Abstract
Seeds of several agriculturally important legumes are rich sources of the only halogenated plant hormone, 4-chloroindole-3-acetic acid. However, the biosynthesis of this auxin is poorly understood. Here, we show that in pea (Pisum sativum) seeds, 4-chloroindole-3-acetic acid is synthesized via the novel intermediate 4-chloroindole-3-pyruvic acid, which is produced from 4-chlorotryptophan by two aminotransferases, TRYPTOPHAN AMINOTRANSFERASE RELATED1 and TRYPTOPHAN AMINOTRANSFERASE RELATED2. We characterize a tar2 mutant, obtained by Targeting Induced Local Lesions in Genomes, the seeds of which contain dramatically reduced 4-chloroindole-3-acetic acid levels as they mature. We also show that the widespread auxin, indole-3-acetic acid, is synthesized by a parallel pathway in pea.
The chlorinated auxin, 4-chloroindole-3-acetic acid (4-Cl-IAA), is found in certain higher plants and is more active in some bioassays than the most widespread endogenous auxin, indole-3-acetic acid (IAA; Reinecke, 1999). In seeds of some key legumes, such as Lens culinaris, Lathyrus latifolius, Vicia faba, and pea (Pisum sativum), the levels of 4-Cl-IAA are among the highest reported for any auxin in plant tissues (Reinecke, 1999). In pea, it has been suggested that 4-Cl-IAA moves from young seeds into the pod, where it is required for normal pod elongation (Reinecke, 1999; Ozga et al., 2009), but no mutant has been available to aid our understanding of the biosynthesis and roles of 4-Cl-IAA, and there is limited evidence on the origin of this compound (Reinecke, 1999).
Despite more than 70 years of intensive investigation (Normanly, 2010; Zhao, 2010), it is only recently that we have begun to fully understand auxin biosynthesis, even in Arabidopsis (Arabidopsis thaliana). In the past, four Trp-dependent IAA biosynthetic pathways have been proposed (Normanly, 2010; Zhao, 2010), but it now appears that one pathway, involving indole-3-pyruvic acid (IPyA), predominates in Arabidopsis (Mashiguchi et al., 2011; Stepanova et al., 2011; Won et al., 2011; Kriechbaumer et al., 2012). Until recently, it was thought that the enzymes involved in converting Trp to IPyA, the TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS (TAA) family (Stepanova et al., 2008; Tao et al., 2008), acted in a separate pathway from the YUCCA (YUC) family (Zhao et al., 2001; Zhao, 2010). The YUCs were originally thought to function in the tryptamine pathway, converting tryptamine to N-hydroxytryptamine (Zhao et al., 2001; Expósito-Rodríguez et al., 2007; LeClere et al., 2010). However, the original biochemical function of the YUCs was called into question (Tivendale et al., 2010), and it has now been reported that YUCs act downstream of the TAA family, converting IPyA to IAA (Mashiguchi et al., 2011; Stepanova et al., 2011; Won et al., 2011; Kriechbaumer et al., 2012). Nevertheless, Arabidopsis is known to utilize Brassicaceae-specific biosynthetic pathways (Sugawara et al., 2009), and the IPyA pathway has yet to be demonstrated definitively in species other than Arabidopsis. A potential co-ortholog has been isolated from maize (Zea mays; Phillips et al., 2011), but its enzymatic activity has not been reported. In this study, we examine the biosynthesis of 4-Cl-IAA and IAA in pea seeds, presenting evidence for the importance of the pathway through IPyA and its chlorinated analog in these organs.
RESULTS
In Vitro Analysis of PsTAR1 and PsTAR2
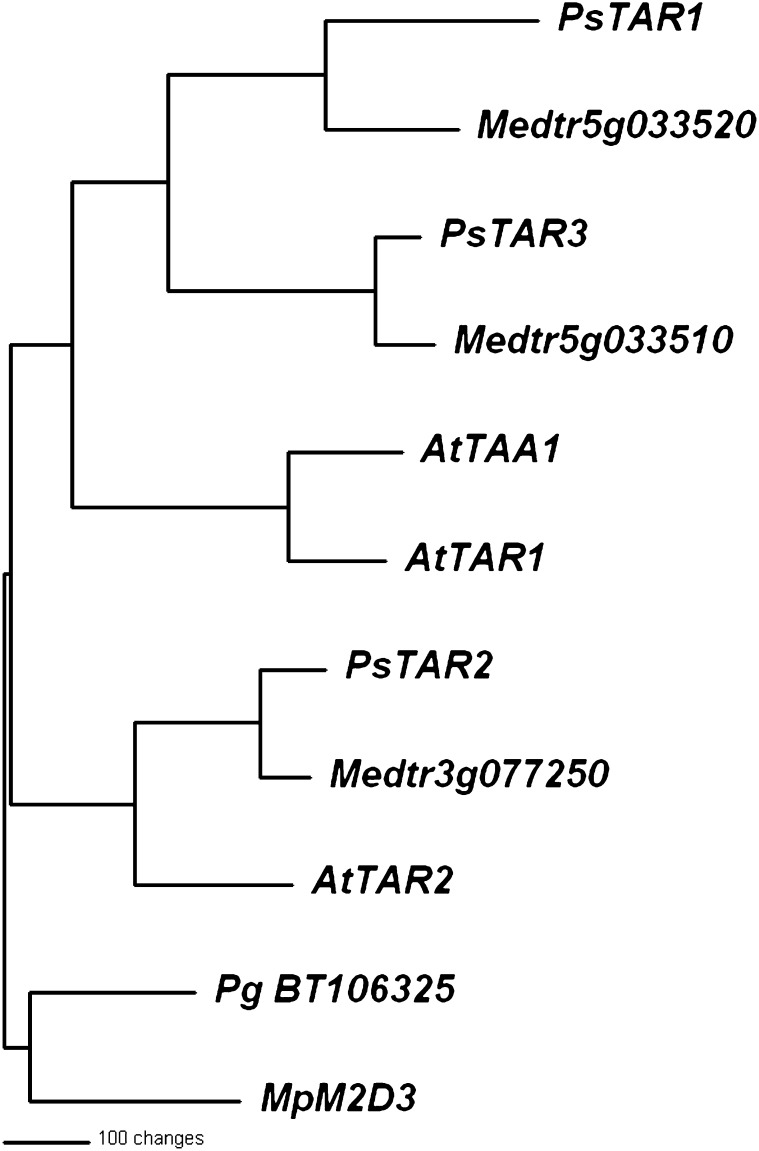
Since evidence for the involvement of Trp aminotransferases in auxin biosynthesis is accumulating (Stepanova et al., 2008; Tao et al., 2008; Chourey et al., 2010; Phillips et al., 2011), we isolated from pea three Trp aminotransferase genes, which we named TRYPTOPHAN AMINOTRANSFERASE RELATED1 (PsTAR1), PsTAR2, and PsTAR3. Their inferred phylogenetic relationships are shown in Figure 1. The sequences of PsTAR1 and PsTAR3 are very similar to each other and 51% homologous to AtTAA1 at the protein level. However, PsTAR2 is more closely aligned to the AtTAR2 (56% identity).
Figure 1.
Inferred phylogenetic relationship of the three pea TAR genes, PsTAR1 (JN990988), PsTAR2 (JN990989), and PsTAR3 (JN990990), with other TAR-like genes. The phylogram was generated as described previously (Tivendale et al., 2010). Included were AtTAA1 (At1g70560), AtTAR1 (At1g23320), and AtTAR2 (At4g24670) from Arabidopsis and Medicago TAR-like sequences Medtr5g033510, Medtr5g033520, and Medtr3g077250. The Medicago sequences were obtained by a BLAST search of the International Medicago Genome Annotation Group (IMGAG) database version 3.5 at the M. truncatula Sequencing Resources (www.medicagohapmap.org/?genome) and correspond to Mt5g034880, Mt5g034890, and Mt3g114550, respectively, from version 3.0 of the IMGAG database. Sequences from the gymnosperm Picea glauca BT106325 and the liverwort M. polymorpha MpM2D3 AF542555_1 were chosen as the outgroup from the more comprehensive phylogram of Phillips et al. (2011).
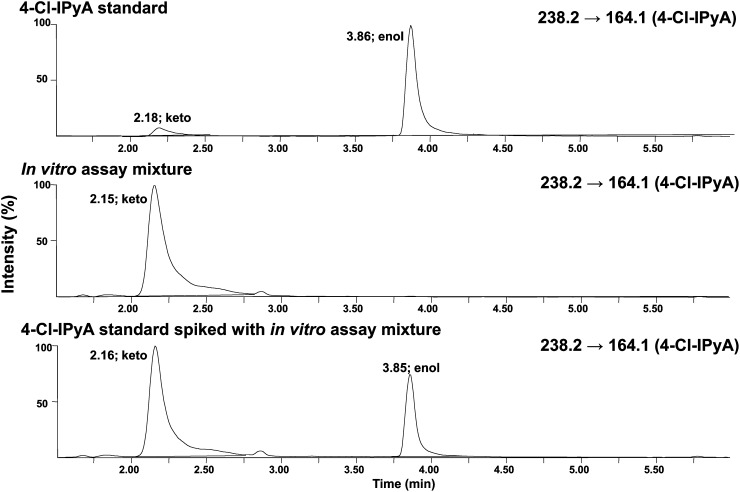
To determine the biochemical function of PsTAR1 and PsTAR2, the corresponding coding sequences were expressed in Escherichia coli, and the resulting recombinant maltose-binding protein (MBP) fusion proteins were purified using affinity chromatography. As in Stepanova et al. (2008) and Mashiguchi et al. (2011), the MBP tag was not cleaved from the protein of interest prior to in vitro analysis. After incubation with potential substrates, analysis by ultra-performance liquid chromatography (UPLC)-mass spectrometry (MS) revealed that both PsTAR1 and PsTAR2 produced 4-chloroindole-3-pyruvic acid (4-Cl-IPyA) when 4-chlorotryptophan (4-Cl-Trp) was supplied and IPyA when Trp was supplied (Supplemental Fig. S1). The identities of the products were confirmed by comparison of retention times (RTs) and mass spectra with those of authentic 4-Cl-IPyA (Fig. 2) and IPyA (Supplemental Fig. S1). The novel compound, 4-Cl-IPyA, was synthesized in our laboratory. These results show that both proteins have aminotransferase activity for Trp and its chlorinated analog, 4-Cl-Trp.
Figure 2.
UPLC-MS chromatograms (MRM mode; acetic acid program) of 4-Cl-IPyA standard (top channel), an in vitro assay mixture using PsTAR1 as the enzyme and 4-Cl-Trp as the substrate (middle channel), and a mixture of the two samples (bottom channel). Mixing the two samples showed coelution of the product with the standard. Given the relative signal strengths of the keto and enol tautomers in the mixture, compared with the individual samples, the mixture represents approximately a 1:1 ratio of the two samples. Identical mass transitions and coelution of the product with the standard confirm its identity as keto 4-Cl-IPyA.
4-Cl-IAA and IAA were also detected in the above in vitro assays, and to determine if this was due to enzymatic conversion or physicochemical breakdown, we conducted in vitro assays using 4-Cl-IPyA and IPyA as substrates. UPLC-MS analysis of reaction mixtures showed no difference in 4-Cl-IAA or IAA production between the tests and controls, thereby indicating that the 4-Cl-IAA and IAA detected after feeds of 4-Cl-Trp and Trp, respectively, were due to physicochemical, rather than enzymatic, conversion of the initial enzyme products, 4-Cl-IPyA and IPyA.
Keto-Enol Tautomerization of IPyA and 4-Cl-IPyA
IPyA, like all compounds containing a ketone functionality, exists as an equilibrium between two tautomers: keto and enol (Supplemental Fig. S2; Schwarz, 1961). In the case of IPyA, it might be predicted that the enol tautomer would be more stable, as it increases the conjugation of the molecule. To better understand the behavior of this compound in auxin biosynthesis, we characterized it in vitro.
We analyzed authentic IPyA by 1H- and 13C-NMR and UPLC-MS. NMR analyses of IPyA in a variety of solvents (at concentrations greater than 10 mm), including CD3OD (1H-NMR, 300 MHz δ: 6.92 [s, 1H], 7.09 [m, 2H], 7.36 [d, J = 4.2 Hz, 1H], 7.68 [d, J = 7.5 Hz, 1H], 7.93 [s, 1H]; 13C-NMR, 75 MHz δ: 104.4 [CH], 110.2 [C], 111.2 [CH], 117.8 [CH], 119.5 [CH], 121.7 [CH], 127.1 [C], 127.4 [CH], 136.2 [C], 137.5 [C], 137.4 [C]), [D6]dimethyl sulfoxide (1H-NMR, 300 MHz δ: 6.76 [s, 1H], 7.08 [m, 2H], 7.38 [d, J = 8.1 Hz, 1H], 7.68 [d, J = 7.8 Hz, 1H], 7.84 [s, 1H], 8.73 [s, 1H], 11.39 [s, 1H]), and aqueous solutions with various pH values (neutral to 13) all indicated the presence of the IPyA enol tautomer only, along with small amounts of breakdown products. When synthetic IPyA was dissolved in methanol and analyzed by UPLC-MS (multiple reaction monitoring [MRM] mode; acetic acid program), one peak dominated the IPyA channels and the breakdown products IAA and indole-3-carboxaldehyde were observed in their respective channels (data not shown). The NMR data indicate that this peak was IPyA in the enol tautomer.
However, when we dissolved IPyA in KH2PO4/K2HPO4 buffer (greater than 10 mm; pH 6.5 or 8.5) and heated to 40°C for 3 h (roughly mimicking the in vitro assay conditions), water-suppression NMR revealed primarily the keto tautomer of IPyA with a small amount of the enol form (1H-NMR, 400 MHz δ: 4.1 [s, 2H] 7.0–7.2 [m, 3H], 7.4 [d of d, J = 8.23 Hz, 8.45 Hz 2H]; 13C-NMR, 100 MHz δ: 40.9 [CH2], 110.9 [C], 117.1 [CH], 123.7 [CH], 124.6 [CH], 127.1 [CH], 130.5 [CH], 132.0 [C], 141.4 [C], 175.7 [C], 209.5 [C]). This treatment of IPyA also changed its behavior on UPLC-MS (acetic acid program). Under these conditions, IPyA (shown by NMR to be the keto form) eluted earlier than IAA, whereas the enol tautomer eluted later than IAA. There were substantial similarities in the tandem MS spectra produced from the solution in CD3OD and the solution in KH2PO4/K2HPO4 buffer (pH 8.5), both consistent with IPyA (keto: mass-to-charge ratio [m/z]: 115 [8%], 118 [8%], 130 [100%], 142 [8%], 158 [63%]; enol: m/z: 103 [6%], 130 [100%], 144 [7%], 158 [82%]). As further evidence that the early peak represented IPyA, we performed a multiplicity-edited 13C-1H heteronuclear single quantum coherence (HSQC) NMR experiment on an IPyA sample prepared in KH2PO4/K2HPO4 buffer (pH 8.5 in water) and heated to 40°C for 3 h. This revealed the presence of five aromatic CH groups and one aliphatic CH2 (Supplemental Fig. S3). These findings indicate that the early peak was the keto tautomer of IPyA. Interestingly, at lower concentrations, mixtures of the tautomers were observed by UPLC-MS, sometimes approximating to 1:1 (Supplemental Fig. S4, top channel). Our analyses show that the relative proportions of the IPyA tautomers can be controlled using temperature and pH, and the two forms are readily resolved by UPLC; the same was found for 4-Cl-IPyA (Supplemental Fig. S4).
In functional assays of PsTAR1 and PsTAR2 with Trp or 4-Cl-Trp as the substrate, both keto and enol tautomers of IPyA and 4-Cl-IPyA were detected as products by MS, but the keto form predominated (Fig. 2; Supplemental Fig. S1). However, direct monitoring of endogenous 4-Cl-IPyA and IPyA extracted from pea seeds yielded evidence for the keto tautomers only (at low levels; Supplemental Fig. S4). The evidence presented above supports a role for keto IPyA in IAA biosynthesis in pea.
Analysis of PsTAR1 and PsTAR2 Expression Patterns and Auxin Levels during Seed Development
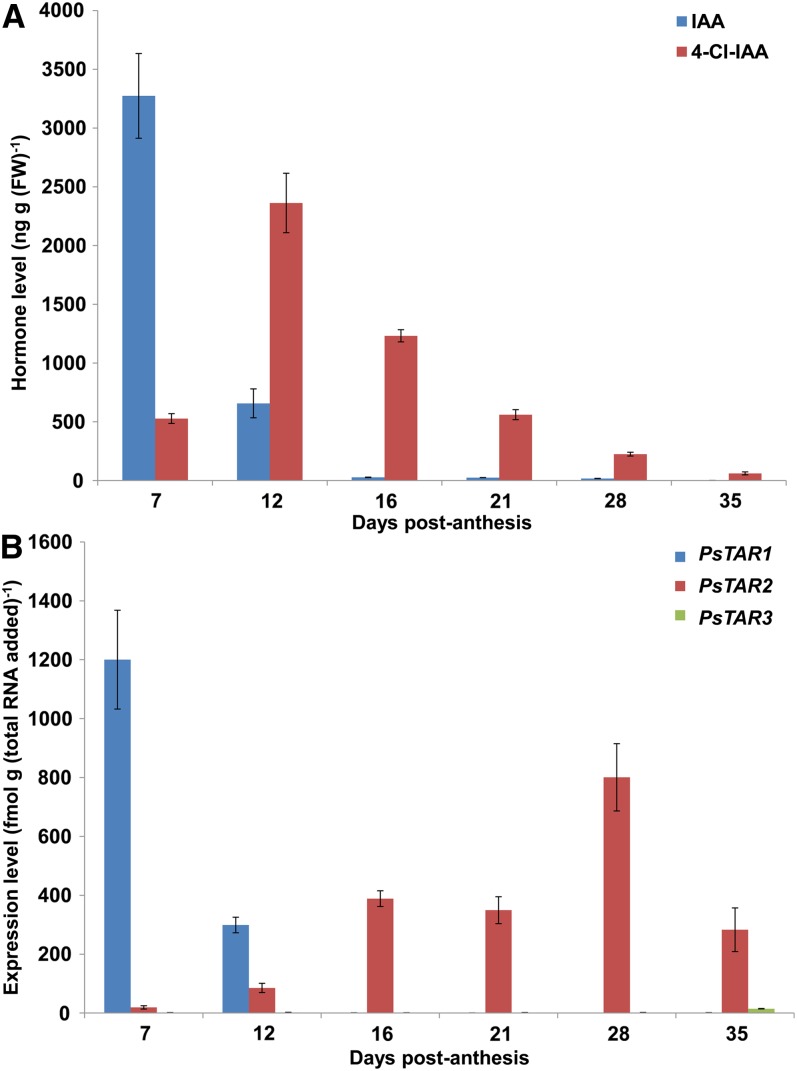
We next measured auxin levels, and the expression of PsTAR1, PsTAR2, and PsTAR3, over the course of seed development. UPLC-MS analyses showed that the level of 4-Cl-IAA in developing seeds increased dramatically from 7 to 12 d post anthesis (DPA) and then steadily declined until the completion of seed development (Fig. 3A). IAA levels were initially high but decreased markedly from 7 to 16 DPA and remained low thereafter (Fig. 3A). Quantitative real-time PCR showed that PsTAR1 is strongly expressed early in seed development (7 DPA), when IAA levels are maximal, and PsTAR2 is strongly expressed at later stages (16–28 DPA), when 4-Cl-IAA levels are high, although past their peak (Fig. 3B).
Figure 3.
IAA and 4-Cl-IAA levels (A) and PsTAR1, PsTAR2, and PsTAR3 transcript levels (B) over the course of seed development in line 107. Shown are means ± se (n = 3 for auxin and n = 4 for transcript levels). Mean seed fresh weights for each time point, in order of increasing age, were 3.8 ± 0.4, 54 ± 4, 199 ± 10, 293 ± 8, 432 ± 27, and 459 ± 12 mg. FW, Fresh weight.
Effects of the tar2 Mutation on Auxin Levels
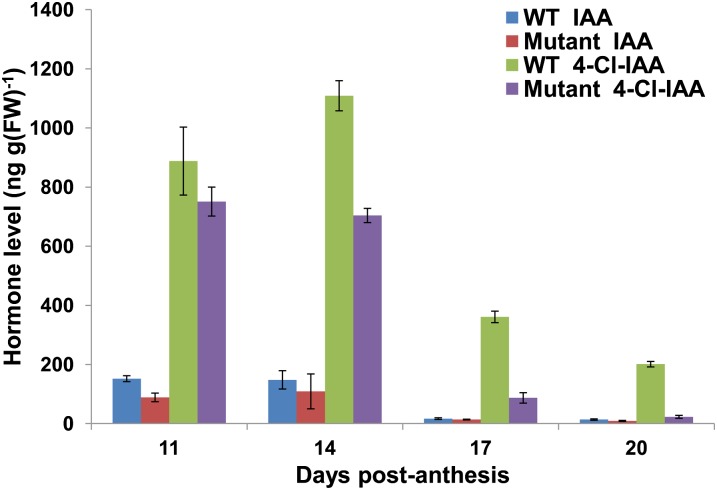
To investigate the role of aminotransferases in 4-Cl-IAA biosynthesis in vivo, we utilized a Targeting Induced Local Lesions IN Genomes (TILLING) population to identify mutants affected in the PsTAR2 gene. A single tar2 knockout mutant was isolated on the genetic background Cameor. The stop codon in this mutant results in a protein truncated prior to the catalytic Lys that is common to all pyridoxal-5-phosphate (PLP)-dependent aminotransferases. The tar2 mutation did not substantially affect auxin levels during early stages of seed development (Fig. 4). However, seeds of the mutant contained much less 4-Cl-IAA than the wild type at the later stages (P < 0.001); by 20 DPA, the reduction was approximately 90%. Effects of the tar2 mutation on the content of IAA, the minor auxin at later stages, were relatively small. The large reduction in 4-Cl-IAA at the later stages indicates the importance of the 4-Cl-IPyA pathway for 4-Cl-IAA biosynthesis in seeds. 4-Cl-Trp was identified by UPLC-MS in seed extracts, and we found that at later stages of seed development, the extracts from mutant seeds contained significantly more 4-Cl-Trp and Trp than did extracts from wild-type seeds (Supplemental Fig. S5).
Figure 4.
IAA and 4-Cl-IAA content of wild-type (WT) and tar2 mutant seeds. Shown are means ± se (n = 3). Wild-type seed fresh weights were as follows: 30 ± 1 mg (11 DPA), 114 ± 5 mg (14 DPA), 252 ± 17 mg (17 DPA), and 339 ± 7 mg (20 DPA); tar2 seed fresh weights were as follows: 41 ± 8 mg (11 DPA), 125 ± 5 mg (14 DPA), 260 ± 5 mg (17 DPA), and 339 ± 13 mg (20 DPA). All plants were on a tall background. FW, Fresh weight.
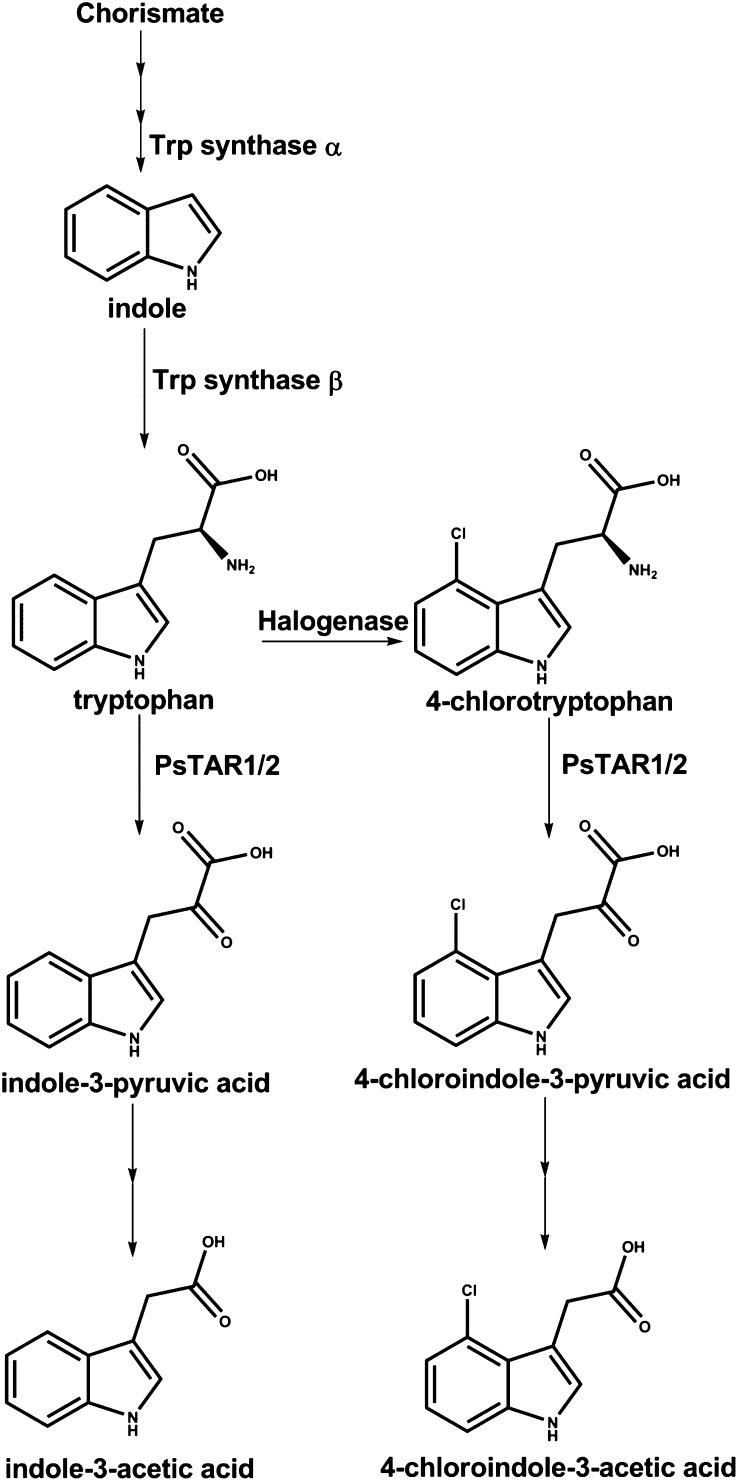
Taken together, the in vitro and in vivo analyses described above allow several conclusions to be drawn. In early seed development, pea seeds contain high levels of IAA and comparatively low levels of 4-Cl-IAA, and at this stage, PsTAR1 is far more strongly expressed than PsTAR2 (Fig. 3). In late seed development, the situation is reversed (high 4-Cl-IAA, low IAA, strong PsTAR2 expression, and comparatively weak PsTAR1 expression). Our evidence is consistent with a role for both PsTAR1 and PsTAR2 in maintaining levels of IAA and 4-Cl-IAA throughout the course of seed development.
Labeled-Precursor Feeding Studies
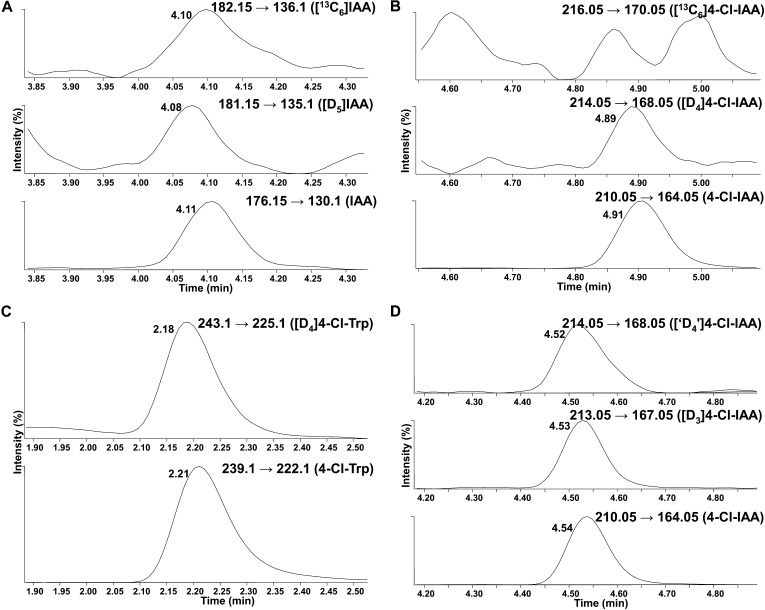
To investigate auxin biosynthesis in young pea seeds, where PsTAR2 does not strongly affect auxin levels, we injected labeled intermediates into seeds at the liquid endosperm stage. We first tested the possibility that 4-Cl-IAA is synthesized directly from IAA by injecting a mixture of [13C6]IAA (20 ng) and [D5]Trp (5 µg) into developing pea seeds (approximately 70 mg). The amount of [13C6]IAA injected was low to avoid a physiologically unrealistic situation. Incorporation of the D5 label from Trp into IAA was observed (Fig. 5A), and the IAA conjugate, indole-3-acetyl-Asp, contained 13C label (data not shown), indicating that IAA metabolism was occurring during the feeding period, but 4-Cl-IAA was not diluted with 13C label (Fig. 5B), indicating that it is not IAA itself that becomes chlorinated. In the same experiment, D4 label (from the [D5]Trp) was detected in both 4-Cl-Trp and 4-Cl-IAA (Fig. 5, B and C), indicating that Trp is a point of chlorination and that the biosynthesis of 4-Cl-IAA continues parallel to that of IAA (Fig. 6). Consistent with the theory that Trp is a point of chlorination, when deuterated 4-Cl-Trp was injected, label incorporation into 4-Cl-IAA was observed (Fig. 5D). It cannot be excluded, however, that chlorination also occurs at a stage prior to Trp.
Figure 5.
UPLC-MS chromatograms (MRM mode; acetic acid program) obtained from extracts of pea seeds that had been previously injected with a mixture of [D5]Trp and [13C6]IAA (A–C) or [D4]4-Cl-Trp (D). On UPLC, the RT of a deuterated species is earlier than the RT for the endogenous species; 13C-labeled species have the same RT as their endogenous counterparts (on the UPLC-MS system used, within 0.01 min). A, IAA became enriched with D5 label (middle channel; RT = 4.08) after injection of [D5]Trp; the endogenous IAA (bottom channel; RT = 4.11) and the injected [13C6]IAA (top channel; RT = 4.10) were also detected. B, 4-Cl-IAA also became enriched with D4 label (middle channel; RT = 4.89) from the injected [D5]Trp (one deuterium is replaced with a chlorine atom), but the 13C6 label from the injected [13C6]IAA was not incorporated into 4-Cl-IAA (top channel; the peaks observed in this channel did not have the correct RT for [13C6]4-Cl-IAA). C, 4-Cl-Trp became enriched with D4 label (top channel; RT = 2.18) from the injected [D5]Trp. D, In a separate experiment, 4-Cl-IAA became enriched with deuterium label (top two channels) after deuterated 4-Cl-Trp was injected; endogenous 4-Cl-IAA was also detected (bottom channel). The [D4]4-Cl-IAA signal detected in this experiment contained a small contribution from [37Cl,D2]4-Cl-IAA.
Figure 6.
Proposed parallel IAA and 4-Cl-IAA biosynthetic pathways in pea seeds, based on this study and previous evidence (Manabe et al., 1999; Reinecke, 1999; Quittenden et al., 2009; Tivendale et al., 2010; Zhao, 2010; Mashiguchi et al., 2011; Stepanova et al., 2011; Won et al., 2011; Kriechbaumer et al., 2012). IAA does not become chlorinated, and Trp is a point of chlorination.
Furthermore, after injections of deuterated Trp, we did not detect, by UPLC-MS, a labeled form of another putative intermediate, indole-3-acetamide (IAM; data not shown); endogenous IAM was similarly below the limit of detection (Supplemental Fig. S6). These results indicate that auxin biosynthesis does not occur via this intermediate in these organs.
DISCUSSION
In this study, we provide evidence for the biosynthesis of 4-Cl-IAA via the 4-chlorinated version of the IPyA pathway. We have isolated three new genes from pea, PsTAR1 to 3 which are homologous to the AtTAA family (Stepanova et al., 2008; Tao et al., 2008), and vt2 and ZmTAR1 from maize (Chourey et al., 2010; Phillips et al., 2011). We have shown, by in vitro assays, that they have aminotransferase activity using either Trp or its 4-chlorinated analog. The novel intermediate, 4-Cl-IPyA, was detected as a product of 4-Cl-Trp, and its identity was confirmed by comparison with 4-Cl-IPyA synthesized in our laboratory. In these assays, 4-Cl-IAA and IAA were also detected, but further analysis showed that this was merely due to nonenzymatic conversion from the initial products, 4-Cl-IPyA and IPyA to 4-Cl-IAA and IAA, respectively. However, the conversion of IPyA to IAA is reportedly enzymatic in Arabidopsis (Mashiguchi et al., 2011; Stepanova et al., 2011; Won et al., 2011; Kriechbaumer et al., 2012). Interestingly, in the functional assays, both the enol and keto tautomers of IPyA and 4-Cl-IPyA were detected as products, but the keto form predominated. In contrast, on the basis of liquid chromatography RTs relative to that of IAA, previous aminotransferase assays appear to have yielded mainly the enol form of IPyA (He et al., 2011). Tao et al. (2008), Stepanova et al. (2008), and He et al. (2011) all report the production of IPyA from in vitro Trp aminotransferase assays, but not the phenomenon of tautomerization. However, an unidentified broad peak is visible in the UV chromatograms presented by Tao et al. (2008) and He et al. (2011), which might represent the keto tautomer of IPyA. Nevertheless, the in vitro assays were conducted under conditions that favor the keto form, as indicated by our NMR analyses. The issue of which form is predominantly produced by aminotransferases remains to be resolved, but our in vivo evidence indicates that in pea seeds, both 4-Cl-IPyA and IPyA are present primarily as their respective keto tautomers.
It appears that in pea seeds, IAA and 4-Cl-IAA are synthesized in parallel via the IPyA pathway and its 4-chlorinated version (Fig. 6). On the basis of gene expression patterns, PsTAR1 is a key enzyme during the early stages of seed development, whereas the dramatic effect of tar2 on 4-Cl-IAA levels in maturing seeds is consistent with an important role for PsTAR2 in the later stages. We found no evidence that IAA itself becomes chlorinated, and metabolism studies indicated that Trp (and possibly earlier precursors) become(s) chlorinated and that the resulting 4-Cl-Trp is subsequently converted to 4-Cl-IAA. Compound-based studies reported here and previously (Quittenden etal., 2009; Tivendale et al., 2010) indicate that Trp (and, by implication, 4-Cl-Trp) is not converted to auxin in pea seeds via tryptamine, IAM, or indole-3-acetaldoxime.
There is currently a renewed focus on auxin biosynthesis, due to reports that the YUC proteins do not operate in the tryptamine pathway (Tivendale et al., 2010) but rather the IPyA pathway (Mashiguchi et al., 2011; Stepanova et al., 2011; Won et al., 2011; Kriechbaumer et al., 2012), as suggested previously by Strader and Bartel (2008). However, Mano and Nemoto (2012) recently noted that functional activity for Trp aminotransferases from the IPyA pathway has been demonstrated only for Arabidopsis genes and implied that the IPyA pathway might be restricted to the Brassicaceae. Our evidence from metabolism and genetic studies indicates that the IPyA pathway does operate, and indeed can predominate, in other species.
MATERIALS AND METHODS
Chemicals
The following compounds were obtained from commercial sources: [13C6]IAA (Cambridge Isotope Laboratories), 4-Cl-Trp (Amatek Chemical), Trp (Sigma-Aldrich), and IPyA (Sigma-Aldrich). The deuterated 4-Cl-IAA was supplied by Prof. Jerry Cohen (Department of Horticultural Science, University of Minnesota). All other compounds were synthesized in our laboratory as described previously (Quittenden et al., 2009) or below.
Isolation and Cloning of PsTAR1, PsTAR2, and PsTAR3
Pea (Pisum sativum) cDNA was synthesized with oligo(dT)20 primer (SuperScript III; Invitrogen) from RNA extracted from apical portions of 4-week-old seedlings and immature pea seeds (RNeasy plant mini kit with on-column DNase digestion; Qiagen). Degenerate primers were designed from blocks generated by CODEHOP (Rose et al., 1998) from conserved regions in Trp aminotransferase-like sequences in a range of species, including Arabidopsis (Arabidopsis thaliana), Medicago truncatula, Oryza sativa, Zea mays, Marchantia polymorpha, and Physcomitrella patens. These degenerate primers were then modified to reflect the TAR-like M. truncatula nucleotide sequences (Medtr5g033520.1, Medtr5g033510.1, and Medtr3g077250.1) obtained by a BLAST search from the International Medicago Genome Annotation Group database at M. truncatula Sequencing Resources (www.medicagohapmap.org/?genome).
PCR with Advantage 2 polymerase (Clontech) resulted in PCR fragments that were purified (Wizard SV Gel and PCR cleanup system; Promega), ligated into pGEM-T-easy, and transformed into JM109 competent Escherichia coli (Promega). Plasmid DNA was isolated from individual colonies (Wizard plus SV minipreps; Promega) and sequenced by Macrogen. Full-length sequences were obtained by 5′ and 3′ RACE (SMARTScribe Reverse Transcriptase; Clontech). A proofreading polymerase (Velocity DNA polymerase; Bioline) with specific primers amplified the full-length coding region of the three PsTAR genes, which were cloned into pGEM-T-easy as above.
PsTAR2 was transferred from pGEM-T-easy to pMAL-c5X (New England Biolabs [NEB]) by NotI (NEB) restriction digestion, dephosphorylation (Antarctic phosphatase; NEB), agarose gel purification (Wizard SV Gel and PCR cleanup system; Promega), and ligation (T4 DNA ligase; Promega).
PsTAR1 was inserted into pMAL-c5X vector by ligation of a PCR fragment amplified (Velocity DNA polymerase; Bioline) from cDNA prepared from RNA extracted from immature pea seeds with the forward primer at the start codon and the reverse primer containing the SbfI restriction site, after digestion of the vector with SbfI and XmnI (NEB). The construct was then transformed into JM109 E. coli (Promega), and the resulting plasmids were sequenced to check that they were in frame and free of PCR-induced errors (Macrogen).
Expression and Purification of Recombinant PsTAR2 and PsTAR1
The following procedure (adapted from the NEB pMAL Protein Fusion and Purification Instruction Manual) was used for protein expression and purification.
A starter culture was prepared by inoculating superoptimal broth with catabolite repression (15 mL) containing ampicillin (0.29 mm) with E. coli, known to contain PsTAR2 in frame in the vector pMAL. A “vector-only” starter culture was also prepared. The two starter cultures were incubated at 37°C for 16 h with shaking, after which the optical density at 600 nm (OD600) was determined by UV-visible spectrophotometry with superoptimal broth with catabolite repression as a blank. Ten milliliters of each starter culture was diluted to 1.0 L with sterile terrific broth containing ampicillin (0.29 mm). These broths were incubated at 37°C with shaking for 4 h, when the OD600 was approximately 0.5 (terrific broth blank). The mixtures were then cooled to 30°C, and isopropyl β-d-1-thiogalactopyranoside (filter sterilized) was added to each one to a final concentration of 300 µm, along with PLP (filter sterilized) to a final concentration of 200 µm. The resulting broth was incubated for 20 h with shaking at 23°C to 30°C.
The flasks were then briefly cooled on ice, and the OD600 of each sample was determined (greater than 0.5; blank: isopropyl β-d-1-thiogalactopyranoside and PLP in terrific broth). Aliquots (0.5 mL) from each sample were taken for SDS-PAGE. The remainders of each sample were centrifuged at 4.0 × 10,000g for 30 min at 4°C, after which the supernatants were discarded. The pellets were resuspended in lysis buffer (15 mL; 50 mm K2HPO4/KH2PO4 buffer [pH 8.5], 1 mm dithiothreitol, 1 mm EDTA, 0.2 mm PLP, and 0.5 m phenylmethylsulfonyl fluoride [added just before use]), and the resulting broth was cooled overnight to −20°C.
The samples were thawed in cold water, sonicated in 15-s bursts for a total of 4 min, and centrifuged at 4°C and 2.0 × 100,000g for 20 min. The supernatant (crude extract) was kept at −20°C until purification.
PsTAR2-MBP fusion and MBP were purified as follows. The column was poured and then rinsed with water (120 mL), SDS (45 mL; 0.1%, v/v), cold water (60 mL), and finally column buffer (75 mL; 100 mm K2HPO4/KH2PO4 [pH 8.5]) at a flow rate of 5 mL min−1. The crude extract, diluted 1:6 with lysis buffer, was loaded onto the column at a flow rate of 3 mL min−1. The column was then washed with cold washing buffer (50 mm K2HPO4/KH2PO4 buffer [pH 8.5], 0.2 mm PLP, and 0.5 mm phenylmethylsulfonyl fluoride [added just before use]; 180 mL, flow rate of 5 mL min−1). The protein was eluted with elution buffer (10 mm maltose in washing buffer; 15 mL) followed by column buffer (15 mL). Fractions (2.5 mL) were collected, and the total protein concentration was determined by spectrophotometry (Thermo Scientific NanoDrop 8000); the fractions were stored at −70°C until needed.
PsTAR1 was obtained using the same procedure.
In Vitro Assays Using PsTAR2 and PsTAR1
The following procedure was adapted from Stepanova et al. (2008) and Tao et al. (2008). Fusion protein tags are not typically required to be cleaved from the protein of interest for in vitro assays (Zhao et al., 2001; Stepanova et al., 2008; Mashiguchi et al., 2011).
For in vitro assays where Trp or 4-Cl-Trp was the substrate, each reaction tube contained purified recombinant protein (0.20 µg µL−1), substrate (34 ng µL−1), sodium pyruvate (4.8 µg µL−1), PLP (1.2 mm), and maltose (3.8 mm) in K2HPO4/KH2PO4 buffer (10 mm; pH 8.5). For in vitro assays where IPyA or 4-Cl-IPyA was the substrate, each reaction tube contained purified recombinant protein (0.20 µg µL−1), substrate (34 ng µL−1), sodium pyruvate (43.1 mm), PLP (1.2 mm), and maltose (4.3 mm) in K2HPO4/KH2PO4 buffer (10 mm; pH 6.5). For all functional assays, the total reaction volume was 174 µL. “No-protein” controls were prepared in a similar manner, except that the protein solution was replaced with buffer. All samples were incubated at 37°C for 3 h, after which the reaction was stopped by the addition of acetic acid in methanol (5%, v/v) to give an 80:19:1 buffer:methanol:acetic acid mixture. These mixtures were then analyzed by UPLC-MS.
UPLC-MS
Samples were analyzed using a Waters Acquity H-series UPLC device coupled to a Waters Xevo triple quadrupole mass spectrometer. A Waters Acquity UPLC BEH C18 column (2.1 mm × 100 mm × 1.7 µm) was used. There were two solvent combinations: (1) 5 mm ammonium acetate (pH 5.4; solvent A) and acetonitrile (solvent B); and (2) 1% (v/v) acetic acid in water (solvent A) and acetonitrile (solvent B).
The UPLC program was 80% A:20% B to 50% A:50% B at 4.5 min, and this was followed by immediate reequilibration to starting conditions for 3 min. The flow rate was 0.35 mL min−1, the column was held at 35°C, and the sample compartment was at 6°C. Approximate RTs using solvent combination 1 were 0.89 min for Trp, 1.0 min for keto-IPyA, 1.2 min for enol-IPyA, 1.3 min for both IAA and [13C6]IAA, 1.1 min for 4-Cl-Trp, 1.3 min for keto-4-Cl-IPyA, 2.0 min for enol-4-Cl-IPyA, and 2.2 min for 4-Cl-IAA and [D3]4-Cl-IAA. RTs using solvent combination 2 were approximately 1.7 min for keto-IPyA, 3.5 min for enol-IPyA, 2.7 min for IAA, 2.2 min for keto-4-Cl-IPyA, 3.9 min for enol-4-Cl-IPyA, and 3.3 min for 4-Cl-IAA.
For improved separation of interfering peaks for IAM analyses, solvent combination 2 and a slower gradient were employed; this was a linear gradient from 99% A:1% B to 80% A:20% B at 6 min, followed by a linear gradient to 40% A:60% B at 12 min, followed by immediate reequilibration to starting conditions for 3 min.
The mass spectrometer was operated in positive ion electrospray mode with a needle voltage of 2.8 kV, and MRM was used to detect all analytes. The ion source temperature was 130°C, the desolvation gas was N2 at 950 L h−1, the cone gas flow was 100 L h−1, and the desolvation temperature was 450°C. Data were processed using MassLynx software.
For Trp, IAA, IPyA, and IAM, all MRM transitions were monitored in a single time window with a dwell time of 92 ms per channel. The channels were as follows: for IAA, m/z 176.15 to 130.1; for [13C6]IAA, m/z 182.15 to 136.1 (cone voltage, 18 V; collision energy [CE], 18 V); for Trp, m/z 205.2 to 146.1 (cone voltage, 17 V; CE, 17 V) and m/z 205.2 to 188.1 (cone voltage, 17 V; CE, 10 V); IPyA, m/z 204.2 to 130.1 (cone voltage, 18 V; CE, 22 V) and m/z 204.2 to 158.1 (cone voltage, 18 V; CE, 22 V); for IAM, m/z 175.1 to 130.1 and m/z 181.1 to 135.1 (cone voltage, 24 V; CE, 15 V).
The chlorinated analogs were analyzed in three overlapping time windows. The first window from 0 to 1.6 min was for 4-Cl-Trp (dwell time, 95 ms per channel), the second window from 0 to 6 min was for 4-Cl-IPyA (dwell time, 66 ms per channel), and the third window from 1.6 to 6 min was for 4-Cl-IAA and [D4]4-Cl-IAA (dwell time, 66 ms per channel). MRM channels for the chlorinated species were as follows: 4-35Cl-Trp, m/z 239.2 to 222.2; 4-37Cl-Trp, m/z 241.2 to 224.2; [D3]4-Cl-Trp, m/z 242.2 to 225.2 and m/z 242.2 to 224.2 (cone voltage, 19 V; CE, 12 V); 4-35Cl-IPyA, m/z 238.2 to 164.1 and 238.2 to 192.1; 4-37Cl-IPyA, m/z 240.2 to 166.1 (cone voltage, 20 V; CE, 24 V); 4-35Cl-IAA, m/z 210.05 to 164.05; [D2]4-35Cl-IAA and 4-37Cl-IAA, m/z 212.05 to 166.05; [D3]4-35Cl-IAA, m/z 213.05 to 167.05 (cone voltage, 20 V; CE, 18 V).
When tandem MS was required, the cone voltage and CE were 20 V.
Plant Material
Wild-type (TAR2) pea plants used for this study were the Hobart tall (LE) line 107 (derived from cv Torsdag) and the dwarf (le-1) line Cameor. All plants were grown as described previously (Jager et al., 2005).
Quantitative Real-Time PCR
At a range of developmental stages, three or four seeds from multiple pods were harvested and immersed in liquid N2, for each of four replicates. Comparable seeds from the same pods were harvested for analysis of auxin content (Fig. 3).
The seeds were ground frozen, and total RNA was extracted from approximately 100 mg of ground tissue (RNeasy plant mini kit; Qiagen) with on-column DNase digestion (RNase-free DNase set; Qiagen). For later-stage seeds, the ground tissue was divided among four or five QIAshredder columns and the flow-through was combined to one RNA column, to avoid overloading one shredder with contaminants that can reduce yield and damage the RNA.
Spectrophotometer measurements (Thermo Scientific NanoDrop 8000) showed the RNA to be of good quality, with a clear peak at 260 nm, 260:280 ratio of 2, and 260:230 ratio above 1.5 (260:230 mean of 2.17, median of 2.30, and sd of 0.30).
In a single batch, cDNA was synthesized from 0.5 µg of total RNA using reverse transcriptase primer mix (QuantiTect reverse transcription kit; Qiagen) with integrated removal of genomic DNA. Controls lacking reverse transcriptase enzyme were prepared to monitor for contamination with genomic DNA.
First-strand cDNA was diluted 6-fold, and 3 µL was used in each quantitative real-time PCR using SYBR Green chemistry (SensiMix SYBR; Bioline). Samples were set up with a CAS-1200N robotic liquid-handling system (Corbett Research) and run for 50 or 55 cycles in a Rotor-Gene RG3000A Dual-Channel machine (Qiagen). Quantitative PCR primer sequences are presented in Supplemental Table S1. Two technical replicates and four biological replicates were performed for each sample. The concentrations were calculated relative to a curve containing seven serial 10-fold dilution points of cloned cDNA plasmids containing the full-length gene of interest created in the same quantitative PCR run. Reaction efficiencies obtained [E = 10(−1/slope) − 1] were between 0.99 and 1.20, and the correlation coefficient of the standard curve (r2) was greater than 0.99. The temperatures at which the PCR products melted were measured to check for primer specificity, and these were all found to be within 0.2 sd.
PsActin and PsPP2a housekeeper genes were not useful in the comparison of seeds of different ages, as there is elevated expression of these genes in the rapidly growing embryo around contact point. To test that the same quantity of total RNA was added to each cDNA synthesis reaction and, hence, quantitative PCR, the pea 18s ribosomal RNA (rRNA) levels were utilized as before (Ozga et al., 2003). The coefficient of variation of the 18S rRNA threshold cycle among all the samples was low (less than 4%); therefore, the target amplicon mRNA values were not normalized to the 18S signal (Ozga et al., 2003).
Quantification of IAA, 4-Cl-IAA, Trp, and 4-Cl-Trp
Seeds were harvested, weighed, homogenized, and extracted as before (Tivendale et al., 2010). After filtering, internal standards ([13C6]IAA and [D4]4-Cl-IAA) were added, and aliquots were purified using a Sep-Pak cartridge. Smaller aliquots were taken for the determination of Trp and 4-Cl-Trp; the internal standards were [D4]Trp and [D3]4-Cl-Trp. All samples were analyzed by UPLC-MS (MRM).
TILLING
PsTAR2 mutants were identified from an ethyl methanesulfonate mutant collection of 4,800 pea lines using TILLING screening as described by Dalmais et al. (2008). Details of all primer sequences are given in Supplemental Table S2. The nature of the mutations was identified by sequencing. PsTAR2 genomic sequence and TILLING mutations were integrated in UTILLdb (http://urgv.evry.inra.fr/UTILLdb).
The tar2 mutant (line 918), with a stop codon encoding a protein truncated prior to the catalytic Lys common to all PLP-dependent aminotransferases, was obtained among 13 mutations identified in the TILLING screen and was backcrossed four times to the parental cv Cameor (dwarf; le-1). Line 918 was also crossed to the Hobart tall line 107, and single plant selection was carried out for three generations to give tall TAR2 and tar2 plants on a similar background. Seeds from these plants were harvested for the quantification of auxins and auxin precursors.
Genotyping
DNA from plants at the seedling stage was amplified with PsTAR2-specific primers (Phire Plant Direct kit; Finnzymes); the restriction enzyme BseRI (NEB) cut the mutant allele. Alternatively, when they failed to emerge from the soil, the genotype was determined by digging up the seed, removing the testa, and extracting DNA from the cotyledons/embryo (DNeasy plant kit; Qiagen), PCR amplification (Taq DNA polymerase with ThermoPol buffer; NEB), and BseRI (NEB) digestion.
Synthesis of Deuterated 4-Cl-Trp
A suspension of dl-4-Cl-Trp (69 mg) in deuterated HCl in D2O, prepared by the careful addition of thionyl chloride (0.5 mL; Riedel-de Haën) to D2O (4.5 mL), was stirred at room temperature for 4 weeks in a sealed vial. The solvent was removed under reduced pressure, and a fresh portion of deuterated HCl in D2O was added and the suspension was stirred for a further 4 weeks. This process was repeated a second time, before evaporation under reduced pressure, to yield deuterated dl-4-Cl-Trp as the hydrochloride salt. The product was checked for deuterium incorporation by UPLC-MS (D4, 15%; D3, 25%; D2, 33%; D1, 23%; unlabeled, 5%).
Synthesis and Characterization of 4-Cl-IPyA
The 4-Cl-IPyA synthesis method was adapted from Politi et al. (1996). Triethylamine (76 µL) was added to a suspension of dl-4-Cl-Trp (102 mg) in methanol (1.7 mL) under an atmosphere of N2 at room temperature. After stirring for 10 min, pyridine-4-carboxaldehyde (80 µL) was added and the mixture was stirred for a further 10 min, after which ZnCl2 (anhydrous; 42 mg) was added to the mixture, which was stirred for a further 10 min. Total dissolution was achieved when 1,8-diazabicyclo-[5.4.0.]-undec-7-ene (164 µL) was added to the mixture, which was then stirred for a further 3 h under N2, after which it was quickly added drop-wise to HCl (2 m, 5 mL) preheated to 50°C. After 10 min, the temperature was increased to 55°C and the mixture was left for another 25 min, after which it was slowly cooled to room temperature and stood for 16 h. The resulting brown precipitate was collected by vacuum filtration, air dried overnight, and then dried in a desiccator. The precipitate was identified as the target compound (47 mg, 46% yield) by 1H- and 13C-NMR (1H-NMR, CD3OD, 300 MHz δ: 7.06 [m, 2H], 7.33 [doublet of doublets, J = 2.4 Hz, 3.9 Hz, 1H], 7.72 [s, 1H], 8.12 [d, J = 2.1 Hz, 1H], 11.18 [broad singlet, 1H]; 13C-NMR, C2H3O2H, 75 MHz δ: 105.4 [CH], 110.5 [C], 110.4 [CH], 120.9 [CH], 122.1 [CH], 122.9 [C], 125.9 [C], 129.4 [CH], 137.1 [C], 137.8 [C], 167.5 [C]). This compound was characterized by UPLC-MS for comparison with the product of our PsTAR1 and PsTAR2 in vitro assays using 4-Cl-Trp as the substrate (Fig. 2).
Application of Heavy-Isotope-Labeled Intermediates
Solutions of d- or 13C-labeled substrates in water were injected into excised immature pea seeds (30-135 mg), which contained liquid endosperm, using a sterile syringe, as described previously (Tivendale et al., 2010), and left up to 3 h before harvesting. After the appropriate incubation time, seed extracts were prepared in one of two ways: (1) the seeds were quickly frozen in liquid N2 and ground into a powder, which was immersed in methanol at 4°C for 1 h; (2) the seeds were immersed in −20°C 4:1 methanol:water containing butylated hydroxytoluene (1.13 mm) and subsequently extracted as before (Tivendale et al., 2010). After removal of impurities using Waters Sep-Pak Vac RC C18 cartridges, all extracts were analyzed by UPLC-MS.
Sequence data for pea cv Torsdag mRNA can be found in the GenBank/EMBL data libraries under accession numbers JN990988 (PsTAR1), JN990989 (PsTAR2), and JN990990 (PsTAR3). The GenBank accession numbers for genomic nucleotide sequences from cv Cameor are JQ002582 (PsTAR1), JQ002584 (PsTAR2), and JQ002583 (PsTAR3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. UPLC-MS chromatograms of in vitro assay mixtures.
Supplemental Figure S2. Keto-enol tautomerization of IPyA.
Supplemental Figure S3. HSQC NMR spectrum for IPyA.
Supplemental Figure S4. UPLC-MS chromatograms showing endogenous IPyA and 4-Cl-IPyA in pea seeds.
Supplemental Figure S5. Trp and 4-Cl-Trp levels of wild-type (WT) and tar2 seeds.
Supplemental Figure S6. UPLC-MS chromatograms showing lack of endogenous IAM in pea seeds.
Supplemental Table S1. qPCR-specific primers.
Supplemental Table S2. TILLING-specific primers.
Supplementary Material
Acknowledgments
We thank Sam Cook, Ian Cummings, Lauren Febey, Stevie Florent, Jasmine Janes, Claire Sayers, Vincent Walter, James Weller, Tracey Winterbottom, and Yunde Zhao for technical assistance or advice.
Glossary
- 4-Cl-IAA
4-chloroindole-3-acetic acid
- IAA
indole-3-acetic acid
- IPyA
indole-3-pyruvic acid
- UPLC
ultra-performance liquid chromatography
- MS
mass spectrometry
- 4-Cl-IPyA
4-chloroindole-3-pyruvic acid
- 4-Cl-Trp
4-chloro-tryptophan
- RT
retention time
- MRM
multiple reaction monitoring
- m/z
mass-to-charge ratio
- DPA
d post anthesis
- PLP
pyridoxal-5-phosphate
- IAM
indole-3-acetamide
- NEB
New England Biolabs
- OD600
optical density at 600 nm
- CE
collision energy
- TILLING
Target-Induced Local Lesions In Genome
- cDNA
to be defined
References
- Chourey PS, Li QB, Kumar D. (2010) Sugar-hormone cross-talk in seed development: two redundant pathways of IAA biosynthesis are regulated differentially in the invertase-deficient miniature1 (mn1) seed mutant in maize. Mol Plant 3: 1026–1036 [DOI] [PubMed] [Google Scholar]
- Dalmais M, Schmidt J, Le Signor C, Moussy F, Burstin J, Savois V, Aubert G, Brunaud V, de Oliveira Y, Guichard C, et al. (2008) UTILLdb, a Pisum sativum in silico forward and reverse genetics tool. Genome Biol 9: R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Expósito-Rodríguez M, Borges AA, Borges-Pérez A, Hernández M, Pérez JA. (2007) Cloning and biochemical characterisation of ToFZY, a tomato gene encoding a flavin monooxygenase involved in a tryptophan-dependent auxin biosynthesis pathway. J Plant Growth Regul 26: 329–340 [Google Scholar]
- He W, Brumos J, Li H, Ji Y, Ke M, Gong X, Zeng Q, Li W, Zhang X, An F, et al. (2011) A small-molecule screen identifies l-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 23: 3944–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager CE, Symons GM, Ross JJ, Smith JJ, Reid JB. (2005) The brassinosteroid growth response in pea is not mediated by changes in gibberellin content. Planta 221: 141–148 [DOI] [PubMed] [Google Scholar]
- Kriechbaumer V, Wang P, Hawes C, Abell BM. (2012) Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartmentation. Plant J 70: 292–302 [DOI] [PubMed] [Google Scholar]
- LeClere S, Schmelz EA, Chourey PS. (2010) Sugar levels regulate tryptophan-dependent auxin biosynthesis in developing maize kernels. Plant Physiol 153: 306–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manabe K, Higashi M, Hatori M, Sakagami Y. (1999) Biosynthetic studies on chlorinated indoles in Vicia faba. Plant Growth Regul 27: 15–19 [Google Scholar]
- Mano Y, Nemoto K. (2012) The pathway of auxin biosynthesis in plants. J Exp Bot 63: 2853–2872 [DOI] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, et al. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA 108: 18512–18517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J. (2010) Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb Perspect Biol 2: a001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozga JA, Reinecke DM, Ayele BT, Ngo P, Nadeau C, Wickramarathna AD. (2009) Developmental and hormonal regulation of gibberellin biosynthesis and catabolism in pea fruit. Plant Physiol 150: 448–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozga JA, Yu J, Reinecke DM. (2003) Pollination-, development-, and auxin-specific regulation of gibberellin 3β-hydroxylase gene expression in pea fruit and seeds. Plant Physiol 131: 1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips K, Skirpan A, Christensen A, Slewinski T, Hudson C, Barazesh S, Cohen JD, Malcomber S, McSteen P. (2011) vanishing tassel2 endcodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell 53: 550–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politi V, De Luca G, Di Stazio G, Materazzi M, inventors. November 20, 1996. 3-Indolepyruvic acid derivatives: their method of production and therapeutic use. European Patent No. EP0421946
- Quittenden LJ, Davies NW, Smith JA, Molesworth PP, Tivendale ND, Ross JJ. (2009) Auxin biosynthesis in pea: characterization of the tryptamine pathway. Plant Physiol 151: 1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinecke DM. (1999) 4-Chloroindole-3-acetic acid and plant growth. Plant Growth Regul 27: 3–13 [Google Scholar]
- Rose TM, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S. (1998) Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res 26: 1628–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz K. (1961) Separation of enol and keto tautomers of aromatic pyruvic acids by paper chromatography. Arch Biochem Biophys 92: 168–175 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Doležal K, Schlereth A, Jürgens G, Alonso JM. (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Robles LM, Novak O, He W, Guo H, Ljung K, Alonso JM. (2011) The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23: 3961–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Bartel B. (2008) A new path to auxin. Nat Chem Biol 4: 337–339 [DOI] [PubMed] [Google Scholar]
- Sugawara S, Hishiyama S, Jikumaru Y, Hanada A, Nishimura T, Koshiba T, Zhao Y, Kamiya Y, Kasahara H. (2009) Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc Natl Acad Sci USA 106: 5430–5435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tivendale ND, Davies NW, Molesworth PP, Davidson SE, Smith JA, Lowe EK, Reid JB, Ross JJ. (2010) Reassessing the role of N-hydroxytryptamine in auxin biosynthesis. Plant Physiol 154: 1957–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, Kasahara H, Kamiya Y, Chory J, Zhao Y. (2011) Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci USA 108: 18518–18523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2010) Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol 61: 49–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J. (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.