Abstract
Many species in Rosaceae, Solanaceae, and Plantaginaceae exhibit S-RNase-based self-incompatibility (SI). In this system, the pistil and pollen specificities are determined by S-RNase and the S locus F-box protein, respectively. The pollen S determinant F-box protein in Prunus (Rosaceae) is referred to by two different terms, SFB (for S-haplotype-specific F-box protein) and SLF (for S locus F box), whereas it is called SLF in Solanaceae and Plantaginaceae. Prunus SFB is thought to be a molecule indispensable for its cognate S-RNase to exert cytotoxicity and to arrest pollen tube growth in incompatible reactions. Although recent studies have demonstrated the molecular function of SCFSLF in the SI reaction of Solanaceae and Plantaginaceae, how SFB participates in the Prunus SI mechanism remains to be elucidated. Here we report the identification of sweet cherry (Prunus avium) SFB (PavSFB)-interacting Skp1-like1 (PavSSK1) using a yeast (Saccharomyces cerevisiae) two-hybrid screening against the pollen cDNA library. Phylogenetic analysis showed that PavSSK1 belongs to the same clade as Antirrhinum hispanicum SLF-interacting Skp1-like1 and Petunia hybrida SLF-interacting Skp1-like1 (PhSSK1). In yeast, PavSSK1 interacted not only with PavSFBs from different S haplotypes and Cullin1-likes (PavCul1s), but also with S-locus F-box-likes. A pull-down assay confirmed the interactions between PavSSK1 and PavSFB and between PavSSK1 and PavCul1s. These results collectively indicate that PavSSK1 could be a functional component of the SCF complex and that PavSFB may function as a component of the SCF complex. We discuss the molecular function of PavSFB in self-/nonself-recognition in the gametophytic SI of Prunus.
Gametophytic self-incompatibility (GSI) is one of the most common reproductive systems in flowering plants, and it prevents inbreeding and promotes outcrossing by enabling the pistil to reject pollen from genetically related individuals (de Nettancourt, 2001; Franklin-Tong, 2008). In most cases, self-incompatibility (SI) is genetically controlled by a single polymorphic S locus, which harbors pistil S and pollen S determinant genes. The GSI system in many species of Rosaceae, Solanaceae, and Plantaginaceae recruits the extracellular cytotoxic RNase, S-RNase, as the pistil S determinant; this is referred to as S-RNase-based GSI (McClure, 2009). Recently, Rubiaceae was also shown to have the S-RNase-based GSI system (Nowak et al., 2011). Phylogenetic analyses of S-RNase and related sequences suggested that the S-RNase-based GSI system had a single evolutionary origin about 120 million years ago (Igic and Kohn, 2001; Steinbachs and Holsinger, 2002; Vieira et al., 2008a; Nowak et al., 2011).
F-box protein genes encoding a subunit of the SCF complex are commonly identified as the pollen S determinants in S-RNase-based GSI. Several lines of evidence suggest that the S-haplotype-specific F-box gene (SFB) or the S locus F-box gene (SLF) is the pollen S in Prunus (Rosaceae), whereas SLFs have been shown to control pollen specificity in Solanaceae and Plantaginaceae (Lai et al., 2002; Entani et al., 2003; Ushijima et al., 2003; Wang et al., 2003; Yamane et al., 2003b; Qiao et al., 2004a; Sijacic et al., 2004; Sonneveld et al., 2005; Kubo et al., 2010). Although Prunus pollen S has been referred to by two different terms, we use SFB in this article because it distinguishes the Prunus system from other systems, and recent studies have used SFB (Newbigin et al., 2008; Sassa et al., 2010; Tao and Iezzoni, 2010). S locus F-box brothers (SFBBs) have been reported to be pollen S candidates in the tribe Pyreae of the Rosaceae (Sassa et al., 2007; Okada et al., 2008, 2011; Minamikawa et al., 2010; Francheschi et al., 2011; Kakui et al., 2011). The SCF complex, which is made up of an F-box protein, Skp1, Cullin1 (Cul1), and Rbx1, is one of the Really Interesting New Gene-type E3 ubiquitin ligases, which polyubiquitinates substrate proteins to be degraded by the ubiquitin proteasome system. In the SCF complex, the F-box protein determines substrate specificity, Skp1 serves as an adaptor to connect the variable F-box protein to Cul1, and Cul1 forms a core catalytic scaffold with Rbx1 (Zheng et al., 2002; Deshaies and Joazeiro, 2009).
Inhibition of self-pollen tube growth in S-RNase-based GSI occurs in the transmitting tissue of the style, where S-RNase is secreted into the extracellular matrix. Because microscopic observations of pollen tube growth in Solanaceae showed that the pollen tube uptakes both self- and nonself-S-RNases, self-/nonself-recognition has been assumed to occur inside the pollen tube cell (Luu et al., 2000; Goldraij et al., 2006).
Two models, the compartmentalization model and the degradation model, have been proposed to explain self-pollen-specific rejection in Solanaceae, although these models are not necessarily mutually exclusive. Goldraij et al. (2006) proposed the compartment model based on the microscopic observations of pollen tube growth in Nicotiana. In this model, S-RNase that is taken up nonspecifically into the luminal compartments of the pollen endomembrane system remains stably sequestered in the vacuoles of compatible pollen tubes and therefore does not exert its cytotoxicity. In incompatible pollen tubes, however, the vacuoles break down and cytotoxic S-RNase is released into the cytosol. The degradation model was developed based on the expected biochemical functions of the pollen S determinant as the F-box protein. In this model, the SCF complex containing the F-box pollen S protein is supposed to polyubiquitinate all nonself-S-RNases for degradation, but it specifically interacts with its cognate S-RNase, leaving it active and leading to the arrest of self-pollen tube growth (Zhang et al., 2009; Meng et al., 2011). Data supporting the degradation model has been obtained from a series of biochemical experiments conducted with Antirrhinum and Petunia. Formation of SCFSLF with SLF-interacting Skp1-like1 (SSK1) and/or S-RNase binding protein1 in Antirrhinum and Petunia has been observed in vitro (Hua and Kao, 2006; Hua et al., 2007; Kubo et al., 2010). Furthermore, higher SLF affinity to nonself-S-RNase than to self-S-RNase and selective polyubiquitination of nonself-S-RNase have also been reported (Qiao et al., 2004b; Hua and Kao, 2006; Hua et al., 2007; Kubo et al., 2010). All of these observations are compatible with the degradation model.
The SI recognition system in Prunus (Rosaceae) was later shown to be incompatible with the degradation model. According to the degradation model, the defective pollen S that loses its function would result in SI and cross-incompatibility because pollen S is assumed to be involved in detoxification of the presumably cytotoxic S-RNase (Golz et al., 2001; Meng et al., 2011). Consistent with the degradation model, mutations that disrupt the pollen S (SLF) function in Solanaceae and Plantaginaceae have not been found (Meng et al., 2011). However, dysfunction of the pollen S (SFB) leads to self-compatibility (SC) in Prunus (Ushijima et al., 2004; Sonneveld et al., 2005). Therefore, SFB is thought to be an indispensable component for its cognate S-RNase to exert cytotoxicity and to arrest pollen tube growth in incompatibility reactions (Tao and Iezzoni, 2010).
In addition to the distinct outcome of the pollen S mutation in Prunus, differences between Prunus and the other plant species showing S-RNase-based GSI (including Solanaceae, Plantaginaceae, and Rosaceae tribe Pyreae) were also observed in the SI/SC reaction in heterodiallelic pollen. Breakdown of the incompatibility reaction in heterodiallelic pollen, called competitive interaction, is absent in Prunus, although it is well documented in other plants with S-RNase-based GSI (Entani et al., 1999; Yamane et al., 2003a; Hauck et al., 2006; Adachi et al., 2009; Xue et al., 2009; Tsukamoto et al., 2010). Competitive interaction can be explained by the degradation model, whereas it is incompatible with the one-allele match model that has been proposed to explain the SI reaction in Prunus (Hua and Kao, 2006; Hauck et al., 2006).
Distinct functions of Prunus SFB are also indicated by the phylogenetic analyses of the pollen S and pollen S-like F box. In the phylogenetic tree, the Prunus SFB clade diverged from the clade containing SLF of Solanaceae and Plantaginaceae and SFBB of Pyreae (Sassa et al., 2007; Matsumoto et al., 2008; Vieira et al., 2009; Minamikawa et al., 2010). As data have accumulated for the distinctiveness of Prunus SI, pollen S of Prunus has been assumed to be involved in the release of S-RNase cytotoxicity rather than the avoidance of S-RNase cytotoxicity, a presumed pollen S function in the degradation model (Tao and Iezzoni, 2010). In Prunus, a hypothetical general inhibitor (GI) has been proposed to detoxify S-RNase cytotoxicity unless affected by a cognate SFB in an unknown mechanism (Luu et al., 2001; Tao and Iezzoni, 2010). Because Prunus SFB appears to have a distinct function from SLF of Solanaceae and Plantaginaceae, the biochemical properties of SFB should be investigated thoroughly.
This article reports the identification and characterization of sweet cherry (Prunus avium) SFB-interacting Skp1-like1 (PavSSK1), which was isolated by yeast (Saccharomyces cerevisiae) two-hybrid (Y2H) screening against the sweet cherry pollen cDNA library. Y2H analysis showed that PavSSK1 interacted with PavSFBs from different S haplotypes and sweet cherry Cul1-like proteins (PavCul1s). A glutathione S-transferase (GST) pull-down assay confirmed the interactions between PavSSK1-PavSFB and PavSSK1-PavCul1s. These results collectively indicate that PavSSK1 could be a functional component of the SCF complex and that PavSFB may function as a component of the SCF complex. We discuss the molecular function of PavSFB in self-/nonself-recognition in the GSI of Prunus.
RESULTS
Identification of the Skp1-Like Protein Interacting with PavSFB F-Box Domain
Y2H screening against the sweet cherry pollen cDNA library was performed using the N-terminal region of PavSFB-S6 (48 amino acids), which contains the F-box domain, as bait to isolate partner molecules of PavSFB in the SCFSFB complex. We isolated 29 positive clones from 3.5 × 105 cells. Of these, 16 clones were derived from an identical gene. The gene was named sweet cherry PavSSK1, because it showed high similarity to Ricinus communis putative Skp1 (E value; 7e-36). DNA sequences of all 16 clones were found in the full-length coding sequence (CDS) of PavSSK1. Interaction between the full-length PavSSK1 and the N-terminal region of PavSFB-S6 containing the F-box motif was confirmed in the second round of the Y2H assay using the SD/-adenine/-His/-Leu/-Trp plate (data not shown). On the contrary, the N-terminal region of PavSFB-S3 and PavSFB-S6 showed no interaction with the sweet cherry Skp1-like1 protein (PavPSK1) that was isolated from pollen cDNA using degenerate PCR based on DNA sequences in Arabidopsis (Arabidopsis thaliana) Skp1-like genes reported to be under strong purifying selection, i.e. ASK1, ASK2, and ASK4 (data not shown; Kong et al., 2004, 2007).
Molecular Characterization of PavSSK1
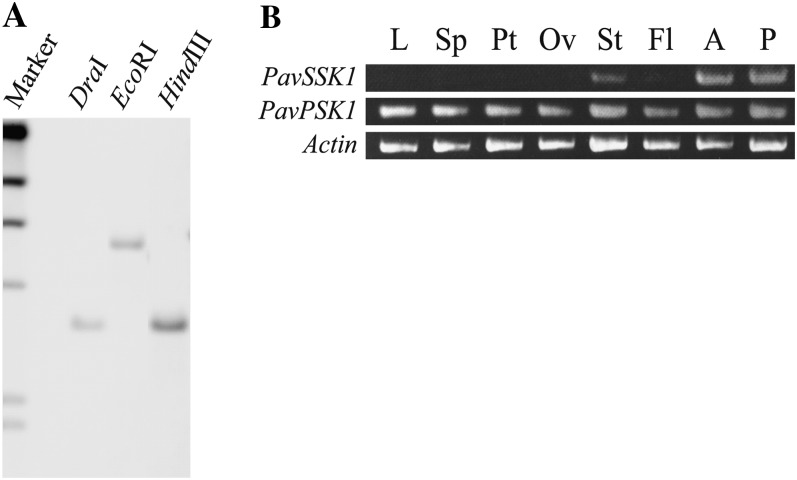
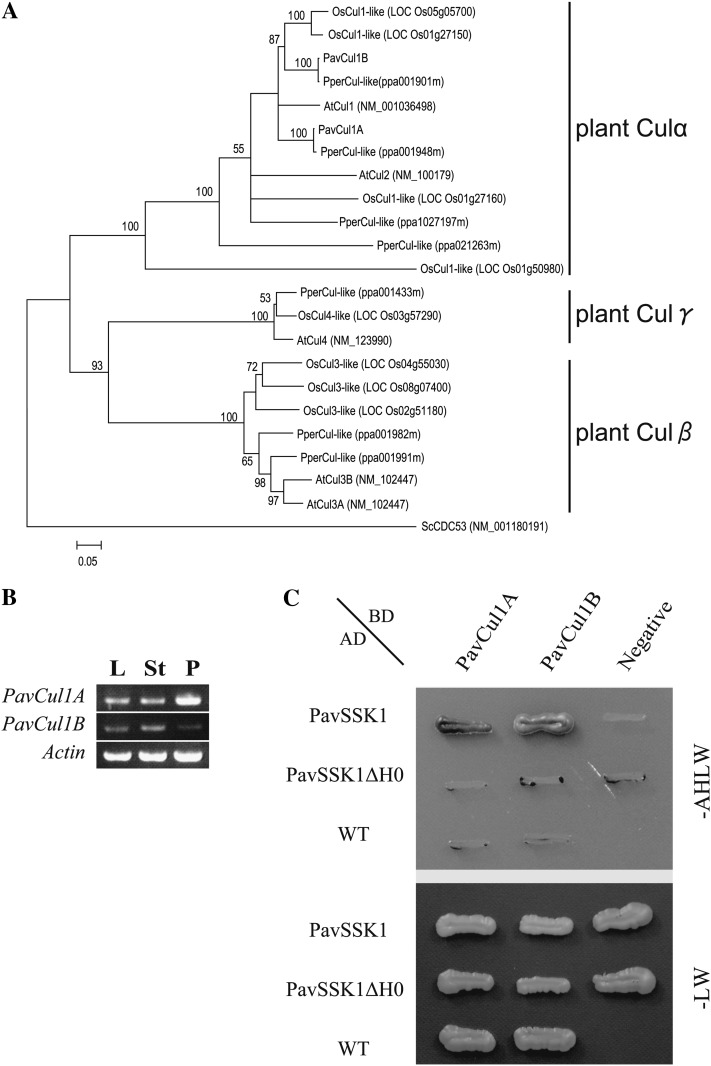
PavSSK1 was confirmed to be a single-copy gene in the genome of sweet cherry by DNA-blot analysis (Fig. 1A). Comparison between the genomic and mRNA sequences of PavSSK1 revealed that PavSSK1 had no intron in its CDS and untranslated region, whereas Antirrhinum hispanicum SLF-interacting Skp1-like1 (AhSSK1) was reported to have >1.5 kb of introns at its 3′ untranslated region and yield transcript variants resulting from alternative splicing (Huang et al., 2006). PavSSK1 was expressed strongly in anthers and pollen, weakly in styles, and not at all in other floral organs or leaves (Fig. 1B). Because Skp1-like genes belong to a multigene family, we sequenced the amplified bands to confirm that they were from PavSSK1.
Figure 1.
PavSSK1 was a single-copy gene and expressed in several floral organs. A, Genomic DNA-blot analysis using the PavSSK1 probe. Genomic DNA from sweet cherry cv Satonishiki was digested with DraI, EcoRI, or HindIII and probed with the PavSSK1 probe. B, PavSSK1 and PavPSK1 expression patterns were examined by RT-PCR. Total RNAs from various organs were used as templates for cDNA synthesis and RT-PCR with gene-specific primer pairs. L, Leaves; Sp, sepals; Pt, petals; Ov, ovaries; St, styles; Fl, filaments; A, anthers; P, pollen.
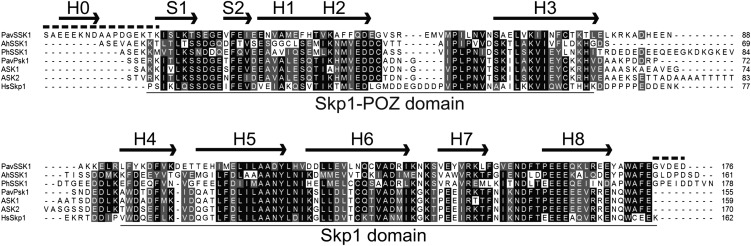
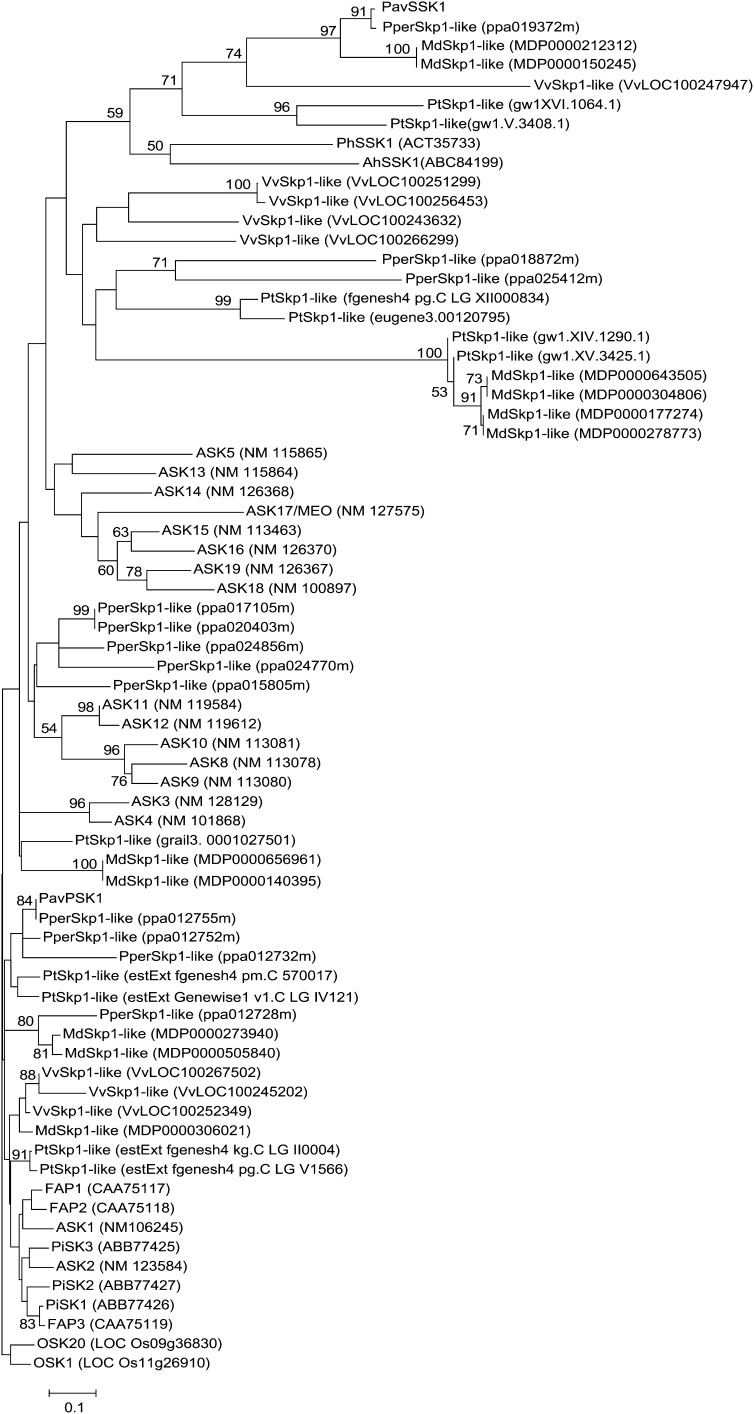
The canonical Skp1 comprises 150 to 200 amino acid residues and contains an Skp1-POZ domain, which interacts with Cul1 at the N terminus, and an Skp1 domain, which interacts with F-box domains at the C terminus. The tertiary structure of human Skp1 in the SCF complex consists of eight helices (H1–H8) and three sheets (S1–S3; Schulman et al., 2000). H2, S3, and H5 form the interface that interacts with Cul1, whereas H5 to H8 constitute the interface that interacts with the F-box protein (Schulman et al., 2000; Zheng et al., 2002). The putative amino acid sequences of PavSSK1 comprised 177 residues and contained both the Skp1-POZ and Skp1 domains (Fig. 2). The predicted secondary structure of PavSSK1 showed the presence of not only the typical helix and sheet structures, but also the N-terminal disordered helix (H0) and the C-terminal tail commonly present in AhSSK1 and PhSSK1 (Fig. 2; Huang et al., 2006; Zhao et al., 2010). PavSSK1 showed relatively high amino acid sequence similarity with AhSSK1 (55.3%) and PhSSK1 (50.8%). In the neighbor-joining (NJ) tree, PavSSK was classified in the same clade as AhSSK1 and PhSSK1 (Fig. 3; Supplemental Fig. S1).
Figure 2.
Sequence alignment of SSK1s and typical Skp1-like proteins. The putative amino acid sequence of PavSSK1 was aligned with that of AhSSK1 (ABC84199) from Antirrhinum, PhSSK1 (ACT35733) from P. hybrida, ASK1 (NM_106245) and ASK2 (NM_123584) from Arabidopsis, PavPSK1 (from this study) from sweet cherry, and human HsSkp1 (P63208). The N-terminal Met residue was excluded because its excision was predicted by the TermiNator (http://www.isv.cnrs-gif.fr/terminator3/index.html; Frottin et al., 2006; Martinez et al., 2008). Black and gray blocks indicate identical and similar amino acid residues, respectively. Top arrows and dashed lines represent the secondary structure and the intrinsically disordered regions of PavSSK1, respectively. H, α-helix; S, β-sheet.
Figure 3.
NJ tree constructed using aligned deduced amino acid sequences of PavSSK1 and other plant Skp1-like proteins. Skp1-like proteins shorter than about 200 amino acid residues that possessed both Skp1 and Skp1-POZ domains were used to construct a NJ tree. The deduced amino acid sequences were from Arabidopsis (ASK1-19), grapevine (Vitis vinifera; VvSkp1-likes), poplar (Populus spp.; PtSkp1-likes), apple (Malus domestica; MdSkp1-likes), peach (PperSkp1-likes), Antirrhinum (AhSSK1 and FAP1-3), Petunia (PhSSK1 and PiSKP1-3), and rice (OSK1 and OSK20). The NJ tree was generated with 1,000 bootstrap replicates. OSK1 and OSK20, which had the best blast scores among OSKs when PavSSK1 was used as a query, were used as outgroups to construct the NJ tree.
The N-Terminal Disordered Helix of PavSSK1 Enhances Its Interaction with F-Box Protein in Yeast
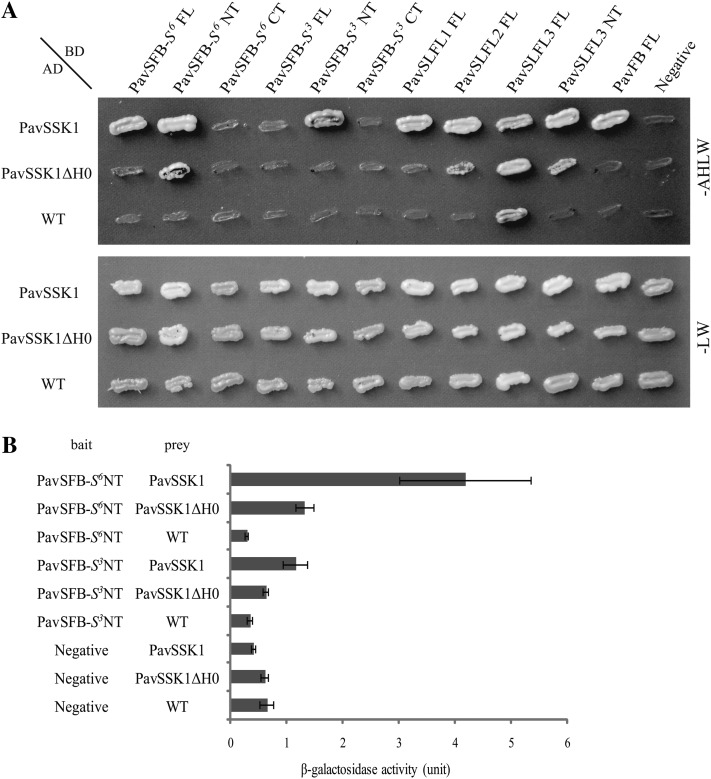
A Y2H assay was performed using various constructs designed from PavSSK1, PavSFB-s3, and PavSFB-S6 to investigate the interaction between PavSSK1 and PavSFB (Fig. 4A). PavSSK1 interacted with the full-length PavSFB-S6 and its N-terminal region that contained the F-box domain, whereas PavSSK1 did not interact with the C-terminal region of PavSFB-S6 that lacked the F-box domain. Although PavSSK1 interacted with the N-terminal region of PavSFB-S3, no interaction was detected with the full-length and C-terminal regions of PavSFB-S3. PavSSK1 lacking the specific N-terminal helix (PavSSK1ΔH0) did not interact with the full-length PavSFB-S6 or the N-terminal region of PavSFB-S3 (Fig. 4A). The β-galactosidase activity assay confirmed that the F-box domain of PavSFB more strongly interacted with the full-length PavSSK1 than with PavSSK1ΔH0 in both S haplotypes tested (Fig. 4B).
Figure 4.
Y2H analysis to investigate interactions between PavSSK1 and Prunus F-box proteins. A, A colony of AH109 cotransformants was grown on nonselective (-LW) and selective (-AHLW) media for 7 d at 25°C. The empty pAD-GAL4-2.1 vector (WT) and the empty pGBKT7 vector (Negative) were used as negative controls. B, β-Galactosidase activity was detected in a liquid assay using o-nitrophenyl-β-d-galactopyranoside as a substrate. The activity is given as the mean ± sd of five independent transformants.
We also examined the interactions between PavSSK1 and four other pollen-expressed F-box proteins, S-locus F-box-like1 to -3 (PavSLFL1–3) and Prunus avium F-box protein (PavFB), to investigate the specificity of PavSSK1/F-box interactions. Prunus SLFL1–3 are located at the flanking region of the Prunus S locus and show low sequence identity with each other and with SFB (Entani et al., 2003; Ushijima et al., 2003, 2004; Matsumoto et al., 2008; Vieira et al., 2008b). Although Prunus armeniaca F-box (ParFB) was reported to show higher sequence identity to Prunus SFB than SLFLs, there is no linkage between the FB and the S loci (Vilanova et al., 2006). The PavFB cloned in this study showed very high amino acid sequence similarity to ParFB (96.1%), high sequence similarity to PavSFB-S3 (74.8%) and PavSFB-S6 (76.6%), and low sequence similarity to PavSLFL1-3-S4 (approximately 35.0%). The Y2H assay was conducted using full-length CDSs of these F-box protein genes and the N-terminal sequence of SLFL3, the full-length sequence of which showed autoactivation of reporter genes in yeasts (Fig. 4A). All of these F-box proteins exhibited strong interactions with full-length PavSSK1 and weaker or no interactions with PavSSK1ΔH0 in yeast (Fig. 4A).
PavSSK1 Interacts with PavCul1s in Yeast
To ascertain whether PavSSK1 acts as a component of the SCF complex, we attempted to isolate sweet cherry Cul1-like genes and examine the interaction between PavSSK1 and PavCul1. Six peach (Prunus persica) Cul-like CDSs were retrieved from the peach genome database, four of which clustered with Arabidopsis Cul α-group genes in the NJ tree. Because Arabidopsis Cul α-group proteins act as components of the SCF complex (Fig. 5A; Supplemental Fig. S2; Risseeuw et al., 2003; Marín, 2009), we selected two of the four genes, excluding ppa1027197m and ppa021263m, for further analyses. ppa1027197m was excluded because it lacked the N-terminal region binding to Skp1, whereas ppa021263m was excluded because it showed much lower amino acid sequence similarity (62.3%) with Arabidopsis Cul1 than did ppa001948m and ppa001901m (>92.7%). Both homologs of ppa001948m and ppa001901m were confirmed to be expressed in sweet cherry pollen (Fig. 5B). Full-length homologous DNA sequences of ppa001948m and ppa001901m were cloned from sweet cherry pollen cDNA and named PavCul1-likeA (PavCul1A) and PavCul1B, respectively. In the Y2H assays, both PavCul1A and PavCul1B interacted with PavSSK1 but not with PavSSK1ΔH0 (Fig. 5C).
Figure 5.
Identification of the pollen-expressed sweet cherry Cul1s. A, An NJ tree was constructed based on aligned deduced amino acid sequences of Prunus Cul-likes. Deduced amino acid sequences were from peach (PperCul-likes), sweet cherry (PavCul1A and PavCul1B), Arabidopsis (AtCul1-4), and rice (OsCul1–4-likes). The NJ tree was generated with 1,000 bootstrap replicates. Budding yeast Cul α (ScCDC53) was defined as the outgroup. B, PavCul1A and PavCul1B expression profiles. Total RNAs from leaves (L), styles (St), and pollen (P) were used as a template for cDNA synthesis and RT-PCR with gene-specific primer pairs. C, Y2H analysis to investigate interactions between PavSSK and PavCul1s. A colony of AH109 cotransformants was grown on nonselective (-LW) and selective (-AHLW) media for 10 d at 25°C. The empty pAD-GAL4-2.1 vector (WT) and the empty pGBKT7 vector (Negative) were used as negative controls.
PavSSK1 Interacted with PavSFB and PavCul1s in Pull-Down Assays
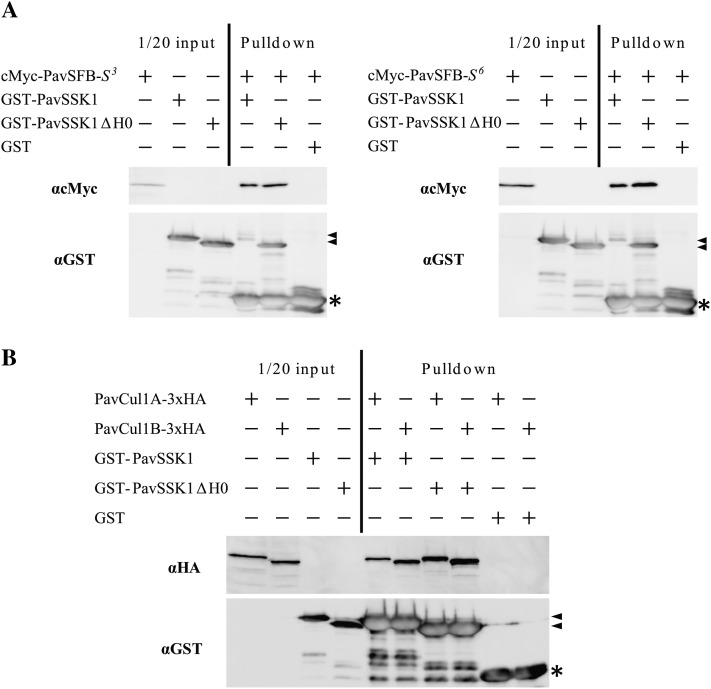
Pull-down assays were carried out using purified GST-PavSSK1 and GST-PavSSK1ΔH0. For the pull-down assay, PavSSK1 fusion protein expressed in Escherichia coli was used after purification, whereas cMyc-tagged PavSFB and 3x hemagglutinin (HA)-tagged PavCul1s were expressed in the cell-free system. Both cMyc-tagged PavSFB-S3 and cMyc-tagged PavSFB-S6 bound not only to GST-PavSSK1, but also to GST-PavSSK1ΔH0 (Fig. 6A). In addition, both 3xHA-tagged PavCul1A and 3xHA-tagged PavCul1B bound to GST-PavSSK1 and GST-PavSSK1ΔH0 (Fig. 6B). PavSSK1ΔH0 bound to PavSFB and PavCul1s in pull-down assays, although PavSSK1ΔH0 did not interact with PavSFB-S3, PavCul1A, and PavCul1B in the Y2H assays (Fig. 6).
Figure 6.
In vitro binding assays of PavSSK1-PavSFB and PavSSK1-PavCul1s. A, The interaction between PavSSK1 and PavSFB was tested. Purified GST-tagged PavSSK1 or GST-tagged PavSSK1ΔH0 was used as bait against crude cell-free expressed cMyc-SFB-S3 or cMyc-SFB-S6. Bound fractions were treated with thrombin before SDS-PAGE to dissociate the GST tag from the GST-fusion proteins. Bound proteins were detected using the anticMyc antibody. GST was used as a negative control. Triangles and asterisks indicate the GST-fusion proteins and the GST fragment, respectively. Left section: assays using cMyc-SFB-S3; right section: assays using cMyc-SFB-S6. B, The interaction between PavSSK1 and PavCul1s was tested. Purified GST-tagged PavSSK1 or GST-tagged PavSSK1ΔH0 was used as bait against crude cell-free expressed PavCul1A-3xHA or PavCul1B-3xHA. Bound proteins were detected using the anti-HA antibody. GST was used as a negative control. Triangles and asterisks indicate the GST-fusion protein and the GST fragment, respectively.
DISCUSSION
The S-RNase-based GSI systems in Rosaceae, Solanaceae, and Plantaginaceae have been shown to use the same kind of molecule, an F-box protein, as the pollen S determinant. The F-box protein generally interacts with a substrate protein and Skp1, which bridges an F-box protein to a Cul1 to form the SCF complex. There are 692 F-box-like genes, 21 Skp1-like genes, and two Cul1-like genes in the genome of Arabidopsis, whereas 779 F-box-like genes, 32 Skp1-like genes, and four Cul1-like genes were present in the genome of rice (Oryza sativa; Kong et al., 2007; Marín, 2009; Xu et al., 2009). Skp1 proteins are important adaptors that allow diverse F-box proteins to function as substrate receptors of an E3 ligase. The solanaceous and plantaginaceous pollen S determinant SLF forms the SCF complex with a Skp1-like protein (AhSSK1 or PhSSK1) and a Cul1-like protein and/or S-RNase binding protein1 (Hua and Kao, 2006; Huang et al., 2006; Zhao et al., 2010). Furthermore, biochemical studies indicated that the SCFSLF functioned as the E3 ligase (Qiao et al., 2004b; Hua and Kao, 2006, 2008). However, whether the rosaceous pollen S determinant forms the SCF complex has yet to be determined. To our knowledge, this study identified the PavSSK1 that binds to PavSFB and PavCul1 for the first time. Although this finding does not conclusively prove that PavSFB forms a functional SCF complex, the identification of PavSSK strongly indicates that PavSFB forms an SCF complex with PavSSK1 and PavCul1. Here, we discuss the characteristics of PavSSK1 and propose a working hypothesis of a possible molecular basis of GSI recognition in Prunus.
PavSSK1 Sequence Features in Comparison with AhSSK1 and PhSSK1
The putative amino acid sequence of PavSSK1 encodes the canonical Skp1 domains, although its secondary structure is unique, especially at its termini (Fig. 2). PavSSK1 appears to have the N-terminal disordered helix and the C-terminal tail that is also found in AhSSK1 and PhSSK1. Furthermore, PavSSK1 showed relatively high amino acid sequence identity with AhSSK1 and PhSSK1 and clustered together with AhSSK1 and PhSSK1 in the NJ tree. All these results indicate that Skp1-like proteins that interact with the pollen S determinant in the S-RNase-based GSI might share a common origin (Huang et al., 2006; Zhao et al., 2010; Fig. 3). Because some of the Skp1-like proteins of Malus also clustered with PavSSK1 in the NJ tree, these Skp1-like proteins may also function as adaptors for pollen S determinants in Malus to form SCF complexes (Fig. 3).
Unlike the shared characteristics of C-terminal SSK1 homologs, the N-terminal disordered helix is unique in PavSSK1 (Fig. 2). The N-terminal disordered helix, H0, was also found in other phylogenetically distant Skp1-like proteins, such as Arabidopsis ASK18, Oryza OSK20, and Malus Skp1-likes MDP000021232, MDP0000150245, MDP0000140395, MDP0000278773, and MDP000017724 (Fig. 2; Supplemental Fig. S1). The H0 of PavSSK1 strengthened its interactions with F-box proteins and with PavCul1s in yeast, although it was not necessary for these interactions in the pull-down assays (Figs. 4–6). This discrepancy might be attributed to the different sensitivity of the two assays. In other words, the Y2H assay we used may not have been sensitive enough to detect the interaction between PavSSK1ΔH0 and PavSFB. In fact, PavSSK1ΔH0 could bind to the F-box domains of some PavSFB constructs in yeast (Fig. 4). Although a disordered region was often reported to be involved in the molecular recognition (Dyson and Wright, 2005), to the best of our knowledge, there has been no prior report on a functional dissection of the N-terminal disordered helix of Skp1-likes.
Tissue-Specific Expression Pattern of PavSSK1
PavSSK1 showed a tissue-specific expression pattern (Fig. 1), as opposed to the constitutive expression patterns across different tissues observed with Skp1-like proteins with general functions, such as ASK1, ASK2, and ASK11 in Arabidopsis (Risseeuw et al., 2003; Kong et al., 2004, 2007; Takahashi et al., 2004). Although Skp1-like proteins bind to the pollen S determinants of Solanaceae and Plantaginaceae, PhSSK1 and AhSSK1, respectively, also showed a tissue-specific expression pattern, their transcription was reported to be confined to pollen (Huang et al., 2006; Zhao et al., 2010). The difference in the transcriptional patterns might reflect the wider range of interactive partners for PavSSK1 than PhSSK1 and AhSSK1. PhSSK1 and AhSSK1 interacted only with SLF among the SLF and SLF-like proteins tested in Y2H assays (Huang et al., 2006; Zhao et al., 2010), but PavSSK1 appeared to interact with pollen-expressed F-box proteins other than PavSFB, such as PavSLFL1-3 and PavFB (Fig. 4A). Sweet cherry SLFL1 was reported to be expressed in pollen, anthers, and styles, whereas ParFB was expressed in pollen, styles, and leaves (Ushijima et al., 2003; Vilanova et al., 2006). PavSSK1 may also serve as an adaptor for SLFLs and FB in tissues other than pollen.
PavSSK1 Can Be an Adaptor for SCFSFB and SCFSLFLs
There have been several suggestions that the recognition mechanism of S-RNase-based GSI in Prunus differed from that in Solanaceae, Plantaginaceae, and even the Rosaceae tribe Pyreae, although SI recognition is commonly assumed to involve the interaction of pollen S and pistil S products that influence S-RNase cytotoxicity (Sassa et al., 2010; Tao and Iezzoni, 2010; Meng et al., 2011). In Prunus, SFB loss-of-function mutations confer SC, whereas mutants of the other plants with dysfunctional SLFs in Solanaceae and Plantaginaceae have not been found thus far, and are thought to lead to either lethality or incompatibility. Consistent with this idea, it has recently been shown that the mutated S4 haplotype lacking SFBB1-S4 (S4sm) of Pyrus pyrifolia had altered SI specificities and S4sm pollen was rejected by pistils with an otherwise compatible S1, whereas it was accepted by other nonself-pistils (Okada et al., 2008; Kakui et al., 2011; Saito et al., 2011). These different outcomes of pollen S dysfunction in Prunus and others, as well as the different outcomes of heterodiallelic pollen behavior, could indicate the presence of an unidentified unique mechanism that inactivates and/or detoxifies S-RNase cytotoxicity in Prunus. Namely, the pollen S determinant in Solanaceae, Plantaginaceae, and Pyreae is thought to be the molecule responsible for the detoxification of S-RNase, whereas Prunus SFB is believed to protect the cognate S-RNase from detoxification by an unknown mechanism or to release cytotoxicity of S-RNase in an S-haplotype-specific manner.
In this study, we showed that PavSSK1 could interact with PavSFB and PavCul1s. These results indicate that PavSSK1 could be a functional Skp1-like component in the SCF complex. Furthermore, sequence analyses of SFB indicated that its F-box motif was under purifying selection, supporting the inference that SFB interacts with Skp1-like protein (Ikeda et al., 2004; Nunes et al., 2006). Nevertheless, these results do not necessarily mean that SFB forms a functional SCF complex. Although most F-box proteins form SCF complexes, several F-box proteins have been reported to form non-SCF complexes that lacked Cul1 and had unknown functions (Hermand, 2006). The non-SCF complex consisted of an F-box protein, a functional Skp1 that normally constitutes the SCF complex with other F-box proteins, and other proteins such as regulator of V-ATPase in vacuolar membrane1 in budding yeast or Pof6 inteactor1in fission yeast (Schizosaccharomyces pombe; Dawson et al., 2008; Jourdain et al., 2009). Therefore, it may be possible that SFB forms a nonfunctional SCF with PavSSK1 to function in unknown mechanisms to protect or release cognate S-RNase cytotoxicity.
Phylogenetic analyses of pollen S-like genes showed that Prunus SLFLs were more closely related to SFBBs of Pyreae than Prunus SFBs were (Sassa et al., 2007; Matsumoto et al., 2008; Minamikawa et al., 2010; Vieira et al., 2009). SFBBs have been assumed to possess a function similar to solanaceous and plantaginaceous SLFs, because SFBB1 of the P. pyrifolia S4 haplotype was shown to be required for pollen tube growth in pistils containing S1-RNase (Okada et al., 2008; Kakui et al., 2011; Saito et al., 2011). These reports, combined with the sequence similarity among PavSSK1, PhSSK1, and AhSSK1, as well as the binding ability of PavSSK1 to PavSLFLs reported here, may support our speculation that Prunus SLFLs may act as the GI to form SCFSLFL to polyubiquitinate S-RNase for degradation (Tao and Matsumoto, 2012). Although we previously reported that a mutation in PavSLFL1 in sweet cherry had no effect on GSI response (Matsumoto et al., 2008), that data only ruled out the possibility that SLFL1 was the sole molecule acting as the GI. The effect of mutation in PavSLFL1 would be masked, if SLFLs work collaboratively, like Petunia SLFs, to detoxify S-RNase (Kubo et al., 2010). How S-RNase cytotoxicity, which is otherwise detoxified by presumed GI, is released by cognate SFB in the Prunus SI reaction is still unclear. Although the possibility that PavSSK1 and SFB form a non-SCF complex cannot be ruled out, the finding that SFB could function as an SCF complex using PavSSK1 as an adopter molecule strongly indicated that the ubiquitin proteasome protein degradation pathway could be involved in the SFB-mediated S-haplotype-specific release of S-RNase cytotoxicity.
MATERIALS AND METHODS
Plant Materials
The sweet cherry (Prunus avium) cultivar Satonishiki (S3S6) was used in this study. Floral organs from young buds and young leaves were collected in spring, frozen in liquid nitrogen, and stored at −80°C until used. Genomic DNA was extracted from leaves using a DNA extraction kit (GE Healthcare), and total RNA was extracted by the cold phenol method as described previously (Tao et al., 1999). Poly(A)+ RNA was isolated using the PolyATract mRNA isolation system (Promega).
Y2H Screening and Assays
Y2H screening was performed using the N-terminal region of PavSFB-S3 that contains the F-box motif as bait. The cDNA library was constructed from pollen poly(A)+ RNA using the BD matchmaker library construction and screening kits (Takara Bio). Yeast (Saccharomyces cerevisiae) cotransformations were performed using yeast strain AH109, according to the manufacturer’s instructions. Approximately 3.5 × 105 transformed cells were grown on SD/-adenine/-His/-Leu/-Trp medium for 7 d at 25°C. Plasmids of the positive clones were extracted, sequenced, and used for further experiments to confirm the interaction.
To construct plasmids for Y2H assays, full-length cDNA for PavSSK1 was cloned and sequenced using the GeneRacer advanced RACE kit (Invitrogen). PavPSK1 fragments were amplified from the pollen cDNA using the degenerate primer pair dePSK Fw (5′-ATH AAR CAY ARG ATH GAR GAY GAY TG-3′) and dePSK Rv (5′-TCY TCY TCT CCT CNG GNG TRA AGT-3′). Based on the partial cDNA sequence for PavPSK1, its full-length sequence was cloned and determined using the GeneRacer kit. Similarly, full-length cDNA sequences for PavFB, PavCul1A, and PavCul1B were cloned by 3′-RACE and 5′-RACE using pollen cDNA as a template. Primers for 3′- and 5′-RACE were designed based on the Genome Database for Rosaceae (http://www.rosaceae.org). The full-length CDS for PavSSK1 and the CDS lacking the N-terminal helix of PavSSK1 were cloned into pAD-GAL4-2.1 (AD; Agilent), whereas the full-length CDSs for PavSFB-S3, PavSFB-S6, PavFB, PavSLFL1-S4, PavSLFL2-S4, PavSLFL3-S4, PavCul1A, and PavCul1B were cloned into pGBKT7 (BD) vector (Takara Bio). In addition to the full-length CDSs, partial CDSs for the N-terminal regions containing the F-box motif of PavSFB-S3, PavSFB-S6, and PavSLFL3-S4 and the partial CDSs for the C-terminal region of PavSFB-S3 and PavSFB-S6 were cloned into pGBKT7 (BD).
For the Y2H assay, AH109 cells containing both AD and BD plasmids were grown on SD/-Leu/-Trp medium for 3 d at 30°C. Six independent clones for each combination were streaked on SD/-adenine/-His/-Leu/-Trp medium containing X-α-gal (Takara Bio) and grown for 7 to 10 d at 25°C to check viability and α-galactosidase activity. For quantitative measurements, β-galactosidase activity was determined using o-nitrophenyl-β-d-galactopyranoside (Sigma-Aldrich) as a substrate according to Miller (1972) as described in the Yeast Protocols Handbook (Takara Bio).
Reverse Transcription-PCR
Total RNA was isolated from leaves, petals, sepals, ovaries, styles, filaments, anthers, and pollen, and treated with DNase I (Invitrogen). cDNA was synthesized from the treated RNA using SuperScript III (Takara Bio) and poly-dT primer. cDNA equivalent to the amount synthesized from 50 ng of total RNA was used for reverse transcription (RT)-PCR with gene-specific primer pairs for PavSSK1 (Fw: 5′-AAG GCG TTC TTC CAA GAC GAA-3′ and Rv: 5′-CTT CAA AAG CCC AAG CAT ACT G-3′), PavPSK1 (Fw: 5′-GAC CGC CCT TCC AAC GAT GA-3′ and Rv: 5′-GCA TTC CAC AGC CAC TAG CTC-3′), peach (Prunus persica) Cul1A (PperCul1A Fw: 5′-TTG GTT TAT CGG GAG GTC-3′ and Rv: 5′-AGC TCC ACT TGA AGC CTG-3′), and PperCul1B (Fw: 5′-GAG ATC TGG TCT ACC AAG AAT TA-3′ and Rv: 5′-CTT TTT CTC CGC CTT CCT-3′). As a reference, RT-PCR for an actin gene was also conducted using the ActF1 and ActR1 primers (Ushijima et al., 2003).
DNA-Blot Analysis
DNA-blot analysis was conducted as previously described (Matsumoto et al., 2008). Genomic DNA (6 μg) was digested with DraI, HindIII, or EcoRI, separated in an agarose gel, and transferred onto a nylon membrane. The membrane was probed with the digoxigenin-labeled full-length PavSSK1 probe and washed under low-stringency conditions (twice in 2× SSC, 0.1% SDS for 5 min at room temperature, and then twice in 0.1× SSC, 0.1% SDS for 15 min at 60°C). The hybridization signals were visualized using the LAS-3000 system (Fujifilm).
Construction and Screening of Fosmid Libraries
A fosmid library was constructed from genomic DNA using the CopyControl fosmid library production kit (Epicentre) as previously described (Matsumoto et al., 2008). Clones containing PavSSK1 were obtained by screening using digoxigenin-labeled full-length PavSSK1 probes. The positive clones were used as templates to determine the genomic sequence of PavSSK1.
GST Pull-Down Assay
Full-length CDSs and partial CDSs lacking the N-terminal helix of PavSSK1 were cloned into pGEX-KG (GE Healthcare) to produce recombinant proteins fused with GST, named GST-PavSSK1 and PavSSK1ΔH0. GST, GST-PavSSK1, and GST-PavSSK1ΔH0 were introduced to BL21 Codon Plus (DE3) Escherichia coli (Agilent). The transformed E. coli were grown for 24 h at 18°C and lysed with lysozyme in phosphate-buffered saline (KCl−) containing 0.5% Triton X-100 and 1 mm dithiothreitol (DTT). Recombinant fusion proteins in the clarified lysate were bound to Glutathione Sepharose 4B (GE Healthcare) for purification. Bound recombinant proteins were eluted with 50 mm Tris-HCl buffer (pH 9.0) containing 50 mm reduced glutathione. Then the buffer was exchanged with 50 mm HEPES buffer containing 150 mm NaCl and 1 mm DTT using an Amicon Ultra-4 (Millipore).
Full-length CDS for PavSFB-S3 and PavSFB-S6 were cloned into pGBKT7 and used to produce recombinant fusion proteins, cMyc-PavSFB-S3 and cMyc-PavSFB-S6. Full-length CDS for PavCul1A and PavCul1B were fused with 3xHA tag sequence at the 3′ terminus and cloned into pGADT7 (Invitrogen) to produce recombinant fusion proteins, PavCul1A-3xHA and PavCul1B-3xHA. The recombinant fusion proteins cMyc-PavSFB-S3, cMyc-PavSFB-S6, PavCul1A-3xHA, and PavCul1B-3xHA were expressed using the TNT T7 coupled wheat germ extract system (Promega) and used for GST pull-down assays.
In the GST pull-down assays, 20 μg of GST-fusion proteins were incubated with 50 μL TNT reaction mixtures for PavCul1 fusion proteins or 25 μL TNT reaction mixtures for PavSFB fusion proteins in 50 mm HEPES (pH 7.0) buffer containing 150 mm NaCl, 1 mm DTT, and 0.1% Triton X-100 for 30 min at 18°C. After incubation, 20 μL Glutathione Sepharose 4B was added and samples were further incubated for 6 h at 4°C. The beads were washed four times with 50 mm HEPES (pH 7.0) buffer containing 150 mm NaCl, 1 mm DTT, and 0.1% Triton X-100 and once with 50 mm Tris-HCl (pH 8.5) buffer containing 150 mm NaCl, 1 mm DTT, and 2 mm CaCl2. Pulled-down PavCul1 fusion proteins were eluted with SDS-loading buffer (0.2 m Tris-HCl [pH 6.8], 2% SDS, 0.85 m 2-mercaptoethanol, 10% glycerol). PavSFB fusion proteins were digested with 3 units of thrombin (GE Healthcare) for 2 h at 22°C and then eluted with 2× SDS-loading buffer, because GST-SSK, which was equivalent in size to cMyc-SFB, blocked the detection of the cMyc signal on the blot. Eluted proteins were separated in 10% SDS-polyacrylamide gels, blotted onto polyvinylidene difluoride membranes (Millipore), and probed with anticMyc monoclonal antibodies (Nacalai Tesque), anti-HA polyclonal antibodies (Takara Bio), or anti-GST monoclonal antibodies (Nacalai Tesque). Immune complex signals were detected using the ECL advance western blotting detection kit (GE Healthcare) and the LAS-3000 system.
Sequence Analysis
Protein motifs were searched using the National Center for Biotechnology Information Conserved Domain Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). Secondary structures were predicted using the Quick Phyre program (http://www.sbg.bio.ic.ac.uk/~phyre) and intrinsically disordered regions were predicted using the DISOPRED server (Ward et al., 2004). Phylogenetic trees were constructed using the predicted amino acid sequences from genes annotated as belonging to the Skp1 family or the genes retrieved when Arabidopsis (Arabidopsis thaliana) Cul1-Cul5 was used as the query in the Genome Database for Rosaceae (http://www.rosaceae.org). The deduced amino acid sequences of Skp1-family genes or those of Cul-like genes were aligned using MEGA version 4.0 (Tamura et al., 2007) and adjusted manually. The alignments were used to generate a phylogenetic tree using the NJ method with 1,000 bootstrap replicates using MEGA version 4.0.
The accession numbers for the sequences described in this article are JQ322646 (PavSSK1), JQ322647 (PavPSK1), JQ322648 (PavFB), JQ322649 (PavCul1A), and JQ322650 (PavCul1B).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of PavSSK1 and other plant Skp1-like proteins.
Supplemental Figure S2. Alignment of Prunus Cul-likes and other plant Cul-likes.
Supplementary Material
Glossary
- SI
self incompatibility
- GSI
gametophytic self incompatibility
- SC
self compatibility
- GI
general inhibitor
- Y2H
yeast two-hybrid
- CDS
coding sequence
- NJ
neighbor-joining
- RT
reverse transcription
- DTT
dithiothreitol
References
- Adachi Y, Komori S, Hoshikawa Y, Tanaka N, Abe K, Bessho H, Watanabe M, Suzuki A. (2009) Characteristics of fruiting and pollen tube growth of apple autotetraploid cultivars showing self-compatibility. J Jpn Soc Hortic Sci 78: 402–409 [Google Scholar]
- Dawson K, Toone WM, Jones N, Wilkinson CR. (2008) Loss of regulators of vacuolar ATPase function and ceramide synthesis results in multidrug sensitivity in Schizosaccharomyces pombe. Eukaryot Cell 7: 926–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nettancourt D. (2001) Incompatibility and Incongruity in Wild and Cultivated Plants. Springer, Berlin.
- Deshaies RJ, Joazeiro CAP. (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78: 399–434 [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. (2005) Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6: 197–208 [DOI] [PubMed] [Google Scholar]
- Entani T, Iwano M, Shiba H, Che FS, Isogai A, Takayama S. (2003) Comparative analysis of the self-incompatibility (S-) locus region of Prunus mume: identification of a pollen-expressed F-box gene with allelic diversity. Genes Cells 8: 203–213 [DOI] [PubMed] [Google Scholar]
- Entani T, Takayama S, Iwano M, Shiba H, Ches FS, Isogai A. (1999) Relationship between polyploidy and pollen self-incompatibility phenotype in Petunia hybrida Vilm Bios. Biotech Biochem 63: 1882–1888 [DOI] [PubMed] [Google Scholar]
- Francheschi P, Pierantoni L, Dondini L, Grandi M, Sansavini S, Sanzol J. (2011) Evaluation of candidate F-box genes for the pollen S of gametophytic self-incompatibility in the Pyrinae (Rosaceae) on the basis of their phylogenomic context. Tree Genet Genomes 7: 663–683 [Google Scholar]
- Franklin-Tong VE. (2008) Self-Incompatibility in Flowering Plants. Springer, Berlin.
- Frottin F, Martinez A, Peynot P, Mitra S, Holz RC, Giglione C, Meinnel T. (2006) The proteomics of N-terminal methionine cleavage. Mol Cell Proteomics 5: 2336–2349 [DOI] [PubMed] [Google Scholar]
- Goldraij A, Kondo K, Lee CB, Hancock CN, Sivaguru M, Vazquez-Santana S, Kim S, Phillips TE, Cruz-Garcia F, McClure BA. (2006) Compartmentalization of S-RNase and HT-B degradation in self-incompatible Nicotiana. Nature 439: 805–810 [DOI] [PubMed] [Google Scholar]
- Golz JF, Oh HY, Su V, Kusaba M, Newbigin E. (2001) Genetic analysis of Nicotiana pollen-part mutants is consistent with the presence of an S-ribonuclease inhibitor at the S locus. Proc Natl Acad Sci USA 98: 15372–15376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck NR, Yamane H, Tao R, Iezzoni AF. (2006) Accumulation of nonfunctional S-haplotypes results in the breakdown of gametophytic self-incompatibility in tetraploid Prunus. Genetics 172: 1191–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermand D. (2006) F-box proteins: more than baits for the SCF? Cell Div 1: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Kao TH. (2006) Identification and characterization of components of a putative petunia S-locus F-box-containing E3 ligase complex involved in S-RNase-based self-incompatibility. Plant Cell 18: 2531–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Kao TH. (2008) Identification of major lysine residues of S(3)-RNase of Petunia inflata involved in ubiquitin-26S proteasome-mediated degradation in vitro. Plant J 54: 1094–1104 [DOI] [PubMed] [Google Scholar]
- Hua Z, Meng XY, Kao TH. (2007) Comparison of Petunia inflata S-locus F-box protein (Pi SLF) with Pi SLF like proteins reveals its unique function in S-RNase based self-incompatibility. Plant Cell 19: 3593–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zhao L, Yang Q, Xue Y. (2006) AhSSK1, a novel SKP1-like protein that interacts with the S-locus F-box protein SLF. Plant J 46: 780–793 [DOI] [PubMed] [Google Scholar]
- Igic B, Kohn JR. (2001) Evolutionary relationships among self-incompatibility RNases. Proc Natl Acad Sci USA 98: 13167–13171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Igic B, Ushijima K, Yamane H, Hauck NR, Nakano R, Sassa H, Iezzoni AF, Kohn JR, Tao R. (2004) Primary structural features of the S haplotype-specific F-box protein, SFB, in Prunus. Sex Plant Reprod 16: 235–243 [Google Scholar]
- Jourdain I, Spielewoy N, Thompson J, Dhut S, Yates JR, Toda T. (2009) Identification of a conserved F-box protein 6 interactor essential for endocytosis and cytokinesis in fission yeast. Biochem J 420: 169–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakui H, Kato M, Ushijima K, Kitaguchi M, Kato S, Sassa H. (2011) Sequence divergence and loss-of-function phenotypes of S locus F-box brothers genes are consistent with non-self recognition by multiple pollen determinants in self-incompatibility of Japanese pear (Pyrus pyrifolia). Plant J 68: 1028–1038 [DOI] [PubMed] [Google Scholar]
- Kong H, Landherr LL, Frohlich MW, Leebens-Mack J, Ma H, dePamphilis CW. (2007) Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J 50: 873–885 [DOI] [PubMed] [Google Scholar]
- Kong H, Leebens-Mack J, Ni W, dePamphilis CW, Ma H. (2004) Highly heterogeneous rates of evolution in the SKP1 gene family in plants and animals: functional and evolutionary implications. Mol Biol Evol 21: 117–128 [DOI] [PubMed] [Google Scholar]
- Kubo K, Entani T, Takara A, Wang N, Fields AM, Hua Z, Toyoda M, Kawashima S, Ando T, Isogai A, et al. (2010) Collaborative non-self recognition system in S-RNase-based self-incompatibility. Science 330: 796–799 [DOI] [PubMed] [Google Scholar]
- Lai Z, Ma W, Han B, Liang L, Zhang Y, Hong G, Xue Y. (2002) An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol Biol 50: 29–42 [DOI] [PubMed] [Google Scholar]
- Luu DT, Qin X, Laublin G, Yang Q, Morse D, Cappadocia M. (2001) Rejection of S-heteroallelic pollen by a dual-specific s-RNase in Solanum chacoense predicts a multimeric SI pollen component. Genetics 159: 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu DT, Qin X, Morse D, Cappadocia M. (2000) S-RNase uptake by compatible pollen tubes in gametophytic self-incompatibility. Nature 407: 649–651 [DOI] [PubMed] [Google Scholar]
- Marín I. (2009) Diversification of the cullin family. BMC Evol Biol 9: 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A, Traverso JA, Valot B, Ferro M, Espagne C, Ephritikhine G, Zivy M, Giglione C, Meinnel T. (2008) Extent of N-terminal modifications in cytosolic proteins from eukaryotes. Proteomics 8: 2809–2831 [DOI] [PubMed] [Google Scholar]
- Matsumoto D, Yamane H, Tao R. (2008) Characterization of SLFL1, a pollen-expressed F-box gene located in the Prunus S locus. Sex Plant Reprod 21: 113–121 [Google Scholar]
- McClure BA. (2009) Darwin’s foundation for investigating self-incompatibility and the progress toward a physiological model for S-RNase-based SI. J Exp Bot 60: 1069–1081 [DOI] [PubMed] [Google Scholar]
- Meng X, Sun P, Kao TH. (2011) S-RNase-based self-incompatibility in Petunia inflata. Ann Bot (Lond) 108: 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Minamikawa M, Kakui H, Wang S, Kotoda N, Kikuchi S, Koba T, Sassa H. (2010) Apple S locus region represents a large cluster of related, polymorphic and pollen-specific F-box genes. Plant Mol Biol 74: 143–154 [DOI] [PubMed] [Google Scholar]
- Newbigin E, Paape T, Kohn JR. (2008) RNase-based self-incompatibility: puzzled by pollen S. Plant Cell 20: 2286–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak MD, Davis AP, Anthony F, Yoder AD. (2011) Expression and trans-specific polymorphism of self-incompatibility RNases in coffea (Rubiaceae). PLoS ONE 6: e21019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes MDS, Santos RAM, Ferreira SM, Vieira J, Vieira CP. (2006) Variability patterns and positively selected sites at the gametophytic self-incompatibility pollen SFB gene in a wild self-incompatible Prunus spinosa (Rosaceae) population. New Phytol 172: 577–587 [DOI] [PubMed] [Google Scholar]
- Okada K, Tonaka N, Moriya Y, Norioka N, Sawamura Y, Matsumoto T, Nakanishi T, Takasaki-Yasuda T. (2008) Deletion of a 236 kb region around S4-RNase in a stylar-part mutant S 4sm-haplotype of Japanese pear. Plant Mol Biol 66: 389–400 [DOI] [PubMed] [Google Scholar]
- Okada K, Tonaka N, Taguchi T, Ichikawa T, Sawamura Y, Nakanishi T, Takasaki-Yasuda T. (2011) Related polymorphic F-box protein genes between haplotypes clustering in the BAC contig sequences around the S-RNase of Japanese pear. J Exp Bot 62: 1887–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Wang F, Zhao L, Zhou JL, Lai Z, Zhang YS, Robbins TP, Xue Y. (2004a) The F-box protein AhSLF-S2 controls the pollen function of S-RNase-based self-incompatibility. Plant Cell 16: 2307–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Wang H, Zhao L, Zhou J, Huang J, Zhang Y, Xue Y. (2004b) The F-box protein AhSLF-S2 physically interacts with S-RNases that may be inhibited by the ubiquitin/26S proteasome pathway of protein degradation during compatible pollination in Antirrhinum. Plant Cell 16: 582–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risseeuw EP, Daskalchuk TE, Banks TW, Liu E, Cotelesage J, Hellmann H, Estelle M, Somers DE, Crosby WL. (2003) Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J 34: 753–767 [DOI] [PubMed] [Google Scholar]
- Saito T, Sato Y, Sawamura Y, Shoda M, Takasaki-Yasuda T, Kotobuki K. (December 9, 2011) Dual recognition of S1 and S4 pistils by S4sm pollen in self-incompatibility of Japanese pear (Pyrus pyrifolia Nakai). Tree Genet Genomes http://dx.doi.org/10.1007/s11295-011-0456-5 [Google Scholar]
- Sassa H, Kakui H, Minamikawa M. (2010) Pollen-expressed F-box gene family and mechanism of S-RNase-based gametophytic self-incompatibility (GSI) in Rosaceae. Sex Plant Reprod 23: 39–43 [DOI] [PubMed] [Google Scholar]
- Sassa H, Kakui H, Miyamoto M, Suzuki Y, Hanada T, Ushijima K, Kusaba M, Hirano H, Koba T. (2007) S locus F-box brothers: multiple and pollen-specific F-box genes with S haplotype-specific polymorphisms in apple and Japanese pear. Genetics 175: 1869–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, Elledge SJ, Harper JW, Pagano M, Pavletich NP. (2000) Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408: 381–386 [DOI] [PubMed] [Google Scholar]
- Sijacic P, Wang X, Skirpan AL, Wang Y, Dowd PE, McCubbin AG, Huang S, Kao TH. (2004) Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature 429: 302–305 [DOI] [PubMed] [Google Scholar]
- Sonneveld T, Tobutt KR, Vaughan SP, Robbins TP. (2005) Loss of pollen-S function in two self-compatible selections of Prunus avium is associated with deletion/mutation of an S haplotype-specific F-box gene. Plant Cell 17: 37–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbachs JE, Holsinger KE. (2002) S-RNase-mediated gametophytic self-incompatibility is ancestral in eudicots. Mol Biol Evol 19: 825–829 [DOI] [PubMed] [Google Scholar]
- Takahashi N, Kuroda H, Kuromori T, Hirayama T, Seki M, Shinozaki K, Shimada H, Matsui M. (2004) Expression and interaction analysis of Arabidopsis Skp1-related genes. Plant Cell Physiol 45: 83–91 [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tao R, Iezzoni AF. (2010) The S-RNase-based gametophytic self-incompatibility system in Prunus exhibits distinct genetic and molecular feature. Sci Hortic (Amsterdam) 124: 423–433 [Google Scholar]
- Tao R, Matsumoto D. (2012) S locus mutation and self-compatibility in stone fruits. Acta Hortic (in press) [Google Scholar]
- Tao R, Yamane H, Sugiura A, Murayama H, Sassa H, Mori H. (1999) Molecular typing of S-alleles through identification, characterization and cDNA cloning for S-RNases in sweet cherry. J Am Soc Hortic Sci 124: 224–233 [Google Scholar]
- Tsukamoto T, Hauck NR, Tao R, Jiang N, Iezzoni AF. (2010) Molecular and genetic analyses of four nonfunctional S haplotype variants derived from a common ancestral S haplotype identified in sour cherry (Prunus cerasus L.). Genetics 184: 411–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima K, Sassa H, Dandekar AM, Gradziel TM, Tao R, Hirano H. (2003) Structural and transcriptional analysis of the self-incompatibility locus of almond: identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell 15: 771–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima K, Yamane H, Watari A, Kakehi E, Ikeda K, Hauck NR, Iezzoni AF, Tao R. (2004) The S haplotype-specific F-box protein gene, SFB, is defective in self-compatible haplotypes of Prunus avium and P. mume. Plant J 39: 573–586 [DOI] [PubMed] [Google Scholar]
- Vieira J, Fonseca NA, Vieira CP. (2008a) An S-RNase-based gametophytic self-incompatibility system evolved only once in eudicots. J Mol Evol 67: 179–190 [DOI] [PubMed] [Google Scholar]
- Vieira J, Fonseca NA, Vieira CP. (2009) RNase-based gametophytic self-incompatibility evolution: questioning the hypothesis of multiple independent recruitments of the S-pollen gene. J Mol Evol 69: 32–41 [DOI] [PubMed] [Google Scholar]
- Vieira J, Teles E, Santos RAM, Vieira CP. (2008b) Recombination at Prunus S-locus region SLFL1 gene. Genetics 180: 483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilanova S, Badenes ML, Burgos L, Martínez-Calvo J, Llácer G, Romero C. (2006) Self-compatibility of two apricot selections is associated with two pollen-part mutations of different nature. Plant Physiol 142: 629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang X, McCubbin AG, Kao TH. (2003) Genetic mapping and molecular characterization of the self-incompatibility (S) locus in Petunia inflata. Plant Mol Biol 53: 565–580 [DOI] [PubMed] [Google Scholar]
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. (2004) Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol 337: 635–645 [DOI] [PubMed] [Google Scholar]
- Xu G, Ma H, Nei M, Kong H. (2009) Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification. Proc Natl Acad Sci USA 106: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zhang Y, Yang Q, Li Q, Cheng Z, Dickinson HG. (2009) Genetic features of a pollen-part mutation suggest an inhibitory role for the Antirrhinum pollen self-incompatibility determinant. Plant Mol Biol 70: 499–509 [DOI] [PubMed] [Google Scholar]
- Yamane H, Ikeda K, Hauck NR, Iezzoni AF, Tao R. (2003a) Self-incompatibility (S) locus region of the mutated S6-haplotype of sour cherry (Prunus cerasus) contains a functional pollen S allele and a non-functional pistil S allele. J Exp Bot 54: 2431–2437 [DOI] [PubMed] [Google Scholar]
- Yamane H, Ikeda K, Ushijima K, Sassa H, Tao R. (2003b) A pollen-expressed gene for a novel protein with an F-box motif that is very tightly linked to a gene for S-RNase in two species of cherry, Prunus cerasus and P. avium. Plant Cell Physiol 44: 764–769 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhao Z, Xue Y. (2009) Roles of proteolysis in plant self-incompatibility. Annu Rev Plant Biol 60: 21–42 [DOI] [PubMed] [Google Scholar]
- Zhao L, Huang J, Zhao Z, Li Q, Sims TL, Xue Y. (2010) The Skp1-like protein SSK1 is required for cross-pollen compatibility in S-RNase-based self-incompatibility. Plant J 62: 52–63 [DOI] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. (2002) Structure of the Cul1-Rbx1-SKp1-F-boxSkp2 SCF ubiquitin ligase complex. Science 416: 703–709 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.