Abstract
A major challenge is to unravel how genes interact and are regulated to exert specific biological functions. The integration of genome-wide functional genomics data, followed by the construction of gene networks, provides a powerful approach to identify functional gene modules. Large-scale expression data, functional gene annotations, experimental protein-protein interactions, and transcription factor-target interactions were integrated to delineate modules in Arabidopsis (Arabidopsis thaliana). The different experimental input data sets showed little overlap, demonstrating the advantage of combining multiple data types to study gene function and regulation. In the set of 1,563 modules covering 13,142 genes, most modules displayed strong coexpression, but functional and cis-regulatory coherence was less prevalent. Highly connected hub genes showed a significant enrichment toward embryo lethality and evidence for cross talk between different biological processes. Comparative analysis revealed that 58% of the modules showed conserved coexpression across multiple plants. Using module-based functional predictions, 5,562 genes were annotated, and an evaluation experiment disclosed that, based on 197 recently experimentally characterized genes, 38.1% of these functions could be inferred through the module context. Examples of confirmed genes of unknown function related to cell wall biogenesis, xylem and phloem pattern formation, cell cycle, hormone stimulus, and circadian rhythm highlight the potential to identify new gene functions. The module-based predictions offer new biological hypotheses for functionally unknown genes in Arabidopsis (1,701 genes) and six other plant species (43,621 genes). Furthermore, the inferred modules provide new insights into the conservation of coexpression and coregulation as well as a starting point for comparative functional annotation.
The sequencing of Arabidopsis (Arabidopsis thaliana) and the emergence of high-throughput functional genomics techniques like microarrays, systematic T-DNA knockout screens, and protein-protein interaction (PPI) mapping have enabled the development of integrative approaches to study gene function and regulation. One of the major challenges of computational biology is the integration and exploitation of genome-wide data sets such as transcriptome and interactome data, metabolomics and other “-omics” data, and large-scale phenotyping (Brady and Provart, 2009). Data integration is often performed through gene network analysis (Lee et al., 2010; Kourmpetis et al., 2011), and the resulting networks can increase our knowledge of functional gene relationships and the interplay of different types of interactions. However, to study specific biological processes, networks are frequently studied through gene modules (Aoki et al., 2007). From a practical point of view, modules are typically identified as highly connected subgraphs within the network (Barabási and Oltvai, 2004). Depending on the type of interaction data, different types of modules are defined, and examples in Arabidopsis include coexpression modules (Lisso et al., 2005; Horan et al., 2008; Atias et al., 2009), protein complexes (Geisler-Lee et al., 2007; Boruc et al., 2010; De Bodt et al., 2010; Arabidopsis Interactome Mapping Consortium, 2011), and modules grouping genes that are regulated by the same transcription factor (TF; Ferrier et al., 2011). Genes can be part of different (sometimes overlapping) modules, while modules can be involved in different biological processes. As a consequence, gene networks are frequently highly connected, revealing the pleiotropic roles of different genes. Consequently, the module context can be explored to identify genes that are present in many different modules and that have a functional association with many other genes (hub genes; Barabási and Oltvai, 2004). These hub genes represent important components of biological systems and can provide cross talk between different processes.
Modules based on expression data are typically inferred through the clustering of genes with similar expression profiles. Most often, each gene pair receives an expression similarity measure, and this coexpression information is used to detect highly connected subgraphs in the coexpression network, representing modules. Although numerous expression network analyses have been performed in Arabidopsis, some studies focused on a specific process using guide genes (genes known to function in the process) to draw new hypotheses about the functional interplay between functionally known and unknown genes based on guilt by association (Lisso et al., 2005; Persson et al., 2005; Wei et al., 2006). Other studies employed module delineation and guilt by association on a genome-wide scale to predict gene functions (Wolfe et al., 2005; Vandepoele et al., 2006, 2009; Ma and Bohnert, 2007; Horan et al., 2008; Atias et al., 2009). From a regulatory point of view, module genes are often regulated by multiple cis-regulatory elements (referred to as motifs) organized into cis-regulatory modules (not to be confused with the gene module; Michael et al., 2008). Therefore, coexpression modules are often used to investigate the cis-regulatory elements controlling the genes within the modules using known DNA motifs or de novo motif finding (Tompa et al., 2005; Ma and Bohnert, 2007; Vandepoele et al., 2009).
A disadvantage of coexpression analysis is the false assumption that coexpressed genes are de facto coregulated (Stuart et al., 2003). The emergence of chromatin immunoprecipitation (ChIP) allows the direct profiling of the regions bound by a TF and the detection of TF target genes. The technique can be applied in a genome-wide fashion when followed by a whole-genome tiling array (ChIP-chip) or deep sequencing (ChIP-Seq; Ferrier et al., 2011). The ChIP technique provides a snapshot of the regulatory binding state of the genome by cross-linking all proteins to nearby bound DNA. In Arabidopsis, ChIP-chip/Seq has been applied to a range of TFs, primarily those active in flowering and development. Because of the static nature of a ChIP experiment (it is a snapshot of the biological state), the genome-wide profiling of TF-binding sites is often combined with differential expression analysis in a knockout (Lee et al., 2007; Morohashi and Grotewold, 2009; Busch et al., 2010; Yant et al., 2010) or an inducible overexpression line (Thibaud-Nissen et al., 2006; Kaufmann et al., 2009, 2010; Mathieu et al., 2009). By combining these two data types, TF target interactions can be viewed with respect to the expression of both the TF and the target, thus transforming the static ChIP image to a set of dynamic transcriptional modules.
A third type of module is based on PPI networks. Although there have been several PPI studies in Arabidopsis, their main focus lay in building the interactome rather than on breaking down the network to the module level (Fujikawa and Kato, 2007; Geisler-Lee et al., 2007; Van Leene et al., 2007; Arabidopsis Interactome Mapping Consortium, 2011; Li et al., 2011). Studies that did explore the network module contexts found modules recapitulating known biological functions and also suggesting new biological hypotheses for several plant-specific genes, often through the integration with expression data (De Bodt et al., 2009, 2010; Boruc et al., 2010; Arabidopsis Interactome Mapping Consortium, 2011).
Although several plant studies performed some kind of data integration when delineating gene modules, the number of data types is often limited. Recently, a few Arabidopsis studies have been published reporting large networks for function prediction based on multiple data types. These networks were built combining expression and PPI data with sequence data (Kourmpetis et al., 2011), genetic and physical interaction data (Warde-Farley et al., 2010), phylogenetic profiles and gene location (Bradford et al., 2010), and the integration of functional genomics, proteomics, and comparative genomics data sets (Lee et al., 2010). Apart from studying gene modules in one species, recent studies have applied comparisons across species to identify conserved gene coexpression in plants (Ficklin and Feltus, 2011; Movahedi et al., 2011; Mutwil et al., 2011). The analysis of coexpression networks between more distantly related species exploits the assumption that predicted gene function associations, occurring by chance within one organism, will not be conserved in a multispecies context. Consequently, the analysis of conserved modules with specific functions provides an invaluable approach for biological gene discovery in model species and for the translation of new gene functions into species with agricultural or economical value (Movahedi et al., 2012).
In this study, we investigated how Arabidopsis genes are organized into gene modules based on four different data types (Gene Ontology [GO], PPI, ChIP, and AraNet) and studied the functional and regulatory properties of these modules. Furthermore, module evolution was examined by the integration of orthologous sequences and expression data of six related plant species. Overall, our results revealed that currently available experimental data sources are highly complementary, different functional categories show varying levels of regulatory complexity, a large number of Arabidopsis gene modules are conserved in other plant species, and conserved modules provide a valuable source to study gene functions.
RESULTS
Construction of Arabidopsis Gene Modules Using Experimental and Computational Gene Associations
Based on an ensemble of primary data sets covering TF target interactions from AtRegNet (Palaniswamy et al., 2006), probabilistic gene-gene associations from AraNet (Lee et al., 2010), nonelectronic gene-GO annotations from The Arabidopsis Information Resource (TAIR; Berardini et al., 2004), and PPIs from CORNET (De Bodt et al., 2010), functional gene modules were delineated in Arabidopsis (Table I). To assemble a set of high-quality gene associations, the GO, PPI, and TF target data were filtered to only contain experimental information (see “Materials and Methods”). In contrast, the AraNet data are an integration of 24 distinct types of gene associations (e.g. coexpression, PPI, shared protein domains, similarity in phylogenetic profile, orthology) including both experimental and computational observations. In total, the final input data set covered 22,492 unique genes and more than 1 million interactions, with the largest fraction coming from the AraNet network. Nearly all gene associations were unique to one input data type, with the fraction of unique associations ranging from 75% for PPI to 99% for AraNet and TF targets (Table I).
Table I. Overview of the primary data sets and delineated modules with their properties.
| Data Type | Primary Data Sets |
Modules |
||||
|---|---|---|---|---|---|---|
| No. of Genes | No. of Associations (% Unique)a | No. of Genes | No. of Modules (% Unique)b | Functional Enrichmentc | Motif Enrichmentd | |
| PPI | 3,194 | 7,210 (75%) | 597 | 72 (95%) | 51 | 43 |
| AraNet | 19,647 | 1,062,222 (99%) | 6,377 | 419 (99%) | 116 | 172 |
| TF targets | 9,422 | 13,037 (99%) | 5,127 | 518 (96%) | 51 | 224 |
| GO | 6,588 | 89,100 (n.a.) | 7,750 | 1,105 (99%) | 943 | 341 |
| Total | 22,492 | 1,089,661 | 13,428 | 2,114 | 1,161 | |
| Nonredundant modules | 13,142 | 1,563 | 676 | 772 | ||
Percentage of associations unique for this data type. As GO does not consist of pairwise gene-gene associations, no unique fraction is reported. n.a., Not available.
Percentage of modules unique for this data type (based on the output of detecting redundant modules across different input data types).
Calculated for the nonredundant modules only.
To delineate gene modules from the different gene association data sets, two clustering strategies were applied (Fig. 1). First, for the TF targets and GO data, expression information was integrated to cluster genes into modules (expression-based clustering; see “Materials and Methods”). This was done because the TF target ChIP data provide a static image of genome-wide TF binding, and as a consequence, TF target genes do not necessarily form functionally coherent modules. By integrating expression data, these static images are converted into spatiotemporal TF target maps. Similarly, GO categories do not represent functionally coherent gene modules (Vandepoele et al., 2009). Therefore, per GO category, genes with nonelectronic GO annotations were used as prior information to guide the creation of coexpression clusters using different expression compendia from CORNET (De Bodt et al., 2010). Genes used as guides are referred to as seed genes in the remainder of this paper. Different Arabidopsis expression compendia (see “Materials and Methods”) were used because the degree of coexpression can be influenced by the specific expression data used (Usadel et al., 2009). Therefore, genes from GO categories were clustered using the compendium in which the coexpression was highest, measured by expression coherence (EC). EC is a measure for the amount of expression similarity within a set of genes for a given expression compendium (see “Materials and Methods”). All GO categories across the three GO hierarchies “Biological Process” (BP), “Molecular Function,” and “Cellular Component” were used as sources for seed genes to build modules of different specificity (i.e. general versus very specific processes). As many genes in Arabidopsis have not yet been functionally annotated, many GO categories are incomplete. To overcome this problem, GO category-based seed sets were expanded with genes showing high coexpression with the seed genes prior to the clustering (multi-query seed expansion [MQSE]; see “Materials and Methods”). Since different TFs can regulate the same gene and genes can be associated with multiple GO categories, genes can belong to more than one resulting module. Second, PPI and AraNet gene associations were clustered based on the connectivity of the genes in their respective input networks without linking to expression data (referred to as connectivity-based clustering; see “Materials and Methods”). As a consequence, highly connected subgraphs were identified in both networks to delineate PPI and AraNet modules, respectively.
Figure 1.
Delineation of functional gene modules. A, Four different primary data sets were processed to extract functional gene modules, resulting in 1,563 nonredundant modules. Data types are in roman font, and methods are in italic font. B, Biological properties (functional coherence, EC, and cis-regulatory coherence) of gene modules were characterized. Dotted lines indicate gene-GO associations and nonsignificant PCCs for the functional coherence and the EC panels, respectively. In the cis-regulatory coherence panel, the blue triangle represents an enriched motif.
All modules from the different input data types (PPI, 72; AraNet, 419; TF targets, 518; GO, 1,105) were compiled into one final data set covering 2,114 coexpression modules derived from GO, transcriptional modules derived from TF targets, PPI modules derived from the PPI network, and AraNet modules. To determine the extent to which the different data sets complement each other, the overlap between the different data types was assessed (see “Materials and Methods”). On the level of gene content, 40% of the genes in the modules are present in more than one input data type (Fig. 2A). However, the overlap based on the gene-gene associations both in the input (Table I) and the module associations was drastically smaller, with only 3% of the gene pairs within a module having support by more than one primary data type (Fig. 2B). After removing redundant modules based on the number of shared genes (see “Materials and Methods”), the final data set consisted of 1,563 modules comprising 13,142 genes (63% of all genes on the ATH1 microarray). Based on the redundant modules, the low overlap between different data types was confirmed, as most modules (1,556 of 1,563) could only be found through a single data type (Fig. 2C). Examples of modules confirmed by multiple data types (seven) include genes related to amino acid metabolism and transport (for modules and gene sets discussed throughout the article, see Supplemental Table S1). The majority of modules contained five to 10 genes (50%), while larger module sizes were increasingly less frequent (Fig. 2D). These observations were in line with the notion of a hierarchical structure of biological networks, where smaller and more specific clusters reside within larger and more general clusters (Mutwil et al., 2010).
Figure 2.
Basic properties of the derived functional gene modules. A, Number of different module types per gene. B, Number of different input data types per module edge. C, Overlap between the different types of modules. D, Gene size distribution for the set of 1,563 nonredundant gene modules.
Functional, Expression, and cis-Regulatory Coherence of Plant Modules
Based on the gene modules inferred through the different primary data types, we next sought to characterize different biological properties. The investigated properties describe the level of coexpression among the genes in a module, whether the module genes are potentially regulated by the same TF, and whether a specific function or biological process can be linked to a module (Table I; Fig. 3). An additional Web site (http://bioinformatics.psb.ugent.be/cig_data/plant_modules/) is available to browse modules, genes, coexpression information, primary gene associations, functional annotations, and motifs.
Figure 3.
Functional, expression, and cis-regulatory coherence. A, Comparison of EC scores between the modules from different input data types. The EC scores are shown for both the general compendium (dotted lines) and the compendium showing the maximum EC (solid lines). The vertical dotted line indicates the threshold for significant EC. B, GO-BP and motif enrichment statistics for the modules delineated using the different input data types.
For each module, the level of coexpression was determined using EC. To minimize the possible influence of the specific expression data set used to determine the level of coexpression, EC scores were initially calculated for each module based on a global compendium and other specific compendia, and only the maximum EC score was retained for further analysis. Note that for GO and TF targets, these compendia correspond with the expression data used to delineate the modules. Overall, for the nonredundant set of 1,563 modules, the median EC score was above 50%, indicating that coexpression is an important property of most modules (Fig. 3A). Comparing the maximal EC scores for modules derived from different primary data types revealed that coexpression levels were also high for PPI and AraNet modules (98.6% and 88.5% show significant EC), despite the fact that expression information was not directly integrated during the module delineation. At the 10% EC threshold, which corresponds with P ≤ 0.02 (based on randomized gene modules; see “Materials and Methods”), the difference between the EC scores from the global and specific expression compendia was the largest for the TF target modules.
To assess the cis-regulatory module properties (cis-regulatory coherence), de novo motif finding was performed to identify putative TF binding sites in the 1-kb promoters of the genes. The motif finding was performed with the complementary tools Weeder and MotifSampler (Pavesi et al., 2001; Thijs et al., 2002; Tompa et al., 2005). To discard potentially false motifs, enrichment analysis was performed and only motifs showing significant enrichment within a module were retained (q ≤ 0.01). Redundant motifs within modules were removed based on sequence similarity and gene-motif occurrences (see “Materials and Methods”), resulting in 1,544 different motifs in the modules. MotifSampler and Weeder exclusively supported 1,190 (77.1%) and 285 (18.5%) motifs, respectively, while 69 (4.5%) motifs were supported by both tools, emphasizing their complementarity. To validate the reliability of motifs found by only one tool, the overlap of motifs found by MotifSampler or Weeder was compared with a set of 515 known motifs from PLACE (Higo et al., 1999) and AGRIS (Palaniswamy et al., 2006). Of the 1,544 de novo motif instances in modules, 528 corresponded to a known motif. For these 528 known motif instances, 408 (77.3%) and 71 (13.4%) were found uniquely by MotifSampler and Weeder, respectively, and 49 (9.3%) were retrieved by both tools. In addition, both methods reported a similar but complementary fraction of known motifs (MotifSampler, 408 of 1,190 [34.3%]; Weeder, 71 of 285 [24.9%]) among their total number of reported motifs. To facilitate downstream analysis, the combined set of de novo motifs and known motifs from PLACE and AGRIS was grouped into 813 motif families based on sequence similarity (see “Materials and Methods”). Within these de novo motif families, 65 contained a known motif while 748 families contained purely de novo motifs. Finally, the cis-regulatory coherence was defined as the fraction of modules with at least one enriched motif (Fig. 3B). The cis-regulatory coherence scores ranged from 40% (AraNet, 172 of 419; TF target, 224 of 502) to 60% (PPI, 43 of 72; GO, 341 of 579). In total, 49.4% of the nonredundant set of modules contained at least one motif (772 of 1,563). A weak but significant (r2 = 0.03; P < 1.42e-11) relation was found for the number of different motif families in one module in the function of EC. Apart from the cis-regulatory coherence analysis, these motifs provide an important resource to annotate and map specific TF target interactions at the module level.
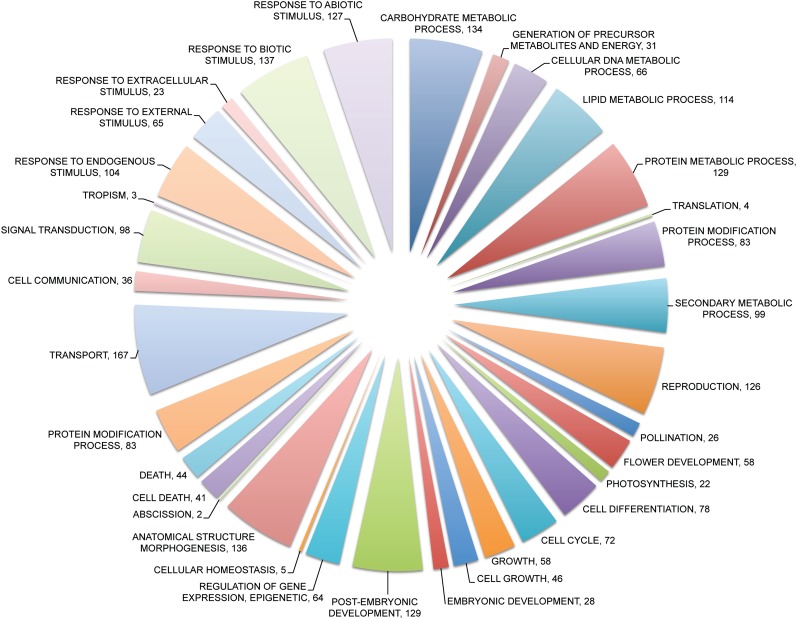
The functional coherence was determined by GO enrichment analysis for nonelectronic biological process annotations and enrichment for genes associated with embryo lethality. Information about genes involved in embryo lethality was based on the SeedGenes database (Meinke et al., 2008). The functional coherence revealed large differences between modules from the different primary data types (Table I; Fig. 3B). As expected, the GO modules showed the highest functional coherence (80% of the modules). While for AraNet and PPI, 27% and 72%, respectively, of the modules showed functional coherence, the TF target data had the lowest functional coherence (10% of the modules). Overall, 40% of the modules could be linked to a significantly enriched biological process or embryo lethality, while 98% of the modules contained one or more genes with a known experimental annotation. To obtain an overview of the different biological processes in which the modules were involved, the module predictions were categorized according to their GO slim terms (Fig. 4). Control experiments indicated that there were no significant enrichments toward any GO category in either the complete set of input genes or the complete set of resulting modules.
Figure 4.
Overview of GO-BP slim biological processes in which modules were predicted to be involved. Modules with multiple GO-BP annotations can be present in different GO slim categories.
Hub Genes and the Organization of Transcriptional Regulation in Arabidopsis
Genes can have pleiotropic roles and thus can be involved in multiple processes or modules. Because of the different input data types and the way different GO categories were used to guide module detection using MQSE, genes can occur in multiple although nonredundant modules. Hub genes (Barabási and Oltvai, 2004) were identified as genes that are present in a large number of modules and are possibly providing cross talk between the different biological processes they are involved in. The number of modules per gene ranged from one to 26, following a power law, making the gene-module associations a scale-free network (Fig. 5A; Supplemental Fig. S1). Genes present in more than 10 modules (116 genes; top 5%) were extracted as hub genes, and a functional enrichment analysis revealed that these genes are involved in immune response, photosynthesis, cell cycle, and carbohydrate metabolism (Supplemental Table S1; Supplemental Fig. S2A), which is in accordance with earlier studies (Mao et al., 2009; Chao et al., 2011). Among the hub genes, we found MEK1 (mitogen-activated protein kinase kinase), MPK11 and MPK4 (mitogen-activated protein kinase), SOLUBLE N-ETHYLMALEIMIDE-SENSITIVE FACTOR ADAPTOR PROTEIN33 (SNARE), RAB GTPASE HOMOLOGH1C (RAB GTPase), and EXTRA-LARGE GTP-BINDING PROTEIN2 (GTP-binding protein), revealing that several hub genes are involved in signal transduction. Evidence for cross talk mediated by hub genes was found for chromatin modification and development through the genes CYTOCHROME P450, FAMILY 71, AT5G63960, and FUSED. Light response and photosynthesis were found to be coupled through the genes Light-harvesting-like3:1, FRUCTOSE-BISPHOSPHATE ALDOLASE1, ISOPRENOID F, and 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE. Finally, SYNTAXIN OF PLANTS121 (SYP121), SYP122, ARABIDOPSIS PHYTOALEXIN DEFICIENT4, NECROTIC SPOTTED LESIONS1, AVRPPHB SUSCEPTIBLE3, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN70 (TF), JASMONATE-ZIM-DOMAIN PROTEIN1, ARABIDOPSIS NONEXPRESSER OF PR GENES1, RADICAL-INDUCED CELL DEATH1, CHITINASE-LIKE PROTEIN1, and AT1G15430 (based on module-based GO prediction) describe the cross-link between the response to biotic/abiotic stimuli and hormone signaling through jasmonic acid (JA) and salicylic acid (SA). In addition, hub genes are also 3-fold enriched for embryo-lethal genes, confirming the relationship between network connectivity and essentiality (Mutwil et al., 2010).
Figure 5.
Regulatory complexity of genes in modules. A, Number of modules in which a gene is present. Asterisks denote values higher than zero. B, Number of motifs per module. C, Number of motifs per gene promoter. D, Regulatory complexity, defined as a combination of the number of modules in which a gene is present and the number of motifs in its promoter. All 13,142 genes are included, and the number of genes at each coordinate is given as a colored size scale. The gray circle indicates the average regulatory complexity for all 13,142 genes. The dotted line is the function f(x) = x.
Besides investigating gene-module organization, the organization of motifs was also examined at the module and gene level. On the module level, the number of motifs ranged from zero to eight (Fig. 5B), and modules regulated by five or more motifs (approximately 2%; Supplemental Table S1) are involved in processes associated with flower development, protein synthesis, and stimulus responses. On the gene level, the number of motifs per gene varied from zero to 26 (Fig. 5C). Genes are mostly regulated by one to five motifs, but approximately 2% are regulated by more than 10 motifs. These highly regulated genes are involved in cell cycle, systemic acquired response, and SA signaling (Supplemental Table S1).
To define the regulatory complexity of a gene, the number of modules and the number of motifs were combined in one plot (Fig. 5D). A gene is considered complexly regulated when present in multiple modules and harboring multiple motifs. A significant positive correlation was found between the number of motifs and the number of modules (adjusted r2 = 0.40; P ≤ 2.2e-16). Whereas for the GO-BP slim main category BP the linear fit followed the 1:1 line, not all genes follow this strict “one module, one motif” principle. Examining the module-motif relationships for different GO-BP slim subcategories revealed processes where genes were present in many modules but without being regulated by many motifs. This indicates that, based on the number of motifs, hub genes are not necessarily regulated by many TFs (Supplemental Fig. S2B). Carbohydrate metabolism, lipid metabolism, secondary metabolism, photosynthesis, cellular homeostasis, and generation of precursor metabolites and energy consistently showed a linear fit with a less steep slope, indicating more modules than motifs. In contrast, DNA metabolism and cell cycle showed a steeper slope than the main BP category, indicating more motifs than modules and combinatorial regulation.
When isolating the top 200 genes based on regulatory complexity (i.e. genes with a high number of modules and motifs), functional enrichments were found related to immune response, stress response, and cell cycle (Supplemental Table S1).
Conservation of Gene Modules in Other Plants
Based on the inferred Arabidopsis modules and their different biological properties, we next characterized if these modules are conserved in other plant species, since it has been shown that dynamic properties are primarily conserved at the module level (Zinman et al., 2011). The evolution of functional gene modules was examined using conservation of coexpression (EC) and conservation of regulatory DNA motifs (cis-regulatory coherence) based on orthologous genes in the dicots soybean (Glycine max), Medicago truncatula, poplar (Populus trichocarpa), and grapevine (Vitis vinifera) and the monocots maize (Zea mays) and rice (Oryza sativa ssp. japonica). Orthologous modules were delineated using the PLAZA integrative orthology approach, which infers orthologous genes using complementary detection methods (i.e. phylogenetic trees, OrthoMCL families, and Best-Hits-and-Inparalog families), which are considered as evidence (Van Bel et al., 2012). For each Arabidopsis gene, the orthologous gene(s) with the greatest evidence was retained in each of the sampled species (Supplemental Table S2). Orthologous modules were subsequently constructed by grouping the orthologous genes based on the Arabidopsis modules. Despite the potential problem of modules expanding significantly in size due to one-to-many orthology relationships, the applied ensemble approach retrieved one-to-one orthologs for on average (over the six species) 67% of the genes.
To study coexpression conservation, EC scores were calculated in the six species using publicly available microarray data (see “Materials and Methods”). For gene pairs with multiple orthologs, coexpression was considered present when at least one orthologous gene pair showing significant coexpression was found. Orthologous genes missing from the microarray were not taken into account. EC values of orthologous modules with less than five genes on the microarray of the respective species were marked as missing to distinguish them from zero values. The EC scores were compared with those of a set of random modules with the same gene size distribution (Supplemental Fig. S3A), and based on these background scores, 910 modules (58%) with EC of 10% or greater in three or more species showed significant conservation of coexpression (P ≤ 0.025; Supplemental Table S1). These conserved modules comprised a wide range of functions and biological processes, while modules with ultraconserved coexpression (i.e. EC > 10% in seven species, 92 modules; Supplemental Table S1) showed enrichments for processes linked with energy metabolism (e.g. NADPH metabolism, photosynthesis, and starch biosynthesis).
For the set of modules with significant coexpression conservation in other plants, the conservation of cis-regulatory coherence was investigated, since conservation of both properties would strongly indicate the conservation of regulation. To measure motif conservation, enrichment analysis for each of the motifs present in the original Arabidopsis modules was conducted in each of the species based on the promoter sequences of genes in the orthologous modules (Supplemental Fig. S3B). Fifty-five percent of modules with conserved coexpression (500 of 910 modules) had at least one enriched motif in Arabidopsis, and based on the comparative motif analysis, we were able to confirm motif enrichment for 27.4% of these modules in at least one other species (137 of 500 modules; Supplemental Table S1). Four modules exhibited both expression and motif conservation in all seven species. These were involved in ribosome assembly, DNA modification, and response pathways and harbored motifs such as SORLIP2 (for sequences overrepresented in light-induced promoter 1), SITEIIATCYTC, TELOBOX, UP1/2, BS1EGCCR, E2F, ABRE, and G-box. In contrast, 42% of modules without coexpression conservation had at least one motif in Arabidopsis (272 of 653 modules), but for only 5% of those modules, the motif enrichment was conserved (20 of 272 modules). This result showed that modules with conserved coexpression in other species are 4-fold enriched in motif conservation compared with modules lacking conserved coexpression. The modules with conserved motif enrichment harbor 90 motif families (5% of all motif families), of which 67 represent new motifs and 23 were previously known. A detailed map associating motifs with specific functional categories is shown in Supplemental Figure S4.
Module-Based Functional Annotation of Unknown Plant Genes
Complementary to the cross-species analysis of different regulatory module properties, the conserved module contexts provide a promising resource for hypothesis-driven gene discovery in other plant species. The Arabidopsis sequencing project was succeeded by the Arabidopsis 2010 program, of which the goal was the annotation of all Arabidopsis genes by 2010 (http://www.arabidopsis.org/portals/masc/FG_projects.jsp). Despite many efforts based on forward and reverse genetics as well as computational predictions, functional annotation is still lacking for many genes. Although advanced computational gene function prediction tools have been developed (Lee et al., 2010; Bassel et al., 2011), our main intention was to investigate how the integrated gene associations could lead to new functional hypotheses.
Since the initial download of the GO data for the module delineation (hereafter referred to as “data freeze”), 2,940 genes belonging to the gene modules have received new experimental GO-BP annotations. Since these gene-GO associations were not available at the time of the module delineation, they form an ideal basis to evaluate the module-based gene function predictions inferred through the integration of the different primary gene associations. These new associations can be categorized into three groups: (1) genes that had no GO information from any hierarchy in the input data; (2) genes that had no GO information with nonelectronic evidence tags in the BP hierarchy; and (3) other experimental BP genes that had nonelectronic BP information available, which was not linked to the new experimental association. To evaluate our module-based function predictions, very general categories were not taken into account to avoid an overestimation of the number of true positives (see “Materials and Methods”). Results showed that out of the 2,940 genes with a new experimental GO-BP, 1,460 genes were assigned to modules with GO-BP enrichment, and 29.7% (434) of those had a correct GO-BP inferred through the modules (Table II; Supplemental Table S3). For the 197 functionally unknown genes from category 1, this percentage was 38.1%. Conversely, from the perspective of the modules, 5,562 genes received a new module-based GO-BP prediction, of which 434 genes had their prediction confirmed by a new experimental GO annotation (7.8%; Table III). Based on the fraction of true positives for the functionally unknown genes from category 1, this would suggest that more than 2,000 genes (38.1% of 5,562) can be correctly characterized based on the functional coherence of the modules. The results for the different categories are presented in more detail in Tables II and III. All new module-based Arabidopsis functional annotations were submitted to TAIR.
Table II. Comparison of 2,940 genes having new experimental GO-BP annotations (of which 1,460 are present in modules) with the module-based function predictions.
| Genes | Unknowna |
Unknown Experimental BPb |
Other Experimental BPc |
Total |
||||
|---|---|---|---|---|---|---|---|---|
| No. Predicted | No. Confirmedd | No. Predicted | No. Confirmed | No. Predicted | No. Confirmed | No. Predicted | No. Confirmed | |
| All Genese | 197 | 75 (38.1) | 255 | 108 (42.4) | 1,008 | 251 (24.9) | 1,460 | 434 (29.7) |
| Conserved | 166 | 65 (39.2) | 195 | 80 (41) | 871 | 215 (24.7) | 1,232 | 360 (29.2) |
| Not conserved | 48 | 10 (20.8) | 83 | 31 (37.3) | 315 | 52 (16.5) | 446 | 93 (20.9) |
No GO information from any hierarchy in the input data.
Nonelectronic GO information is available in the BP hierarchy, which is not linked to the new experimental association.
Numbers in parentheses represent percentages of confirmed genes (number confirmed/number predicted).
Genes that were present in both conserved and nonconserved modules could gain a prediction by both. The total of genes in conserved and nonconserved modules is the set of unique genes from these two sets.
Table III. Comparison of 5,562 module-based function predictions with new experimental GO-BP annotations.
| Genes | Unknowna |
Unknown Experimental BPb |
Other Experimental BPc |
Total |
||||
|---|---|---|---|---|---|---|---|---|
| No. Predicted | No. Confirmedd | No. Predicted | No. Confirmed | No. Predicted | No. Confirmed | No. Predicted | No. Confirmed | |
| All genese | 2,241 | 75 (3.3) | 2,386 | 108 (4.5) | 935 | 251 (26.8) | 5,562 | 434 (7.8) |
| Conserved | 1,826 | 65 (3.6) | 1,926 | 80 (4.2) | 818 | 215 (26.3) | 4,570 | 360 (7.9) |
| Not conserved | 565 | 10 (1.8) | 645 | 31 (4.8) | 260 | 52 (20) | 1,470 | 93 (6.3) |
No GO information from any hierarchy in the input data.
Nonelectronic GO information is available in the BP hierarchy, which is not linked to the new experimental association.
Numbers in parentheses represent percentages of confirmed genes (number confirmed/number predicted).
Genes that were present in both conserved and nonconserved modules could gain a prediction by both. The total of genes in conserved and nonconserved modules is the set of unique genes from these two sets.
Despite the increasing number of genes receiving experimental GO-BP annotations during the last decades, still 7,233 Arabidopsis genes exist for which no GO-BP information is available (neither experimental nor electronic information in any GO hierarchy; Supplemental Table S1). From these functionally unknown genes, 3,553 genes were assigned to a module, of which 68% (2,419 of 3,553) were part of a module that showed expression conservation (Table IV). Based on a functional enrichment analysis using GO or embryo-lethal genes, a functional annotation could be associated to 1,701 genes. The fraction of modules containing genes of unknown function and having enrichment-based functional predictions was roughly two times higher for conserved modules compared with modules lacking expression conservation (1,435 of 2,419 and 266 of 1,134, respectively). The newly annotated genes in the coexpression conserved modules represented a wide range of biological processes, as can be seen in Supplemental Figure S5. Based on gene orthology in the significantly coexpression conserved modules, 43,621 genes with unknown experimental GO-BP in other plants could be assigned a function.
Table IV. Module-based annotation of genes for which the GO-BP is currently unknown using experimental GO and embryo lethality data.
| Genes | No. of Genes of Unknown Functiona | Module-Based Annotation |
||
|---|---|---|---|---|
| No. of Genes with GO Enrichment | No. of Genes Predicted with Embryo Lethality | Total No. of Genes with Functional Prediction (Uniqueb) | ||
| Modules | 3,553 | 1,680 | 281 | 1,701 |
| Conserved | 2,419 | 1,418 | 275 | 1,435 |
| Not conserved | 1,134 | 262 | 6 | 266 |
| Not in modules | 3,680 | |||
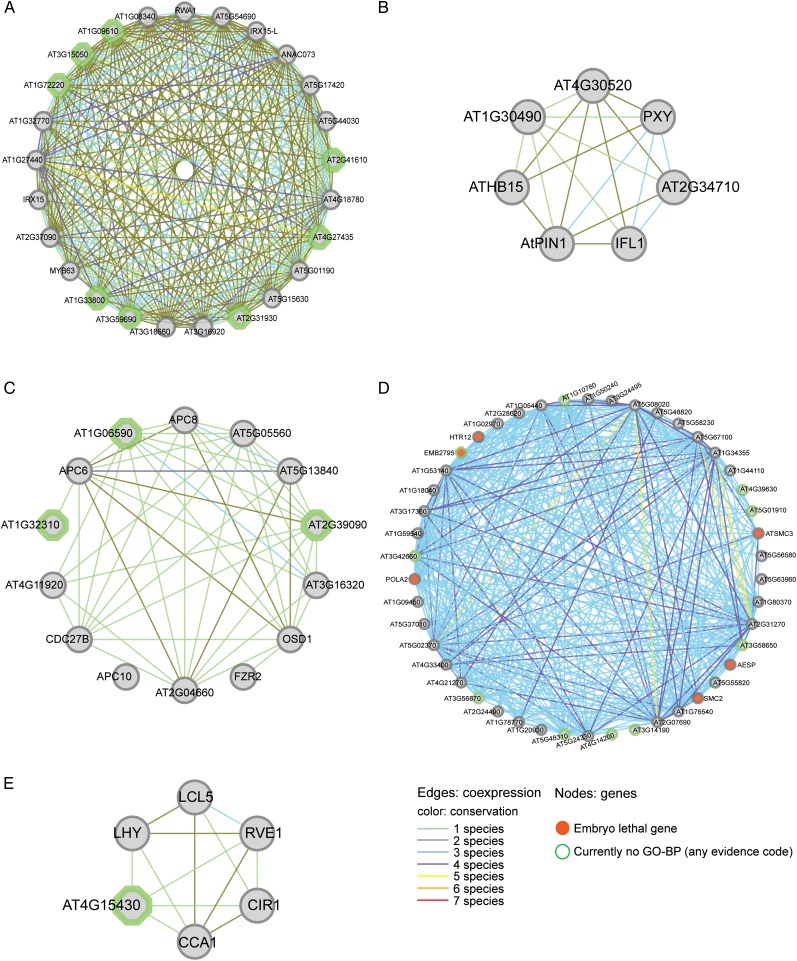
The following paragraphs report a number of examples of module-based gene function predictions that correspond with recent experimental work, which can be explored using the additional data Web site (http://bioinformatics.psb.ugent.be/cig_data/plant_modules/). The first module, MQSE_BP_GO:0006030_3 (Fig. 6A), is derived from the GO term “chitin metabolic process” and also includes some PPIs, TF targets, and AraNet edges. The module contains five true-positive genes, MYB DOMAIN PROTEIN63 (MYB63; Zhou et al., 2009), IRREGULAR XYLEM15 (IRX15) and IRX15-L (Brown et al., 2011), NAC DOMAIN CONTAINING PROTEIN73 (ANAC073) (Zhong et al., 2008), and REDUCED WALL ACETYLATION1 (RWA1) (Lee et al., 2011), all of which were correctly predicted to be involved in cell wall biogenesis. MYB63 and ANAC073 are a MYB and a NAC TF, respectively, and whereas MYB63 was known to be involved in JA/SA response pathways (Yanhui et al., 2006), no BP information was known for ANAC073. In contrast, RWA1, IRX15, and IRX15-L were completely unknown (no GO in any hierarchy). Additionally, eight currently functionally unknown genes (AT2G41610, AT2G31930, AT1G09610, IQ-DOMAIN10 (IQD10), AT1G72220, AT1G33800, IQD13, and AT4G27435) are present in the module. Furthermore, the genes reported in the module correspond with those found by Persson et al. (2005) in their study of cell wall biogenesis. Out of the four genes that were tested by mutant analysis in their investigation, IRX8 was present as a seed gene in the input data, but CHITINASE-LIKE PROTEIN2 and AT4G27435 were added by the MQSE methodology (AT5G03170 was not present in the module). In addition, looking at the 25 highest ranked genes with CELLULOSE SYNTHASEA4 (CESA4), CESA7, and CESA8 (including the four tested genes), we observed four genes that were seed genes and 10 genes that were added to our module by MQSE.
Figure 6.
Example of a delineated module with true-positive genes. A, Cell wall biogenesis. B, Xylem and phloem pattern formation. C, DNA endoreduplication. D, Cell cycle regulation. Edges with EC conservation in less than three species are hidden. E, Response to GA. Modules can be explored in detail using the additional data Web site.
The second module originated from the GO category “meristem initiation” (MQSE_BP_GO:0010014_1; Fig. 6B). The true-positive gene in this module is PHLOEM INTERCALATED WITH XYLEM, which had only a computational BP annotation related to protein amino acid phosphorylation. Based on the module, the gene was predicted to be involved in xylem and phloem pattern formation, which has recently been annotated by an experimental GO annotation (Hirakawa et al., 2008). The module contains multiple genes known to be involved in xylem and phloem pattern formation, including ARABIDOPSIS THALIANA PIN-FORMED1, INTERFASCICULAR FIBERLESS1, and ATHB15. All genes in the module have experimental associations with meristem-related processes, which refers to the formation of phloem and xylem out of cambium cells (meristematic tissue).
The third module, PPI_14 (Fig. 6C), is based on the experimental PPI network, but many edges are supported by AraNet as well. This PPI module contains 14 genes and is predicted to be involved in DNA endoreduplication, the process of continued DNA replication without mitosis in order to support cell growth. Genes AT1G32310, AT1G06590, and OMISSION OF SECOND DIVISION1 were unknown, but AT1G06590 has recently been experimentally validated (Quimbaya et al., 2012). Experiments have shown that a hemizygous mutant line of this gene has an endoreduplication index (the mean number of endoreduplication cycles) significantly different from wild-type plants. Genes in the module with a known link to endoreduplication were ANAPHASE-PROMOTING COMPLEX SUBUNIT8 (APC8), APC6, FIZZY-RELATED2, CDC27B, and APC10.
The fourth module (MQSE_BP_GO:0051726_1; Fig. 6D) was identified based on the GO term “regulation of cell cycle” and includes the functionally unknown genes AT5G48310, AT3G56870, AT3G14190, AT1G10780, AT2G32590, AT3G42660, AT3G56870, AT4G14200, AT3G58650, AT5G01910, and AT4G39630. Given the strong coexpression in the entire module (EC = 0.97) and the conservation of the coexpression (in all six species but M. truncatula), there is strong evidence that these genes are involved in cell cycle regulation as well. A large fraction of the genes are coregulated by the E2F TRANSCRIPTION FACTOR A (E2FA)-DPA TF complex. An essential role in cell division coincides with the observed embryo lethality of the module genes CENTROMERIC HISTONE H3, EMBRYO DEFECTIVE2795, DNA POLYMERASE ALPHA2, STRUCTURAL MAINTENANCE OF CHROMOSOMES2, HOMOLOG OF SEPARASE, and STRUCTURAL MAINTENANCE OF CHROMOSOME3. The prediction for AT5G55820, which was only known to be functionally involved in embryo sac and seed development, is supported by additional InterPro domain evidence, as it contains the “inner centromere protein, ARK-binding domain.” This domain is involved in the coordination of chromosome segregation during cell division in yeast (Leverson et al., 2002), thus linking it to the cell cycle. Furthermore, the de novo motif discovery retrieved enriched motifs with an E2F core (TCCCGC).
The last module, MQSE_BP_GO:0009739_3, is delineated from the GO category “response to GA stimulus” (Fig. 6E) and has some AraNet and PPI edges as well. The functional prediction of the module yielded the GO terms “response to GA” as well as “response to salt stress and hormones (auxin, JA, SA, and abscisic acid).” However, the module also showed enrichment toward “circadian rhythm” and “long-day photoperiodism, flowering” (GO:0007623 and GO:0048574, respectively). These two predictions are particularly interesting, as the module contains LHY-CCA1-LIKE5 (LCL5), encoding a MYB family TF that was only known to be involved in response to hormone stimuli but has recently been experimentally assigned to both “photoperiodism, flowering” and “circadian rhythm” (Farinas and Mas, 2011). Next to the newly assigned MYB LCL5, the module contains two more MYB TF genes (REVEILLE1 [RVE1] and CIRCADIAN1 [CIR1]). Although the MYB TF gene RVE1 had a GO-BP association based on a traceable author statement, the annotation “regulation of cellular transcription” (GO:0045449) was far from specific. Together with the unknown gene AT4G15430, the module thus provides a strong prediction for two functionally unknown genes. CIR1, LATE ELONGATED HYPOCOTYL, and CIRCADIAN CLOCK ASSOCIATED1 were the known circadian regulators on which the module prediction was based. The module is enriched for the motif sTsAGCCACwAn, which contains the SORLIP1 core (CCAC) described in PLACE, which is a phytochrome A-induced motif. Finally, given the enrichments for genes responsive to a GA stimulus and circadian clock genes, this module reaffirms the cross talk between both processes reported by Arana et al. (2011).
DISCUSSION
To delineate a wide range of gene modules, an ensemble of input data types was assembled based on experimental gene associations (GO, PPI, and TF targets) and AraNet. Although the different combined data sets comprised more than 1 million gene associations, the overlap between individual data sets was surprisingly low. This observation was confirmed by the large fraction of unique associations per primary data type and the low overlap in gene content between the modules before redundancy removal, indicating the advantage of combining different experimental data sources. Based on a set of 2,355 Arabidopsis proteins, Lysenko et al. (2011) also reported that the integration of multiple data sets, apart from sequence-based gene functions, was beneficial for the functional annotation of modules inferred using graph-based clustering. In addition, their data revealed that, despite the integration of experimental data sources, only a limited number of all Arabidopsis genes could be embedded into an integrative network. Complementary to network construction methods that start from a limited number of experimentally characterized genes, other studies have applied clustering tools on large expression compendia to identify gene modules at a genome-wide scale (Ma and Bohnert, 2007; Horan et al., 2008; Atias et al., 2009). Although including more genes, these approaches typically yield a limited number of functional modules, as functional gene information is mostly incorporated during postprocessing to link modules to specific biological processes (Atias et al., 2009; Mutwil et al., 2010). To circumvent this problem, we developed the MQSE method to use genes with nonelectronic GO annotations as guide genes to define coexpression modules. While guide gene approaches are typically applied for the analysis of a specific process (Lisso et al., 2005; Persson et al., 2005; Wei et al., 2006), the integration of all GO categories resulted in a set of modules covering a wide range of processes in Arabidopsis (Fig. 4). Although Cho et al. (2011) also integrated different GO annotations during the delineation of yeast modules, as far as we are aware, this approach has not been applied to plants. GO-based clustering without any modification to the gene sets would result in many missing genes due to the incomplete functional annotation of the Arabidopsis genome and the low EC in some categories. To overcome this problem, the guide gene MQSE strategy allowed us to fine-tune the GO seed sets prior to expression clustering by identifying strongly coexpressed seeds and by adding more than 1,000 genes with highly similar expression profiles. Whereas MQSE is related to the multi-experiment matrix (MEM) method of Adler et al. (2009), MEM uses one gene as seed, while our approach can integrate multiple seed genes. This is a significant improvement, since it allows the analysis of coregulation within a process of interest. Second, whereas the output of MEM is a ranked list of genes that are coexpressed with the query gene, there is no determination of an optimal set of coexpressed genes. In contrast, MQSE returns the optimal set of coexpressed genes using a rank-based enrichment score.
Based on the EC scores and the percentage of modules for which a regulatory DNA motif could be found (50%), it is clear that coexpression and coregulation are two important factors to ensure the proper functioning of genes. Remarkably, PPI is the second best data type when considering expression and cis-regulatory coherence, indicating that interacting genes are also frequently coregulated. Conversely, the cis-regulatory coherence of the TF target data were not higher than in other data sets, supporting the concerns about the specificity of ChIP data sets, as many reported TF targets do not correlate with each other at the expression level (Ferrier et al., 2011). However, the EC of the TF target data set was influenced most by different expression compendia, suggesting differences in the condition specificity for the different target genes (Fig. 3A). The analyzed module properties indicate that GO combined with coexpression and PPI data are the most suited to delineate functionally and regulatory coherent modules. The same trend was observed when determining true-positive module-based GO predictions per input type, as true positives were found in 214 (37%) GO, 22 (31%) PPI, 47 (11%) AraNet, and 15 (3%) TF modules. In addition, we observed that highly integrative approaches, such as AraNet, yielded many modules lacking functional coherence and that more than 1,000 conserved gene modules were found, based on one of the other primary data type.
On the organizational level, it is clear that, as for other biological networks, most genes are present in a few modules while a limited number of hub genes exists. On the regulatory level, a similar pattern was observed, with most modules and genes containing a limited number of motifs. The maximum number of 26 motifs per gene is high but in line with a recent estimation of the number of binding sites per gene being, based on available Arabidopsis Chip-Seq studies, up to 75 binding events per gene (Ferrier et al., 2011). Although it is currently unclear whether this pattern holds for all genes, this estimate provides an experimental indication that complex regulation, as indicated by our modules, will be true for some genes. The variation in regulatory complexity for different GO-BP slim categories confirms that function, apart from other factors, is an important element contributing to a gene’s regulation (Freeling et al., 2007; Movahedi et al., 2011).
Genome-wide modular approaches have often been used to infer functions for functionally unknown genes. However, to our knowledge, this study is the first one to integrate different functional data types as well as conserved coexpression in seven species (soybean, M truncatula, poplar, grapevine, rice, Arabidopsis, and maize) to characterize new plant gene functions. Whereas integrative approaches have been performed combining heterogeneous data in Arabidopsis (Bradford et al., 2010; Kourmpetis et al., 2011), Mutwil et al. (2011) included cross-species expression information to study gene functions in seven plant species. An important advantage of the module-based approach with respect to function prediction is that homologs are not required for a gene to receive a prediction. In agreement with a recent comparative transcriptomics study reporting conserved modules between maize and rice (Ficklin and Feltus, 2011), we observed that modules showing ultraconserved coexpression primarily cover genes that are related to energy and housekeeping functions, such as photosynthesis, ribosome biogenesis, and translation. However, the 910 modules showing significant coexpression in other angiosperms cover a broad range of biological processes and provide a valuable resource to identify new gene functions and translate biological information from model species to crops. Based on our module-based functional predictions, 5,562 Arabidopsis genes received a functional annotation, and an evaluation experiment showed that, based on a set of previously functionally unknown genes that were recently experimentally characterized, 38.1% of these gene functions could be inferred through the modules. Clearly, the annotation of genes of unknown function seems to benefit from the integration of coexpression conservation, as modules showing conserved coexpression recover almost two times more experimental GO-BP annotations compared with nonconserved modules. However, true-positive annotations could be found in nonconserved modules as well, thus not only providing support for these annotations but also suggesting that high-quality experimental data sets are important to study species-specific or adaptive gene functions. Overall, as a result of the integration of sequence and expression data for six plant species, the module-based predictions offer new biological hypotheses for currently functionally unknown genes in Arabidopsis (1,701 genes) and six other plant species (43,621 genes).
CONCLUSION
We have shown that there is a large discrepancy in the gene associations between the different experimental data sets, being GO, PPI, and TF targets and the AraNet resource, to delineate functional gene modules in Arabidopsis. Results of EC and motif analyses reveal that coexpression among module genes is most prevalent while coregulation and functional coherence is less widespread. By combining the number of modules with that of motifs, we showed that different processes exhibit a different regulatory complexity and that hub genes are enriched for essential genes involved in signaling between different processes. Finally, using module-based functional predictions, 5,562 genes were annotated, and an experiment evaluating recent gene annotations confirmed that a large fraction of the inferred functions are biologically valid. As a result, the presented Arabidopsis modules and their orthologous counterparts in other angiosperms provide an excellent starting point to study gene function and regulation in plants.
MATERIALS AND METHODS
Data Sets
Twelve expression data sets (abiotic stress conditions, biotic stress conditions, developmental stages, flowering tissue, genetic modification, hormone treatment, leaf tissue, root tissue, seed tissue, all stress conditions, whole plants, and AtGenExpress, as well as a general compendium) for Arabidopsis (Arabidopsis thaliana) were retrieved from the CORNET database in November 2010 (De Bodt et al., 2010). The expression data for soybean (Glycine max; 15,753 genes), Medicago truncatula (17,614 genes), poplar (Populus trichocarpa; 28,969 genes), grapevine (Vitis vinifera; 8,255 genes), rice (Oryza sativa ssp. japonica; 34,153 genes), and maize (Zea mays; 10,068 genes) were assembled from the National Center for Biotechnology Information Gene Expression Omnibus (Barrett et al., 2011). Cell intensity files were analyzed using a custom-made chip definition file (at least five probes per probe set) and normalized using the robust multiarray average method (Irizarry et al., 2003). A list of experiments for the different species is given in Supplemental Table S4. Redundant experiments were removed by clustering experiments over genes, and experiments with Pearson correlation coefficient (PCC) of 0.99 or greater were considered redundant. The number of retained experiments was 1,153, 43, 108, 39, 258, and 85 for soybean, M. truncatula, poplar, grapevine, rice, and maize, respectively. AraNet gene associations were retrieved in November 2010 (Lee et al., 2010). GO associations (Ashburner et al., 2000) for Arabidopsis genes were retrieved from the PLAZA 2.0 database in November 2010 (Proost et al., 2009). Genes assigned to a GO term were recursively assigned to all of the GO terms’ parental terms. Only gene-GO associations with nonelectronic evidence codes were taken into account for module delineation: EXP, IDA, IPI, IMP, IGI, IEP, IC, and TAS. The PPI data were downloaded from the CORNET database in November 2010 (De Bodt et al., 2010), and only experimentally identified PPIs were retained. Interaction data of TFs and their targets were retrieved from the AtRegNet database in November 2010 (Palaniswamy et al., 2006). The targets of each TF were divided based on the effect on their expression: activation, repression, and all (this group also contains the genes with unknown effect). Orthologous genes were identified using the integrative orthology method available from PLAZA 2.0 only retaining orthologs with the greatest evidence (Van Bel et al., 2012). The embryo-lethal genes were obtained from the SeedGenes database by selecting for confirmed embryo-defective genes (Meinke et al., 2008).
Module Delineation Using Expression- and Connectivity-Based Clustering
Both connectivity-based clustering and expression-based clustering were performed with a Perl implementation of the graph-based Cluster Affinity Search Technique (CAST) algorithm (Ben-Dor et al., 1999). Connectivity-based clustering was directly applied to the PPI and AraNet input gene associations and was optimized by selecting the threshold that maximized the largest number of genes assigned to modules and the number of modules with GO functional enrichment (PPI, 0.5; AraNet, 0.33).
Expression-based clustering was performed using a relative PCC threshold (95th percentile) based on a set of 10,000 random gene pairs specific to each expression compendium. Clustering was optimized for each set of genes (either a set of TF target genes or a set of genes with a common GO annotation) by prior selection of the CORNET expression compendium with the best EC for the given set of genes. The minimum and maximum clustering size was set at five and 100, respectively.
The GO seed genes were submitted to the MQSE approach prior to clustering. The MQSE approach adds new genes that show significant coexpression while also removing seed genes that do not coexpress coherently with the other seed genes. The decision of which genes to add and which genes to remove is based on a rank statistic that incorporates the number of coexpressed seed genes, the sd of the expression profile of the coexpressed seed genes, and the median rank toward all seed genes (Supplemental Protocol S1). The final expanded gene set is defined by selecting the top set of ranked genes yielding the highest significant enrichment toward seed genes. Subsequently, these expanded gene sets are clustered using CAST, after which only clusters with enrichment toward the initial seed genes are retained, to ensure retention of the initial functional category (hypergeometric distribution; P ≤ 0.05).
To identify and remove redundant modules within and across the different data types, the gene overlap between all modules was assessed using the Jaccard score. In cases where one module was completely embedded in the other, the overlap score was set at 1. Based on all pairwise overlap scores, modules were clustered by CAST using a score cutoff of 0.85. As a result, overlapping modules were grouped in a cluster of similar modules and the most highly connected module in each cluster was assigned as being the representative (i.e. the module with the highest average overlap in the cluster of similar modules).
In order to make the module information publicly available, an additional data Web site was developed (http://bioinformatics.psb.ugent.be/cig_data/plant_modules/). From the start page, all genes, modules, and GO categories from the module data set can be queried. Results include the modules and their genes, regulatory DNA motifs, comparative coexpression results, and visualizations of the modules based on either the comparative coexpression links or the input data gene associations. Bulk downloads are also available.
EC
The EC for a set of N genes was calculated as the fraction of all possible N × (N − 1)/2 gene pairs with a PCC higher than or equal to the threshold value defined for that compendium (Pilpel et al., 2001). The P value for an EC threshold of 10% in Arabidopsis modules was estimated at P ≤ 0.02 based on 960,000 random modules with a size distribution identical to the real data set.
Gene Functional Annotation
GO enrichment analysis was based on the same GO data set as for the module delineation (described in “Data Sets” above). Enrichment of a GO category in a module was calculated as the ratio of the module frequency over the genome-wide frequency. The enrichment values were validated statistically using the hypergeometric distribution and adjusted using false discovery rate (FDR) correction for multiple hypotheses testing (Storey and Tibshirani, 2003). The significance level was set at 0.01, and at least two genes in the cluster had to be associated with the GO label before a GO was assigned to a module. Due to this stringent threshold, some GO modules determined by MQSE lack enrichment in the final set of nonredundant modules. Enrichment toward embryo-lethal genes was performed similarly.
Motif Finding
De novo motifs were identified using MotifSampler (default settings) followed by MotifRanking (default settings; Thijs et al., 2002) and Weeder (default settings; Pavesi et al., 2001) for word sizes ranging from six to 12 on the 1-kbpromoter (sequence upstream of start codon, based on TAIR9), taking both strands into account. MotifSampler was run with a third-order background model based on all Arabidopsis promoters from PLAZA 2.0. Weeder motifs were transformed to position weight matrices (PWMs) based on their reported frequency matrix. Motif enrichment was determined for each motif based on genome-wide promoter mapping of their PWMs using MotifLocator (default settings; Thijs et al., 2002). Enrichment was defined as the ratio of the module frequency over the genome-wide frequency, and enrichment values were statistically evaluated using the hypergeometric distribution adjusted by the FDR correction for multiple hypothesis testing (Storey and Tibshirani, 2003). Only significantly enriched motifs with a corrected P ≤ 0.01 were retained. To determine motif representatives (and remove redundancy) within each module, motifs were clustered based on sequence similarity and gene-motif occurrences. To compare sequence similarity, motif PWMs were transformed into vectors, and for each pair of motifs, the PCC between the vectors was determined using a sliding window while retaining a minimum overlap of six nucleotides. Subsequently, the motifs were clustered using a PCC threshold of 0.75. The results of the sequence-based clustering were submitted to occurrence-based clustering, based on the method described by Xie et al. (2005). Based on these results, a set of nonredundant motifs was defined for each module, and motifs with a similar sequence but residing in a distinct set of genes, were considered as distinct motifs. Known motifs were extracted from AGRIS (Palaniswamy et al., 2006) and PLACE (Higo et al., 1999), and the redundancy was removed similarly as for the modules.
Motif conservation was determined by mapping the PWMs on the 1-kb promoters (both strands) of the different species with MotifLocator. For each species, backgrounds of the third order were built based on all 1-kb promoters (PLAZA 2.0). For each module, the enrichment was determined in each species based on the occurrences in the orthologous module and the genome-wide occurrences. P values for enrichment were calculated based on the hypergeometric distribution and corrected by FDR.
Motif annotation was performed by integrating the module functional annotation and the coexpression conservation. For each motif family, the motif instances across different modules were used to translate the functional annotation of the module to the motif family. Furthermore, each motif family annotation obtained in this manner was weighted by the expression conservation of the module. When multiple modules supported the association between GO and the motif family, the expression conservation was averaged over the different modules. The motif-GO network was created using Cytoscape (Shannon et al., 2003) and reduced by retaining the most specific GO nodes (and discarding related but less significant nodes).
Functional Prediction of Genes of Unknown Function
To validate module-based GO predictions, an updated GO gene association file was downloaded from TAIR on January 20, 2012. All associations with nonelectronic evidence tags that were created after the input data freeze were compared with the module-based predictions. Note that some new experimental gene associations were derived from publications prior to the data freeze. A prediction was called true positive if and only if the most specific common parent between the prediction and the new experimental association was more specific than any existing experimental GO-BP term. If the most specific common parent was a general term (GO:0008150, GO:0051704, GO:0009987, GO:0008152, GO:0044237, GO:0044238, GO:0050794, GO:0044260, GO:0043170, GO:0044249, GO:0050789, GO:0034645, GO:0010468, GO:0031326, GO:0010556, GO:0051171, GO:0009889, GO:0080090, GO:0019222, GO:0060255, GO:0065007, GO:0031323, GO:0009058, GO:0006139, GO:0009059, GO:0034641, GO:0044267), it was not considered a true positive. The different categories for a true-positive prediction listed in Table II are visualized in Supplemental Figures S6 and S7. The categories “unknown” and “unknown experimental BP” were the same from the perspective of the true-positive determination, as in both cases there were no existing GO-BP categories in the input data (only nonelectronic evidence GO-BPs were selected for input data). These scenarios are depicted in Supplemental Figure S6. The third category, “other experimental BP,” describes genes that had GO-BP annotations with experimental evidence codes but for which the true-positive prediction was not linked to the existing annotations (Supplemental Fig. S7). As such, the predictions were not a consequence of the existing nonelectronic annotations.
Genes that did not have GO-BP associations with nonelectronic evidence types in the updated GO association file were selected as currently unknown. The functional prediction was based on the enrichments for GO-BP categories and embryo-lethal genes. Orthologous genes without nonelectronic GO-BP associations were assigned the functional prediction of the Arabidopsis module if and only if these modules had a significant EC conservation as well as a significant EC in the respective species.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Number of modules per gene.
Supplemental Figure S2. Functional enrichment of hub genes, and regulatory complexity of different biological processes.
Supplemental Figure S3. Conservation of EC and motif enrichment across the green plant lineage.
Supplemental Figure S4. Motif-GO map based on coexpression conservation.
Supplemental Figure S5. GO slim overview of GO functional predictions for 3,553 genes currently without GO-BP.
Supplemental Figure S6. True-positive gene annotation prediction for AT1G73805.
Supplemental Figure S7. True-positive gene annotation prediction for AT1G70940.
Supplemental Table S1. Modules and genes discussed throughout the analyses.
Supplemental Table S2. Orthologous genes.
Supplemental Table S3. True-positive annotations.
Supplemental Table S4. Overview of the CEL files used for comparative expression analysis.
Supplemental Protocol S1. MQSE: optimizing a set of seed genes prior to clustering.
Supplementary Material
Acknowledgments
We thank Sara Movahedi for microarray normalization, Michiel Van Bel for technical assistance with the additional Web site, Stefanie de Bodt for critical reading of the manuscript and providing the CORNET data, Annick Bleys for help in preparing the manuscript, and Yves Van de Peer for general support.
Glossary
- PPI
protein-protein interaction
- TF
transcription factor
- ChIP
chromatin immunoprecipitation
- GO
Gene Ontology
- TAIR
The Arabidopsis Information Resource
- EC
expression coherence
- BP
Biological Process
- MQSE
multi-query seed expansion
- JA
jasmonic acid
- SA
salicylic acid
- MEM
multi-experiment matrix
- PCC
Pearson correlation coefficient
- CAST
Cluster Affinity Search Technique
- FDR
false discovery rate
- PWMs
position weight matrices
References
- Adler P, Kolde R, Kull M, Tkachenko A, Peterson H, Reimand J, Vilo J. (2009) Mining for coexpression across hundreds of datasets using novel rank aggregation and visualization methods. Genome Biol 10: R139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Ogata Y, Shibata D. (2007) Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol 48: 381–390 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana MV, Marín-de la Rosa N, Maloof JN, Blázquez MA, Alabadí D. (2011) Circadian oscillation of gibberellin signaling in Arabidopsis. Proc Natl Acad Sci USA 108: 9292–9297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000) Gene Ontology: tool for the unification of biology. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atias O, Chor B, Chamovitz DA. (2009) Large-scale analysis of Arabidopsis transcription reveals a basal co-regulation network. BMC Syst Biol 3: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabási A-L, Oltvai ZN. (2004) Network biology: understanding the cell’s functional organization. Nat Rev Genet 5: 101–113 [DOI] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, et al. (2011) NCBI GEO: archive for functional genomics data sets—10 years on. Nucleic Acids Res 39: D1005–D1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel GW, Glaab E, Marquez J, Holdsworth MJ, Bacardit J. (2011) Functional network construction in Arabidopsis using rule-based machine learning on large-scale data sets. Plant Cell 23: 3101–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Dor A, Shamir R, Yakhini Z. (1999) Clustering gene expression patterns. J Comput Biol 6: 281–297 [DOI] [PubMed] [Google Scholar]
- Berardini TZ, Mundodi S, Reiser L, Huala E, Garcia-Hernandez M, Zhang P, Mueller LA, Yoon J, Doyle A, Lander G, et al. (2004) Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol 135: 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boruc J, Van den Daele H, Hollunder J, Rombauts S, Mylle E, Hilson P, Inzé D, De Veylder L, Russinova E. (2010) Functional modules in the Arabidopsis core cell cycle binary protein-protein interaction network. Plant Cell 22: 1264–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford JR, Needham CJ, Tedder P, Care MA, Bulpitt AJ, Westhead DR. (2010) GO-At: in silico prediction of gene function in Arabidopsis thaliana by combining heterogeneous data. Plant J 61: 713–721 [DOI] [PubMed] [Google Scholar]
- Brady SM, Provart NJ. (2009) Web-queryable large-scale data sets for hypothesis generation in plant biology. Plant Cell 21: 1034–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D, Wightman R, Zhang Z, Gomez LD, Atanassov I, Bukowski J-P, Tryfona T, McQueen-Mason SJ, Dupree P, Turner S. (2011) Arabidopsis genes IRREGULAR XYLEM (IRX15) and IRX15L encode DUF579-containing proteins that are essential for normal xylan deposition in the secondary cell wall. Plant J 66: 401–413 [DOI] [PubMed] [Google Scholar]
- Busch W, Miotk A, Ariel FD, Zhao Z, Forner J, Daum G, Suzaki T, Schuster C, Schultheiss SJ, Leibfried A, et al. (2010) Transcriptional control of a plant stem cell niche. Dev Cell 18: 841–853 [DOI] [PubMed] [Google Scholar]
- Chao WS, Foley ME, Doğramacı M, Anderson JV, Horvath DP. (2011) Alternating temperature breaks dormancy in leafy spurge seeds and impacts signaling networks associated with HY5. Funct Integr Genomics 11: 637–649 [DOI] [PubMed] [Google Scholar]
- Cho J-H, Wang K, Galas DJ. (2011) An integrative approach to inferring biologically meaningful gene modules. BMC Syst Biol 5: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S, Carvajal D, Hollunder J, Van den Cruyce J, Movahedi S, Inzé D. (2010) CORNET: a user-friendly tool for data mining and integration. Plant Physiol 152: 1167–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S, Proost S, Vandepoele K, Rouzé P, Van de Peer Y. (2009) Predicting protein-protein interactions in Arabidopsis thaliana through integration of orthology, gene ontology and co-expression. BMC Genomics 10: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinas B, Mas P. (2011) Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J 66: 318–329 [DOI] [PubMed] [Google Scholar]
- Ferrier T, Matus JT, Jin J, Riechmann JL. (2011) Arabidopsis paves the way: genomic and network analyses in crops. Curr Opin Biotechnol 22: 260–270 [DOI] [PubMed] [Google Scholar]
- Ficklin SP, Feltus FA. (2011) Gene coexpression network alignment and conservation of gene modules between two grass species: maize and rice. Plant Physiol 156: 1244–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M, Rapaka L, Lyons E, Pedersen B, Thomas BC. (2007) G-boxes, bigfoot genes, and environmental response: characterization of intragenomic conserved noncoding sequences in Arabidopsis. Plant Cell 19: 1441–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikawa Y, Kato N. (2007) Split luciferase complementation assay to study protein-protein interactions in Arabidopsis protoplasts. Plant J 52: 185–195 [DOI] [PubMed] [Google Scholar]
- Geisler-Lee J, O’Toole N, Ammar R, Provart NJ, Millar AH, Geisler M. (2007) A predicted interactome for Arabidopsis. Plant Physiol 145: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y, Shinohara H, Kondo Y, Inoue A, Nakanomyo I, Ogawa M, Sawa S, Ohashi-Ito K, Matsubayashi Y, Fukuda H. (2008) Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc Natl Acad Sci USA 105: 15208–15213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan K, Jang C, Bailey-Serres J, Mittler R, Shelton C, Harper JF, Zhu J-K, Cushman JC, Gollery M, Girke T. (2008) Annotating genes of known and unknown function by large-scale coexpression analysis. Plant Physiol 147: 41–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Muiño JM, Jauregui R, Airoldi CA, Smaczniak C, Krajewski P, Angenent GC. (2009) Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol 7: e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Wellmer F, Muiño JM, Ferrier T, Wuest SE, Kumar V, Serrano-Mislata A, Madueño F, Krajewski P, Meyerowitz EM, et al. (2010) Orchestration of floral initiation by APETALA1. Science 328: 85–89 [DOI] [PubMed] [Google Scholar]
- Kourmpetis YAI, van Dijk ADJ, van Ham RCHJ, ter Braak CJF. (2011) Genome-wide computational function prediction of Arabidopsis proteins by integration of multiple data sources. Plant Physiol 155: 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Teng Q, Zhong R, Ye Z-H. (2011) The four Arabidopsis reduced wall acetylation genes are expressed in secondary wall-containing cells and required for the acetylation of xylan. Plant Cell Physiol 52: 1289–1301 [DOI] [PubMed] [Google Scholar]
- Lee I, Ambaru B, Thakkar P, Marcotte EM, Rhee SY. (2010) Rational association of genes with traits using a genome-scale gene network for Arabidopsis thaliana. Nat Biotechnol 28: 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW. (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverson JD, Huang HK, Forsburg SL, Hunter T. (2002) The Schizosaccharomyces pombe aurora-related kinase Ark1 interacts with the inner centromere protein Pic1 and mediates chromosome segregation and cytokinesis. Mol Biol Cell 13: 1132–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JF, Bush J, Xiong Y, Li L, McCormack M. (2011) Large-scale protein-protein interaction analysis in Arabidopsis mesophyll protoplasts by split firefly luciferase complementation. PLoS ONE 6: e27364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisso J, Steinhauser D, Altmann T, Kopka J, Müssig C. (2005) Identification of brassinosteroid-related genes by means of transcript co-response analyses. Nucleic Acids Res 33: 2685–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysenko A, Defoin-Platel M, Hassani-Pak K, Taubert J, Hodgman C, Rawlings CJ, Saqi M. (2011) Assessing the functional coherence of modules found in multiple-evidence networks from Arabidopsis. BMC Bioinformatics 12: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Bohnert HJ. (2007) Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression. Genome Biol 8: R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L, Van Hemert JL, Dash S, Dickerson JA. (2009) Arabidopsis gene co-expression network and its functional modules. BMC Bioinformatics 10: 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Yant LJ, Mürdter F, Küttner F, Schmid M. (2009) Repression of flowering by the miR172 target SMZ. PLoS Biol 7: e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke D, Muralla R, Sweeney C, Dickerman A. (2008) Identifying essential genes in Arabidopsis thaliana. Trends Plant Sci 13: 483–491 [DOI] [PubMed] [Google Scholar]
- Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, Hazen SP, Shen R, Priest HD, Sullivan CM, et al. (2008) Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet 4: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K, Grotewold E. (2009) A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet 5: e1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movahedi S, Van Bel M, Heyndrickx KS, Vandepoele K. (May 10, 2012) Comparative co-expression analysis in plant biology. Plant Cell Environ http://dx.doi.org/10.1111/j.1365-3040.2012.02517.x [DOI] [PubMed] [Google Scholar]
- Movahedi S, Van de Peer Y, Vandepoele K. (2011) Comparative network analysis reveals that tissue specificity and gene function are important factors influencing the mode of expression evolution in Arabidopsis and rice. Plant Physiol 156: 1316–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil M, Klie S, Tohge T, Giorgi FM, Wilkins O, Campbell MM, Fernie AR, Usadel B, Nikoloski Z, Persson S. (2011) PlaNet: combined sequence and expression comparisons across plant networks derived from seven species. Plant Cell 23: 895–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil M, Usadel B, Schütte M, Loraine A, Ebenhöh O, Persson S. (2010) Assembly of an interactive correlation network for the Arabidopsis genome using a novel heuristic clustering algorithm. Plant Physiol 152: 29–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaniswamy SK, James S, Sun H, Lamb RS, Davuluri RV, Grotewold E. (2006) AGRIS and AtRegNet: a platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant Physiol 140: 818–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi G, Mauri G, Pesole G. (2001) An algorithm for finding signals of unknown length in DNA sequences. Bioinformatics (Suppl 1) 17: S207–S214 [DOI] [PubMed] [Google Scholar]
- Persson S, Wei H, Milne J, Page GP, Somerville CR. (2005) Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA 102: 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilpel Y, Sudarsanam P, Church GM. (2001) Identifying regulatory networks by combinatorial analysis of promoter elements. Nat Genet 29: 153–159 [DOI] [PubMed] [Google Scholar]
- Proost S, Van Bel M, Sterck L, Billiau K, Van Parys T, Van de Peer Y, Vandepoele K. (2009) PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell 21: 3718–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quimbaya M, Vandepoele K, Raspé E, Matthijs M, Dhondt S, Beemster GT, Berx G, De Veylder L. (2012) Identification of putative cancer genes through data integration and comparative genomics between plants and humans. Cell Mol Life Sci 69: 2041–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JM, Segal E, Koller D, Kim SK. (2003) A gene-coexpression network for global discovery of conserved genetic modules. Science 302: 249–255 [DOI] [PubMed] [Google Scholar]
- Thibaud-Nissen F, Wu H, Richmond T, Redman JC, Johnson C, Green R, Arias J, Town CD. (2006) Development of Arabidopsis whole-genome microarrays and their application to the discovery of binding sites for the TGA2 transcription factor in salicylic acid-treated plants. Plant J 47: 152–162 [DOI] [PubMed] [Google Scholar]
- Thijs G, Marchal K, Lescot M, Rombauts S, De Moor B, Rouzé P, Moreau Y. (2002) A Gibbs sampling method to detect overrepresented motifs in the upstream regions of coexpressed genes. J Comput Biol 9: 447–464 [DOI] [PubMed] [Google Scholar]
- Tompa M, Li N, Bailey TL, Church GM, De Moor B, Eskin E, Favorov AV, Frith MC, Fu Y, Kent WJ, et al. (2005) Assessing computational tools for the discovery of transcription factor binding sites. Nat Biotechnol 23: 137–144 [DOI] [PubMed] [Google Scholar]
- Usadel B, Obayashi T, Mutwil M, Giorgi FM, Bassel GW, Tanimoto M, Chow A, Steinhauser D, Persson S, Provart NJ. (2009) Co-expression tools for plant biology: opportunities for hypothesis generation and caveats. Plant Cell Environ 32: 1633–1651 [DOI] [PubMed] [Google Scholar]
- Van Bel M, Proost S, Wischnitzki E, Movahedi S, Scheerlinck C, Van de Peer Y, Vandepoele K. (2012) Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol 158: 590–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Casneuf T, Van de Peer Y. (2006) Identification of novel regulatory modules in dicotyledonous plants using expression data and comparative genomics. Genome Biol 7: R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Quimbaya M, Casneuf T, De Veylder L, Van de Peer Y. (2009) Unraveling transcriptional control in Arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol 150: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J, Stals H, Eeckhout D, Persiau G, Van De Slijke E, Van Isterdael G, De Clercq A, Bonnet E, Laukens K, Remmerie N, et al. (2007) A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis thaliana. Mol Cell Proteomics 6: 1226–1238 [DOI] [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. (2010) The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38: W214–W220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Persson S, Mehta T, Srinivasasainagendra V, Chen L, Page GP, Somerville C, Loraine A. (2006) Transcriptional coordination of the metabolic network in Arabidopsis. Plant Physiol 142: 762–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe CJ, Kohane IS, Butte AJ. (2005) Systematic survey reveals general applicability of “guilt-by-association” within gene coexpression networks. BMC Bioinformatics 6: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. (2005) Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434: 338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, Zhiqiang L, Yunfei Z, Xiaoxiao W, Xiaoming Q, et al. (2006) The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol 60: 107–124 [DOI] [PubMed] [Google Scholar]
- Yant L, Mathieu J, Dinh TT, Ott F, Lanz C, Wollmann H, Chen X, Schmid M. (2010) Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22: 2156–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye Z-H. (2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20: 2763–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lee C, Zhong R, Ye Z-H. (2009) MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21: 248–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinman GE, Zhong S, Bar-Joseph Z. (2011) Biological interaction networks are conserved at the module level. BMC Syst Biol 5: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.