Abstract
The protein kinase AvrPto-dependent Pto-interacting protein3 (Adi3) is a known suppressor of cell death, and loss of its function has been correlated with cell death induction during the tomato (Solanum lycopersicum) resistance response to its pathogen Pseudomonas syringae pv tomato. However, Adi3 downstream interactors that may play a role in cell death regulation have not been identified. We used a yeast two-hybrid screen to identify the plant SnRK1 (for Sucrose non-Fermenting-1-Related Protein Kinase1) protein as an Adi3-interacting protein. SnRK1 functions as a regulator of carbon metabolism and responses to biotic and abiotic stresses. SnRK1 exists in a heterotrimeric complex with a catalytic α-subunit (SnRK1), a substrate-interacting β-subunit, and a regulatory γ-subunit. Here, we show that Adi3 interacts with, but does not phosphorylate, the SnRK1 α-subunit. The ability of Adi3 to phosphorylate the four identified tomato β-subunits was also examined, and it was found that only the Galactose Metabolism83 (Gal83) β-subunit was phosphorylated by Adi3. This phosphorylation site on Gal83 was identified as serine-26 using a mutational approach and mass spectrometry. In vivo expression of Gal83 indicates that it contains multiple phosphorylation sites, one of which is serine-26. An active SnRK1 complex containing Gal83 as the β-subunit and sucrose nonfermenting4 as the γ-subunit was constructed to examine functional aspects of the Adi3 interaction with SnRK1 and Gal83. These assays revealed that Adi3 is capable of suppressing the kinase activity of the SnRK1 complex through Gal83 phosphorylation plus the interaction with SnRK1 and suggested that this function may be related to the cell death suppression activity of Adi3.
Programmed cell death (PCD) is a genetically encoded, highly regulated process in multiple-cell and single-cell eukaryotic organisms (Lam, 2004; Brownlee, 2008; Deponte, 2008; Lane, 2008) and bacteria (Engelberg-Kulka et al., 2006; Lane, 2008). In multicellular organisms, PCD often occurs during developmental processes, imparting a positive effect by killing specific cells in the organ connected with the process (Lam, 2004). Without PCD, proper development is not achieved. In plants, flower and embryo development, seed coat formation, senescence, leaf shape formation, xylem formation, and resistance to pathogens all involve PCD (Lam, 2004). Thus, PCD plays a central role in many aspects of the maturation and survival of plants.
Despite the many processes in plants that require PCD, the identification of genes and signaling pathways involved in plant PCD has been difficult compared with mammalian systems (Lam et al., 2001; Hoeberichts and Woltering, 2003; Lam, 2004, 2008). However, in the past decade, the number of genes identified to be involved in plant PCD control has increased substantially. Just to name a few, these genes range from the plant homologs for the mammalian PCD regulators Bax inhibitor-1 and the Bcl-2 associated athanogene (BAG) proteins (Doukhanina et al., 2006; Watanabe and Lam, 2009) to mitogen-activated protein kinases and transcription factors (Zhang and Klessig, 2001; Ren et al., 2002; Pedley and Martin, 2005; Kaneda et al., 2009; Phan et al., 2011). However, the signaling pathways associated with the proteins encoded by these genes remain, for the most part, unresolved.
One of the plant genes identified to control PCD encodes the AGC Ser/Thr protein kinase AvrPto-dependent Pto-interacting protein3 (Adi3) from tomato (Solanum lycopersicum), which functions as a cell death suppressor (Devarenne et al., 2006; Ek-Ramos et al., 2010). Adi3 was first characterized for its role in the hypersensitive response cell death induced during the resistance of tomato to its bacterial pathogen Pseudomonas syringae pv tomato (Pst; Bogdanove and Martin, 2000; Devarenne et al., 2006). We have used a variety of methods to analyze Adi3 cell death suppression (CDS) activity and parts of the Adi3 signaling pathway. A loss of Adi3 function by virus-induced gene silencing causes spontaneous cell death lesions to form on leaves and stems, ultimately leading to death of the plant (Devarenne et al., 2006). Adi3 can prevent cell death when overexpressed in the presence of PCD-inducing conditions. 3-Phosphoinositide-dependent protein kinase1 (Pdk1) is the upstream kinase that phosphorylates Adi3 at Ser-539 for the activation of its CDS activity (Devarenne et al., 2006; Ek-Ramos et al., 2010). The phosphorylation of Adi3 by Pdk1 also directs Adi3 to the nucleus, where its CDS activity is manifested (Ek-Ramos et al., 2010). The prevention of nuclear entry eliminates Adi3 CDS and, thus, induces cell death (Ek-Ramos et al., 2010). Thus, it is hypothesized that prevention of Adi3 signaling and CDS, possibly by inhibiting nuclear entry, may be a mechanism by which cell death is initiated for PCD-requiring situations such as the hypersensitive response to Pst (Ek-Ramos et al., 2010).
Our studies on Adi3 CDS show remarkable similarity to the mammalian cell death (apoptosis) suppressor protein kinase B (PKB; a.k.a. Akt). PKB is also an AGC Ser/Thr kinase that is phosphorylated by Pdk1 for activation and nuclear localization, where PKB CDS activity occurs (Vivanco and Sawyers, 2002; Scheid et al., 2005; Miyamoto et al., 2009). Loss of PKB function by knockout or elimination of kinase activity causes spontaneous cell death, and the PKB knockout is lethal (Dudek et al., 1997; Chen et al., 2001; Luo et al., 2003). PKB can prevent cell death when overexpressed in the presence of PCD-inducing conditions (Arico et al., 2002; Kulp et al., 2004; Zhu et al., 2004). These similarities in function, cell localization, and signaling between Adi3 and PKB suggest that Adi3 may be the functional homolog of PKB in plants (Devarenne et al., 2006; Ek-Ramos et al., 2010).
While many PKB substrates for CDS are known (Carnero, 2010), substrates for Adi3 have yet to be identified. Thus, we initiated a yeast two-hybrid (Y2H) screen to identify potential substrates of Adi3. This screen identified the α-subunit of the sucrose nonfermenting 1-related protein kinase 1 (SnRK1) complex as an Adi3 interactor. SnRK1 is the plant homolog of the conserved protein complex known as sucrose nonfermenting 1 (Snf1) in yeast and AMP-activated protein kinase (AMPK) in mammals. These protein complexes regulate carbon metabolism and metabolic stress responses (i.e. low Glc or starvation; Halford et al., 2003; Polge and Thomas, 2007; Halford and Hey, 2009; Hey et al., 2010). Snf1 controls the shift from Glc fermentation to the aerobic use of alternate carbon sources (Polge and Thomas, 2007). AMPK is activated under low-ATP/Glc conditions and represses ATP-consuming pathways while activating ATP-producing pathways (Polge and Thomas, 2007; Halford and Hey, 2009). SnRK1 activates starch mobilization and metabolism under low-Glc conditions such as during darkness (Halford et al., 2003; Polge and Thomas, 2007; Halford and Hey, 2009; Hey et al., 2010). Additionally, SnRK1 regulates metabolism in response to environmental stresses such as pathogen attack and herbivory, flooding, and cell death during development (Sreenivasulu et al., 2006; Coello et al., 2010; Hey et al., 2010; Cho et al., 2012). Thus, SnRK1 appears to be a key regulator connecting metabolism and stress responses in plants (Halford and Hey, 2009; Hey et al., 2010).
The SnRK1 (and Snf1/AMPK) complex exists as a heterotrimer of an α-subunit Ser/Thr kinase called SnRK1, one of several possible β-subunits (SNF1-interacting protein1, Sip2, or Galactose Metabolism83 [Gal83] in yeast) and a γ-subunit called sucrose nonfermenting4 (Snf4) (Halford and Hey, 2009; Coello et al., 2010). The γ-subunit has been shown to regulate kinase activity of the complex (Jiang and Carlson, 1996), whereas the β-subunits regulate complex substrate specificity and cellular localization (Mitchelhill et al., 1997; Vincent and Carlson, 1999; Vincent et al., 2001; Warden et al., 2001). The signaling mechanisms exerted on the β-subunits for controlling function are not fully understood. But at least for yeast Sip1 and the human β-subunit AMPKβ1, phosphorylation appears to be involved in controlling β-subunit function (Warden et al., 2001; Hedbacker et al., 2004). Recently, two kinases have been shown to phosphorylate yeast Gal83, but a connection to function has not been shown (Mangat et al., 2010), and it has yet to be shown that a plant β-subunit is phosphorylated. Here, we present data showing that Adi3 interacts with the tomato SnRK1 α-subunit and the Gal83 β-subunit, that Adi3 can only phosphorylate Gal83, and that Adi3 can inhibit the kinase activity of the SnRK1 complex.
RESULTS
Identification of SnRK1 as an Adi3-Interacting Protein
In an effort to identify Adi3-interacting proteins, we carried out a Y2H screen using a cDNA prey library that has been used previously to identify proteins that interact with the tomato resistance protein kinase Pto (Zhou et al., 1995). Approximately 15 million yeast transformants were screened for Adi3-interacting proteins using selection on Leu− plates, and 1,366 transformants were followed up in a LacZ screen. The prey inserts from 85 random positive clones were sequenced and screened against GenBank by BLAST for identification. Of these clones, SnRK, encoding the α-subunit of the SnRK1 protein complex, was identified four times. The SnRK insert in the prey library was a partial open reading frame (ORF), and a full-length ORF was identified by searching the tomato EST database (http://solgenomics.net/) by BLAST with the SnRK Y2H fragment. Unigene SGN-U564382 was identified as containing a full-length SnRK ORF, and this sequence was amplified from tomato leaf tissue RNA by reverse transcription (RT)-PCR. A BLAST search against GenBank with the full-length SnRK sequence showed that it was identical to a previously identified tomato SnRK cDNA (Bradford et al., 2003). In Arabidopsis (Arabidopsis thaliana), SnRK proteins are separated into three distinct families, SnRK1, SnRK2, and SnRK3 (Halford and Hey, 2009). BLAST and alignment comparison of the protein encoded by the SnRK sequence cloned here with members of the Arabidopsis SnRK (AtSnRK) family indicated that it belongs to the SnRK1 family (Supplemental Fig. S1). The tomato gene identified here will be referred to as SlSnRK1 throughout this study.
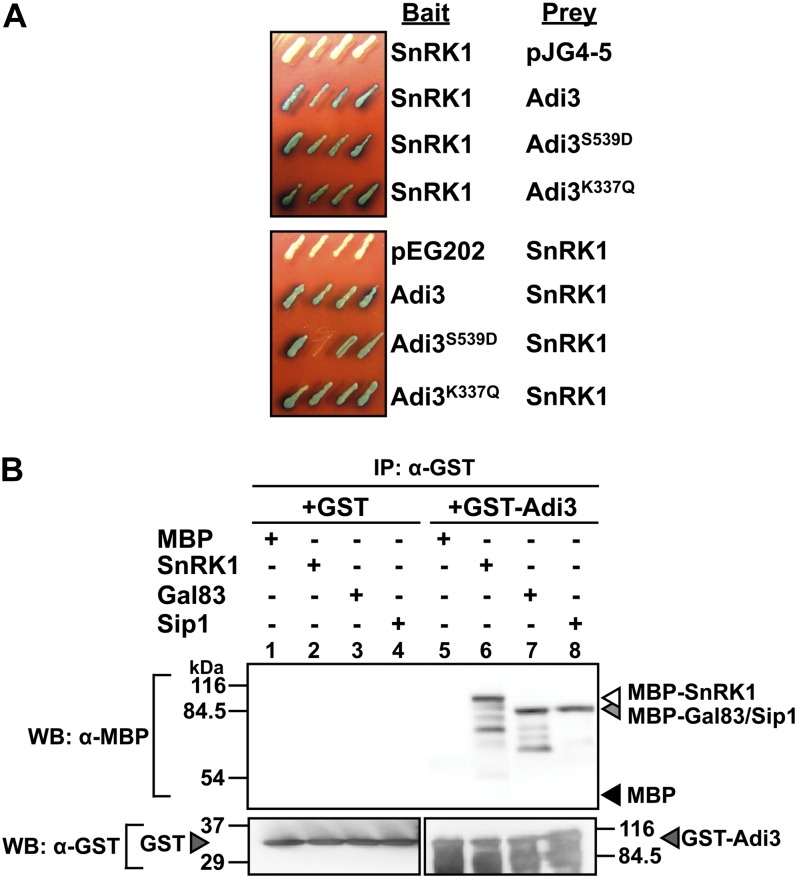
The full-length SlSnRK1 ORF was used to confirm the Y2H interaction with Adi3 and to test the interaction with kinase activity mutants of Adi3. SlSnRK1 does not autoactivate in the Y2H assay when expressed from either the prey or bait vector (Fig. 1A). Our previous studies have shown that mutation of the Pdk1 phosphorylation site on Adi3 (Ser-539) to Asp (Adi3S539D) confers constitutive kinase activity on Adi3 and that mutation of Lys-337 to Gln (Adi3K337Q) in the ATP-binding pocket eliminates Adi3 kinase activity (Devarenne et al., 2006). The interaction of SlSnRK1 with Adi3 was not abolished by either of these Adi3 kinase activity mutants (Fig. 1A). This was the case whether the proteins were in the bait or prey vector (Fig. 1A), suggesting that kinase activity is not required for this interaction. The SlSnRK1 and Adi3 interaction was also tested by immunoprecipitation. Glutathione S-transferase (GST)-Adi3 immunoprecipitated with an α-GST antibody was not capable of pulling down maltose binding protein (MBP) but was capable of pulling down MBP-SlSnRK1 (Fig. 1B, compare lanes 5 and 6).
Figure 1.
Adi3 interaction with SnRK1 complex members. A, Adi3 and SnRK1 interact in the Y2H assay. The indicated bait and prey constructs were expressed in yeast and tested for expression of the LacZ gene on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside plates (blue = interaction). B, Adi3 interacts with SnRK1 complex members by immunoprecipitation (IP). Top panels, GST or a GST-Adi3 fusion protein was incubated at 4°C for 1 h with MBP fusion proteins of SnRK1, Gal83, or Sip1, immunoprecipitated with an α-GST antibody, and the proteins associated with GST-Adi3 were analyzed by α-MBP western blot (WB). Bottom panels, a one-tenth aliquot of each MBP fusion protein was analyzed by α-MBP western blot for a loading control. [See online article for color version of this figure.]
Adi3 Also Interacts with Two SlSnRK1 β-Subunits
We also tested if Adi3 could interact with two of the previously identified SlSnRK1 β-subunits. First, cDNAs for these two tomato β-subunits, SlGal83 and SlSip1 (Bradford et al., 2003), were cloned. The reported SlGal83 sequence is not a full-length ORF and is missing a portion of the 5′ end (Bradford et al., 2003). Thus, we used the tomato EST and genomic databases to identify the remaining 5′ end of the SlGal83 sequence and to make sure that the published SlSip1 sequence contained the full-length ORF. A BLAST search of the tomato ESTs with the published SlGal83 sequence identified a full-length ORF within unigene SGN-U564868, which indicated that the published SlGal83 sequence was missing 51 bp from the 5′ end, or 17 N-terminal amino acids (Supplemental Fig. S2). The original SlGal83 sequence also had a misidentification of nucleotide 58 as guanine, whereas EST and genomic sequence indicate that nucleotide 58 is a cytosine (data not shown). The full-length SlGal83 ORF was amplified by PCR from Sol Genomics Network (SGN) clone cTOF-18-D18.
A BLAST search with the published SlSip1 sequence (Bradford et al., 2003) against the tomato genomic database identified the SlSip1 gene within genomic locus AC186291.2. The deduced ORF from this genomic sequence was longer than the published sequence and indicated that the published SlSip1 ORF was missing 177 bp of 5′ sequence, or 59 N-terminal amino acids (Supplemental Figs. S2 and S3A). The full-length ORF sequence of SlSip1 was cloned by RT-PCR based on the deduced ORF sequence, confirming the presence of this transcript in tomato (Supplemental Fig. S3A). Both of these cloned full-length tomato sequences were used for all subsequent studies reported here.
The interaction of SlGal83 and SlSip1 with Adi3 was tested by α-GST immunoprecipitation as with SlSnRK1. The results indicated that both β-subunits were capable of interacting with Adi3 (Fig. 1B, lanes 7 and 8). For reasons that will become apparent below, we made SlGal83 the main subject of our research and have shown that Adi3 also interacts with SlGal83 in the Y2H assay (Supplemental Fig. S3B). These results indicated that Adi3 is capable of interacting with several members of the SlSnRK1 complex.
Adi3 Phosphorylates SlGal83
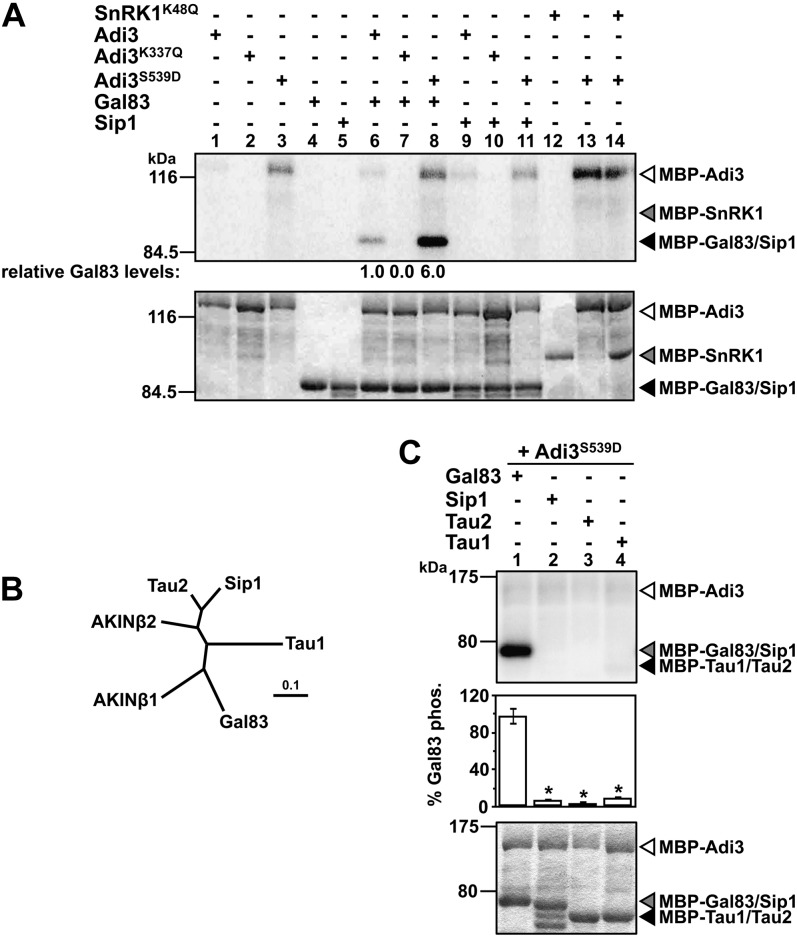
Because Adi3 interacts with the SlSnRK1 α-subunit, it is possible that Adi3 acts as an upstream activator of SlSnRK1. Thus, we analyzed Adi3 kinase activity toward SlSnRK1. First, a kinase-inactive SlSnRK1 was generated by mutating Lys-48 to Gln, giving SlSnRK1K48Q (Fig. 2, lane 12). This Lys corresponds to the invariant Lys-45 in AMPK required for ATP binding (Dyck et al., 1996; Supplemental Fig. S1). Phosphorylation of SlSnRK1K48Q by the constitutively active Adi3S539D was not seen (Fig. 2A, lane 14), suggesting that Adi3 is not an upstream activator of SlSnRK1.
Figure 2.
Adi3 phosphorylates Gal83. In A and C, top panels show phosphor images and bottom panels show Coomassie blue-stained gels. Quantity One software was used to normalize the phosphorylation levels to the protein levels in each assay. A, Analysis of SnRK1 α- and β-subunit phosphorylation by Adi3. Kinase-active and -inactive MBP-Adi3 proteins were tested for phosphorylation of MBP-Gal83, MBP-Sip1, and kinase-inactive MBP-SnRK1K48Q using [γ-32P]ATP in in vitro kinase assays. Gal83 phosphorylation values are reported as a percentage of wild-type Adi3 phosphorylation of Gal83 and are representative of two independent experiments. B, Phylogenetic relationship between tomato and Arabidopsis β-subunits. Proteins were aligned using ClustalW (Larkin et al., 2007), and the tree produced was analyzed using TreeView (Page, 1996). The scale bar indicates the number of amino acid substitutions per site. C, Adi3 only phosphorylates the Gal83 β-subunit. Kinase-active MBP-Adi3S539D was tested for phosphorylation of MBP-Gal83, MBP-Sip1 MBP-Tau1, and MBP-Tau2 as in A. Values are averages of three independent experiments. Error bars represent se. Asterisks indicate significant decreases in β-subunit phosphorylation as compared with Gal83 phosphorylation (Student’s t test, P < 0.01).
Because Adi3 can interact with SlGal83 and SlSip1, it is possible that Adi3 can phosphorylate these β-subunits. The β-subunits from yeast and mammals are known to be phosphorylated (Mitchelhill et al., 1997; Warden et al., 2001; Mangat et al., 2010), whereas phosphorylation of the plant β-subunits has not been reported to date. Thus, the ability of Adi3 to phosphorylate the SlGal83 and/or SlSip1 β-subunits was examined. Kinase assays showed that both wild-type Adi3 and constitutively active Adi3S539D were able to phosphorylate SlGal83, with Adi3S539D phosphorylating SlGal83 approximately six times more than the wild type (Fig. 2A, compare lanes 6 and 8). Interestingly, neither form of Adi3 was capable of phosphorylating SlSip1 (Fig. 2A, lanes 9–11), even though Adi3 can interact with SlSip1 (Fig. 1B, lane 8). This would suggest that there is some catalytic specificity of Adi3 toward SlGal83 over that of SlSip1.
The phosphorylation of SlGal83 but not SlSip1 by Adi3 led us to search the tomato genome for additional SlSnRK1 β-subunits that may be phosphorylated by Adi3. The SlGal83 sequence was used to search the tomato genome by BLAST against the SGN Tomato Combined Database (whole genome, bacterial artificial chromosome, and unigene sequences), and two additional sequences with high similarity to SlSnRK1 β-subunits were discovered and termed Tomato SnRK1 Associated β-subunit1 (Tau1) and Tau2 (Supplemental Fig. S2). Additionally, BLAST of the Tau1 and Tau2 proteins against GenBank returned the Arabidopsis β-subunit AKINβ2 (E values of 1E-90 and 3E-136, respectively) as a top hit, suggesting that these proteins are SnRK1 β-subunits. The Tau1 and Tau2 cDNAs were amplified from leaf RNA by RT-PCR (Supplemental Fig. S4), and the proteins derived from these ORFs appear to be more related to SlSip1 and the Arabidopsis β-subunit AKINβ2 than to SlGal83 (Fig. 2B). Next, the phosphorylation of Tau1 and Tau2 by Adi3S539D was tested using in vitro kinase assays, which showed that Adi3 did not phosphorylate Tau1 or Tau2 to a significant level and only phosphorylated SlGal83 (Fig. 2C).
Because Adi3 only phosphorylates SlGal83 and not the other β-subunits, we focused on SlGal83 and confirmed that it is a functional SnRK1 β-subunit using yeast complementation that was not done in the initial SlGal83 study (Bradford et al., 2003). In yeast, the Snf1 complex functions to allow growth on alternative carbon sources such as Suc (Carlson et al., 1981; Polge and Thomas, 2007), and loss of the three yeast β-subunits (ScSip1, ScSip2, and ScGal83; sip1Δsip2Δgal83Δ yeast) does not allow for growth on Suc (Schmidt and McCartney, 2000). Complementation of sip1Δsip2Δgal83Δ cells and restoration of growth on Suc can be accomplished by introducing any one of the β-subunits (Schmidt and McCartney, 2000). Individually, each of the Arabidopsis β-subunits (AKINβ1, AKINβ2, and AKINβ3) is also capable of complementing the sip1Δsip2Δgal83Δ cells (Gissot et al., 2004; Polge et al., 2008). We carried out this assay and showed that SlGal83-GFP was capable of restoring sip1Δsip2Δgal83Δ growth on Suc, confirming complementation (Supplemental Fig. S5A). As an additional confirmation of SlGal83 complementation of sip1Δsip2Δgal83Δ yeast, we tested for the restoration of invertase activity, which is regulated by the Snf1 complex under low-Glc conditions (Carlson et al., 1984). Our results show that SlGal83-GFP was able to restore basal and low-Glc-induced invertase activity to sip1Δsip2Δgal83Δ yeast (Supplemental Fig. S5B). These studies confirm SlGal83 as a true SnRK1 β-subunit and that SlGal83-GFP is functional in vivo.
Identification of Ser-26 as the Adi3 Phosphorylation Site on SlGal83
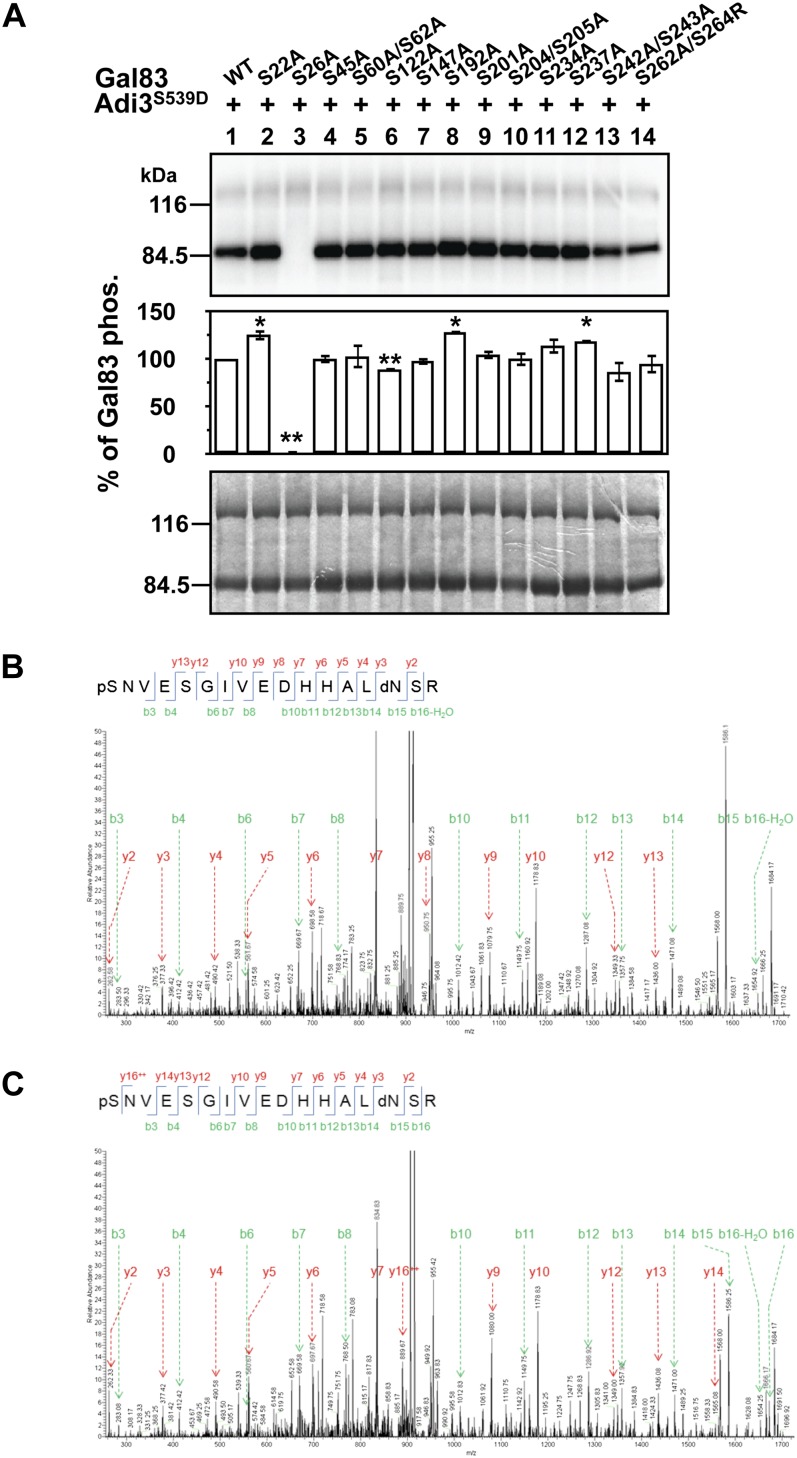
In an effort to identify the SlGal83 residue phosphorylated by Adi3, we carried out a kinase assay screen of several SlGal83 Ser mutants. Within the SlGal83 protein, there are 28 Ser amino acids (Supplemental Fig. S2), 17 of which were mutated to Ala and tested for loss of phosphorylation by Adi3 using in vitro kinase assays. Once the assays were completed, the SlGal83 phosphorylation levels were normalized to the SlGal83 and Adi3 protein levels in each assay, and the amount of SlGal83 phosphorylation was expressed as a percentage of wild-type SlGal83 phosphorylation. The results indicate that while many of the mutations slightly increased or decreased the ability of Adi3 to phosphorylate SlGal83, only the Ser-26A (S26A) mutation completely eliminated phosphorylation by Adi3 (Fig. 3A, lane 3). There are eight Thr residues in SlGal83 (Supplemental Fig. S2). Ala mutation of one Thr residue did not eliminate Adi3 phosphorylation (data not shown) and the remaining seven Thr residues were not tested because S26A was a complete knockout of Adi3 phosphorylation of SlGal83 (Fig. 3A). These results indicate that while Adi3 can interact with several members of the SlSnRK1 complex, it can only phosphorylate SlGal83. The β-subunit protein alignment indicates that SlSip1, Tau1, and Tau2 do not contain a Ser corresponding to SlGal83 Ser-26 (possibly marginally conserved in Tau2; Supplemental Fig. S2), supporting the inability of Adi3 to phosphorylate these proteins.
Figure 3.
Adi3 phosphorylates Gal83 at Ser-26. A, Adi3 phosphorylates Ser-26 of Gal83. Kinase-active MBP-Adi3S539D was used to phosphorylate the indicated MBP-Gal83 Ser-to-Ala mutants using [γ-32P]ATP in in vitro kinase assays. Quantity One software was used to normalize the phosphorylation levels to the protein levels in each assay. Gal83 phosphorylation values are reported as a percentage of wild-type (WT) Gal83 phosphorylation and are averages of three independent experiments. Error bars represent se. Asterisks indicate significant increases (*) or decreases (**) in phosphorylation of Gal83 Ser-to-Ala mutants compared with wild-type Gal83 phosphorylation (Student’s t test, P < 0.05). Top and middle panels show phosphor images, and the bottom panel shows a Coomassie blue-stained gel. B, MS identification of Gal83 Ser-26 in vitro phosphorylation by Adi3. In vitro Adi3-phosphorylated Gal83-MBP as in A was digested with trypsin, passed over an immobilized metal affinity chromatography column, and eluted peptides were analyzed by MS/MS. C, MS identification of Gal83 Ser-26 in vivo phosphorylation. Gal83-GFP was expressed in tomato protoplasts, α-GFP was immunoprecipitated, the protein was digested with trypsin and passed over an immobilized metal affinity chromatography column, and the eluted peptides were analyzed by MS/MS. dN, Deaminated Asn; pS, phospho-Ser. [See online article for color version of this figure.]
Phosphorylation of SlGal83 Ser-26 was confirmed by mass spectrometry (MS) analysis. Trypsin digestion of SlGal83 will produce two possible peptides containing Ser-26, SNVESGIVEDHHALNSR and RSNVESGIVEDHHALNSR (with Ser-26 underlined), and tandem mass spectrometry (MS/MS) analysis of in vitro Adi3-phosphorylated, trypsin-digested SlGal83 identified Ser-26 phosphorylation in both peptides (Fig. 3B; Supplemental Fig. S6A). The in vivo phosphorylation of Ser-26 was also analyzed by first expressing SlGal83-GFP in tomato protoplasts and immunoprecipitating the protein with an α-GFP antibody (Supplemental Fig. S6, B and C). The trypsin-digested protein was analyzed by MS/MS, and Ser-26 phosphorylation was identified in the SNVESGIVEDHHALNSR peptide (Fig. 3C) but not the RSNVESGIVEDHHALNSR peptide. These data indicate that Adi3 phosphorylates SlGa83 Ser-26 in vitro and support the possibility that Adi3 also performs this phosphorylation event in vivo.
An additional SlGal83 phosphorylation site was identified in vitro and in vivo in peptide RSNVEpSGIVEDHHALNSR corresponding to Ser-30 (Supplemental Fig. S7, A and B), suggesting that Adi3 may also phosphorylate Ser-30. The Ser-30-to-Ala mutation was not initially tested, as shown in Figure 3A. So the SlGal83S30A protein was produced and tested for loss of Adi3 phosphorylation as in Figure 3A. The results indicate that the S30A mutation does not significantly reduce the SlGal83 phosphorylation by Adi3 in vitro (Supplemental Fig. S7C). While Adi3 could be responsible for this phosphorylation event in vivo, it remains to be positively determined. It should be noted that for both the in vitro and in vivo MS/MS analyses, peptides with the Ser-26 phosphorylation were approximately twice as prevalent as those with Ser-30 phosphorylation, and no peptides were found with both Ser-26 and Ser-30 phosphorylation.
Tomato Gal83 Is Phosphorylated in Vivo
In order to analyze the in vivo phosphorylation status of SlGal83, we used an alteration to the standard SDS-PAGE by adjusting the ratio of bis-acrylamide to acrylamide. This method has been used to distinguish different phosphorylation states of yeast phosphatidylinositol 4-kinase (Demmel et al., 2008). SlGal83-GFP transgenic Arabidopsis plants were created, and the SlGal83-GFP protein was analyzed by α-GFP western blot using increasing ratios of bis-acrylamide to acrylamide. The 1:200 bis-acrylamide:acrylamide ratio was capable of separating five different forms of SlGal83-GFP, and two of these forms are lost when expressing the SlGal83S26A-GFP protein (Supplemental Fig. S8). This would suggest that the 1:200 SDS-PAGE/α-GFP western blot can be used to effectively separate and identify different modified forms of SlGal83.
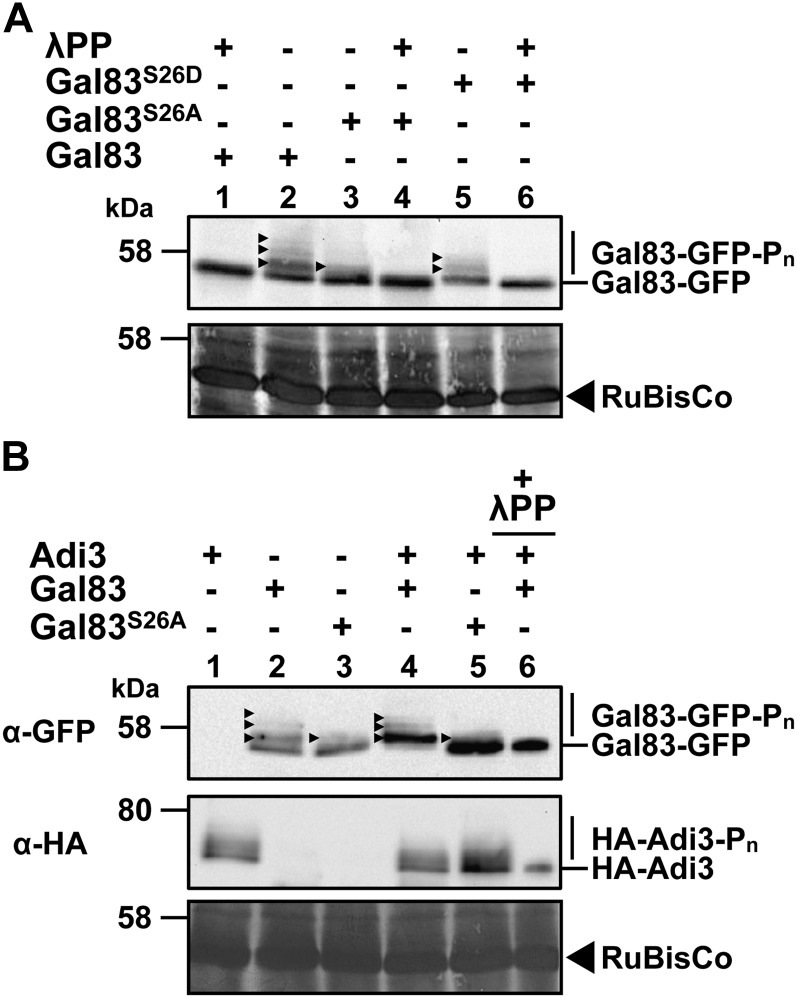
Next, the in vivo phosphorylation status of SlGal83 as expressed in tomato was analyzed. SlGal83-GFP was expressed in protoplasts, an extract was made, the extract was treated with λ phosphatase, and SlGal83-GFP was analyzed using 1:200 gels/α-GFP western blot. In the presence of λ phosphatase, SlGal83-GFP appeared as a single band (Fig. 4A, lane 1). However, in the absence of λ phosphatase, SlGal83-GFP appeared as at least four distinct protein bands, and by comparison with the λ phosphatase treatment, this can be interpreted as one unphosphorylated form and three phosphorylated forms of SlGal83-GFP (Fig. 4A, lane 2).
Figure 4.
In vivo phosphorylation status of Gal83. Proteins were separated by SDS-PAGE using a 1:500 bis-acrylamide:acrylamide ratio followed by α-GFP or α-HA western blot. A, Gal83 is phosphorylated in tomato protoplasts. Total protein extracts from Gal83-GFP-expressing protoplasts were treated with and without λ phosphatase (λPP) and analyzed by α-GFP western blot. The arrowheads indicate different Gal83-GFP phosphorylated forms. B, Adi3 phosphorylates Gal83 in vivo. Protoplasts expressing the indicated combinations of HA-Adi3 and Gal83-GFP were analyzed by α-GFP for analysis of the phosphorylation status of Gal83-GFP and α-HA western blot.
The contribution of Ser-26 phosphorylation to the SlGal83 phosphorylated protein bands was analyzed by mutating SlGal83 Ser-26 to the nonphosphorylatable Ala (SlGal83S26A) and the phosphomimetic Asp (SlGal83S26D). Expression of the GFP fusions of both of these proteins in tomato protoplasts appeared to reduce the number of SlGal83-GFP phosphorylated forms: SlGal83S26A only had one phosphoprotein band (Fig. 4A, lane 3), whereas SlGal83S26D showed a reduction of one phosphoprotein band (Fig. 4A, lane 5). The phosphoprotein bands for both SlGal83S26A and SlGal83S26D can be removed by λ phosphatase treatment (Fig. 4A, lanes 4 and 6, respectively). This would suggest that Ser-26 phosphorylation contributes to the in vivo phosphorylation status of SlGal83.
Adi3 Phosphorylates SlGal83 in Vivo
We looked for evidence that SlGal83 Ser-26 is phosphorylated in vivo by Adi3 using a coexpression approach. SlGal83-GFP, SlGal83S26A-GFP, and hemagglutinin (HA)-Adi3 were coexpressed in tomato protoplasts, and the banding pattern of phosphorylated SlGal83-GFP was analyzed by 1:200 gels/α-GFP western blot. In the absence of HA-Adi3, SlGal83-GFP and SlGal83S26A-GFP appeared as was seen in Figure 4A (Fig. 4B, lanes 2 and 3). However, in the presence of HA-Adi3, wild-type SlGal83-GFP protein appeared to shift upward (Fig. 4B, lane 4). Treatment of this sample with λ phosphatase reduced SlGal83-GFP to a single nonphosphorylated protein band (Fig. 4B, lane 6, compare with lane 4). In the presence of HA-Adi3, the SlGal83S26A-GFP protein appeared similar to that without HA-Adi3 (Fig. 4B, lane 5). Taken together, these data would suggest that SlGa83 is phosphorylated by Adi3 in vivo. Additionally, it was seen that HA-Adi3 exists as several phosphoprotein bands that can be reduced to a single band with λ phosphatase treatment (Fig. 4B, middle panel).
Functional Analysis of SlGal83 Ser-26 Phosphorylation
In order to begin to analyze possible roles for Adi3 phosphorylation of SlGal83, we first utilized the sip1Δsip2Δgal83Δ yeast complementation assay. The ability of the SlGal83S26A-GFP and SlGal83S26D-GFP proteins to complement the sip1Δsip2Δgal83Δ cells was tested, and the results indicate that these proteins complement to an extent similar to that of wild-type SlGal83-GFP (Supplemental Fig. S5A). This suggests that Adi3 phosphorylation of SlGal83 may not affect the function, at least in a heterologous system, of controlling growth on alternate carbon sources.
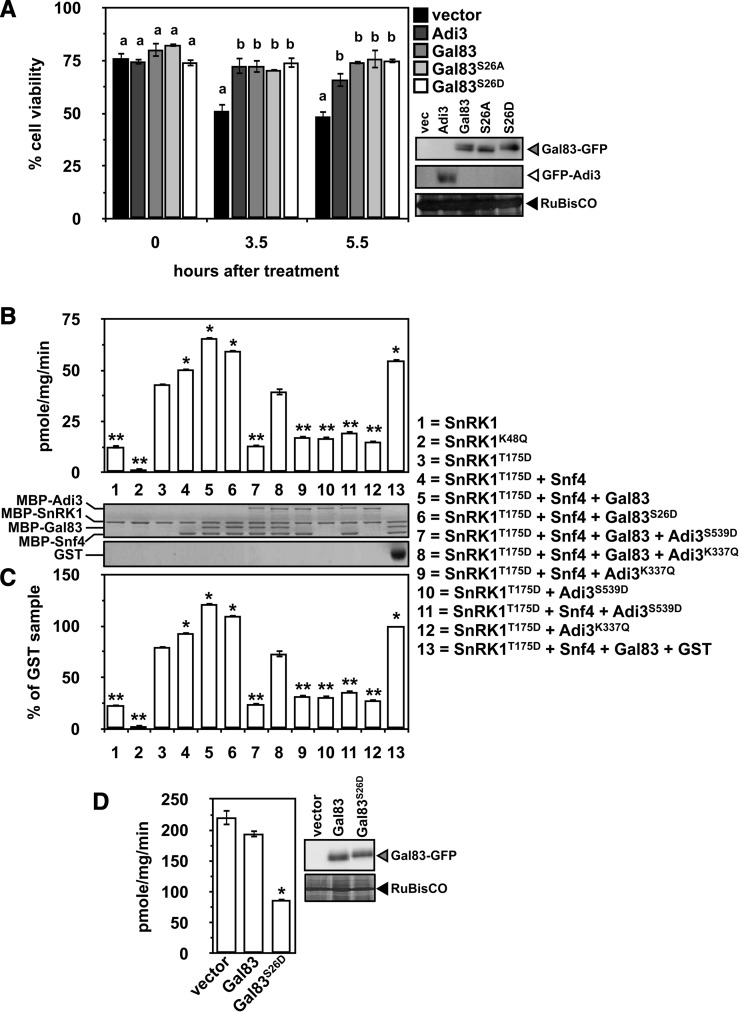
Given the role of Adi3 in the suppression of cell death (Devarenne et al., 2006; Ek-Ramos et al., 2010) and that Adi3 can phosphorylate SlGal83, the ability of SlGal83 and its Ser-26 phosphorylation mutants to suppress cell death was analyzed in tomato cells. It is known that high levels of NaCl are capable of inducing cell death in plants (Katsuhara and Kawasaki, 1996; Lin et al., 2006; Tuteja, 2007; Jiang et al., 2008; Affenzeller et al., 2009; Banu et al., 2009; Chen et al., 2009; Wang et al., 2010), and a functional Snf1 complex has been shown to be required for yeast cell survival in the presence of high NaCl (Hong and Carlson, 2007). We expressed SlGal83-GFP, SlGal83S26A-GFP, SlGal83S26D-GFP, and GFP-Adi3 in tomato protoplasts, treated them with 200 mm NaCl, and measured cell viability over a 5.5-h time course. Both Adi3 and SlGal83 were capable of CDS activity and provided increased cell viability in response to NaCl compared with the vector-transformed sample (Fig. 5A). The SlGal83 Ser-26 phosphorylation mutants did not confer increased or decreased cell viability over wild-type SlGal83 (Fig. 5D). These results indicated that SlGal83 does have a role in cell death suppression, but phosphorylation of Ser-26 may not play a role in controlling SlGal83 CDS activity.
Figure 5.
Functional analysis of Gal83 Ser-26 phosphorylation mutants. A, Gal83 confers cell viability in the presence of high NaCl. Tomato protoplasts expressing GFP-Adi3 or the indicated Gal83-GFP constructs for 18 h were treated with 200 mm NaCl, and cell viability was determined by Evans blue staining over a 5.5-h time course. Values are averages of at least three independent experiments. Error bars represent se. Data analysis was carried out using Duncan’s multiple-range test. Samples with the same letter above the bars are not significantly different (P < 0.05). Protein expression detected by α-GFP western blot is shown on the right. B, SnRK1 substrate phosphorylation with the Gal83S26D mutant and Adi3. Kinase-active and -inactive MBP-SnRK1 proteins were tested for phosphorylation of the SAMS peptide in combination with MBP-Snf4, MBP-Gal83, and MBP-Adi3 using [γ-32P]ATP in in vitro kinase assays. Values are shown as pmol phosphate incorporated mg−1 SnRK1 protein min−1 and are averages of three independent experiments. Error bars represent se. Asterisks indicate significant increases (*) or decreases (**) in SAMS phosphorylation as compared with phosphorylation by SnRK1T175D alone (Student’s t test, P < 0.01). The SDS-PAGE gel shows proteins put into the assay. C, Expression of the data in B as a percentage of the GST sample (column 13). All other information is as in B. D, SAMS phosphorylation by protoplast extracts expressing SlGal83. The indicated SlGal83-GFP proteins were expressed in protoplasts for 16 h, and an extract was made and tested for phosphorylation of SAMS as in B. Values are averages of three independent experiments, and error bars represent se. The asterisk indicates a significant decrease in SAMS phosphorylation as compared with the SlGal83 sample (Student’s t test, P < 0.01).
Next, the effect of SlGal83 phosphorylation on SlSnRK1 complex kinase activity was tested. In order to carry out these assays, an in vitro active SnRK complex must be assembled. Thus, the SlSnRK complex members studied here were analyzed for the formation of an active complex by testing kinase activity against the AMPK/SnRK1 SAMS peptide substrate (HMRSAMSGLHLVKRR; with the phosphorylation site underlined; Halford et al., 2003). We also cloned the tomato cDNA for Snf4, which encodes the γ-subunit of the SlSnRK complex (Bradford et al., 2003), for inclusion in the kinase assays. The α-subunit SlSnRK1 by itself showed limited SAMS phosphorylation (Fig. 5B, column 1). The phosphomimetic mutation of SlSnRK1 Thr-175 (SlSnRK1T175D), which corresponds to the identified phosphorylation activation site in AMPK (Thr-172) and spinach (Spinacia oleracea) and Arabidopsis SnRK1 (Thr-175; Hawley et al., 1996; Sugden et al., 1999; Crozet et al., 2010; Supplemental Fig. S1), conferred an increase in SAMS phosphorylation (Fig. 5B, column 3). The addition of SlSnf4 marginally but significantly increased SlSnRK1T175D SAMS phosphorylation (Fig. 5B, column 4). Inclusion of all complex subunits (SlSnRK1, SlSnf4, SlGal83) imparted a greater increase in SlSnRK1T175D SAMS phosphorylation (Fig. 5B, column 5). These assays show that the SlSnRK1 subunits constitute a functional complex. To the best of our knowledge, this is the first report of reconstituting an active plant SnRK complex in vitro.
The contribution of SlGal83 Ser-26 phosphorylation toward SlSnRK1 kinase activity on the SAMS peptide was analyzed by including the SlGal83S26D protein in the complex or adding Adi3 to the complex. The results show that SlGal83S26D conferred a slight yet statistically significant decrease in SlSnRK1 SAMS phosphorylation (Fig. 5B, column 6), whereas the addition of Adi3S539D to the assay drastically lowered the phosphorylation of SAMS to a level close to that of SlSnRK1 alone (Fig. 5B, column 7). This drop in SAMS phosphorylation appears to partially depend on Adi3 kinase activity, as inclusion of the kinase-inactive Adi3K337Q restored activity of the complex similar to SlSnRK1T175D alone but not to the level of the full complex (Fig. 5B, column 8). This would suggest that even though Adi3 does not phosphorylate SlSnRK1 (Fig. 2A, lane 14), it may inhibit SnKR1 kinase activity through their interaction. This appears to be the case, because kinase-active Adi3S539D or kinase-inactive Adi3K337Q reduced SAMS phosphorylation by SlSnRKT175D and SlSnRKT175D + Snf4 close to the level of SlSnRK1 alone (Fig. 5B, columns 9–12). In order to analyze if the drop in complex kinase activity in the presence of Adi3 is due to an additional protein in the assay, the analysis was repeated with the addition of GST protein. This assay had strong kinase activity, but not to the level of the full complex (Fig. 5B, column 13). This would suggest that some loss of kinase activity in the presence of Adi3 could be due to the addition of another protein. To take this into account, the values in Figure 5B were normalized to that of the assay in the presence of GST (i.e. the GST sample was set as 100% and the other samples were expressed as a percentage of this value). Figure 5C shows that when expressed in this manner, the trends do not change.
We extended the SnRK1 SAMS phosphorylation assays to a more in vivo approach by expressing SlGal83-GFP or SlGal83S26D-GFP in tomato protoplasts, making extracts of these cells, and using the extracts to phosphorylate the SAMS peptide. We found that the extract from SlGal83S26D-GFP-expressing cells had greatly reduced SAMS phosphorylation compared with expression of SlGal83-GFP (Fig. 5D). This reduction in SAMS phosphorylation is much lower than what was seen for the in vitro assay (Fig. 5, B and C), suggesting that a more in vivo context is needed to better realize the effects of Ser-26 phosphorylation. Taken together, these kinase assay data suggest that the Adi3 interaction with the SlSnRK complex has the ability to inhibit the kinase activity of the complex. This may be mediated through two mechanisms, phosphorylation of SlGal83 and interaction with SlSnRK1.
DISCUSSION
In this study, we present evidence for the interaction of Adi3 with the SnRK complex in tomato. Our finding that Adi3 can only phosphorylate the Gal83 SnRK β-subunit out of the four β-subunits identified in tomato has far-reaching implications, because Snf1/AMPK/SnRK1 β-subunits control the cellular localization and substrate specificity of the complex (Mitchelhill et al., 1997; Vincent and Carlson, 1999; Vincent et al., 2001; Warden et al., 2001). Additionally, β-subunit phosphorylation has been associated with the regulation of some of these β-subunit functions (Mitchelhill et al., 1997; Warden et al., 2001; Hedbacker et al., 2004; Mangat et al., 2010), and the SnRK complex appears to link signaling connected with metabolism and stresses (Halford and Hey, 2009; Hey et al., 2010). Given the role of Adi3 in cell death control, our studies add additional evidence for the connection of SnRK with stress signaling. Alternatively, Adi3 may also be involved in the direct regulation of metabolism through its interactions with the SlSnRK1 complex.
A Role for Adi3 Phosphorylation in Regulating SnRK Complex Kinase Activity?
We have shown that Adi3 phosphorylates SlGal83 at Ser-26 (Fig. 2) and explored the functional relevance of this phosphorylation event. While several β-subunits have been shown to be phosphorylated in yeast and mammals (Warden et al., 2001; Hedbacker et al., 2004; Mangat et al., 2010), our studies appear to be the first report of phosphorylation for a plant β-subunit. An in vitro functional analysis showed that kinase-active Adi3 has drastic effects on the kinase activity of the SnRK1 complex. If Adi3 phosphorylation of SlGal83 at Ser-26 is controlling this large decrease in SnRK1 complex activity, the SlGal83S26D protein should also confer a decrease in kinase activity. A large reduction in SAMS phosphorylation in the presence of SlGal83S26D was seen in vivo (Fig. 5D) but was much less drastic in vitro (Fig. 5C), suggesting that there is an in vivo role for Ser-26 phosphorylation in controlling SnRK1 kinase activity.
Interestingly, the restoration of SAMS phosphorylation when including kinase-inactive Adi3K337Qwould suggest that Adi3 kinase activity is at least partially required for this large inhibition of SnRK1 activity in vitro and may suggest additional Adi3 phosphorylation sites on SlGal83 for controlling activity. It is possible that Ser-30 is one of these sites, because we identified phosphorylation of this SlGal83 residue both in vitro and in vivo by MS analysis. However, the inability of Adi3 to phosphorylate this site in vitro raises doubt about the role of Ser-30 phosphorylation. Additionally, the complete loss of Adi3 phosphorylation of the SlGal83S26A protein suggests that Ser-26 is the only Adi3 phosphorylation site on Gal83. Thus, the requirement of Adi3 kinase activity in the suppression of SnRK1 substrate phosphorylation still remains to be fully resolved.
While many studies have shown that phosphorylation of α-subunits controls complex kinase activity in yeast, mammals, and plants (Hong et al., 2003; Nath et al., 2003; Sutherland et al., 2003; Woods et al., 2003, 2005; Hawley et al., 2005; Hurley et al., 2005; Shen and Hanley-Bowdoin, 2006; Hey et al., 2007; Shen et al., 2009), only one previous study has shown that phosphorylation of a β-subunit affects complex kinase activity. A phosphorylation mutant of the human AMPKβ1 β-subunit reduced AMPK complex kinase activity by 60% (Warden et al., 2001). Thus, control of complex kinase activity by β-subunit phosphorylation may be more common than previously thought. This could be supported by determining if phosphorylation of the conserved Ser-26 residue in the Arabidopsis Gal83 homolog, AKINβ1, affects AtSnRK1 complex kinase activity.
Our results also suggest that the interaction of Adi3 with SlSnRK1 is capable of suppressing SlSnRK1 kinase activity (Fig. 5B). In the absence of SlGal83, the kinase-active or -inactive forms of Adi3 are capable of suppressing SlSnRK1 kinase activity (Fig. 5B). This apparently contradicts the finding that the kinase-inactive Adi3 can restore the activity of the complex in the presence of SlGal83. However, these results may indicate that the Adi3/SlGal83 interaction affects the ability of Adi3 to fully interact with and inhibit SlSnRK1. Eliminating SlGal83 from the assay would then allow for full interaction between Adi3 and SlSnRK1 and stronger activity inhibition. The interaction of Adi3 with SlSnRK1 may be inhibiting the ability of SlSnRK1 to bind the SAMS substrate or even ATP.
These data also help to explain the detection of SlSnRK1 as an Adi3 Y2H interactor even though Adi3 does not phosphorylate SlSnRK1 as well as shed light on the biological significance for this interaction. Given the role of Adi3 in the host response to Pst and the function of SnRK1 in stress signaling, Adi3 may be directing reallocation of cellular energy reserves by modulating SlSnRK1 kinase activity during the resistance response of tomato to Pst. Studies using Nicotiana attenuata show that photosynthate is reallocated to the roots in response to herbivore attack through the down-regulation of SnRK β-subunit expression (Schwachtje et al., 2006). Our results indicate that SlGal83 phosphorylation at Ser-26 functions as an inhibitor of SlSnRK kinase activity. Down-regulation of this β-subunit may thus play a role in facilitating the activation of SlSnRK1 and the metabolic modifications required to respond to pathogens. Phosphorylation of SlGal83 by Adi3 offers an additional layer of control over SlSnRK activity, a specificity required given the involvement of this complex in regulating metabolic responses to several environmental stresses (Hong and Carlson, 2007; Hey et al., 2010; Cho et al., 2012).
Multiple Roles for β-Subunit Phosphorylation
Snf1/AMPK/SnRK1 β-subunits appear to be phosphorylated on several amino acids, and our studies also support phosphorylation at several residues on SlGal83. Expression of SlGal83-GFP in plant cells showed the existence of multiple phosphorylated protein bands based on our λ phosphatase treatments, one of which contains Ser-26 phosphorylation (Fig. 4). One of these phosphorylated bands may also contain Ser-30 phosphorylation. This and the identity of any additional SlGal83 phosphorylation sites remain to be determined. Multiple phosphorylation sites have been found for other β-subunits. Mass spectral analysis of human AMPKβ1 isolated from COS cells identified phosphorylation at Ser-24/25, Ser-108, and Ser-182, but the responsible kinase has not been identified (Mitchelhill et al., 1997). Phosphorylation of AMPKβ1 Ser-24/25 and Ser-182, but not Ser-108, appears to prevent nuclear localization (Warden et al., 2001). ScGal83 is phosphorylated by both the α-subunit Snf1 and Casein Kinase2 (CK2), and while the exact sites of phosphorylation have not been identified, they are predicted to be Ser-64 or Ser-65 for Snf1 and Ser-87, Thr-90, or Ser-93 for CK2 (Mangat et al., 2010). The role for Snf1/CK2 phosphorylation of ScGal83 is not clear, because deletion of the region containing both the Snf1 and CK2 phosphorylation sites did not affect Glc-regulated Snf1 function (Mangat et al., 2010). The situation for ScSip1 is similar. Protein kinase A (PKA) has been shown to be required for retaining ScSip1 cytoplasmic localization under high-Glc conditions (Hedbacker et al., 2004). However, mutation of four potential PKA phosphorylation sites did not affect ScSip1 cellular localization (Hedbacker et al., 2004).
Taken together, it appears that the role of Snf1/AMPK/SnRK1 β-subunit phosphorylation is not fully understood and will be an important area of research for the future. From our studies, the full role of SlGal83 Ser-26 phosphorylation by Adi3 is not clear. It appears to have only a minor role in controlling SlSnRK1 complex kinase activity. So, additional functions attributable to this phosphorylation event will be important to identify in the future. Given the role of β-subunits in controlling Snf1/AMPK/SnRK1 complex localization and phosphorylation playing a role in this function, it will be important to examine the contribution of phosphorylation by Adi3 in controlling SlGal83 cellular localization. Consequently, the full extent of the SlGal83 Ser-26 phosphorylation event by Adi3 remains to be determined.
Is There a Link between Cell Death Control and Metabolism?
An important aspect of PCD is the reallocation of cellular resources such as proteins and sugars. This is particularly true of the cell death that occurs during leaf senescence (van Doorn and Woltering, 2004, 2008; Guiboileau et al., 2010). In fact, the reuse of cellular materials was suggested as early as 1891 from the examination of cell death associated with xylem development (Lange, 1891). Thus, it may not be surprising that a gene controlling PCD would also be able to regulate how cells utilize and/or mobilize energy sources. This appears to be the case for mammalian PKB. While it is well known that PKB suppresses cell death by phosphorylating and inactivating proapoptotic proteins or activating antiapoptotic proteins (Luo et al., 2003; Carnero, 2010), PKB also functions in the regulation of metabolism through the control of glycolytic enzymes and Glc uptake (Plas and Thompson, 2002; Carnero, 2010). Such a connection between a specific plant gene controlling cell death and metabolism has been indirect at best. Our previous studies have shown that there are many striking activity and cellular localization similarities between Adi3 and PKB (Devarenne et al., 2006; Ek-Ramos et al., 2010). The studies presented here showing Adi3 inhibition of SlSnRK1 complex kinase activity add one additional similarity between Adi3 and PKB, because PKB is known to modulate AMPK activity. While PKB and AMPK do not directly interact with each other, there is substantial cross talk between the pathways. For example, activation of PKB has been shown to down-regulate AMPK activity and thus a decrease in AMP/ATP cellular ratios (Kovacic et al., 2003; Hahn-Windgassen et al., 2005). Conversely, activation of AMPK has been shown to inactivate PKB-regulated glycolysis (Grabacka and Reiss, 2008). Combining our current and previous Adi3 studies raises the possibility that Adi3 functions similarly to PKB in cell death and metabolism control. Further studies on the role of Adi3 association with the SlSnRK1 complex, especially the phosphorylation of Gal83, will be required to fully understand if there is a connection between Adi3-mediated cell death control and SlSnRK1 metabolism control.
MATERIALS AND METHODS
Cloning of Tomato SnRK1, Gal83, Sip1, Tau1, Tau2, and Snf4
All primers and restriction sites used in this study for ORF amplification, cloning, and mutagenesis are listed in Supplemental Table S1, and the primers used to amplify all genes were designed using sequence data obtained from the SGN databases (http://solgenomics.net/). The ORFs for SlSnRK1, SlSip1, SlSnf4, SlTau1, and SlTau2 were obtained by RT-PCR using cDNA generated with SuperScript III (Invitrogen) from tomato (Solanum lycopersicum) total RNA isolated from 4-week-old leaves. Primers used to amplify SlSnRK1 (accession no. AF143743) were based on the unigene SGN-U564382. The cDNA for SlSip1 (accession no. AF322108) in unigene SGN-U575258 and reported by Bradford et al. (2003) appeared to lack a portion of the 5′ end of the cDNA when compared with homologous β-subunits from yeast and Arabidopsis (Arabidopsis thaliana). Consequently, the tomato genome sequence was searched on SGN for the SlSip1 gene using unigene SGN-U575258. An SlSip1 gene was found in genomic sequence SL2.31ch05:63330625.0.63325020, and primers were designed based on this sequence to amplify the ORF by RT-PCR. The reported SlGal83 cDNA (accession no. AY245177) lacked the 5′ end, and the full-length cDNA was identified in unigene SGN-U564868. Primers based on this unigene were used to amplify the ORF by PCR using SGN EST clone cTOF-18-D18 as a template. The Tau1 (accession no. JQ846034) and Tau2 (accession no. JQ846035) ORFs were isolated using primer sequences based on the unigenes U571217 and U565213, respectively. The SlSnf4 ORF (accession no. AF419320) was amplified by PCR using the published sequence (Bradford et al., 2003). Mutagenesis of SlSnRK1 and SlGal83 was performed using Pfu Turbo Polymerase (Stratagene) and the primer pairs listed in Supplemental Table S1. Cloning of Adi3 and its kinase activity mutants was described previously (Devarenne et al., 2006).
Recombinant Protein Expression and Purification
The ORFs for SlSnRK1, SlGal83, and SlSip1 were cloned as N-terminal MBP fusions into pMAL-c2 vector (New England Biolabs). Recombinant proteins were expressed in Escherichia coli BL21 Star (DE3) as described previously (Devarenne et al., 2006) and purified using maltose-binding resin (New England Biolabs) manufacturer protocols. For GST-Adi3, the Adi3 ORF was cloned into the pGEX-4T N-terminal GST fusion vector (GE Healthcare), and protein was expressed and purified as recommended by the manufacturer. After elution, all fusion proteins were concentrated using Amicon Ultra centrifugal filters (Millipore) and added to buffer for final concentrations of 50% glycerol, 50 mm Tris-HCl, pH 7.5, 0.5 mm EDTA, and 100 mm NaCl. Protein concentrations were quantified using the Bio-Rad Protein Assay Kit before storage at −20C.
Y2H Assay
Y2H assays where conducted using pEG202 for the bait vector and pJG4-5 for the prey vector as described previously (Devarenne et al., 2006). Constructs were transformed into yeast strain EGY48 containing the pSH18-34 reporter vector and analyzed for LacZ gene expression on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside-containing plates. Protein expression was confirmed by western blot. All other procedures for the Y2H assays and Y2H library screen for identifying Adi3 interactors followed standard procedures as described previously (Golemis et al., 2008).
Yeast Complementation and Invertase Assays
The ORF for SlGal83 and its Ser-26 mutants were fused to a C-terminal eGFP tag under the control of the Glyceraldehyde-3-Phosphate Dehydrogenase promoter in the modified vector MBB263. The yeast β-subunit knockout strain MCY4040 (MATα sip1Δ::KanMX6 sip2Δ3::LEU2 Gal83::TRP1 his3-Δ200 leu2-3,112 trp1Δ1 ura3-52 lys2-801; Vincent et al., 2001) was transformed with the SlGal83 constructs using the standard lithium acetate/polyethylene glycol method. Transformants were screened on plates of complete minimal medium with 2% Glc and lacking Leu, Trp, and uracil. Recovered colonies were grown in liquid complete minimal 2% Glc medium for 48 h, and 5-fold serial dilutions were spotted on selective medium supplemented with either 2% Glc or 2% Suc and incubated at 30°C for 2 d (2% Glc) or 6 to 7 d (2% Suc). Invertase assays were performed as reported previously (Celenza and Carlson, 1989; Bradford et al., 2003). Invertase activity of derepressed (0.05% Glc) and Glc-repressed (2% Glc) cells was estimated as a measure of the amount of Suc metabolized into Glc using the Glc (GO) assay kit (Sigma) as described by the manufacturer.
Pull-Down Assays
Immobilized glutathione beads (Thermo Scientific) were equilibrated by washing three times with 200 μL of binding buffer (50 mm NaCl, 50 mm Tris-HCl, pH 7.5, 0.1% Triton X-100, and 5 mm EDTA). For each pull down, 1 μg of either GST or GST-Adi3 and equivalent protein amounts of MBP, MBP-Gal83, MBP-Sip1, and MBP-SnRK1 were mixed in a final volume of 30 μL. Samples were incubated for 15 min at room temperature followed by the addition of buffer-preequilibrated glutathione resin to each sample and incubation for 1 h at 4°C on an orbital shaker. The resin with bound proteins was pelleted by centrifugation at 100g and washed five times with 200 μL of wash buffer (500 mm NaCl, 50 mm Tris-HCl, pH 7.5, 0.1% Triton X-100, and 5 mm EDTA). Bound proteins were eluted using 1× SDS-PAGE sample buffer, resolved by 12% SDS-PAGE, and analyzed by western blotting using α-GST (Santa Cruz Biotechnology) at 1:15,000 and α-MBP (New England BioLabs) at 1:5,000 for pull downs or 1:10,000 for loading controls.
Kinase Assays
In vitro kinase assays were done in 30-μL reactions in Adi3 kinase buffer (10 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 1 mm dithiothreitol [DTT], and 20 μm ATP) or SlSnRK1 kinase buffer (10 mm Tris-HCl, pH 8, 10 mm MnCl2 or MgCl2, 1 mm DTT, and 20 μm ATP). SlSnRK1 autophosphorylation appeared to be slightly stronger using MnCl2 and, therefore, was used for all SlSnRK1 autophosphorylation assays. However, SlSnRK1 substrate phosphorylation was comparable using MnCl2 or MgCl2 as a cofactor. Therefore, MgCl2 was used for all SlSnRK1 substrate phosphorylation experiments. Adi3 substrate phosphorylation assays contained 5 μg of purified MBP-Adi3 or MBP-Adi3S539D and 2 μg of MBP-Gal83, MBP-Gal83 mutants, or MBP-Sip1. For SlSnRK1 kinase assays, 3 μg of MBP-SnRK1, MBP-SnRK1K48Q, or MBP-SnRK1T175D was used. Reactions were initiated upon the addition of 0.25 μCi of [γ-32P]ATP (6,000 Ci mmol−1; Perkin-Elmer) per sample and were incubated for 15 min at room temperature for Adi3 or for 30 min at 30°C for SlSnRK1. Reactions were terminated by the addition of 4× SDS-PAGE sample buffer, and samples were resolved by 7.5% SDS-PAGE. Sample radioactive incorporation imaging and quantification were done with a phosphor imager (Bio-Rad Molecular Imager).
SAMS peptide (HMRSAMSGLHLVKRR) phosphorylation assays were performed as described previously (Davies et al., 1989). Assay conditions for SlSnRK1 phosphorylation of the SAMS peptide were as for the SlSnRK1 substrate phosphorylation assays above plus 100 μm SAMS peptide (AnaSpec). Reactions were spotted on phosphocellulose P81 paper (Whatman), washed three times in 1% H3PO4 and once in acetone, the paper was dried, and the incorporated radioactivity was counted using a Beckman LS5000TA scintillation counter. For SAMS phosphorylation with protoplast lysates, 4 × 105 tomato protoplasts expressing empty pTEX vector, SlGal83-GFP, and SlGal83S26D-GFP were lysed by vortexing in a buffer containing 50 mm Tris, pH 8.0, 1 mm EDTA, 50 mm NaCl, 8% glycerol, 5 mm DTT, 2% plant protease inhibitor cocktail (Sigma), and 2% plant phosphatase inhibitor cocktail (Sigma). Extracts were cleared by centrifugation at 4°C and 13,000g for 10 min. Protein concentration was estimated as described above, and lysates were adjusted to equal protein concentrations with lysis buffer. Reactions were done as described above but using a buffer containing 40 mm HEPES-KOH, pH 7.6, 10 mm MgCl2, 1 mm DTT, 200 μm ATP, 2 μCi of [γ-32P]ATP, and 100 μm SAMS peptide (buffer adapted from Fragoso et al. [2009]). Reactions were initiated with the addition of 8 μL of the protein extract. Phosphate incorporation was analyzed as described above, and the remaining lysates were used for α-GFP western blotting to evaluate the expression efficiency of the proteins.
MS
For sample preparation, Coomassie blue-stained gel bands were in-gel digested with trypsin overnight, and phosphopeptides were enriched using a NuTip metal oxide phosphoprotein enrichment kit according to the manufacturer’s instructions (Glygen).
For liquid chromatography-MS/MS analysis, phosphopeptides were injected onto a capillary trap (LC Packings PepMap) and desalted for 5 min with 0.1% (v/v) acetic acid at a flow rate of 3 µL min−1. The samples were loaded onto an LC Packings C18 PepMap nanoflow HPLC column. The elution gradient of the HPLC column started at 97% solvent A and 3% solvent B and finished at 60% solvent A and 40% solvent B for 30 min. Solvent A consisted of 0.1% (v/v) acetic acid, 3% (v/v) acetonitrile, and 96.9% (v/v) water. Solvent B consisted of 0.1% (v/v) acetic acid, 96.9% (v/v) acetonitrile, and 3% (v/v) water. Liquid chromatography-MS/MS analysis was carried out on a LTQ Orbitrap XL mass spectrometer (Thermo Scientific). The instrument under Xcalibur 2.07 with LTQ Orbitrap Tune Plus 2.55 software was operated in the data-dependent mode to automatically switch between MS and MS/MS acquisition. Survey scan MS spectra (from mass-to-charge ratio [m/z] 300 to 2,000) were acquired in the Orbitrap mass spectrometer with resolution of R = 60,000 at m/z 400. During collision-induced dissociation, if a phosphate neutral loss of 98, 49, 32.66, and 24.5 m/z below the precursor ion mass was detected, there was an additional activation of all four neutral loss m/z values. This multistage activation was repeated for the top five ions in a data-dependent manner. Dynamic exclusion was set to 60 s. Typical mass spectrometric conditions include a spray voltage of 2.2 kV, no sheath and auxiliary gas flow, a heated capillary temperature of 200°C, a capillary voltage of 44 V, a tube lens voltage of 165 V, an ion isolation width of 1.0 m/z, and a normalized collision-induced dissociation collision energy of 35% for MS/MS in the LTQ. The ion selection threshold was 500 counts for MS/MS. The mass spectrometer calibration was performed according to the manufacturer’s guidelines using a mixture of SDS, sodium taurocholate, the MRFA peptide, and Ultramark.
For the protein-search algorithm, all MS/MS spectra were analyzed using Mascot (Matrix Science; version 2.2.2). Mascot was set up to search a current Arabidopsis database assuming the digestion enzyme trypsin. Mascot was searched with a fragment ion mass tolerance of 0.50 D and a parent ion tolerance of 10 ppm. Iodoacetamide derivative of Cys, deamidation of Asn and Gln, oxidation of Met, and phosphorylation of Ser, Thr, and Tyr are specified as variable modifications. The MS/MS spectra of the identified phosphorylated peptides were manually inspected to ensure confidence in phosphorylation site assignment.
Phosphatase Treatment
Gal83-GFP proteins were expressed in tomato protoplasts from pTEX for 22 h and lysed in ice-cold extraction buffer containing 50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 0.1% Triton X-100, 2 mm DTT, 2.5% plant protease inhibitor cocktail (Sigma), and 6 μm epoxymicin (Enzo Life Sciences). Lysates were split into two fractions: one for phosphatase treatment and one for a no-treatment control. Both fractions were adjusted to 3 mm MnCl2 in λ phosphatase buffer (50 mm HEPES, pH 7.5, 100 mm NaCl, 2 mm DTT, and 0.01% Brij-35) in a final volume of 100 μL. The no-treatment fraction was additionally adjusted to 2% phosphatase inhibitors (Sigma; phosphatase inhibitor cocktail 1). Reactions were started with the addition of 800 units of λ phosphatase (New England Biolabs), incubated at 30°C for 30 min, and reactions were terminated by the addition of 1× SDS-PAGE sample buffer. Samples were then resolved by 7.5% SDS-PAGE with a 1:200 bis-acrylamide:acrylamide ratio and analyzed by α-GFP western blotting.
Protoplast Protein Expression and Cell Death Assays
The ORFs for Gal83 and Gal83Ser-26 were cloned into the BamHI and SalI restriction sites of pTEX-eGFP (Ek-Ramos et al., 2010) to yield an in-frame C-terminal Gal83-GFP fusion under the control of the 35S promoter. Cloning of Adi3 into pTEX-eGFP for an N-terminal tagged GFP-Adi3 was described previously (Ek-Ramos et al., 2010). The resulting constructs were purified using CsCl gradient centrifugation. Protoplasts were isolated from expanded leaves of 4-week-old PtoR tomato plants and transformed as reported previously (Devarenne et al., 2006; Ek-Ramos et al., 2010) using 8 × 105 protoplasts and 25 μg of plasmid DNA. For NaCl-induced cell death experiments, transformed protoplasts expressing proteins for 18 h were suspended in 200 μL of WI buffer (0.5 m mannitol, 4 mm MES, pH 5.7, and 20 mm KCl) with or without 200 mm NaCl, incubated in the dark at 25°C, and aliquots taken over a 5.5-h time course. Cell viability was estimated by treating 30-μL protoplast aliquots with 0.05% Evans blue for 5 min and counting a minimum of 200 cells as described previously (Devarenne et al., 2006; Ek-Ramos et al., 2010). Cell viability estimates are a measurement of at least three independent transformation experiments. Protein expression was confirmed by western blot with 4 × 105 transformed protoplast resuspended in 1× SDS-PAGE sample buffer and boiled at 95°C for 5 min. GFP fusion proteins were detected with a horseradish peroxidase-conjugated α-GFP antibody (Santa Cruz Biotechnology) at 1:1,000.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers JF895513 (SlGal83), JF8955212 (SlSip1), JQ846034 (SlTau1), and JQ846035 (SlTau2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of SnRK proteins from tomato and Arabidopsis.
Supplemental Figure S2. Alignment of SnRK complex β-subunits.
Supplemental Figure S3. RT-PCR amplification of SlSip1 and Adi3/SlGal83 Y2H interaction.
Supplemental Figure S4. RT-PCR amplification of SlTau1 and SlTau2.
Supplemental Figure S5. SlGal83 complementation of sip1Δsip2Δgal83Δ yeast.
Supplemental Figure S6. MS identification of SlGal83 Ser-26 phosphorylation and α-GFP immunoprecipitation of SlGal83-GFP.
Supplemental Figure S7. MS identification of SlGal83 Ser-30 phosphorylation.
Supplemental Figure S8. Separation of SlGal83-GFP phosphoproteins by SDS-PAGE with varying bis-acrylamide:acrylamide ratios.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank the other members of the Devarenne laboratory for comments and constructive discussions. The yeast β-subunit triple knockout strain MCY4040 was kindly provided by Marian Carlson (Columbia University Medical Center). The yeast vector MBB263 was kindly provided by Mary Bryk (Texas A&M University). We also thank Al Burlingame (University of California, San Francisco) and Zhiyong Wang (Carnegie Institution for Science) for allowing access to their mass spectrometers.
Glossary
- PCD
programmed cell death
- Pst
Pseudomonas syringae pv tomato
- CDS
cell death suppression
- Y2H
yeast two-hybrid
- ORF
open reading frame
- RT
reverse transcription
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- SGN
Sol Genomics Network
- DTT
dithiothreitol
- m/z
mass-to-charge ratio
- HA
hemagglutinin
- cDNA
to be defined
References
- Affenzeller MJ, Darehshouri A, Andosch A, Lütz C, Lütz-Meindl U. (2009) Salt stress-induced cell death in the unicellular green alga Micrasterias denticulata. J Exp Bot 60: 939–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arico S, Pattingre S, Bauvy C, Gane P, Barbat A, Codogno P, Ogier-Denis E. (2002) Celecoxib induces apoptosis by inhibiting 3-phosphoinositide-dependent protein kinase-1 activity in the human colon cancer HT-29 cell line. J Biol Chem 277: 27613–27621 [DOI] [PubMed] [Google Scholar]
- Banu NA, Hoque A, Watanabe-Sugimoto M, Matsuoka K, Nakamura Y, Shimoishi Y, Murata Y. (2009) Proline and glycinebetaine induce antioxidant defense gene expression and suppress cell death in cultured tobacco cells under salt stress. J Plant Physiol 166: 146–156 [DOI] [PubMed] [Google Scholar]
- Bogdanove AJ, Martin GB. (2000) AvrPto-dependent Pto-interacting proteins and AvrPto-interacting proteins in tomato. Proc Natl Acad Sci USA 97: 8836–8840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford KJ, Downie AB, Gee OH, Alvarado V, Yang H, Dahal P. (2003) Abscisic acid and gibberellin differentially regulate expression of genes of the SNF1-related kinase complex in tomato seeds. Plant Physiol 132: 1560–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee C. (2008) Diatom signalling: deadly messages. Curr Biol 18: R518–R519 [DOI] [PubMed] [Google Scholar]
- Carlson M, Osmond BC, Botstein D. (1981) Mutants of yeast defective in sucrose utilization. Genetics 98: 25–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M, Osmond BC, Neigeborn L, Botstein D. (1984) A suppressor of SNF1 mutations causes constitutive high-level invertase synthesis in yeast. Genetics 107: 19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnero A. (2010) The PKB/AKT pathway in cancer. Curr Pharm Des 16: 34–44 [DOI] [PubMed] [Google Scholar]
- Celenza JL, Carlson M. (1989) Mutational analysis of the Saccharomyces cerevisiae SNF1 protein kinase and evidence for functional interaction with the SNF4 protein. Mol Cell Biol 9: 5034–5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WS, Xu PZ, Gottlob K, Chen ML, Sokol K, Shiyanova T, Roninson I, Weng W, Suzuki R, Tobe K, et al. (2001) Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene. Genes Dev 15: 2203–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wang Y, Li J, Jiang A, Cheng Y, Zhang W. (2009) Mitochondrial proteome during salt stress-induced programmed cell death in rice. Plant Physiol Biochem 47: 407–415 [DOI] [PubMed] [Google Scholar]
- Cho YH, Hong JW, Kim EC, Yoo SD. (2012) Regulatory functions of SnRK1 in stress-responsive gene expression and in plant growth and development. Plant Physiol 158: 1955–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coello P, Hey SJ, Halford NG. (2011) The sucrose non-fermenting-1-related (SnRK) family of protein kinases: potential for manipulation to improve stress tolerance and increase yield. J Exp Bot 62: 883–893 [DOI] [PubMed] [Google Scholar]
- Crozet P, Jammes F, Valot B, Ambard-Bretteville F, Nessler S, Hodges M, Vidal J, Thomas M. (2010) Cross-phosphorylation between Arabidopsis thaliana sucrose nonfermenting 1-related protein kinase 1 (AtSnRK1) and its activating kinase (AtSnAK) determines their catalytic activities. J Biol Chem 285: 12071–12077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SP, Carling D, Hardie DG. (1989) Tissue distribution of the AMP-activated protein kinase, and lack of activation by cyclic-AMP-dependent protein kinase, studied using a specific and sensitive peptide assay. Eur J Biochem 186: 123–128 [DOI] [PubMed] [Google Scholar]
- Demmel L, Beck M, Klose C, Schlaitz AL, Gloor Y, Hsu PP, Havlis J, Shevchenko A, Krause E, Kalaidzidis Y, et al. (2008) Nucleocytoplasmic shuttling of the Golgi phosphatidylinositol 4-kinase Pik1 is regulated by 14-3-3 proteins and coordinates Golgi function with cell growth. Mol Biol Cell 19: 1046–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deponte M. (2008) Programmed cell death in protists. Biochim Biophys Acta 1783: 1396–1405 [DOI] [PubMed] [Google Scholar]
- Devarenne TP, Ekengren SK, Pedley KF, Martin GB. (2006) Adi3 is a Pdk1-interacting AGC kinase that negatively regulates plant cell death. EMBO J 25: 255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doukhanina EV, Chen S, van der Zalm E, Godzik A, Reed J, Dickman MB. (2006) Identification and functional characterization of the BAG protein family in Arabidopsis thaliana. J Biol Chem 281: 18793–18801 [DOI] [PubMed] [Google Scholar]
- Dudek H, Datta SR, Franke TF, Birnbaum MJ, Yao R, Cooper GM, Segal RA, Kaplan DR, Greenberg ME. (1997) Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science 275: 661–665 [DOI] [PubMed] [Google Scholar]
- Dyck JR, Gao G, Widmer J, Stapleton D, Fernandez CS, Kemp BE, Witters LA. (1996) Regulation of 5′-AMP-activated protein kinase activity by the noncatalytic β and γ subunits. J Biol Chem 271: 17798–17803 [DOI] [PubMed] [Google Scholar]
- Ek-Ramos MJ, Avila J, Cheng C, Martin GB, Devarenne TP. (2010) The T-loop extension of the tomato protein kinase AvrPto-dependent Pto-interacting protein 3 (Adi3) directs nuclear localization for suppression of plant cell death. J Biol Chem 285: 17584–17594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R. (2006) Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet 2: e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoso S, Espíndola L, Páez-Valencia J, Gamboa A, Camacho Y, Martínez-Barajas E, Coello P. (2009) SnRK1 isoforms AKIN10 and AKIN11 are differentially regulated in Arabidopsis plants under phosphate starvation. Plant Physiol 149: 1906–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gissot L, Polge C, Bouly JP, Lemaitre T, Kreis M, Thomas M. (2004) AKINbeta3, a plant specific SnRK1 protein, is lacking domains present in yeast and mammals non-catalytic beta-subunits. Plant Mol Biol 56: 747–759 [DOI] [PubMed] [Google Scholar]
- Golemis EA, Serebriiskii I, Finley RL, Jr, Kolonin MG, Gyuris J, Brent R. (2008) Interaction trap/two-hybrid system to identify interacting proteins. Curr Protoc Mol Biol Chapter 20: Unit 20.1. [DOI] [PubMed]
- Grabacka M, Reiss K. (2008) Anticancer properties of PPARalpha: effects on cellular metabolism and inflammation. PPAR Res 2008: 930705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiboileau A, Sormani R, Meyer C, Masclaux-Daubresse C. (2010) Senescence and death of plant organs: nutrient recycling and developmental regulation. C R Biol 333: 382–391 [DOI] [PubMed] [Google Scholar]
- Hahn-Windgassen A, Nogueira V, Chen CC, Skeen JE, Sonenberg N, Hay N. (2005) Akt activates the mammalian target of rapamycin by regulating cellular ATP level and AMPK activity. J Biol Chem 280: 32081–32089 [DOI] [PubMed] [Google Scholar]
- Halford NG, Hey S, Jhurreea D, Laurie S, McKibbin RS, Paul M, Zhang Y. (2003) Metabolic signalling and carbon partitioning: role of Snf1-related (SnRK1) protein kinase. J Exp Bot 54: 467–475 [DOI] [PubMed] [Google Scholar]
- Halford NG, Hey SJ. (2009) Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochem J 419: 247–259 [DOI] [PubMed] [Google Scholar]
- Hawley SA, Davison M, Woods A, Davies SP, Beri RK, Carling D, Hardie DG. (1996) Characterization of the AMP-activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phosphorylates AMP-activated protein kinase. J Biol Chem 271: 27879–27887 [DOI] [PubMed] [Google Scholar]
- Hawley SA, Pan DA, Mustard KJ, Ross L, Bain J, Edelman AM, Frenguelli BG, Hardie DG. (2005) Calmodulin-dependent protein kinase kinase-β is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab 2: 9–19 [DOI] [PubMed] [Google Scholar]
- Hedbacker K, Townley R, Carlson M. (2004) Cyclic AMP-dependent protein kinase regulates the subcellular localization of Snf1-Sip1 protein kinase. Mol Cell Biol 24: 1836–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey S, Mayerhofer H, Halford NG, Dickinson JR. (2007) DNA sequences from Arabidopsis, which encode protein kinases and function as upstream regulators of Snf1 in yeast. J Biol Chem 282: 10472–10479 [DOI] [PubMed] [Google Scholar]
- Hey SJ, Byrne E, Halford NG. (2010) The interface between metabolic and stress signalling. Ann Bot (Lond) 105: 197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeberichts FA, Woltering EJ. (2003) Multiple mediators of plant programmed cell death: interplay of conserved cell death mechanisms and plant-specific regulators. Bioessays 25: 47–57 [DOI] [PubMed] [Google Scholar]
- Hong SP, Carlson M. (2007) Regulation of snf1 protein kinase in response to environmental stress. J Biol Chem 282: 16838–16845 [DOI] [PubMed] [Google Scholar]
- Hong SP, Leiper FC, Woods A, Carling D, Carlson M. (2003) Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc Natl Acad Sci USA 100: 8839–8843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley RL, Anderson KA, Franzone JM, Kemp BE, Means AR, Witters LA. (2005) The Ca2+/calmodulin-dependent protein kinase kinases are AMP-activated protein kinase kinases. J Biol Chem 280: 29060–29066 [DOI] [PubMed] [Google Scholar]
- Jiang AL, Cheng Y, Li J, Zhang W. (2008) A zinc-dependent nuclear endonuclease is responsible for DNA laddering during salt-induced programmed cell death in root tip cells of rice. J Plant Physiol 165: 1134–1141 [DOI] [PubMed] [Google Scholar]
- Jiang R, Carlson M. (1996) Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev 10: 3105–3115 [DOI] [PubMed] [Google Scholar]
- Kaneda T, Taga Y, Takai R, Iwano M, Matsui H, Takayama S, Isogai A, Che FS. (2009) The transcription factor OsNAC4 is a key positive regulator of plant hypersensitive cell death. EMBO J 28: 926–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuhara M, Kawasaki T. (1996) Salt stress induced nuclear and DNA degradation in meristematic cells of barley roots. Plant Cell Physiol 37: 169–173 [Google Scholar]
- Kovacic S, Soltys CL, Barr AJ, Shiojima I, Walsh K, Dyck JR. (2003) Akt activity negatively regulates phosphorylation of AMP-activated protein kinase in the heart. J Biol Chem 278: 39422–39427 [DOI] [PubMed] [Google Scholar]
- Kulp SK, Yang Y-T, Hung C-C, Chen K-F, Lai J-P, Tseng P-H, Fowble JW, Ward PJ, Chen C-S. (2004) 3-Phosphoinositide-dependent protein kinase-1/Akt signaling represents a major cyclooxygenase-2-independent target for celecoxib in prostate cancer cells. Cancer Res 64: 1444–1451 [DOI] [PubMed] [Google Scholar]
- Lam E. (2004) Controlled cell death, plant survival and development. Nat Rev Mol Cell Biol 5: 305–315 [DOI] [PubMed] [Google Scholar]
- Lam E. (2008) Programmed cell death: orchestrating an intrinsic suicide program within walls. Crit Rev Plant Sci 27: 413–423 [Google Scholar]
- Lam E, Kato N, Lawton M. (2001) Programmed cell death, mitochondria and the plant hypersensitive response. Nature 411: 848–853 [DOI] [PubMed] [Google Scholar]
- Lane N. (2008) Marine microbiology: origins of death. Nature 453: 583–585 [DOI] [PubMed] [Google Scholar]
- Lange T. (1891) Beiträge zur Kenntniss der Entwicklung der Gefässe und Tracheiden. Flora 74: 391–434 [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lin J, Wang Y, Wang G. (2006) Salt stress-induced programmed cell death in tobacco protoplasts is mediated by reactive oxygen species and mitochondrial permeability transition pore status. J Plant Physiol 163: 731–739 [DOI] [PubMed] [Google Scholar]
- Luo HR, Hattori H, Hossain MA, Hester L, Huang Y, Lee-Kwon W, Donowitz M, Nagata E, Snyder SH. (2003) Akt as a mediator of cell death. Proc Natl Acad Sci USA 100: 11712–11717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangat S, Chandrashekarappa D, McCartney RR, Elbing K, Schmidt MC. (2010) Differential roles of the glycogen-binding domains of β subunits in regulation of the Snf1 kinase complex. Eukaryot Cell 9: 173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchelhill KI, Michell BJ, House CM, Stapleton D, Dyck J, Gamble J, Ullrich C, Witters LA, Kemp BE. (1997) Posttranslational modifications of the 5′-AMP-activated protein kinase β1 subunit. J Biol Chem 272: 24475–24479 [DOI] [PubMed] [Google Scholar]
- Miyamoto S, Rubio M, Sussman MA. (2009) Nuclear and mitochondrial signalling Akts in cardiomyocytes. Cardiovasc Res 82: 272–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath N, McCartney RR, Schmidt MC. (2003) Yeast Pak1 kinase associates with and activates Snf1. Mol Cell Biol 23: 3909–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RD. (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Pedley KF, Martin GB. (2005) Role of mitogen-activated protein kinases in plant immunity. Curr Opin Plant Biol 8: 541–547 [DOI] [PubMed] [Google Scholar]
- Phan HA, Iacuone S, Li SF, Parish RW. (2011) The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 23: 2209–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plas DR, Thompson CB. (2002) Cell metabolism in the regulation of programmed cell death. Trends Endocrinol Metab 13: 75–78 [DOI] [PubMed] [Google Scholar]
- Polge C, Jossier M, Crozet P, Gissot L, Thomas M. (2008) β-Subunits of the SnRK1 complexes share a common ancestral function together with expression and function specificities: physical interaction with nitrate reductase specifically occurs via AKINβ1-subunit. Plant Physiol 148: 1570–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polge C, Thomas M. (2007) SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci 12: 20–28 [DOI] [PubMed] [Google Scholar]
- Ren D, Yang H, Zhang S. (2002) Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J Biol Chem 277: 559–565 [DOI] [PubMed] [Google Scholar]
- Scheid MP, Parsons M, Woodgett JR. (2005) Phosphoinositide-dependent phosphorylation of PDK1 regulates nuclear translocation. Mol Cell Biol 25: 2347–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MC, McCartney RR. (2000) β-Subunits of Snf1 kinase are required for kinase function and substrate definition. EMBO J 19: 4936–4943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwachtje J, Minchin PE, Jahnke S, van Dongen JT, Schittko U, Baldwin IT. (2006) SNF1-related kinases allow plants to tolerate herbivory by allocating carbon to roots. Proc Natl Acad Sci USA 103: 12935–12940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Hanley-Bowdoin L. (2006) Geminivirus infection up-regulates the expression of two Arabidopsis protein kinases related to yeast SNF1- and mammalian AMPK-activating kinases. Plant Physiol 142: 1642–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Reyes MI, Hanley-Bowdoin L. (2009) Arabidopsis protein kinases GRIK1 and GRIK2 specifically activate SnRK1 by phosphorylating its activation loop. Plant Physiol 150: 996–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasulu N, Radchuk V, Strickert M, Miersch O, Weschke W, Wobus U. (2006) Gene expression patterns reveal tissue-specific signaling networks controlling programmed cell death and ABA-regulated maturation in developing barley seeds. Plant J 47: 310–327 [DOI] [PubMed] [Google Scholar]
- Sugden C, Crawford RM, Halford NG, Hardie DG. (1999) Regulation of spinach SNF1-related (SnRK1) kinases by protein kinases and phosphatases is associated with phosphorylation of the T loop and is regulated by 5′-AMP. Plant J 19: 433–439 [DOI] [PubMed] [Google Scholar]
- Sutherland CM, Hawley SA, McCartney RR, Leech A, Stark MJ, Schmidt MC, Hardie DG. (2003) Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr Biol 13: 1299–1305 [DOI] [PubMed] [Google Scholar]
- Tuteja N. (2007) Mechanisms of high salinity tolerance in plants. Methods Enzymol 428: 419–438 [DOI] [PubMed] [Google Scholar]
- van Doorn WG, Woltering EJ. (2004) Senescence and programmed cell death: substance or semantics? J Exp Bot 55: 2147–2153 [DOI] [PubMed] [Google Scholar]
- van Doorn WG, Woltering EJ. (2008) Physiology and molecular biology of petal senescence. J Exp Bot 59: 453–480 [DOI] [PubMed] [Google Scholar]
- Vincent O, Carlson M. (1999) Gal83 mediates the interaction of the Snf1 kinase complex with the transcription activator Sip4. EMBO J 18: 6672–6681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent O, Townley R, Kuchin S, Carlson M. (2001) Subcellular localization of the Snf1 kinase is regulated by specific β subunits and a novel glucose signaling mechanism. Genes Dev 15: 1104–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivanco I, Sawyers CL. (2002) The phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat Rev Cancer 2: 489–501 [DOI] [PubMed] [Google Scholar]
- Wang J, Li X, Liu Y, Zhao X. (2010) Salt stress induces programmed cell death in Thellungiella halophila suspension-cultured cells. J Plant Physiol 167: 1145–1151 [DOI] [PubMed] [Google Scholar]
- Warden SM, Richardson C, O’Donnell J, Jr, Stapleton D, Kemp BE, Witters LA. (2001) Post-translational modifications of the β-1 subunit of AMP-activated protein kinase affect enzyme activity and cellular localization. Biochem J 354: 275–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Lam E. (2009) Bax inhibitor-1, a conserved cell death suppressor, is a key molecular switch downstream from a variety of biotic and abiotic stress signals in plants. Int J Mol Sci 10: 3149–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A, Dickerson K, Heath R, Hong SP, Momcilovic M, Johnstone SR, Carlson M, Carling D. (2005) Ca2+/calmodulin-dependent protein kinase kinase-beta acts upstream of AMP-activated protein kinase in mammalian cells. Cell Metab 2: 21–33 [DOI] [PubMed] [Google Scholar]
- Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LG, Neumann D, Schlattner U, Wallimann T, Carlson M, Carling D. (2003) LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr Biol 13: 2004–2008 [DOI] [PubMed] [Google Scholar]
- Zhang S, Klessig DF. (2001) MAPK cascades in plant defense signaling. Trends Plant Sci 6: 520–527 [DOI] [PubMed] [Google Scholar]
- Zhou J-M, Loh Y-T, Bressan RA, Martin GB. (1995) The tomato gene Pti1 encodes a serine/threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell 83: 925–935 [DOI] [PubMed] [Google Scholar]
- Zhu J, Huang JW, Tseng PH, Yang YT, Fowble J, Shiau CW, Shaw YJ, Kulp SK, Chen CS. (2004) From the cyclooxygenase-2 inhibitor celecoxib to a novel class of 3-phosphoinositide-dependent protein kinase-1 inhibitors. Cancer Res 64: 4309–4318 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.