TTR-52. When TTR-52 (M5) is superposed with the C-terminal seven strands of the PKCα-C2 domain, the main chain R.M.S.D of these two structures is 3.4 Å. We also superposed strands 1, 4, 3, 2, 5, and 6 of TTR-52, PKCα-C2, and transthyretin, which seem to form a topologically similar core common to all three molecules: The R.M.S.D. between TTR-52 and PKCα-C2 is ∼2.49 Å, and that between TTR-52 and transthyretin is ∼2.66 Å (Fig. 1D), suggesting that they all have a similar core. These results imply that TTR-52 and PKCα-C2 may have a similar PtdSer-binding mechanism. This prompted us to explore potential PtdSer-binding sites in TTR-52 based on the PKCα-C2 domain and PtdSer complex structure.
The PKCα-C2 domain binds to PtdSer using two loops located between β2–β3 and β6–β7 at one end of the β sandwich (Verdaguer et al. 1999; Guerrero-Valero et al. 2009). Two calcium ions coordinated by five aspartate residues and the main chain of the other two residues from the loops provide a binding site for the phosphate group of PtdSer. Similarly, the C-terminal region loops in TTR-52 (M5)—L1 and L4—correspond to the PtdSer-binding loops in the PKCα-C2 motif, based on comparison of the structures (Fig. 2A). We thus assumed that residues in the C-terminal region loops of TTR-52 may be involved in metal ion-mediated PtdSer binding.
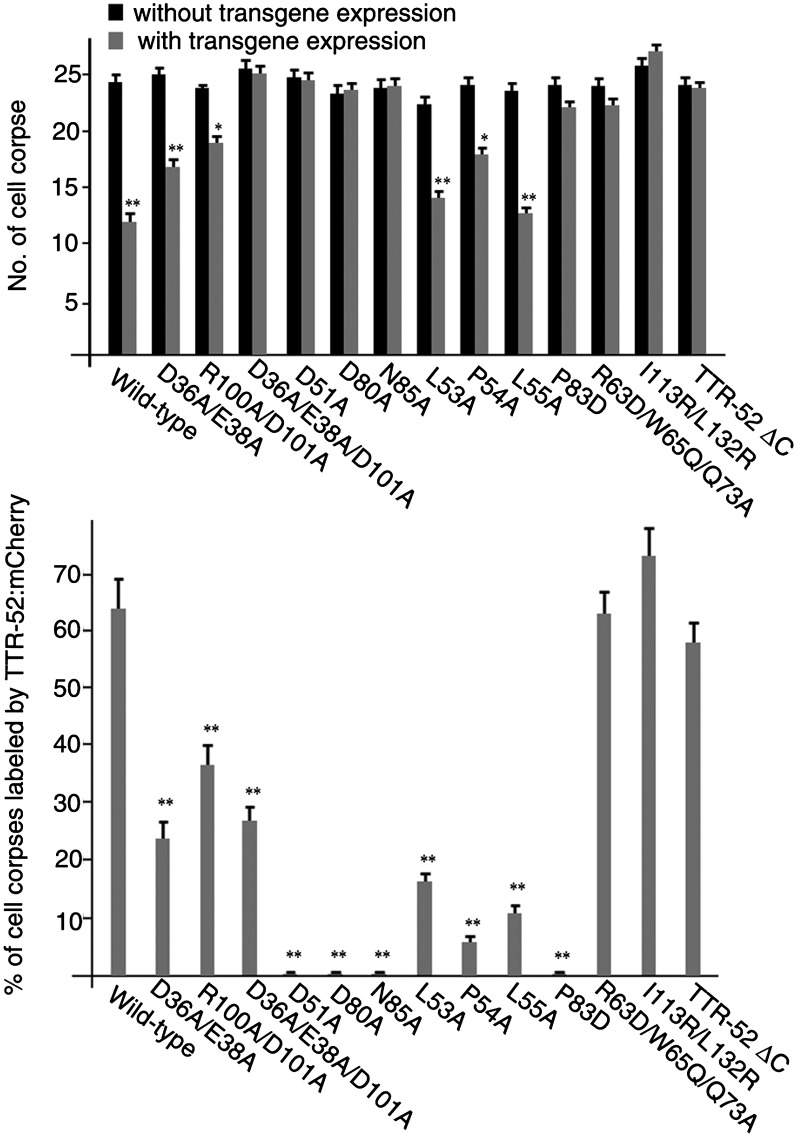
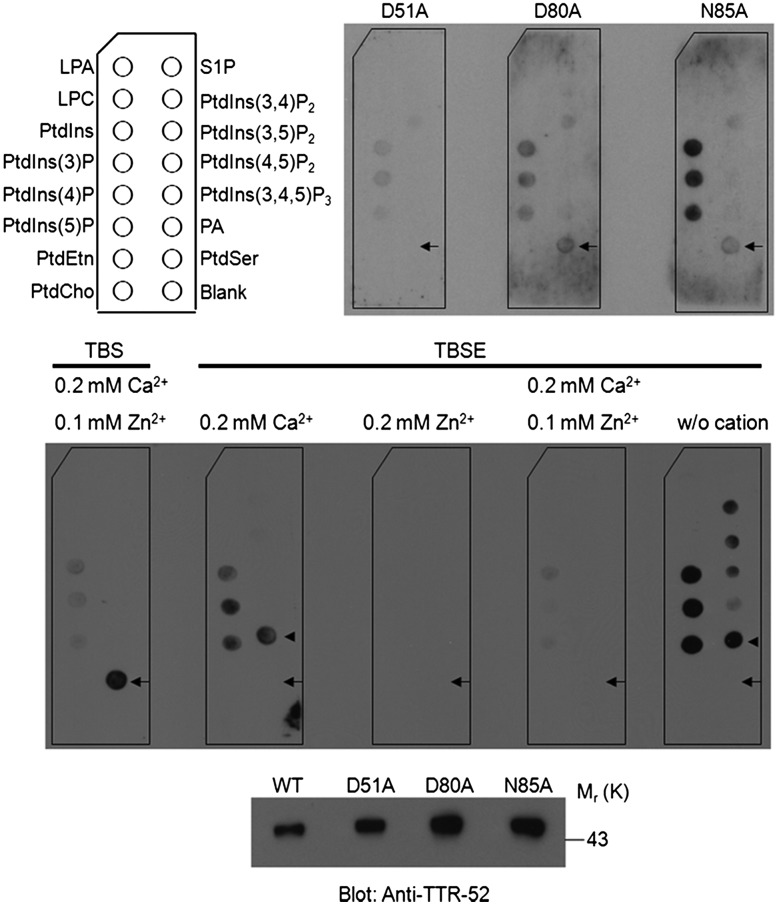
We then studied the residues in the L1 and L4 loops of TTR-52 that may be involved in PtdSer binding in detail. Some negatively charged TTR-52 residues, such as D36 and E38 in L1 and D101 in L4, are similar to aspartate residues D187, D246, and D248 in PKCα-C2 (Verdaguer et al. 1999) and may mediate calcium-dependent PtdSer binding (Fig. 2B). Since PKCα-C2 is shown to bind the phosphatidyl head group of phosphoinositol such as PtdIns(4,5)P2 by basic residues (Guerrero-Valero et al. 2009), we also included one adjacent basic residue, R100 in L4, in the analysis of the PtdSer-binding property of TTR-52. We expressed a series of point mutations of TTR-52 fused to a mCherry fluorescent protein in C. elegans under the control of the C. elegans heat-shock promoters and examined recognition of apoptotic cells by TTR-52 mutant proteins. Wild-type TTR-52::mCherry binds surface-exposed PtdSer on apoptotic cells, forming bright ring-like structures around cell corpses (Fig. 2C). Since these residues were presumed to be required for PtdSer binding, we expected that their mutations would lead to a significant effect on the PtdSer-binding ability of TTR-52. However, we found that mutations D36A/E38A, R100A/D101A, or D36A/E38A/D101A did not completely abolish recognition of apoptotic cells by TTR-52 and that D36A/E38A or R100A/D101A mutations partially rescued the engulfment defect of the ttr-52 deletion mutant tm2078 (Fig. 3; Supplemental Fig. S8). The percentage of cell corpses clustered by TTR-52::mCherry in C. elegans embryos was reduced from 63% (wild-type TTR-52) to 30% (mutant TTR-52) (Fig. 3). We next examined the PtdSer-binding ability of the mutants D36A/E38A, R100A/D101A, and D36A/E38A/R100A/D101A in vitro using membrane lipid strip assay (Wang et al. 2010). The recombinant wild-type and mutant TTR-52-mCherry-Flag proteins were purified from 293T cells and used for PtdSer binding. We found that all of these three proteins retain the PtdSer-binding specificity (Supplemental Fig. S4). Consistent with this, D36A/E38A/R100A/D101A mutant protein expressed in yeast cells can still associate with cell membranes (Supplemental Fig. S5). These results therefore indicate that these residues in the C-terminal region are not absolutely required for TTR-52's binding to PtdSer.
Figure 3.
Cell corpse recognition and rescue activity of TTR-52 mutants. Wild-type and mutant TTR-52-mCherry were expressed in ttr-52 (tm2078) mutants controlled by C. elegans heat-shock promoters. After heat-shock treatment, the number of cell corpses (top) and the clustering of TTR-52::mCherry around dying cells (bottom) were scored in 1.5-fold stage embryos. Two independent transgenic lines were scored (n ≥ 15). Error bars represent the SEM. (**) P < 0.0001; (*) P < 0.01.
In the structure of TTR-52, in addition to the C-terminal negatively charged loops, we noticed that the closely adjacent loops L2 and L3 in the N-terminal region of TTR-52 are also negatively charged, suggesting that they may form a metal ion-binding site similar to that in other PtdSer-binding proteins (Fig. 2B). According to a previous report (Wang et al. 2010), TTR-52 (M5) in which residues 50–55 (in loop L2 in our structure) were replaced by six successive alanines fails to recognize apoptotic cells in C. elegans or bind PtdSer in vitro. These results raise another possibility that the N-terminal region (L2, 49–59 amino acids; L3, 79–88 amino acids) is required for PtdSer binding. We made point mutations (D51A, D80A, and N85A) in this region and found that all three point mutations almost completely abolished the clustering of apoptotic cells by TTR-52::mCherry and failed to rescue the cell corpse phenotype of ttr-52 (tm2078) mutants (Figs. 2C, 3). They also showed decreased membrane labeling in yeast cells (Supplemental Fig. S5) and greatly reduced PtdSer binding in a membrane lipid strip assay (Fig. 4). These results suggest that the loops in the N-terminal region, rather than those in the C-terminal region, may play a crucial role in PtdSer binding.
Figure 4.
TTR-52 mutations and cation depletion disrupt PtdSer binding in vitro. (Top panel) Affinity-purified TTR-52-mCherry-Flag binds specifically to PtdSer on a membrane lipid strip, while point mutations D51A, D80A, and N85A greatly decrease the PtdSer-binding ability of TTR-52. (Bottom panel) Amounts of purified TTR-52 proteins used in lipid binding are shown by immunoblotting. (Middle panel) After chelating with 1 mM EDTA, TTR-52 alters the binding affinity of PtdSer to phosphatidic acid, and the ability of TTR-52 to bind to PtdSer was not restored when it was equilibrated in cation solutions. Two independent experiments were performed. (LPA) Lysophosphatidic acid, (LPC) lysophosphatidylcholine, (S1P) sphingosine-1-phosphate, (PA) phosphatidic acid. Arrows indicate PtdSer, and arrowheads indicate PA.
PtdSer is bound in a cation ion-dependent manner in TIM4, Annexin V, and the PKCα-C2 domain. Particularly, PtdSer is bound by the C2 domain between two loops at one end of the β-sandwich structure that mimic the two arms of a clamp (Verdaguer et al. 1999). Although no sequence similarity can be found between the L2 and L3 loops of TTR-52 and those corresponding regions in the C2 domain (Supplemental Fig. S3), the acidic residues in L2 and L3 may mimic the “ion-mediating” PtdSer-binding properties, similar to TIM4, Annexin V, and the PKCα-C2 domain (Supplemental Fig. S6). Therefore, removal of the predicted cation ions bound to TTR-52 may disrupt the binding of this protein to PtdSer. To test this, Flag beads were washed with TBSE buffer (50 mM Tris-HCl at pH 7.4, 150 mM NaCl, 1 mM EDTA) to chelate divalent cations after affinity purification of TTR-52-mCherry-Flag from the culture medium of 293T cells transfected with pCMV-TTR-52-mCherry-Flag. The eluted proteins were assayed for PtdSer binding using a membrane lipid strip. Instead of binding PtdSer, cation-free TTR-52 showed strong binding activity toward phosphatidic acid. The binding of TTR-52 to PtdSer was not restored even when the TTR-52 protein solution was equilibrated in buffers containing 0.2 mM Ca2+, 0.2 mM Zn2+, or 0.2 mM Ca2+/0.1 mM Zn2+ (Fig. 4), suggesting that removal of cation ions by EDTA may alter the local conformation of the cation ion-binding loops, which is required for specific binding of TTR-52 to PtdSer and is not recovered efficiently by adding back cation ions.
In addition to the negatively charged residues, we also studied the functions of the continuous stretches of hydrophobic residues on the tips of L2 and L3: 53LPL55 and 81FGPI84. Point mutations L53A, P54A, and L55A in L2 had a much lower ability to recognize dying cells in vivo compared with the wild type, although they were able to partially (P54A) or fully (L53A and L55A) rescue the ttr-52 (tm2078) mutant phenotype (Fig. 3). However, point mutation P83D in L3 had a severe phenotype; i.e., it could not form a ring around dying cells (Fig. 2C) and could not rescue the ttr-52 (tm2078) mutant phenotype (Fig. 3). It is important to note that there are no hydrophobic residues corresponding to those mentioned above on the tips of L2 and L3 in the PKCα-C2 domain. TIM4 has also been reported previously to use hydrophobic residues on the tips of loops to interact with the cell membrane (Santiago et al. 2007). We propose that the hydrophobic residues on the tips of the PtdSer-binding loops in TTR-52 may have a similar function; these hydrophobic residues may be located in close contact with the membranes of dying cells. These results indicate that PtdSer binding and membrane insertion are both required for the recognition of dying cells.
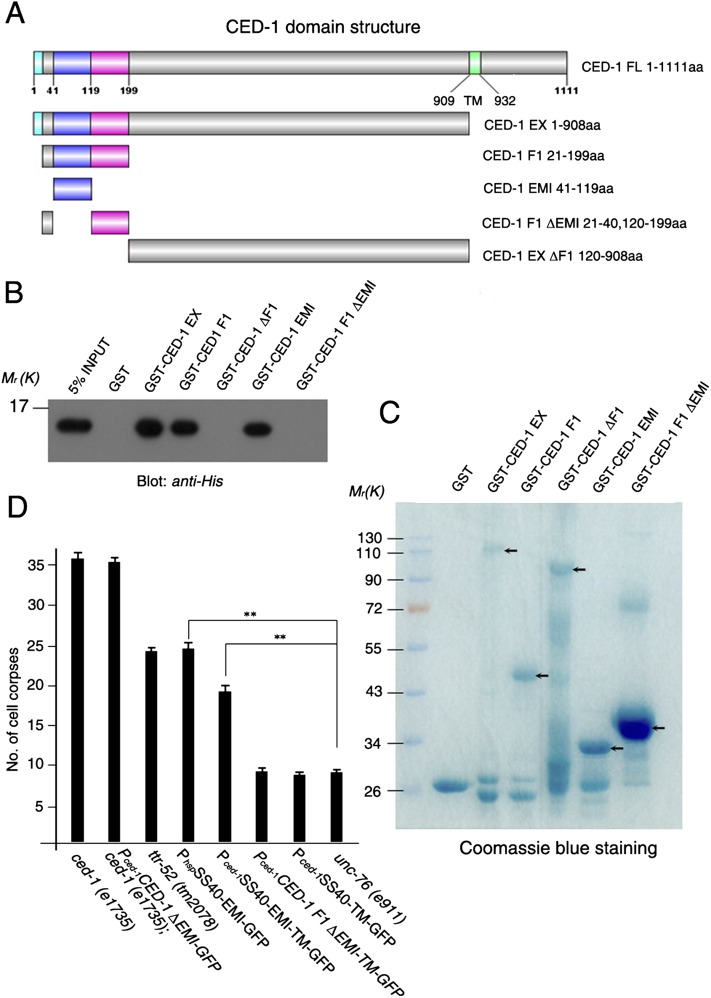
Interactions between TTR-52 and CED-1
As we showed previously, TTR-52 binds to the extracellular domain of CED-1 (Wang et al. 2010). Here, we generated different truncations of CED-1 and analyzed their interactions with TTR-52 by an in vitro pull-down assay (Fig. 5A–C). The EMI domain at the N terminus of CED-1 was found to be sufficient for TTR-52 binding (Fig. 5). We further tested the importance of the EMI domain for CED-1 function in vivo and found that a ced-1 truncation without the EMI domain (Pced-1CED-1ΔEMI-GFP) could not rescue the mutant phenotype of ced-1 (e1735), a null allele of ced-1 (Fig. 5D; Zhou et al. 2001). We then overexpressed the EMI domain under the control of heat-shock promoters (PhspSS40-EMI-GFP, secretion form of EMI-GFP) or the ced-1 promoter (Pced-1SS40-EMI-TM-GFP, membrane-anchored EMI by CED-1 transmembrane domain) in a wild-type unc-76 (e911) worm and found that they caused a dominant-negative effect; i.e., transgenic worms showed a cell corpse engulfment defect similar to that of the ttr-52 mutant (Fig. 5D). Overexpression of the F1 fragment of CED-1 lacking the EMI domain (Pced-1CED-1 F1 ΔEMI-TM-GFP) or the first 20 amino acids prior to the EMI domain (Pced-1SS40-TM-GFP) did not cause similar effects (Fig. 5D). These results suggest that TTR-52 interacts and functions via the CED-1 EMI domain.
Figure 5.
Interactions between TTR-52 and CED-1. (A) The CED-1 extracellular domain (CED-1 EX) was truncated to four fragments, and their interactions with TTR-52 were examined by pull-down assays in vitro. (B,C) Purified GST, GST-CED-1 EX, GST-CED-1 F1, GST-CED-1 ΔF1, GST-CED-1 EMI, and GST-CED-1 F1 ΔEMI were immobilized on glutathione-agarose beads, then incubated with TTR-52-His6. The bound proteins were resolved on a 15% SDS-PAGE gel and revealed by immunoblotting with an antibody against a six-histidine tag. Purified GST fusion proteins were stained with Coomassie blue. Three independent experiments were performed and gave identical results. (D) The EMI domain is essential for CED-1's function in cell corpse engulfment. Transgenic worms were generated by expressing either CED-1 without the EMI domain (Pced-1CED-1 ΔEMI-GFP) in ced-1 (e1735) mutant worms or the secreted CED-1 EMI domain (PhspSS40-EMI-GFP), the membrane-anchored EMI domain (Pced-1SS40-EMI-TM-GFP), CED-1 fragment 1 without the EMI domain (Pced-1CED-1 F1 ΔEMI-TM-GFP), or the CED-1 transmembrane domain fused with GFP (Pced-1SS40-TM-GFP) in unc-76 (e911) worms. The number of cell corpses was scored in fourfold stage embryos in ced-1 (e1735) and ced-1 (e1735);Pced-1CED-1 ΔEMI-GFP worms and in 1.5-fold stage embryos in other transgenic worms (two independent transgenic lines were scored; for each line, n ≥ 15). Error bars represent the SEM. (**) P < 0.0001.
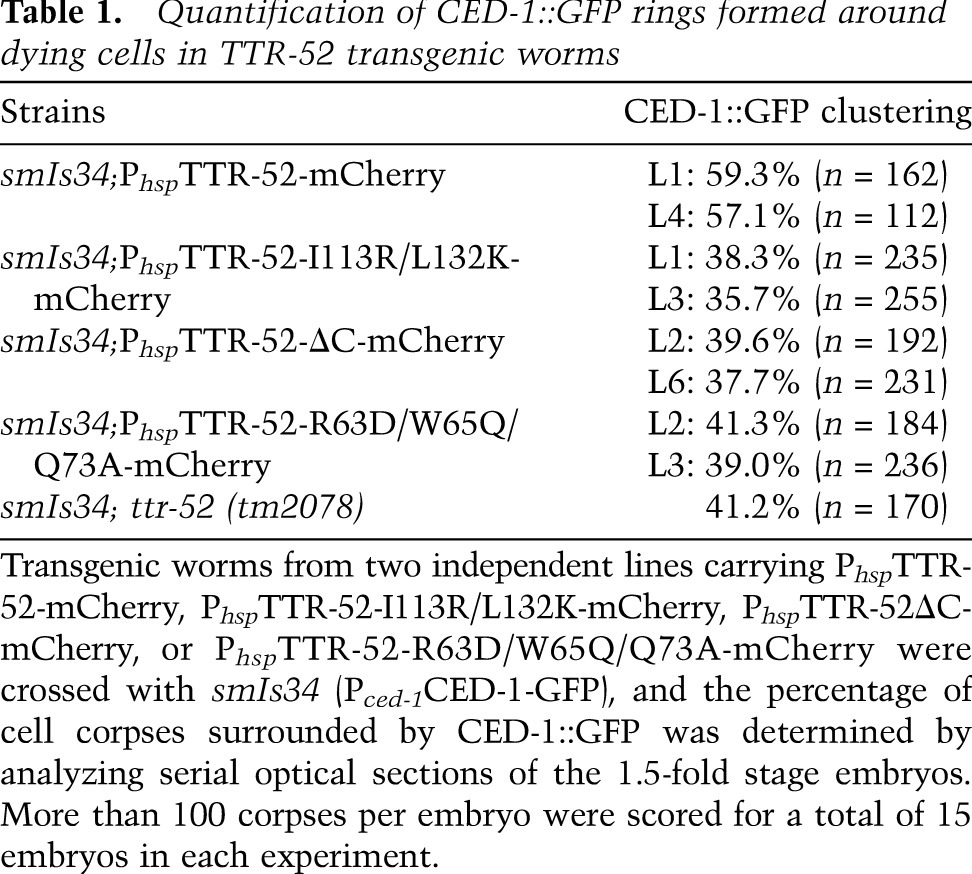
We next investigated the potential CED-1-binding sites on TTR-52. TTR-52 folds into an open β-barrel conformation where strands 6 and 7 form the edges around the opening. Together with the C-terminal tail, the C terminus forms a highly hydrophobic region (Fig. 1B). Some of the hydrophobic residues from this region in our structure are exposed to the solvent. This architecture implies that strands 6 and 7 plus the C-terminal end may be involved in CED-1 binding. We made a double point mutation, I113R/L132K, and a small deletion, 131YLEDY135 (TTR-52 ΔC), in the C terminus of TTR-52, both of which recognized cell corpses normally but failed to rescue the persistent cell corpse phenotype of ttr-52 (tm2078) mutants (Figs. 2C, 3). TTR-52 interacts with the CED-1 receptor to induce its recognition of apoptotic cells (Wang et al. 2010). The percentage of cell corpses clustered by CED-1::GFP in wild-type embryos was 60%, while that in ttr-52 (tm2078) mutants was reduced to 41% (Table 1). We found that expression of TTR-52-I113R/L132K or TTR-52 ΔC failed to rescue the decreased clustering of CED-1 around apoptotic cells in ttr-52 (tm2078) embryos and that <40% of cell corpses were labeled by CED-1::GFP (Table 1). These results indicate that the C-terminal residues of TTR-52 are important for mediating recognition and binding of apoptotic cells by CED-1.
Table 1.
Quantification of CED-1::GFP rings formed around dying cells in TTR-52 transgenic worms
TTR-52 dimer formation is important for its function
TTR-52 (M5) was found to exist as a dimer in our structure, consistent with the behavior of wild-type proteins expressed in 293T cells. Both wild-type TTR-52 proteins purified from the medium of 293T cells transfected with pCMV-TTR-52-Flag and TTR-52 (M5) purified from E. coli were present as dimers in gel filtration chromatography experiments (Fig. 1; Supplemental Fig. S2). In our structure, adjacent TTR-52 (M5) molecules interact back to back to form a butterfly-like dimer structure (Fig. 6A). The interaction surface of the two monomers is ∼700 Å2 in total, as determined by PISA (Krissinel and Henrick 2007). The interaction of adjacent TTR-52 monomers is mainly mediated by residues D58, R63, W65, Q73, and T75 on the third and fourth strands (Fig. 6A). In the structure of TTR-52 (M5), there are salt bridges between the side chains of R63 from one monomer (monomer A) and of D58′ from the other monomer (monomer B, in which residues are indicated with a prime). Three pairs of hydrogen bonds—Q73/Q73′ formed by side chains, and R63/R63′ and W65/T75′ formed by main chains—are also present.
To explore whether the TTR-52 dimer conformation is required for its function, a triple-point mutant R63D/W65Q/Q73A was constructed based on TTR-52 (M5). The purified triple-mutant protein was observed as a monomer in gel filtration experiments (Fig. 1A). To further verify this, cross-linking (Fig. 6B) and sedimentation velocity analytical ultracentrifugation (SV-AUC) (Supplemental Fig. S7) experiments were also performed. Consistent results were obtained using these different methods. Although the triple mutant based on wild-type TTR-52 was not tested here, as it does not fold well in E. coli and has an extraordinarily low level of expression in 293T cells, we presume that the R63/W65/Q73 triple mutation would disrupt its dimer formation.
This triple mutant (PhspTTR-52-R63D/W65Q/Q73A::mCherry) was also expressed in worms to determine whether dimer formation is required for TTR-52 function in vivo. This mutant form of TTR-52::mCherry was still able to form rings on the surface of dying cells with a normal efficiency, but failed to rescue the persistent cell corpse phenotype of ttr-52 (tm2078) (Figs. 2C, 3). We found that the clustering of CED-1::GFP around dying cells was still defective in ttr-52(tm2078) embryos expressing TTR-52-R63D/W65Q/Q73A::mCherry, with only 40% of cell corpses labeled by CED-1::GFP, indicating that interactions between TTR-52 molecules is important for inducing cell corpse recognition by CED-1 (Table 1).
Discussion
In a previous study (Wang et al. 2010), we reported that TTR-52 functions as a bridging molecule that binds PtdSer exposed on the surface of dying cells and the extracellular domain of CED-1 receptor on engulfing cells, thus cross-linking dying cells to engulfing cells and promoting cell corpse engulfment. In this study, we demonstrate how TTR-52 acts in this process by structural and cell biological studies.
While many methods are available for obtaining sufficient amounts of soluble proteins for crystallographic studies (Pantazatos et al. 2004; Keenan et al. 2005), they are not effective in all cases, and new methods are still required. Here, we used alanine-block scanning, a mutagenesis technique that has been broadly applied in protein functional analysis, for recombinant protein optimization and crystallization trials. One mutant of TTR-52—TTR-52 (M5), in which six consecutive residues (50EDSLPL55) were mutated to six alanines—gave well-diffracted crystals, and its structure was determined. We also attempted to purify protein from mutants in which fewer residues in these regions were mutated, but without success (data not shown). We hypothesize that the success of this mutation could be due to its location on a loop in which there are several hydrophobic residues. These hydrophobic residues would lead to the aggregation of this protein when expressed in E. coli. Thus, when this loop was substituted by a stable helix, the folding process for this mutant protein may have been much easier. Thus, we demonstrate here that alanine-block mutagenesis can be successfully applied as a new method in protein crystallization.
The TTR-52 (M5) monomer has an open β-barrel architecture with seven strands connected by five loops. Our data demonstrate that loops L2 and L3 in the N-terminal region are responsible for PtdSer binding because mutations in this region (D51A, D80A, and N85A) greatly reduce its binding affinity to PtdSer in vitro and apoptotic cells in vivo (Figs. 2C, 4; Supplemental Fig. S5) and abolish its function in apoptotic cell engulfment in vivo (Fig. 3). Since many residues in this cleft are negatively charged acidic amino acids, we suspected that cations may mediate PtdSer binding as in other PtdSer-binding proteins, such as TIM4, Annexin V, and the PKCα-C2 domain. However, in spite of its high resolution, we did not find a divalent ion in the TTR-52 (M5) crystal structure, either in the electron density map or by anomalous signal screening, likely due to mutations in predicted cation-binding sites. We therefore verified this hypothesis by chelating divalent cations with EDTA using affinity-purified TTR-52-mCherry-Flag and found that the cation-free protein lost its ability to bind PtdSer, which was not restored when the protein was equilibrated with divalent cations (Fig. 4), implicating its importance in stabilizing protein structure. Although we showed the importance of divalent cations to TTR-52, we were not able to determine which cations are involved. We predicted that calcium is important because of the structural similarity between TTR-52 and the PKCα-C2 domain, in which PtdSer binding has been shown to be calcium-dependent. In contrast to the PKCα-C2 domain, the hydrophobic residues on the tips of loop 2 and loop 3 of TTR-52 are also involved in PtdSer binding; mutations at L53, P54, and L55 in loop 2 and at P83 in loop 3 affected cell corpse recognition, and the latter even abolished TTR-52's function in cell corpse removal (Fig. 3). These hydrophobic residues are predicted to bind directly to the membranes of dying cells, as is the case for TIM4, which is expressed on the surface of antigen-presenting cells and mediates the phagocytosis of apoptotic cells. We propose that these hydrophobic residues may assist the binding of this protein to the surface of dying cells.
The L1 and L4 loops of TTR-52 also contain some negatively charged residues, but our results show that these residues are not essential to the binding of PtdSer by TTR-52 (Supplemental Figs. S4, S5). Combined mutation of these residues has a limited effect on apoptotic cell engulfment in vivo (Fig. 3). No cation ion was observed in this area in the structure. The exact function of this region is thus unclear. Notably, in the structure of synaptotagmin, the prototypical C2 domain-containing protein, there are also some clustered acidic residues in the loop region opposite the PtdSer-binding region (Sutton et al. 1995). This similarity suggests that the additional acidic regions in the two proteins may have important functions.
The presence of a large hydrophobic region in the C terminus of TTR-52—containing strand 6, strand 7, and the C-terminal tail—suggests that it may be the binding site for the extracellular domain of CED-1. Both double mutation of I113R/L132K and deletion of the C-terminal five residues failed to rescue the ttr-52 mutant phenotype (Fig. 3), and the percentage of CED-1::GFP rings around dying cells was greatly decreased when these mutations were expressed (Table 1). This implies that the C-terminal hydrophobic region of TTR-52 may interact with the CED-1 extracellular domain (CED-1 EX), although we could not prove this by using either ex vivo pull-down assays with recombinant proteins purified from E. coli or coimmunoprecipitation in transfected 293T cells. On the other hand, using a truncated protein pull-down assay, we found that the EMI domain in the extracellular region of CED-1 is responsible for its interaction with TTR-52. The EMI domain, which has been reported to be involved in protein–protein interactions (Doliana et al. 2000), is necessary and sufficient for the interaction of CED-1 with TTR-52 in vitro. We further confirmed this result in vivo; overexpression of the EMI domain of CED-1 alone in wild-type worms caused dominant-negative effects, giving a number of corpses in transgenic worms similar to that in the ttr-52 mutant (Fig. 5D).
Unlike other PtdSer-binding proteins, TTR-52 exists as a dimer in both the crystal structure and solution, as confirmed by gel filtration, sedimentation velocity, and cross-linking experiments (Figs. 1A, 6B; Supplemental Fig. S7). When the crucial residues R63, W65, and Q73 between the dimer interface were mutated, TTR-52 (M5) lost its dimer state and behaved as a monomer. When this mutated form of TTR-52 was introduced into worms, it resulted in decreased levels of CED-1::GFP ring formation around dying cells (Table 1) and could not rescue the ttr-52 mutant phenotype, although PtdSer labeling on the surface of dying cells was not affected (Figs. 2C, 3). It is thus clear that the interaction between TTR-52 molecules is important for its function, especially in inducing cell corpse recognition by CED-1. In addition, loss of TTR-52 reduces but does not totally block recognition of apoptotic cells by CED-1 (Wang et al. 2010), suggesting that multiple factors may be involved in mediating cell corpse engulfment through CED-1. It is therefore interesting to determine whether other PtdSer-binding proteins are involved and similar mechanisms are employed.
Based on the structural and functional studies reported here, we propose a model for the role of TTR-52 in mediating cell engulfment. TTR-52 forms a dimer in cells and the dimer provides two possible PtdSer-binding sites. The two PtdSer-binding sites of one TTR-52 dimer mimic the two upper wings of the butterfly-like structure of TTR-52 (Fig. 6A) and extend out from the dimer in the same direction, suggesting that the two PtdSer-binding sites can bind to apoptotic cells simultaneously, likely increasing the binding affinity of this protein to the apoptotic cell membrane. The two large hydrophobic regions in the C terminus of the dimer, mimicking the lower wings of the butterfly, provide binding sites for the EMI domain of CED-1. Thus, the apoptotic cells and phagocytes cells are cross-linked via the bridging molecule.
In summary, we report the first full-length crystal structure of a secreted bridging molecule that can be recruited by PtdSer exposed on the surface of dying cells to trigger CED-1-dependent cell corpse engulfment. These results reveal a potential mechanism of the TTR-52 bridging molecule in cell corpse engulfment in C. elegans. Since the bridging molecule-mediated engulfment phenomenon is conserved in higher organisms, this work provides useful information for studies of engulfment in mammalian cells.
Materials and methods
Plasmid construction
Standard methods of PCR amplification, cloning, and sequencing were used. A detailed description is provided in the Supplemental Material. All constructs generated using PCR-amplified fragments were verified by DNA sequencing.
Protein expression and purification
Plasmid pET21b-TTR-52 ΔN-His was transformed into E. coli strain BL21 (DE3) (Invitrogen). Cells were cultured in LB medium at 37°C with 100 mg/L ampicillin. When the OD600 reached 0.6–0.8, the culture was induced by addition of isopropyl-thio-D-glactosidase (IPTG) (Sigma) to a final concentration of 0.5 mM for 20 h at 16°C. Cells were harvested by centrifugation at 5000 rpm for 10 min. Pellets were resuspended in Tris buffer (20 mM Tris at pH 8.0, 150 mM NaCl) and lysed by sonication. The lysate was separated by centrifugation at 16,000 rpm for 30 min, and the recovered supernatant was applied to a Ni-NTA affinity column (Qiagen), followed by intensive washing with washing buffer (20 mM Tris at pH 8.0, 150 mM NaCl, 50 mM imidazole). Recombinant TTR-52 was eluted from the Ni-NTA affinity column using elution buffer (20 mM Tris at pH 8.0, 150 mM NaCl, 500 mM imidazole) and further purified by gel filtration with a Superdex200 column (GE Healthcare) using Tris buffer as described above on an FPLC protein purification system.
Crystallization and structure determination
The protein was concentrated for crystallization in spin concentrators (Millipore) to a concentration of ∼3–4 mg/mL. SDS-PAGE was used to determine the purity of the proteins. Crystallization trials were set up using crystal screen kits (Hampton Research) using the hanging-drop method by mixing 1 μL of protein solution with an equal volume of a reservoir solution. Crystals of a usable size were obtained in a buffer condition with 0.2 M ammonium sulfate, 0.1 M sodium acetate (pH 4.5), and 35% PEGME2000. PtCl4 salt was soaked into the crystals to obtain heavy atom derivative crystals. Native and derivative data sets were collected on an FR-E SuperBright rotating anode X-ray source equipped with an R-AXIS IV++ imaging plate detector (Rigaku). Data were collected at 150 K and processed using the HKL2000 program suite (Otwinowski and Minor 1997). The platinum sites and initial phases were generated by SHELX (Sheldrick 2008) using the SIRAS method. An automatic program, autoSHARP (Vonrhein et al. 2007), was applied to generate and improve the electron density map. About 90% of the residues were built automatically by ARP/wARP (Morris et al. 2003), and the additional missing residues were added manually in COOT (Emsley et al. 2010). The final model was refined with PHENIX (Adams et al. 2010). In the Ramachandran plot generated using PROCHECK (Laskowski et al. 1993), 95.58% and 4.42% of the amino acids in the final atomic model were in the most favorable and additional allowed regions, respectively. No amino acids were in the disallowed region. The atomic coordinates and diffraction data have been deposited in the Protein Data Bank (3UAF). All structural figures were prepared with COOT and PyMOL (http://www.pymol.org).
Strains and culture conditions
C. elegans strains were maintained using standard methods (Brenner 1974). The N2 Bristol strain was used as the wild-type strain. All of the alleles used in this study have been described previously (Ellis et al. 1991; Riddle et al. 1997) except the ttr-52 (tm2078), which is an engulfment defect mutant in this study (Supplemental Fig. S8).
Quantification of cell corpses
The number of cell corpses in early stage embryos was scored using Nomarski optics as described before (Stanfield and Horvitz 2000).
Fluorescence microscopy and quantification of the CED-1::GFP clustering around apoptotic cells
To score the percentage of cell corpses labeled by CED-1::GFP, Nomarski and fluorescent images of embryos in a 20-μm series (1 μm per section) were captured with a Zeiss Axioimager M1 equipped with epifluorescence and an AxioCam monochrome digital camera. Serial optical sections of embryos were analyzed, first to identify cell corpses by their raised disc-like morphology in 1.5-fold-stage embryos, and then to determine the clustering of CED-1::GFP.
Heat-shock treatment
Animals at various stages were incubated for 1 h at 33°C, followed by recovery at 20°C. Cell corpses and TTR-52 expression were examined 3.5 h after the heat-shock treatment.
PtdSer-binding assays
Wild-type Saccharomyces cerevisiae strain BY4743 (a gift from Y. Zhang, National Institute of Biological Sciences, Beijing, China) was streaked on YPAD plates and cultured at 30°C. p416GPD vectors expressing TTR-52-mCherry, TTR-52 D51A-mCherry, TTR-52 D80A-mCherry, TTR-52 N85A-mCherry, or TTR-52 D36A/E38A/R100A/D101A-mCherry were transformed into yeast cells, followed by colony selection on agar plates with synthetic medium lacking uracil. A single colony was selected and cultured overnight in uracil dropout synthetic medium at 30°C, and the resulting yeast cells were examined with an Axioimager M1 equipped with epifluorescence.
TTR-52-mCherry-Flag, TTR-52-D51A-mCherry-Flag, TTR-52-D80A-mCherry-Flag, and TTR-52-N85A-mCherry-Flag were affinity-purified, respectively, from the culture medium of 293T cells transfected with pCMV-TTR-52-mCherry-Flag or its mutant forms and used for in vitro lipid binding assays. Membrane strips spotted with various phospholipids (P-6001; Echelon Biosciences) were first blocked with buffer B (25 mM Tris-HCl at pH 7.0, 150 mM NaCl, 0.1% Tween 20, 1% dried milk, 0.1 mM ZnSO4, 0.2 mM CaCl2) for 1 h at 22°C and then incubated overnight at 4°C with affinity-purified TTR-52 proteins in buffer B without dried milk. The membrane strip was washed three times in buffer B without dried milk and blotted with the M2 anti-Flag antibody (Sigma). To chelate divalent cations, TTR-52-mCherry-Flag pulled down by the anti-Flag M2 affinity gel (Sigma) was washed with TBSE buffer (50 mM Tris-HCl at pH 7.4, 150 mM NaCl, 1mM EDTA). Proteins were eluted with elution buffer (10 mM Tris-HCl at pH 7.4, 150 mM NaCl, 100 μg/μL Flag peptide [Sigma]) and then aliquoted into four tubes. A final concentration of 0.2 mM Ca2+, 0.2 mM Zn2+, or 0.2 mM Ca2+/0.1 mM Zn2+ was added to three of the tubes. After incubating for 10 min, the protein solution from each of the four tubes was applied to membrane lipid strips. Protein from the same batch, eluted before EDTA chelation, was used in parallel as a control. The amount of protein was quantified by Western blotting with an antibody against TTR-52 (Antibody Production Facility, National Institute of Biological Sciences, Beijing).
GST fusion protein pull-down assays
Purified GST, GST-CED-1 EX, GST-CED-1 F1, GST-CED-1 ΔF1, GST-CED-1 EMI, and GST-CED-1 F1 ΔEMI (1 μg of each) immobilized, respectively, on glutathione-agarose beads (Sigma) were incubated with 2.5 μg TTR-52 ΔN-His6 in a 200-μL reaction volume containing 25 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% Nonidet P40, 10% glycerol, 1 mM phenylmethylsulphonyl fluoride (PMSF), and 5 mM dithiothreitol (DTT) at 4°C. The beads were washed five times with the same buffer, and bound proteins were resolved by 15% SDS-PAGE and detected by Western blotting with an antibody against the hexahistidine tag (Santa Cruz Biotechnology).
Cross-linking assays
Recombinant proteins TTR-52 (M5) and TTR-52 (M5)-R63D/W65Q/Q73A were purified from E. coli as described above. Ethylene glycol bis-succinimidyl succinate (EGS) (1 mM or 5 mM) (Sigma) was added to the recombinant proteins (2 μg) in 15 μL of 1× PBS buffer. Cross-linking reactions were carried out for 30 min at room temperature and stopped by adding EDTA. Samples were separated by 15% SDS-PAGE and stained by Coomassie blue.
Acknowledgments
We thank Jie Zhou and Jian Sun for assistance in crystal data collection and structure determination, Xueyao Jin for technical assistance in cross-linking assays, Xiaoxia Yu for technical assistance in sedimentation velocity analytical ultracentrifugation, and laboratory members in Y.L.'s and X.W.'s laboratories for valuable discussions. We also thank Joy Fleming for critical reading and revision of this manuscript. This work was supported by grants to Y.L. from the Ministry of Science and Technology (863 Project 2006AA02A314; 973 Programs 2007CB914303 and 2011CB910304) and the National Science Foundation of China (30925011, 31030024, and 31021062), to H.L. from the National Science Foundation of China (31000331), and to X.W. from the Ministry of Science and Technology (973 Programs 2010CB835202).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.187815.112.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT 2005. Protein structure prediction servers at University College London. Nucleic Acids Res 33: W36–W38 doi: 10.1093/nar/gki410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut I, Mignotte V, Souchet M, Mornon JP 2003. EMI domains are widespread and reveal the probable orthologs of the Caenorhabditis elegans CED-1 protein. Biochem Biophys Res Commun 300: 619–623 [DOI] [PubMed] [Google Scholar]
- Connelly S, Choi S, Johnson SM, Kelly JW, Wilson IA 2010. Structure-based design of kinetic stabilizers that ameliorate the transthyretin amyloidoses. Curr Opin Struct Biol 20: 54–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietmann S, Park J, Notredame C, Heger A, Lappe M, Holm L 2001. A fully automatic evolutionary classification of protein folds: Dali domain dictionary version 3. Nucleic Acids Res 29: 55–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doliana R, Bot S, Bonaldo P, Colombatti A 2000. EMI, a novel cysteine-rich domain of EMILINs and other extracellular proteins, interacts with the gC1q domains and participates in multimerization. FEBS Lett 484: 164–168 [DOI] [PubMed] [Google Scholar]
- Ellis RE, Jacobson DM, Horvitz HR 1991. Genes required for the engulfment of cell corpses during programmed cell death in Caenorhabditis elegans. Genetics 129: 79–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A 2006. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics 15: 5.6.1–5.6.30 doi: 10.1002/0471250953.bi0506s15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriaud C, Juanhuix J, Gruez A, Lacroix M, Darnault C, Pignol D, Verger D, Fontecilla-Camps JC, Arlaud GJ 2003. The crystal structure of the globular head of complement protein C1q provides a basis for its versatile recognition properties. J Biol Chem 278: 46974–46982 [DOI] [PubMed] [Google Scholar]
- Guerrero-Valero M, Ferrer-Orta C, Querol-Audi J, Marin-Vicente C, Fita I, Gomez-Fernandez JC, Verdaguer N, Corbalan-Garcia S 2009. Structural and mechanistic insights into the association of PKCα-C2 domain to PtdIns(4,5)P2. Proc Natl Acad Sci 106: 6603–6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanayama R, Tanaka M, Miwa K, Shinohara A, Iwamatsu A, Nagata S 2002. Identification of a factor that links apoptotic cells to phagocytes. Nature 417: 182–187 [DOI] [PubMed] [Google Scholar]
- Henson PM, Bratton DL, Fadok VA 2001. Apoptotic cell removal. Curr Biol 11: R795–R805 doi: 10.1016/S0960-9822(01)00474-2 [DOI] [PubMed] [Google Scholar]
- Hornberg A, Eneqvist T, Olofsson A, Lundgren E, Sauer-Eriksson AE 2000. A comparative analysis of 23 structures of the amyloidogenic protein transthyretin. J Mol Biol 302: 649–669 [DOI] [PubMed] [Google Scholar]
- Keenan RJ, Siehl DL, Gorton R, Castle LA 2005. DNA shuffling as a tool for protein crystallization. Proc Natl Acad Sci 102: 8887–8892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N, Karisola P, Pena-Cruz V, Dorfman DM, Jinushi M, Umetsu SE, Butte MJ, Nagumo H, Chernova I, Zhu B, et al. 2007. TIM-1 and TIM-4 glycoproteins bind phosphatidylserine and mediate uptake of apoptotic cells. Immunity 27: 927–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Laskowski AR, Macarthur WM, Moss SD, Thornton MJ 1993. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Liu QA, Hengartner MO 1998. Candidate adaptor protein CED-6 promotes the engulfment of apoptotic cells in C. elegans. Cell 93: 961–972 [DOI] [PubMed] [Google Scholar]
- Morris RJ, Perrakis A, Lamzin VS 2003. ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol 374: 229–244 [DOI] [PubMed] [Google Scholar]
- Nakano T, Ishimoto Y, Kishino J, Umeda M, Inoue K, Nagata K, Ohashi K, Mizuno K, Arita H 1997. Cell adhesion to phosphatidylserine mediated by a product of growth arrest-specific gene 6. J Biol Chem 272: 29411–29414 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pantazatos D, Kim JS, Klock HE, Stevens RC, Wilson IA, Lesley SA, Woods VL Jr 2004. Rapid refinement of crystallographic protein construct definition employing enhanced hydrogen/deuterium exchange MS. Proc Natl Acad Sci 101: 751–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Tosello-Trampont AC, Elliott MR, Lu M, Haney LB, Ma Z, Klibanov AL, Mandell JW, Ravichandran KS 2007. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 450: 430–434 [DOI] [PubMed] [Google Scholar]
- Park SY, Jung MY, Kim HJ, Lee SJ, Kim SY, Lee BH, Kwon TH, Park RW, Kim IS 2008. Rapid cell corpse clearance by stabilin-2, a membrane phosphatidylserine receptor. Cell Death Differ 15: 192–201 [DOI] [PubMed] [Google Scholar]
- Prapunpoj P, Leelawatwattana L 2009. Evolutionary changes to transthyretin: Structure-function relationships. FEBS J 276: 5330–5341 [DOI] [PubMed] [Google Scholar]
- Reddien PW, Horvitz HR 2000. CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nat Cell Biol 2: 131–136 [DOI] [PubMed] [Google Scholar]
- Riddle DL, Blumenthal T, Meyer BJ, Priess JR 1997. Introduction to C. elegans. In C. elegans II, 2nd ed. (ed. DL Riddle et al.), pp. 1–22. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Santiago C, Ballesteros A, Martinez-Munoz L, Mellado M, Kaplan GG, Freeman GJ, Casasnovas JM 2007. Structures of T cell immunoglobulin mucin protein 4 show a metal-ion-dependent ligand binding site where phosphatidylserine binds. Immunity 27: 941–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savill J, Fadok V 2000. Corpse clearance defines the meaning of cell death. Nature 407: 784–788 [DOI] [PubMed] [Google Scholar]
- Savill J, Dransfield I, Gregory C, Haslett C 2002. A blast from the past: Clearance of apoptotic cells regulates immune responses. Nat Rev Immunol 2: 965–975 [DOI] [PubMed] [Google Scholar]
- Schreiber G 2002. The evolutionary and integrative roles of transthyretin in thyroid hormone homeostasis. J Endocrinol 175: 61–73 [DOI] [PubMed] [Google Scholar]
- Shao C, Novakovic VA, Head JF, Seaton BA, Gilbert GE 2008. Crystal structure of lactadherin C2 domain at 1.7A resolution with mutational and computational analyses of its membrane-binding motif. J Biol Chem 283: 7230–7241 [DOI] [PubMed] [Google Scholar]
- Sheldrick GM 2008. A short history of SHELX. Acta Crystallogr A 64: 112–122 [DOI] [PubMed] [Google Scholar]
- Stanfield GM, Horvitz HR 2000. The ced-8 gene controls the timing of programmed cell deaths in C. elegans. Mol Cell 5: 423–433 [DOI] [PubMed] [Google Scholar]
- Su HP, Nakada-Tsukui K, Tosello-Trampont AC, Li Y, Bu G, Henson PM, Ravichandran KS 2002. Interaction of CED-6/GULP, an adapter protein involved in engulfment of apoptotic cells with CED-1 and CD91/low density lipoprotein receptor-related protein (LRP). J Biol Chem 277: 11772–11779 [DOI] [PubMed] [Google Scholar]
- Sutton RB, Davletov BA, Berghuis AM, Sudhof TC, Sprang SR 1995. Structure of the first C2 domain of synaptotagmin I: A novel Ca2+/phospholipid-binding fold. Cell 80: 929–938 [DOI] [PubMed] [Google Scholar]
- Swairjo MA, Concha NO, Kaetzel MA, Dedman JR, Seaton BA 1995. Ca2+-bridging mechanism and phospholipid head group recognition in the membrane-binding protein annexin V. Nat Struct Biol 2: 968–974 [DOI] [PubMed] [Google Scholar]
- Verdaguer N, Corbalan-Garcia S, Ochoa WF, Fita I, Gomez-Fernandez JC 1999. Ca2+ bridges the C2 membrane-binding domain of protein kinase Cα directly to phosphatidylserine. EMBO J 18: 6329–6338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G 2007. Automated structure solution with autoSHARP. Methods Mol Biol 364: 215–230 [DOI] [PubMed] [Google Scholar]
- Wang X, Wu YC, Fadok VA, Lee MC, Gengyo-Ando K, Cheng LC, Ledwich D, Hsu PK, Chen JY, Chou BK, et al. 2003. Cell corpse engulfment mediated by C. elegans phosphatidylserine receptor through CED-5 and CED-12. Science 302: 1563–1566 [DOI] [PubMed] [Google Scholar]
- Wang X, Li W, Zhao D, Liu B, Shi Y, Chen B, Yang H, Guo P, Geng X, Shang Z, et al. 2010. Caenorhabditis elegans transthyretin-like protein TTR-52 mediates recognition of apoptotic cells by the CED-1 phagocyte receptor. Nat Cell Biol 12: 655–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YC, Horvitz HR 1998. The C. elegans cell corpse engulfment gene ced-7 encodes a protein similar to ABC transporters. Cell 93: 951–960 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Hartwieg E, Horvitz HR 2001. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell 104: 43–56 [DOI] [PubMed] [Google Scholar]