Abstract
Chloroplasts in angiosperms contain at least seven nucleus-encoded members of the DEAD box RNA helicase family. Phylogenetic analysis shows that five of these plastid members (RH22, -39, -47, -50, and -58) form a single clade and that RH3 forms a clade with two mitochondrial RH proteins (PMH1 and -2) functioning in intron splicing. The function of chloroplast RH3 in maize (Zea mays; ZmRH3) and Arabidopsis (Arabidopsis thaliana; AtRH3) was determined. ZmRH3 and AtRH3 are both under strong developmental control, and ZmRH3 abundance sharply peaked in the sink-source transition zone of developing maize leaves, coincident with the plastid biogenesis machinery. ZmRH3 coimmunoprecipitated with a specific set of plastid RNAs, including several group II introns, as well as pre23S and 23S ribosomal RNA (rRNA), but not 16S rRNA. Furthermore, ZmRH3 associated with 50S preribosome particles as well as nucleoids. AtRH3 null mutants are embryo lethal, whereas a weak allele (rh3-4) results in pale-green seedlings with defects in splicing of several group II introns and rRNA maturation as well as reduced levels of assembled ribosomes. These results provide strong evidence that RH3 functions in the splicing of group II introns and possibly also contributes to the assembly of the 50S ribosomal particle. Previously, we observed 5- to 10-fold up-regulation of AtRH3 in plastid Caseinolytic protease mutants. The results shown here indicate that AtRH3 up-regulation was not a direct consequence of reduced proteolysis but constituted a compensatory response at both RH3 transcript and protein levels to impaired chloroplast biogenesis; this response demonstrates that cross talk between the chloroplast and the nucleus is used to regulate RH3 levels.
DEAD box RNA helicases are mainly involved in ATP-dependent rearrangement of intermolecular and intramolecular RNA structures or remodeling of ribonucleoprotein complexes and are found in all eukaryotes and most prokaryotes (Cordin et al., 2006; Banroques et al., 2011; Linder and Jankowsky, 2011). DEAD box RNA helicases have been shown to be involved in RNA synthesis, modification, cleavage, and degradation as well as in ribosome biogenesis and translation initiation (Banroques et al., 2011; Linder and Jankowsky, 2011). DEAD box RNA helicase proteins possess conserved ATP-binding and -hydrolyzing domains, RNA-binding motifs, and a characteristic Asp-Glu-Ala-Asp (DEAD) sequence (as part of the Walker B motif; Cordin et al., 2006). However, in some cases, the Ala in the DEAD domain is not conserved (resulting into DExD). These conserved domains are clustered in the central core region that spans 350 to 400 amino acid residues. The N- and C-terminal extensions are highly variable and are thought to provide additional interactions with substrates or cofactors, or to confer additional activities such as protease activity (for NS3, see Cordin et al., 2006). DEAD box RNA helicase proteins belong to the helicase superfamily 2, which also includes DEAH and DExH proteins.
It was reported previously that there are 56 predicted DEAD box RNA helicases (and two pseudogenes) in Arabidopsis (Arabidopsis thaliana; Mingam et al., 2004), 11 of which have a TargetP-predicted chloroplast transit peptide. In this study and in the Plant Proteome Database (http://ppdb.tc.cornell.edu/), we adopt the numbering for the RNA helicases as established for Arabidopsis (Mingam et al., 2004). Mass spectrometry analyses identified six DEAD box helicases in Arabidopsis chloroplasts, namely RH3, RH22, RH26, RH39, RH47, and RH50. These plastid proteins were detected in high-molecular-mass (greater than 0.6–3 MD) chloroplast stromal complexes (Peltier et al., 2006; Olinares et al., 2010). Maize (Zea mays) homologs for these six plastid helicases were found by proteome analysis of maize plastids (Friso et al., 2010; Majeran et al., 2012). Most of these maize proteins were enriched in plastid nucleoids (Majeran et al., 2012) and also in Arabidopsis nucleoids (G. Friso, K. Nishimura, X. Qu, and K.J. van Wijk, unpublished data). The tobacco (Nicotiana tabacum) homolog of RH58, named VDL helicase, was identified in tobacco chloroplasts (Wang et al., 2000) but was not (yet) detected in Arabidopsis, likely because of its low abundance. The maize homolog for RH58 was found in a coimmunoprecipitation (co-IP) with anti-WHY1 serum against maize nucleoids (Majeran et al., 2012), but RH58 abundance levels were very low compared with the other plastid RH helicases.
Mutants for several chloroplast RNA helicases have been characterized to various degrees and exhibit delayed or impaired chloroplast and leaf development and defects in chloroplast RNA metabolism. OsBIRH1, the rice (Oryza sativa) homolog of Arabidopsis RH50, exhibited RNA helicase activities in vitro and helped to confer plant resistance against various stresses (Li et al., 2008). Arabidopsis mutant nara12-1, with reduced accumulation of RH39, showed strongly reduced levels of the photosynthesis machinery due to inefficient translation elongation, most likely due to the loss of RH39-mediated defects in 23S rRNA maturation (formation of the hidden break) affecting the functional state of the ribosome (Nishimura et al., 2010). Complete loss of Arabidopsis RH22 resulted into embryo lethality, whereas reduced RH22 expression resulted in virescent seedlings in which precursors of 23S and 4.5S rRNA (in the 50S particle) accumulated (Chi et al., 2012). It was concluded that RH22 facilitates the assembly of the 50S ribosomal subunit by interacting with ribosomal protein RPL24 and with the region of 23S rRNA encompassing the RPL24 binding site (Chi et al., 2012). Tobacco mutants with loss of the RH58 homolog (VDL helicase) showed pale-green and variegated leaves and abnormal roots and flowers, but no function was determined (Wang et al., 2000).
In this study, we focus on the molecular function of the chloroplast-localized DEAD box RNA helicase AtRH3 in Arabidopsis and its co-orthologs in maize, ZmRH3A and ZmRH3B. Proteome analysis showed that RH3 is the most abundant plastid DEAD box RNA helicase in maize and Arabidopsis (Zybailov et al., 2008; Friso et al., 2010). Our attention was drawn to RH3 for two reasons. First, we found that AtRH3 was strongly (more than 5-fold) up-regulated in the mutants clpr2-1 and clpr4-1, with defects in the chloroplast stroma-localized Caseinolytic protease (Clp) protease core complex (Rudella et al., 2006; Kim et al., 2009; Zybailov et al., 2009). These clpr2-1 and clpr4-1 mutants are chlorophyll-deficient and developmentally delayed mutants and have partial defects in chloroplast rRNA processing (Rudella et al., 2006; Kim et al., 2009; Zybailov et al., 2009), as observed for the Clp core mutant clpr1-1 (Koussevitzky et al., 2007). Second, we detected ZmRH3 in co-IP with several proteins that promote the splicing of chloroplast group II introns (G. Friso, K.P. Watkins, A. Barkan, and K.J. van Wijk, unpublished data). Here, we address the function of RH3 in both maize and Arabidopsis as well as the basis for its up-regulation in Arabidopsis plastid Clp protease mutants. Our results show that RH3 is directly involved in chloroplast intron splicing and possibly also in 50S ribosome biogenesis. RH3 levels peak during early stages of chloroplast biogenesis, consistent with a role in establishing the chloroplast gene expression machinery. Our results suggest that RH3 overaccumulates in Clp mutants not because it is a direct substrate of Clp protease but rather as a consequence of a disruption in chloroplast biogenesis or protein homeostasis. Finally, we use phylogeny and functional domain analysis of the Arabidopsis DEAD box RNA helicase family to compare RH3 with other plastid DEAD box RNA helicases.
RESULTS
Phylogenetic Analysis of Plant DEAD Box RNA Helicases and Architecture of RH3 Orthologs
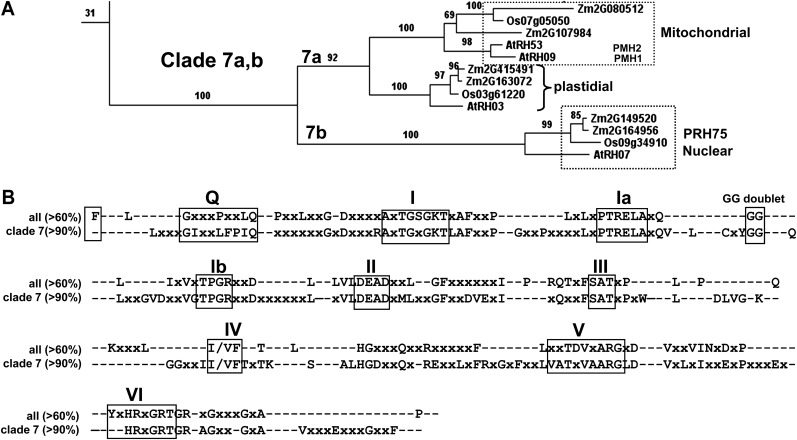
To establish the phylogenetic relationships among maize, rice, and Arabidopsis DEAD box proteins, we created unrooted phylogenetic trees based on the 56 Arabidopsis DEAD box helicases listed by Mingam et al. (2004), 46 related maize and 47 rice sequences, as well as the five known DEAD box RNA helicases from Escherichia coli (for accession numbers, annotations, and clade assignments, see Supplemental Table S1; for all amino acid sequences, see Supplemental Text S1). If there was more than one protein model per gene, we selected the longest protein. Phylogenetic trees were built from alignments based on full-length sequences as well as after removal of the variable N- and C-terminal extensions and removing gaps within the central conserved core helicase region. Sixteen clades could be distinguished for the plant proteins (using a minimal bootstrap value of 50 to define clades), with proteins for each of the three plant species represented in each clade (Supplemental Fig. S1; Supplemental Table S1). The relationship between protein accessions inferred from the various trees was not affected by the removal of gaps and extensions, indicating robustness of these relationships.
The cladogram showed that AtRH3 (At5g26742) has two co-orthologs in maize, ZmRH3A (GRMZM2G415491_P01)and ZmRH3B (GRMZM2G163072_P01), and one rice ortholog (Os03g61220; Fig. 1A; Supplemental Fig. S1). Furthermore, these RH3 orthologs formed a distinct clade (clade 7) with the closely related pair RH9/RH53 and the more distant RH7 (Fig. 1A; Supplemental Fig. S1). AtRH9 (AT3G22310) and AtRH53 (AT3G22330) are mitochondrial proteins (also named PMH1 and PMH2) and were shown to be part of a large complex in the mitochondrial matrix (Matthes et al., 2007). Moreover, PMH2 was shown to be involved in group II intron splicing in mitochondria (Köhler et al., 2010). In the same clade, but more distant, was AtRH7, a demonstrated nuclear protein named PRH75 but with unknown function (Lorković et al., 1997). Furthermore, five of the seven known plastid proteins (RH22, RH39, RH47, RH50, and RH58) formed one clade (clade 8), suggesting a common ancestry and likely functions different from RH3 (Supplemental Fig. S1).
Figure 1.
Phylogenetic and protein domain analyses of the DEAD box RNA helicase family. A, The RH3-containing clade (clade 7) from the phylogenetic tree of all 149 DEAD box helicases in maize, rice, and Arabidopsis and the five E. coli DEAD box RNA helicases (see Supplemental Fig. S1). Bootstrap values are indicated. B, Conserved domains of the maize, rice, and Arabidopsis DEAD box RNA helicase families. The top lines show the conserved motifs (60% minimal conservation threshold) across the 56 Arabidopsis, 46 maize, and 47 rice DEAD box RNA helicases. The bottom lines show the conserved motifs (greater than 90% identity) for the RH3 clade.
To understand the features that are unique to RH3 as compared with other plastid RH proteins, we analyzed the conservation of domains of all 56 Arabidopsis DEAD box RNA helicases and compared that with the conservation in the RH3 clade (Supplemental Table S1). Figure 1B shows the conserved motifs (greater than 60% identity) across all combined maize, rice, and Arabidopsis DEAD box helicases as well as the conserved motifs (greater than 90% identity) for the RH3 clade. The RH3 clade shows the conserved motifs Q, I, Ia, Ib, II, III, IV, V, and VI that are the hallmark for DEAD box RNA helicases (Aubourg et al., 1999; Cordin et al., 2006). Motifs Q, I (Walker A), and II (Walker B; DEAD) are for ATP binding, motif III is for ATP hydrolysis, motifs Ia, Ib, IV, and V are for RNA binding, and motif VI coordinates ATP hydrolysis and RNA unwinding (Cordin et al., 2006). Motif Ia also participates in structural rearrangements upon ATP binding/hydrolysis. An additional recognized motif with unknown function consisting of a single F residue at the beginning of the core domain (Cordin et al., 2006) was conserved (75% identity) across all RH helicases, except for RH3 and its mitochondrial homologs RH9 and RH53. In addition, the RH3 proteins each have a predicted N-terminal chloroplast-targeting sequence and GUCT and 70% Gly/Arg/Ser-enriched domains in the C terminus (Fig. 2A). The GUCT structure has an RNA recognition motif fold (Ohnishi et al., 2009), which is typically found in RNA-binding domains. The five plastid RH proteins in clade 8 (RH22, -39, -47, -50, and -58) lack several conserved motifs (Supplemental Table S1), whereas plastid RH3 and RH26 show full conservation of the DEAD box RNA helicase motifs, suggesting that RH3 and RH26 functions are distinct from the other plastid RH members.
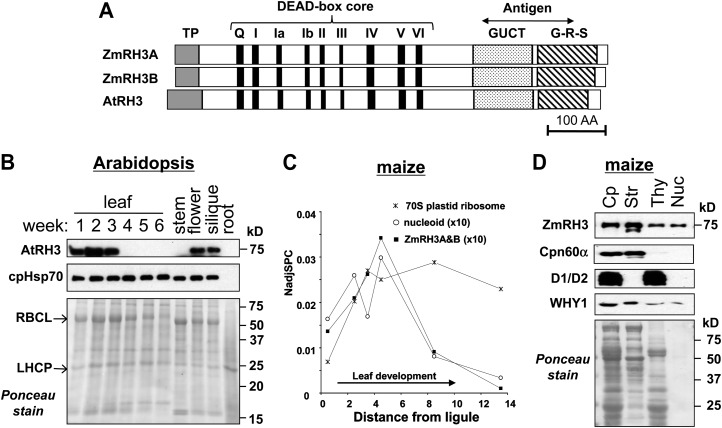
Figure 2.
RH3 proteins in maize and Arabidopsis. A, Domain organization of RH3 proteins. Two maize RH3 proteins, ZmRH3A (GRMZM2G415491_P01) and ZmRH3B (GRMZM2G163072_P01), and the AtRH3 ortholog (At5g26742) are shown. Predicted chloroplast transit peptides (TP) are shown in gray boxes. The conserved motifs in the DEAD box helicase family are indicated by black boxes and numbered (I–VI). The GUCT domain and the C-terminal G-R-S enriched domain are shaded. The region used to generate antisera is indicated. B, AtRH3 protein expresses at an early stage of development. Twenty micrograms of total proteins from green leaves at the stages indicated, or stem, flower, silique, and roots from 6-week-old Arabidopsis seedlings planted on soil, were analyzed by immunoblotting by anti-RH3 and anti-cpHSP70 antisera. The blot stained with Ponceau S is shown in the bottom panel. LHCP, Light-harvesting chlorophyll a/b-binding protein; RbcL, large subunit of Rubisco. C, ZmRH3 shows peak accumulation during the formation of chloroplasts in the sink-source transition zone of developing maize leaves. The plots are based on the identification and quantification of the RH3 homologs by mass spectrometry. For comparison, the sum of all identified plastid ribosomal proteins (53 proteins) and the sum of all identified nucleoid proteins (31 proteins) are shown. Assignment to the nucleoid and original proteomics data were from Majeran et al. (2012). NadjSPC (normalized adjusted spectral counts) is a measure of protein abundance. D, ZmRH3 proteins are localized to the chloroplast stroma, thylakoids, and nucleoids. Chloroplasts (Cp), stroma (Str), and thylakoid (Thy) were subfractionated and applied in equivalent proportions (10 μg of chlorophyll). One microgram of nucleoids (Nuc) from maize chloroplasts was applied. The blot stained with Ponceau S is shown in the bottom panel. CPN60α and D1/D2 were used as markers for the stroma and thylakoids, respectively. WHY1 is the control that localizes to stroma, thylakoids, and nucleoids.
RH3 Accumulates in Stroma and Nucleoids of Green Tissues, with Peak Accumulation during Chloroplast Biogenesis
To examine the temporal and organ expression patterns of AtRH3, we used an antibody raised to a conserved region of ZmRH3 (Fig. 2A and Supplemental Fig. S2). AtRH3 was found to be predominantly expressed in young leaves, peaking in 2-week-old seedlings and decreasing to very low levels in older leaf tissue (Fig. 2B). In addition, the AtRH3 protein was also detected in flowers and siliques, but not in stems or roots, of 6-week-old Arabidopsis plants. Mass spectrometry-based analysis of isolated nucleoids and chloroplasts showed a very strong nucleoid enrichment of ZmRH3 (Majeran et al., 2012) as well as AtRH3 (G. Friso, K. Nishimura, X. Qu, and K.J. van Wijk, unpublished data). Both ZmRH3 homologs were detected by mass spectrometry along the developmental leaf gradient of expanding maize leaves, with expression peaking in the sink-source transition zone (Fig. 2C), together with many other proteins involved in plastid gene expression and protein homeostasis (Majeran et al., 2010), including the nucleoid proteins. In initial accumulation profiles of chloroplast, ribosomal subunits followed ZmRH3 and nucleoids, but accumulation levels of ribosomes remained high and constant in mature tissue, unlike RH3 and (other) nucleoid proteins (Fig. 2C). This is consistent with their respective functions. Western-blot analysis showed that ZmRH3 and DNA/RNA-binding protein ZmWHY1 were present in chloroplast stroma as well as thylakoid membranes, likely reflecting nucleoids associated to the thylakoids (Fig. 2D). Indeed, western blots of nucleoids extracted from the thylakoid membranes identified both ZmRH3 and ZmWHY1 (Fig. 2D), in agreement with previous mass spectrometry analysis of nucleoids (Majeran et al., 2012). We note that the total amount of nucleoid proteins used for the western blot was lower than the other chloroplast fractions, thus underestimating the nucleoid enrichment for ZmRH3 and ZmWHY1. In contrast, the abundant CPN60α chaperone was only found in the stromal fraction (Fig. 2D).
ZmRH3 Associates with Group II Introns and Pre-50S Ribosomal Particles in Vivo
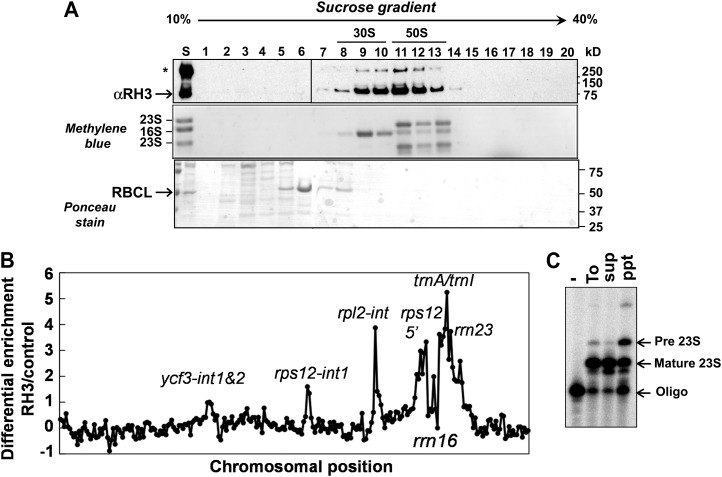
To determine the distribution of the particles with which ZmRH3 associates, maize chloroplast stroma was fractionated by Suc gradient sedimentation under conditions that dissociate 70S ribosomes into 30S and 50S ribosome particles. Immunoblots and RNA staining showed that RH3 primarily accumulated in particles sedimenting between 30S and 50S ribosomal subunits (1–2 MD), consistent with an association with partially assembled 50S ribosomal particles (Fig. 3A).
Figure 3.
RH3 cosediments with pre-50S ribosomal subunits and interacts with specific plastid RNA. A, Maize stroma were sedimented through Suc gradients under the condition that dissociates 30S and 50S subunits. An equal proportion of each fraction was analyzed by probing an immunoblot with anti-RH3 antiserum (top panel). The same blot stained with Ponceau S is shown in the bottom panel and also visualizes the large subunit (RBCL) of the Rubisco holocomplex at 550 kD. RNA extracted from gradients was analyzed by RNA gel blotting and methylene blue staining (middle panel). 16S rRNA marks 30S ribosomal subunits, and 23S rRNA marks 50S ribosomal subunits. B, Identification of RNA ligands of RH3 using the co-IP assay. The RIP-chip assay revealed the enrichment of several RNAs (as indicated) in the RH3 co-IP pellets. C, RH3 associates with the precursor of 23S rRNA determined by poisoned-primer extension assays. Reverse transcriptase reactions were initiated with a radiolabeled primer complementary to the mature and precursor forms of 23S rRNA; a dideoxy nucleotide that terminates the reverse transcription after different distances on mature and precursor forms of 23S was included in the reactions. Pre 23S, Precursor form of 23S rRNA. To, Total; sup, supernatant; ppt, pellet.
To identify chloroplast RNAs that associate with RH3 in vivo, RH3 was coimmunoprecipitated from maize chloroplast extracts. RNAs in the co-IP pellets were identified by hybridization to a tilling microarray of the maize chloroplast genome. Figure 3B shows the degree to which each chloroplast RNA sequence was enriched in the RH3 co-IP pellets, plotted according to chromosomal position, using the well-established RNA immunoprecipitation (RIP)-chip procedure as described by Barkan (2009). RNAs from five loci, rps12, rpl2, trnA, trnI, and rrn23, were strongly enriched in the RH3 co-IP pellet in comparison with a negative control assay. These loci encode two kinds of large and highly structured RNAs: group II introns in the rpl2, trnA, trnI, and rps12 loci and the 23S rRNA (rrn23). A minor enrichment of RNA from the ycf3 locus, which includes two group II introns, was also suggested by these data.
The rrn23, trnI, and tnrA genes are cotranscribed together with rrn16 in the chloroplast rrn operon. The primary transcript is processed via cleaving between coding regions, removal of group II introns in trnI and trnA, and trimming of the termini once ribosome assembly is complete (Stern et al., 2010). Suc gradient fractionation (Fig. 3A) suggested that ZmRH3 is associated primarily with assembling 50S ribosomal subunits (containing incompletely processed 23S rRNA) rather than with mature 50S subunits. To address that possibility, we used poisoned-primer extension to analyze the processing status of the 5′ end of the 23S rRNA in the RH3 co-IP pellets (Fig. 3C). This showed a strong enrichment for incompletely processed 23S rRNA in the RH3 co-IP pellet compared with the supernatant (Fig. 3C). These results, in conjunction with the Suc gradient sedimentation data, suggest that RH3 associates with assembling 50S ribosomal subunits and, potentially, with mature 50S ribosomal particles as well. The coprecipitation with assembling 50S ribosomal subunits could be direct, or it could be due to tethering to as-yet-unprocessed trnI and trnA, whose introns are associated with RH3. However, the cosedimentation of RH3 with pre-50S subunits favors a direct association, as group II intron particles analyzed to date are considerably smaller (approximately 700 kD) than those observed here (Till et al., 2001; Asakura and Barkan, 2007).
AtRH3 Is Required for Embryogenesis, Chloroplast Development, and Photosynthesis
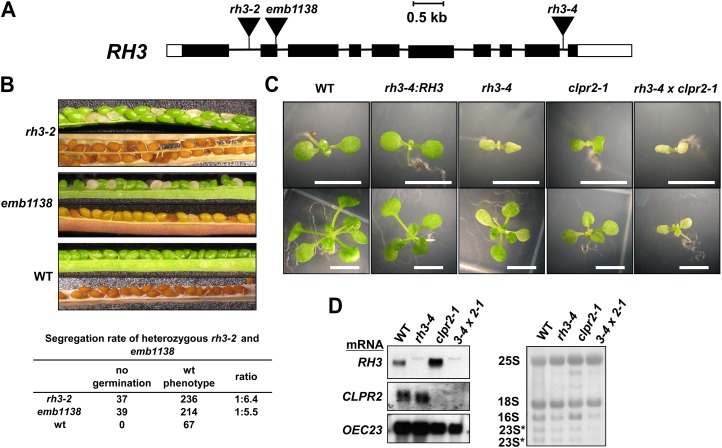
To further assess how RH3 affects the RNAs with which it associates, three T-DNA insertion lines in the single RH3 gene in Arabidopsis were obtained from the SALK and SAIL T-DNA insertion collections (Sessions et al., 2002; Alonso et al., 2003). We verified the positions of the insertions and found that rh3-2, emb1138, and rh3-4 mutants have T-DNA insertions in intron 2, exon 3, and intron 9, respectively (Fig. 4A).
Figure 4.
AtRH3 T-DNA insertion mutants and a double knockdown mutant with rh3-4/clpr2-1. A, T-DNA insertions in AtRH3. Exons and introns are indicated by black rectangles and lines, respectively. 5′ and 3′ untranslated regions are shown as white rectangles. The positions of the T-DNA insertions in emb1138, rh3-2 (SALK_025572), and rh3-4 (SALK_005920) are indicated by triangles. B, Embryo-lethal phenotypes associated with insertions in AtRH3. The green and mature siliques resulting from RH3-2/rh3-2 and Emb1138/emb1138 plants segregated approximately one-third white seeds and brown shriveled seeds, respectively. The shriveled seeds did not germinate. WT, Wild type. C, A rh3-4 homozygote showed a pale-green seedling phenotype, whereas the double knockdown mutant rh3-4/clpr2-1 showed an albino phenotype. Seedlings were grown for 9 d (top row) or 18 d (bottom row) on one-half-strength Murashige and Skoog medium containing 2% Suc. The rh3-4 has white cotyledons at early stages of development and exhibits pale-green true leaves. The double knockdown mutant rh3-4/clpr2-1 showed additive white seedlings. AtRH3 cDNA complemented the rh3-4 phenotype (rh3-4:RH3). Bars = 5 mm. D, RNA gel-blot analysis of RH3, CLPR2, and OEC23 mRNA levels in rh3-4, clpr2-1, and rh3-4/clpr2-1 mutants. Total RNA (6 μg) from leaf samples at leaf stage 1.07 planted on one-half-strength Murashige and Skoog medium with 2% Suc was analyzed by RNA gel blots using RH3, CLPR2, and OEC23 probes (left panels). The same blot stained with methylene blue is shown (right panels).
The different AtRH3 alleles conditioned distinct phenotypes. Self-pollination of heterozygous rh3-2 and emb1138 plants yielded developing siliques with green and white seeds in an approximately 3:1 ratio; these white seeds shriveled after seed maturation (Fig. 4B). We analyzed over 80 progeny of self-pollinated plants for each allele that were germinated on agar plates with Suc at low light intensity (20 µmol photons m−2 s−1) but recovered no homozygous mutants. These results strongly suggest that null mutants for RH3 are embryo lethal (Fig. 4B), consistent with the identification of emb1138 in a large-scale screen for embryo-lethal mutants (Tzafrir et al., 2004). In contrast, an insertion in intron 9 (rh3-4) resulted in seedlings with small chlorotic cotyledons and pale-green leaves with delayed growth (Fig. 4C). Homozygous rh3-4 mutants were able to grow on soil, set viable seeds, and be maintained as a homozygous population. RH3 mRNA was strongly reduced in rh3-4 homozygotes (Fig. 4D), whereas RH3 protein was reduced to approximately 20% of the wild-type level (Fig. 5A). The heteroallelic progeny of a complementation cross between rh3-4 and emb1138 germinated as albino seedlings, which was an intermediate phenotype with respect to those of the parental lines (Supplemental Fig. S3A). Moreover, the AtRH3 complementary DNA (cDNA) complemented the rh3-4 phenotype (Fig. 4C). Collectively, these results confirm that a reduction in RH3 function causes a pale-green seedling phenotype, whereas complete loss leads to embryo lethality.
Figure 5.
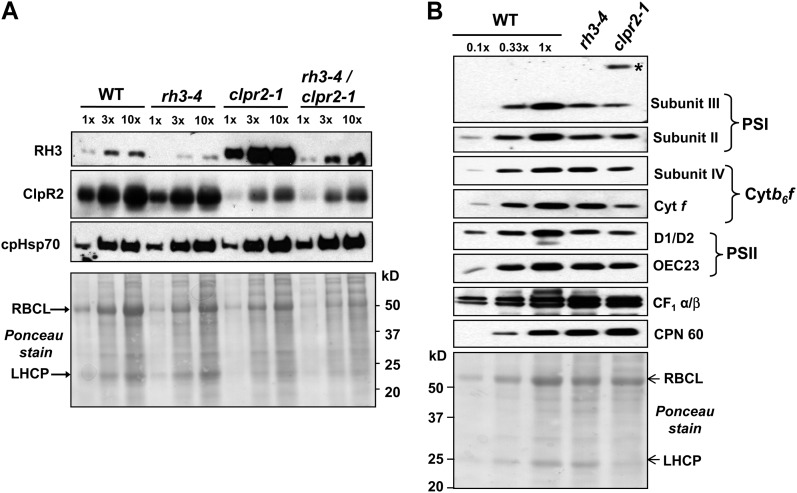
Protein expression patterns in pale-green mutants. A, RH3, ClpR2, and cpHSP70 protein levels in rh3-4, clpr2-1, and rh3-4/clpr2-1 mutants. Total leaf proteins (30 μg of protein or dilutions as indicated) from seedlings at leaf stage 1.07 were analyzed on immunoblots by probing with anti-RH3, anti-ClpR2, and anti-cpHSP70 antisera. The Ponceau S-stained membrane at the bottom shows sample loading and abundance. LHCP, Light-harvesting chlorophyll a/b-binding protein; RBCL, large subunit of Rubisco; WT, wild type. B, Immunoblot analysis of subunits of chloroplast protein complexes in rh3-4 and clpr2-1 mutants. Total leaf extracts (20 μg of protein or dilutions as indicated) from seedlings shown at the top at leaf stage 1.07 planted on one-half-strength Murashige and Skoog medium with 2% Suc were analyzed by immunoblotting probing with antibodies for the proteins named at the right. The blot stained with Ponceau S illustrates sample loading and the abundance of the large subunits of Rubisco.
Immunoblot analysis of total leaf extracts was used to quantify effects on the thylakoid-localized photosynthetic machinery in rh3-4 homozygotes (Fig. 5B). Accumulation of protein subunits of PSI (psaD/subunit II and psaF/subunit III), PSII (D1, D2, and OEC23), as well as the cytochrome b6f (cytb6f) complex (PetD/subunit IV and PetA/cytf) was 2- to 3-fold reduced in rh3-4. In contrast, the α- and β-subunits of the ATP synthase increased severalfold. For comparison, we also included an analysis of clpr2-1 mutant thylakoids. The reduction of PSI, PSII, and cytb6f subunits was 2- to 3-fold stronger than in rh3-4 mutants, consistent with the stronger phenotype of clpr2-1 (Rudella et al., 2006). clpr2-1 accumulated the precursor form of the PSI subunit III (PsaF), as observed previously (Rudella et al., 2006), but rh3-4 did not. Similar to rh4-1, ATP synthase levels increased in clpr2-1. This differential down-regulation was surprising to us, but verification of studies on other chloroplast biogenesis mutants showed that it is frequently observed that the thylakoid ATP synthase is less affected (or even increased), whereas the thylakoid electron transport chain complexes are down-regulated. For instance, the apo1 mutant involved in intron II splicing also shows unchanged levels of ATPase but very strong decreases of PSI, PSII, and the cytb6f complex (Watkins et al., 2011). Arabidopsis lpa2, with defects in PSII assembly, shows a decrease of PSII and PSI but not of ATP synthase (Ma et al., 2007). This shows that chloroplast and thylakoid biogenesis is a complex process. Chloroplast chaperone CPN60 was slightly (approximately 2-fold) up-regulated in both single mutants (Fig. 5B), similar to what we observed previously for clpr2-1 (Rudella et al., 2006).
AtRH3 Is Required for the Splicing of Group II Introns That Coimmunoprecipitated with ZmRH3
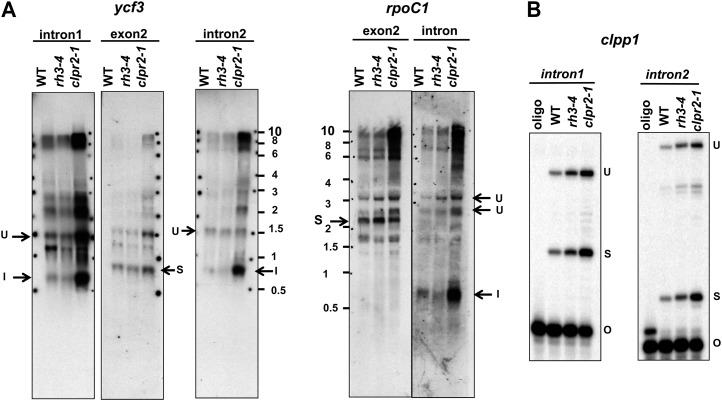
The splicing of each intron whose ortholog coimmunoprecipitated with ZmRH3 (Fig. 4B) was analyzed in the Arabidopsis rh3-4 mutant. For comparative purposes, we included Arabidopsis cfm2-2 mutants that have reduced levels of the plastid-localized splicing factor CFM2 (AT3G01370). cfm2-2 mutants are pale green and have chloroplast biogenesis defects resulting from defects in the splicing of several group II introns, but not in any of the RH3 intron ligands detected here (Asakura and Barkan, 2007). rh3-4 mutants accumulated markedly reduced levels of mature trnA and trnI but accumulated increased levels of their unspliced precursors (Fig. 6A; compare exon and intron levels), indicating a reduction in trnI and trnA splicing efficiency. Poisoned-primer extension assays revealed a clear decrease in the ratio of spliced to unspliced RNA for the trans-spliced intron rps12-int1 (Fig. 6B). The ratio of spliced to unspliced rpl2 and rps12-int2 was also reduced in rh3-4 mutants, but more modestly (Fig. 6B). The fact that effects on the splicing of trnI, trnA, rps12-int1, rps12-int2, and rpl2 were detected even in the weak rh3 mutant allele, in conjunction with the RNA co-IP data, strongly suggests that RH3 directly promotes the splicing of these introns. The splicing of three chloroplast introns that are found in Arabidopsis but not in maize (clpP-int1, clpP-int2, and rpoC1) and introns found in minor peaks in the maize RIP-chip experiments in Figure 3 (ycf3-int1 and ycf3-int2) was unaffected in rh3-4 (Fig. 7). These results suggest that RH3 does not contribute to the splicing of these introns, but analysis of stronger mutant alleles would be required to firmly address this point.
Figure 6.
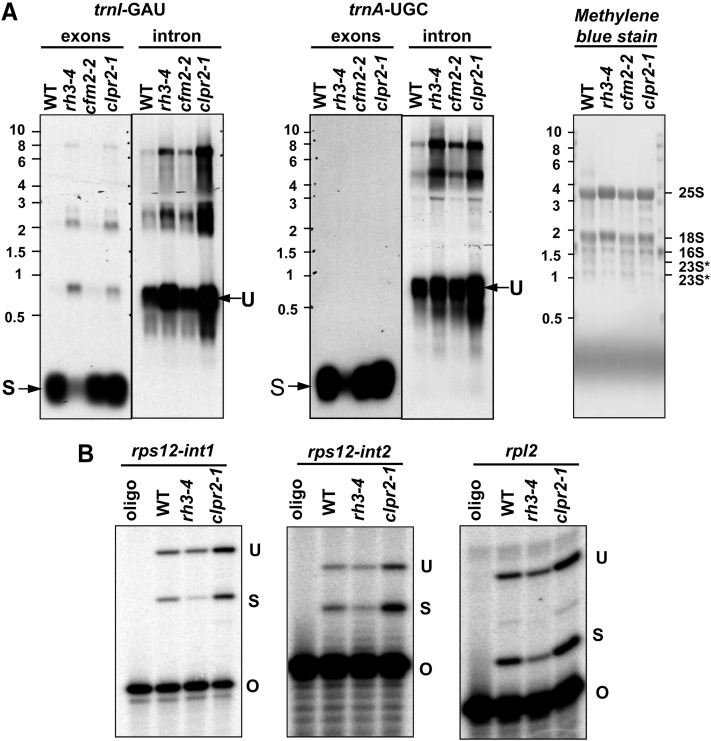
Chloroplast trnA, trnI, and rps12 splicing defects in rh3-4. A, Total leaf RNA (3 μg) from wild-type (WT), rh3-4, cfm2-2, and clpr2-1 seedlings at leaf stage 1.07 was probed with trnI and trnA intron and exon probes. The blot stained with methylene blue is also shown with cytosolic and plastid rRNAs marked. B, Chloroplast rps12-int1 splicing defects in rh3-4. Poisoned-primer extension assays monitored the splicing of rps12-int1, rps12-int2, and rpl2. Total leaf RNA (10 μg) of the wild type, rh3-4, and clpr2-1 was used in reverse transcription reactions using primers mapping several nucleotides downstream of the indicated intron. O, Oligonucleotide; S, spliced; U, unspliced.
Figure 7.
Analysis of ycf3 introns 1 and 2, rpoC intron, and ClpP1 intron 1 and 2 splicing in rh3-4. Total leaf RNA (3 μg) from the wild type (WT), rh3-4, and clpr2-1 planted on one-half-strength Murashige and Skoog medium with 2% Suc until leaf stage 1.07 was analyzed by RNA gel blots (A) or by poisoned-primer extension assays for ClpP intron 1 and intron 2 in rh3-4 (B). The Arabidopsis ClpP gene contains two group II introns, but these are not seen in monocots. I, Intron; O, oligonucleotide; S, spliced; U, unspliced.
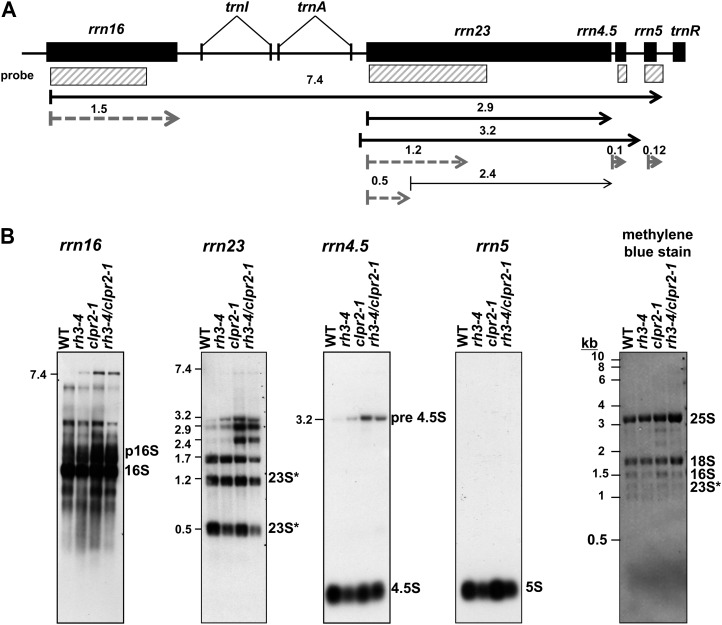
RH3 coimmunoprecipitates immature and mature 23S rRNA (Fig. 3C). This could indicate that RH3 also promotes the assembly of the 50S ribosomal subunit, a defect that is expected to be reflected by a decrease in 23S rRNA processing (for references and discussion, see Nishimura et al., 2010; Chi et al., 2012). To further address the role of RH3 in rRNA metabolism, we analyzed the four rRNAs by RNA gel-blot hybridization (Fig. 8A). We observed that the accumulation of each rRNA in the 50S ribosomal subunit (23S, 5S, and 4.5S) was slightly reduced in rh3-4 mutants, whereas 16S rRNA of the 30S particle was not affected.
Figure 8.
Defects of chloroplast ribosome biogenesis in the rh3-4 and clpr2-1 mutants. A, Chloroplast rRNA operon in Arabidopsis and probes. The rRNA probes used in B are illustrated as white rectangles. The transcripts shown with arrows are based on Bollenbach et al. (2005). Black and dashed lines represent transcripts that were accumulated and reduced in rh3-4, respectively. B, Total leaf RNA (2 μg) from wild-type (WT), rh3-4, clpr2-1, and rh3-4/clpr2-1 seedlings at leaf stage 1.07 planted on one-half-strength Murashige and Skoog medium with 2% Suc was probed with the rRNA probes illustrated as white rectangles in A. The blot stained with methylene blue is shown to illustrate equal loading of the cytosolic rRNAs (25S and 18S). 16S and 23S* bands are plastid rRNA (23S* is a 23S rRNA fragment). p16S corresponds to the 16S precursor.
RH3 and ClpR2 Interact Genetically, But RH3 Is Unlikely To Be a Substrate for the Clp Protease
RH3 was 5- to 10-fold up-regulated in the two ClpPR protease mutants clpr2-1 and clpr4-1 (Kim et al., 2009; Zybailov et al., 2009). This could suggest that RH3 is a substrate of the Clp protease, with the increased accumulation of RH3 due to reduced proteolytic capacity in the chloroplast. Alternatively, the increase in RH3 might be an indirect compensatory or pleiotropic response to the loss of Clp protease capacity. To explore the relationship between RH3 and ClpPR protease, a double homozygous mutant was generated between rh3-4 and clpr2-1 (Fig. 4C; Supplemental Fig. S4). The double-homozygous seedlings were albino and very slow growing; this phenotype was much stronger than the phenotype of either of its parents (Fig. 4C). However, viable seed could eventually be obtained from the homozygous rh3-4 × clpr2-1 double mutants (see “Materials and Methods”).
Immunoblot and RNA gel-blot analyses were carried out to compare levels of RH3 and ClpR2 mRNA (Fig. 4D) and protein (Fig. 5A) in rh3-4 and clpr2-1 single and double mutants. RH3 mRNA was reduced to near undetectable levels in both lines that were homozygous for rh3-4 but was increased severalfold in homozygous clpr2-1 mutants. CLPR2 mRNA levels were unaffected in rh3-4 mutants but strongly reduced in the clpr2-1 background (Fig. 4D). Consistent with our previous mass spectrometry-based analysis (Zybailov et al., 2009), RH3 protein abundance strongly increased (more than 10-fold) in clpr2-1 seedlings (Fig. 5A). Whereas RH3 protein was strongly reduced in the rh3-4 mutant, RH3 protein levels were approximately 2-fold increased in the double mutant as compared with the wild type, indicating that the reduced ClpR2 levels up-regulated RH3 even in the rh3-4 background. The ClpR2 protein level in clpr2-1 was about 20% of the wild type, as reported previously (Rudella et al., 2006), and remained at that level in the double mutant (Fig. 5A). Consistently, ClpR2 protein levels were not significantly affected in the rh3-4 seedlings. Chloroplast cpHSP70 levels were used as an internal control and were slightly increased in the single and double mutants (Fig. 5A).
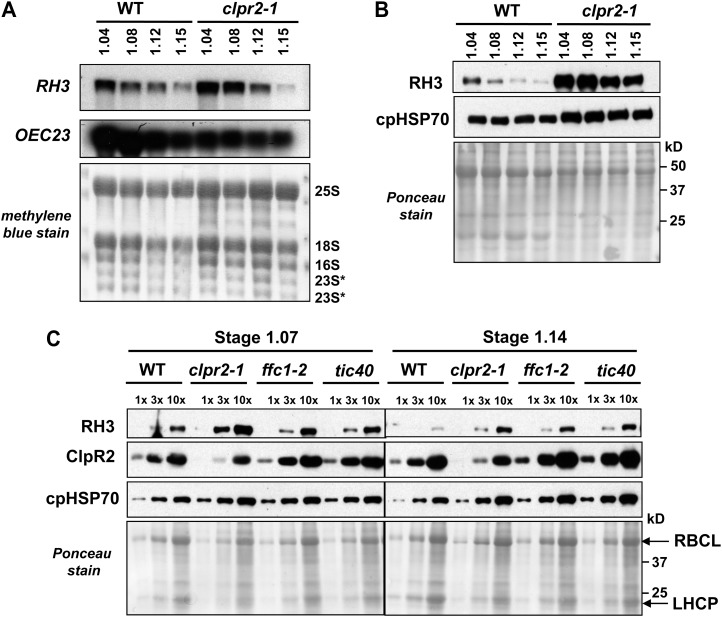
The expression of RH3 is under strong developmental control, with highest expression levels in young, developing leaf tissue (Fig. 2, B–D). To better resolve the developmental kinetics and gain insight into why RH3 protein overaccumulated in the clpr2-1 mutant, we determined RH3 mRNA and protein levels in different leaf rosette developmental stages (stages 1.04–1.15). RH3 mRNA levels decreased during the development of the leaf rosette in both the wild type and clpr2-1 (Fig. 9A); in the oldest rosette, transcript levels were identical in the wild type and clpr2-1. However, RH3 mRNA levels were severalfold higher in the clpr2-1 mutant during early leaf development (stages 1.04 and 1.08), indicating delayed transcriptional down-regulation. A similar but more dramatic pattern was observed at the RH3 protein level (Fig. 9B): RH3 levels clearly decreased with progressive leaf rosette development, but RH3 levels were consistently higher in the clpr2-1 mutant (Fig. 9B).
Figure 9.
RH3 protein and RNA expression patterns in clpr2-1 and the wild type during development. A, Total leaf RNA (6 μg) from the wild type (WT) and clpr2-1 at various developmental vegetative stages planted on soil was analyzed by RNA gel blot and probed with RH3 and OEC23 probes. The blot stained with methylene blue illustrates equal loading. B, Total leaf proteins (30 μg) from wild-type and clpr2-1 seedlings grown on soil at various developmental vegetative stages were tested for the accumulation of RH3 and cpHSP70. The blot stained with Ponceau S illustrates equal loading. C, Total leaf proteins (30 μg) from the wild type and various pale-green mutants (clpr2-1, ffc1-2, and tic40) grown on one-half-strength Murashige and Skoog medium with 2% Suc until leaf stage 1.07 or 1.14 were tested for the accumulation of RH3, ClpR2, and cpHSP70 using western blots. The bottom panel shows the blot stained with Ponceau S. LHCP, Light-harvesting chlorophyll a/b-binding protein; RBCL, large subunit of Rubisco.
To determine whether RH3 up-regulation is specific for ClpPR core protease mutants or is a general result of defects in chloroplast biogenesis, we determined RH3 protein levels for two other pale-green chloroplast mutants, ffc1-2 and tic40, at leaf stages 1.07 and 1.14 (Fig. 9C). ffc1-2 is deficient for chloroplast signal recognition particle subunit SRP54, involved in the targeting of various proteins to the thylakoid membrane (Rutschow et al., 2008). The mutant tic40 is deficient for the inner chloroplast envelope protein TIC40, involved in the import of nucleus-encoded proteins (Kovacheva et al., 2005). At leaf stage 1.07, RH3 levels increased about 2-fold in the ffc1-2 and tic40 mutants, compared with approximately 5-fold in clpr2-1. At leaf stage 1.14, RH3 levels in ffc1-2 and tic40 mutants increased to nearly the level as in the clpr2-1 mutant. Together with the increased RH3 mRNA levels, this indicates that the increase in RH3 levels is not specific to the ClpPR core mutants, even if up-regulation in the early stages of leaf development is stronger in clpr2-1.
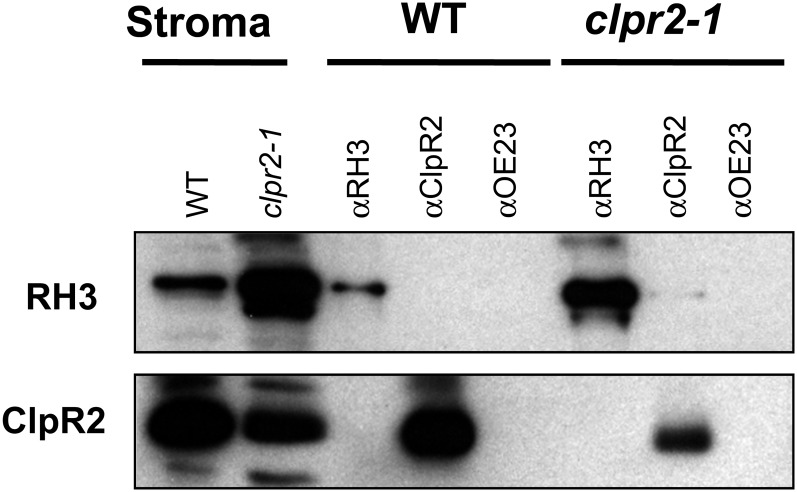
If RH3 were a substrate for degradation by the ClpPR core complex, RH3 or RH3 fragments might be found in association with the ClpPR complex. Therefore, we carried out co-IP experiments against stroma from isolated chloroplasts of wild-type and clpr2-1 plants using our anti-ClpR2 and anti-RH3 antisera. Whereas co-IP using anti-ClpR2 and anti-RH3 successfully precipitated ClpR2 and RH3, respectively, anti-ClpR2 did not coimmunoprecipitate with RH3, nor did anti-RH3 coimmunoprecipitate with ClpR2 (Fig. 10). Furthermore, fractionation of stromal extracts from Arabidopsis wild-type and clpr2-1 mutant plants on Suc gradients identified RH3 in high-molecular-mass fractions over 550 kD but never in 350-kD ClpPR complexes (Supplemental Fig. S5). Similarly, tandem mass spectrometry analysis of tagged ClpPR core complexes, such as His6-tagged ClpPR cores (Rudella et al., 2006) or StrepII-tagged ClpPR cores (Olinares et al., 2011), did not find RH3 protein, unlike other putative substrates (data not shown).
Figure 10.
RH3 and ClpR2 do not coimmunoprecipitate with each other. Arabidopsis stroma from the wild type (WT) or clpr2-1 was used for co-IP with anti-RH3, anti-ClpR2, or anti-OEC23 antiserum. Immunoblots of coimmunoprecipitated proteins were detected by anti-RH3 or anti-ClpR2 antiserum.
Comparison of RNA Metabolism in rh3-4 and clpr2-1 Mutants
So far, we determined that RH3 associates with and promotes the splicing of several group II introns (rps12-int1, rpl2-int, trnA, and trnI) and also coimmunoprecipitates with rrn23 (Fig. 3B). Moreover, we showed that mature forms of rrn4.5 and rrn5 as well as the 0.5-kb fragment of 23S rRNA accumulated at lower levels in rh3-4 (Fig. 8), suggesting a reduced accumulation of the assembled 50S ribosome particles. In contrast to rh3-4, the ratio of spliced to unspliced RNAs from the RH3-dependent introns is nearly normal in clpr2-1 mutants (Figs. 6 and 7). The metabolism of rRNAs also differed between the two mutants: various precursors overaccumulated in the clpr2-1 background, but the fully processed rRNAs accumulated to wild-type levels. Thus, the abundance of assembled 50S ribosomes was unaffected in the clpr2-1 mutant but was reduced in the rh3-4 mutant.
DISCUSSION
RH3 Is a Group II Intron-Splicing Factor in Plastids and May Also Contribute to 50S Ribosomal Subunit Assembly
In this study, we have shown that RH3 is a plastid-localized DEAD box RNA helicase in both maize and Arabidopsis that functions in the splicing of specific group II introns; RH3 is thus part of the network of plastid splice factors (Supplemental Fig. S6). This was based on a number of complementary measurements, namely (1) the interaction of RH3 with a subset of chloroplast type II introns (Fig. 3), and (2) specific splice defects in the rh3-1 mutant (Figs. 6 and 7). Furthermore, sequence analysis of RH3 clearly shows that RH3 contains the conserved domains for ATP-dependent RHA helicase activity and also has the conserved DEAD domain. Thus, our analysis clearly indicates that RH3 belongs to the class of DEAD box RNA helicase proteins. In contrast, some of the other chloroplast helicases do not have a strictly conserved DEAD domain (but have a DEXD domain), whereas others lack some of the key residues involved in ATP binding and hydrolysis. Strikingly, the role of RH3 in intron slicing is also consistent with the phylogenetic analysis of the RH3 family, in that RH3 is part of a clade with two closely related mitochondrial RH proteins (RH9 and RH53; Matthes et al., 2007), one of which (RH53) has been demonstrated to function in intron II splicing in mitochondria (Köhler et al., 2010).
Other results suggest that RH3 may have an additional function in the assembly of 50S ribosomal subunits. First, the abundance of mature 50S ribosomal subunits is reduced in rh3 mutants, as determined by the significant decrease in the accumulation of 23S, 4S, and 5S rRNA but not the 16S RNA of the 30S particle (Fig. 8). In addition, RH3 is found in particles of a suitable size (approximately 45S; Fig. 3A), and it coimmunoprecipitates with pre-23S rRNA (Fig. 3C). Such a role would be in keeping with the fact that other DEAD box proteins promote ribosome assembly in plastids and in other systems (Nishimura et al., 2010; Linder and Jankowsky, 2011; Chi et al., 2012). However, we cannot rule out the possibility that the decrease in ribosome abundance in rh3 mutants is a secondary effect of the splicing defects that impacts several components of the translation machinery (trnI, trnA, and rps12). The only viable rh3 allele was quite weak, and it is possible that a ribosome assembly defect would be revealed more clearly with a stronger (but not total) loss of function. The fact that RH3 is essential for embryogenesis and viability, based on the phenotypes of the two null RH3 alleles, is consistent with the need for functional plastid ribosomes and translation during embryogenesis in Arabidopsis (Stern et al., 2010).
RH3 Increase in Clp Protease Mutants Constitutes a Compensatory Effect and Must Involve Retrograde Signaling from the Plastid to the Nucleus
Both RH3 mRNA transcripts and protein levels of RH3 were severalfold increased in the clpr2-1 backgrounds. The strong increase in RH3 is not specific for clpr2-1 but applies also to pale-green mutants in plastid ClpPR genes, such as clpr4-1 (Kim et al., 2009) and clpp3 (J. Kim, P.D. Olinares, and K.J. van Wijk, unpublished data). Because these ClpPRs are subunits of the same Clp core protease complex, this shows that RH3 levels increase in response to the loss of ClpPR protease capacity. However, we did not find evidence that RH3 increases are directly due to the loss of degradation capacity; rather, our data suggest that the simultaneous increase in RH3 mRNA and protein constitutes a compensatory effect to the Clp mutant phenotype. The increase in RH3 mRNA level suggests a transcriptional response due to the loss of Clp core activity and must involve retrograde signaling from plastid to nucleus; the nature of this signal is still unclear (Sun et al., 2011), but many studies have indicated that inhibition of chloroplast homeostasis does contribute to the signal (Chan et al., 2010; Šimková et al., 2012). Indeed, multiple proteins involved in regulating chloroplast biogenesis and homeostasis, such as nucleoid-associated proteins (e.g. pTAC1/Why1), translation factors (e.g. EF-Tu-1, Ef-Tu-G/Sco1, and EF-Tu-TypeA/BipA), as well as the major chaperones (CPN60, HSP70, HSP90, and ClpB3), were up-regulated in the Clp mutants (Kim et al., 2009; Zybailov et al., 2009), indicative of significant protein stress.
The Difference between RH3 and Other Plastid-Localized DEAD Box Helicases
Out of the seven identified plastid-localized DEAD box RNA helicases (RH3, -22, -26, -39, -47, -50, and -58), only the functions of RH39 and RH22 were previously studied in detail in Arabidopsis (Nishimura et al., 2010; Chi et al., 2012). Our phylogenetic analysis showed that five of these plastid helicases (RH22, -39, -47, -50, and -58) form a separate clade, distinct from the clade with RH3 and its mitochondrial and nuclear homologs, and that RH26 is in a clade with extraplastidic proteins with unknown functions. The five plastid RH proteins in clade 8 lack several conserved motifs likely reducing RNA helicase activity, whereas plastid RH3 and RH26 show full conservation of the DEAD box RNA helicase motifs. Recently, we provided indirect evidence that RNA metabolism and ribosome assembly occur in association with nucleoids (Majeran et al., 2012). The identified plastid RH helicases were found in high-mass complexes (greater than 1 MD) in Arabidopsis stroma (Olinares et al., 2010) or present in maize nucleoids (Majeran et al., 2012), consistent with a role in RNA metabolism and ribosome assembly. Future experiments should focus on the identification of functions of plastid-localized RH26, -47, -50, and -58.
MATERIALS AND METHODS
Phylogenetic and Functional Domain Analysis
Selected proteins were aligned using MUSCLE (http://toolkit.tuebingen.mpg.de/muscle) using 15 to 25 iterations, and the exported aligned sequences in Clustal format were viewed and edited (removal of gaps and variable extensions) in Jalview. Sequences were then converted into PHYLIP format using http://searchlauncher.bcm.tmc.edu/seq-util/readseq.html. Phylogenetic trees were generated (1,000 iterations) using the CIPRES Web portal (http://www.phylo.org/sub_sections/portal/) using the tool RAxML HPC Blackbox, with the general time reversal model selected as the protein substitution matrix. The resulting phylogenetic trees were annotated in FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
Plant Materials and Growth Conditions
Maize (Zea mays inbred line B73) seedlings for stroma and cDNA preparations were grown in soil in a growth chamber under a 16-h-light (28°C, 400 µmol photons m−2 s−1) and 8-h-dark (26°C) cycle. They were harvested between 7 and 10 d after planting and used for preparation. Maize seedlings for chloroplast and nucleoid preparations were grown in a growth chamber under a 12-h-light (31°C, 400 µmol photons m−2 s−1) and 12-h-dark (22°C) cycle. They were harvested 7.5 d after planting for the nucleoid isolation or 8.5 d for chloroplast isolation.
Arabidopsis (Arabidopsis thaliana) T-DNA insertion lines SALK_025572 (rh3-2), SALK_005920 (rh3-4), and CS16011 (emb1138) were obtained from the Salk Institute Genomic Analysis Laboratory. emb1138, SALK_046378 (clpr2-1), SAIL_E67_05 (Atcfm2-2), and ffc1-2 mutants were described previously (Amin et al., 1999; Tzafrir et al., 2004; Asakura and Barkan, 2007; Rutschow et al., 2008). The tic40 mutant (SAIL_92_C10) was described previously (Kovacheva et al., 2005). Arabidopsis plants used for protein, DNA, and RNA extraction were grown on one-half-strength Murashige and Skoog plates supplemented with 2% Suc under a 10-h-light (40 µmol photons m−2 s−1, 23°C) and 14-h-dark (21°C) cycle unless otherwise indicated. Arabidopsis seedlings for stroma, protein, and RNA isolation for different stages and organs (wild type and clpr2-1) were grown on soil under a 10-h-light (100 µmol photons m−2 s−1, 23°C) and 14-h-dark (21°C) cycle and were harvested after 4 weeks (wild type) or 5 to 6 weeks (clpr2-1) after planting. Seeds from the double homozygous clpr2-1 × rh3-4 mutant was obtained by growing seedlings first for 2 months on agar plates supplemented with 2% Suc under short-day and low-light (60 µmol m−2 s−1) conditions followed by transfer to soil and growth for 6 months under long days at low light intensity (20 µmol photons m−2 s−1).
Chloroplast Isolation and Fractionation
Maize stroma for RIP-chip and Suc gradient were isolated as described previously (Voelker and Barkan, 1995). Maize chloroplasts for subfractionation and Arabidopsis stroma were isolated essentially as described (Majeran et al., 2005; Olinares et al., 2010). RNase and DNase treatments of thylakoid membranes were performed as described previously (Prikryl et al., 2008). Chloroplast maize nucleoids were isolated as described (Majeran et al., 2012).
Production of Maize Anti-RH3 and Arabidopsis Anti-ClpR2 Antisera
ZmRH3A (amino acids 531–691) and the C-terminal end of Arabidopsis ClpR2 (amino acids 189–279) were expressed in Escherichia coli as 6× His-tagged fusion proteins from pET-28b(+) or pET21a(+) vectors (Novagen), respectively. A ZmRH3A recombinant protein was expressed in BL21(DE3)Star (Novagen) and induced by the addition of 1 mm isopropyl-1-thio-β-d-galactopyranoside for 2 h. AtClpR2 recombinant protein was expressed in Rosetta (DE3) cells (Novagen) and introduced by the addition of 0.5 mm isopropyl-1-thio-β-d-galactopyranoside for 4 h. Both proteins were recovered in a soluble fraction, purified with nickel-nitrilotriacetic acid agarose beads (Qiagen) according to the manufacturer’s protocol, and equilibrated with phosphate-buffered saline. Polyclonal antisera from 10 mg of antigens were generated in rabbits at the University of Oregon antibody facility for anti-ZmRH3A or at Alfa Diagnostic International for anti-AtClpR2. Antisera were affinity purified against the same antigen that had been used for the immunizations coupled to a HiTrap N-hydroxysuccinimide (NHS) ester-activated column (GE Healthcare Life Science) or cyanogen bromide-activated Sepharose (Sigma).
Protein Extraction and Immunoblot Analysis
Arabidopsis seedlings from six to 10 plants at leaf stage 1.07, unless otherwise mentioned, were ground with a mortar and pestle in liquid nitrogen by adding an extraction buffer (2% SDS, 50 mm Tris-HCl, pH 8.8, and 5 mm EDTA) with protease inhibitors as described above and vortexed for 30 s to solubilize proteins. Insoluble materials were removed by centrifugation (0.8-mL column, 30 μm; Pierce) for 1 min at 10,000 rpm. Protein concentrations were determined by bicinchoninic acid assay (Smith et al., 1985). Samples were denatured in 3× or 2× Laemmli buffer (Laemmli, 1970) and heated at 75°C for 10 min. Proteins were resolved by SDS-PAGE followed by electroblotting to nitrocellulose membranes and then the membranes were stained with Ponceau S (0.3% Ponceau S, 3% TCA). Immunoblotting and enhanced chemiluminescence were performed as described previously (Barkan, 1998). The antisera against pea (Pisum sativum) CPN60α and spinach (Spinacia oleracea) cpHSP70 were kindly provided by Masato Nakai (Osaka University). The antiserum against barley (Hordeum vulgare) PsaF was provided by Hendrik Scheller (Royal Veterinary and Agricultural University, Denmark). The antisera against sorghum (Sorghum bicolor) OEC23 and OEC16, barley PsaD, and maize PetD and WHY1 were described previously (Voelker and Barkan, 1995; Prikryl et al., 2008).
RIP-chip and rrn23 Poisoned-Primer Extension Assays
RIP-chip assay of RNAs that coimmunoprecipitate with RH3 and OEC16 was performed as described previously (Schmitz-Linneweber et al., 2005). Each co-IP used affinity-purified antibodies and 100 µL of stromal extract (approximately 1 mg of stromal protein). A poisoned-primer assay to distinguish mature and precursor forms of 23S rRNA was performed as described previously (Asakura and Barkan, 2006).
Arabidopsis DNA Extraction and PCR Amplification
Arabidopsis DNA isolation was as described previously (Asakura and Barkan, 2006). T-DNA insertions were confirmed by PCR amplification. Primers are provided in Supplemental Table S2.
Arabidopsis RH3 Complementation
A cDNA clone of AtRH3 (stock no. U2189) was obtained from the Arabidopsis Biological Resource Center. A cDNA fragment of AtRH3 was amplified using platinum Pfx DNA polymerase (Invitrogen) with primers YAAtRH3_1-F(start) (5′-CACCATGGCGTCGACGGTAGGAGTT-3′) and YAAtRH3_2247-R(stop) (5′-CTAAAATCCTCTCTTATCAGGAC-3′) and subcloned into a pENTR/D-TOPO vector. The resulting pENTR-RH3 was cut with MluI gel purified, introduced into a binary pEAREYGATE100 vector (Earley et al., 2006) with LR Clonase II enzyme mix (Invitrogen) according to the manufacturer’s instruction, and electroporated into E. coli strain Top10. The resulting pEARLEGATE-AtRH3 clone was electroporated into Agrobacterium tumefaciens strain GV3101. Transformation of the homozygous rh3-4 was carried out using the floral dip method (Clough and Bent, 1998). Transformants were selected on soil with spraying three times with BASTA herbicide with a 3-d interval. T1 and T2 generations of transformants were assayed by PCR genotyping.
Arabidopsis RNA Analysis
RNA was extracted from the Arabidopsis leaf tissues by using Trizol reagent (Invitrogen) according to the manufacturer’s instruction. RNA gel-blot hybridizations and probes for chloroplast ribosomes were as described previously (Zybailov et al., 2009). Other probes were amplified by PCR and reverse transcription-PCR from Arabidopsis genomic DNA or RNA with the primers listed in Supplemental Table S2. Poisoned-primer extension assays were performed using 10 µg of leaf RNA as described previously (Asakura and Barkan, 2006).
Maize Stroma Suc Gradient to Dissociate 50S and 30S Ribosomes
A maize stroma Suc gradient was carried out as described with minor modifications (Barkan et al., 2007). A total of 100 μL of maize stroma (approximately 1 mg of protein) was diluted with 400 µL of dilution buffer (20 mm Tris-HCl, pH 7.8, 100 mm ammonium chloride, and 5 mm 2-mercaptoethanol) to reduce the magnesium concentration to 2 mm in maize stroma. A total of 500 µL of diluted stroma was layered onto a 10% to 40% Suc gradient in dissociation buffer (20 mm Tris-HCl, pH 7.8, 1 mm MgCl2, and 100 mm ammonium chloride) and was ultracentrifuged in a Beckman SW41 rotor at 35,000 rpm for 7 h at 4°C. RNA was extracted from 300 µL of each fraction by adding 50 µL of 5% SDS and 0.2 m EDTA, followed by phenol/chloroform extraction and ethanol precipitation. An equal proportion of each fraction was used for immunoblot and RNA gel-blot analyses.
Arabidopsis Stroma Suc Gradient
A total of 500 μL of Arabidopsis stroma (1 mg of protein) from the wild type and clpr2-1 with 120 units of RNasin (Promega) was fractionated by a 10% to 40% Suc gradient containing KEX buffer [30 mm HEPES-KOH, pH 8, 100 mm KOAc, 10 mm Mg(OAC)2,and 5 mm dithiothreitol] in a Beckman SW40 Ti rotor at 35,000 rpm for 3 h at 4°C. An equal proportion of each fraction was used for immunoblot analysis.
Co-IP
For co-IP for ClpR2 and RH3, 240 μL of Dynabeads protein A (Invitrogen) was washed three times with phosphate-buffered saline, pH 8.0, with 0.1% (v/v) Igepal CA-630. About 400 µL of stroma (390 µg mg−1) for one reaction from wild-type and clpr2-1 Arabidopsis was clarified by centrifugation at top spin for 10 min at 4°C. Twenty microliters of washed Dynabeads was added in the stroma and incubated for 30 min at 4°C, and beads were removed by a DynaMagnet (Invitrogen). Precleared stroma were split into three tubes to 400 µL, and 40 µL was saved as a total sample. Beads were divided into two 60-µL aliquots for wild-type and clpr2-1 stroma and washed once with co-IP buffer (20 mm Tris-HCl, pH 7.0, 150 mm NaCl, 1 mm EDTA, 0.2% Igepal CA-630, and 5 µg mL−1 aprotinin). Precleared stroma aliquots (approximately 400 µL) were incubated with anti-ZmRH3A-Dynabeads or anti-AtClpR2A-Dynabeads in 0.4 mL of co-IP buffer. Samples were incubated for more than 90 min at 4°C and then washed six times with co-IP buffer. Bound proteins were eluted with 40 µL of 1.5× Laemmli buffer without 2-mercaptoethanol at 5 min at 70°C or 95°C, and beads were removed by Dynamagnet. Before running SDS-PAGE gels, 4 µL of 2-mercaptoethanol was added to the samples, heated for 10 min at 75°C, and microfuged for 5 min to remove insoluble materials.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic tree of the RH3 DEAD box helicase family in Arabidopsis, maize, rice, and E. coli based on the core domains with major gaps removed.
Supplemental Figure S2.http://www.plantphysiol.org/cgi/content/full/pp.112.197525/DC1 Multiple sequence alignment of ZmRH3A, ZmRH3B, and AtRH3.
Supplemental Figure S3. An albino seedling phenotype of heteroallelic emb1138/rh3-4.
Supplemental Figure S4. PCR genotype of the double knockdown mutant rh3-4/clpr2-1.
Supplemental Figure S5. RH3 in the wild type and the clpr2-1 mutant is in the same fraction in Suc gradients.
Supplemental Figure S6.http://www.plantphysiol.org/cgi/content/full/pp.112.197525/DC1 RH3 functions and chloroplast splicing factors and their intron target in land plants (updated from Barkan, 2011).
Supplemental Table S1. Protein accession numbers, nomenclature, annotation, and clade assignments for the RH3 DEAD box helicase family in Arabidopsis, maize, and rice used for phylogenetic analysis.
Supplemental Table S2. Primer sequences used for the verification of Arabidopsis T-DNA insertions and RNA analysis.
Supplemental Text S1. All 154 DEAD box RNA helicase sequences (all with DEAD or DExD motifs) used for the phylogenetic analysis.
Supplementary Material
Acknowledgments
We thank Masato Nakai for providing the tic40 mutant and antibodies to CPN60α and cpHSP70 and Hendrik Scheller for antibody to PsaF. We also thank Rena Shimizu, Paul Dominic B. Olinares, Susan Belcher, Rosalind E. Williams-Carrier, Athea Vichas, and Giulia Friso for help.
Glossary
- co-IP
coimmunoprecipitation
- cytb6f
cytochrome b6f
- rRNA
to be defined
- T-DNA
to be defined
- cDNA
to be defined
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Amin P, Sy DA, Pilgrim ML, Parry DH, Nussaume L, Hoffman NE. (1999) Arabidopsis mutants lacking the 43- and 54-kilodalton subunits of the chloroplast signal recognition particle have distinct phenotypes. Plant Physiol 121: 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y, Barkan A. (2006) Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns. Plant Physiol 142: 1656–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y, Barkan A. (2007) A CRM domain protein functions dually in group I and group II intron splicing in land plant chloroplasts. Plant Cell 19: 3864–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubourg S, Kreis M, Lecharny A. (1999) The DEAD box RNA helicase family in Arabidopsis thaliana. Nucleic Acids Res 27: 628–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banroques J, Cordin O, Doère M, Linder P, Tanner NK. (2011) Analyses of the functional regions of DEAD-box RNA “helicases” with deletion and chimera constructs tested in vivo and in vitro. J Mol Biol 413: 451–472 [DOI] [PubMed] [Google Scholar]
- Barkan A. (1998) Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol 297: 38–57 [Google Scholar]
- Barkan A. (2009) Genome-wide analysis of RNA-protein interactions in plants. Methods Mol Biol 553: 13–37 [DOI] [PubMed] [Google Scholar]
- Barkan A. (2011) Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol 155: 1520–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Klipcan L, Ostersetzer O, Kawamura T, Asakura Y, Watkins KP. (2007) The CRM domain: an RNA binding module derived from an ancient ribosome-associated protein. RNA 13: 55–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollenbach TJ, Lange H, Gutierrez R, Erhardt M, Stern DB, Gagliardi D. (2005) RNR1, a 3′-5′ exoribonuclease belonging to the RNR superfamily, catalyzes 3′ maturation of chloroplast ribosomal RNAs in Arabidopsis thaliana. Nucleic Acids Res 33: 2751–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KX, Crisp PA, Estavillo GM, Pogson BJ. (2010) Chloroplast-to-nucleus communication: current knowledge, experimental strategies and relationship to drought stress signaling. Plant Signal Behav 5: 1575–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W, He B, Mao J, Li Q, Ma J, Ji D, Zou M, Zhang L. (2012) The function of RH22, a DEAD RNA helicase, in the biogenesis of the 50S ribosomal subunits of Arabidopsis chloroplasts. Plant Physiol 158: 693–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cordin O, Banroques J, Tanner NK, Linder P. (2006) The DEAD-box protein family of RNA helicases. Gene 367: 17–37 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Friso G, Majeran W, Huang M, Sun Q, van Wijk KJ. (2010) Reconstruction of metabolic pathways, protein expression, and homeostasis machineries across maize bundle sheath and mesophyll chloroplasts: large-scale quantitative proteomics using the first maize genome assembly. Plant Physiol 152: 1219–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Rudella A, Ramirez Rodriguez V, Zybailov B, Olinares PD, van Wijk KJ. (2009) Subunits of the plastid ClpPR protease complex have differential contributions to embryogenesis, plastid biogenesis, and plant development in Arabidopsis. Plant Cell 21: 1669–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler D, Schmidt-Gattung S, Binder S. (2010) The DEAD-box protein PMH2 is required for efficient group II intron splicing in mitochondria of Arabidopsis thaliana. Plant Mol Biol 72: 459–467 [DOI] [PubMed] [Google Scholar]
- Koussevitzky S, Stanne TM, Peto CA, Giap T, Sjögren LL, Zhao Y, Clarke AK, Chory J. (2007) An Arabidopsis thaliana virescent mutant reveals a role for ClpR1 in plastid development. Plant Mol Biol 63: 85–96 [DOI] [PubMed] [Google Scholar]
- Kovacheva S, Bédard J, Patel R, Dudley P, Twell D, Ríos G, Koncz C, Jarvis P. (2005) In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J 41: 412–428 [DOI] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Li DY, Liu HZ, Zhang HJ, Wang XE, Song FM. (2008) OsBIRH1, a DEAD-box RNA helicase with functions in modulating defence responses against pathogen infection and oxidative stress. J Exp Bot 59: 2133–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P, Jankowsky E. (2011) From unwinding to clamping: the DEAD box RNA helicase family. Nat Rev Mol Cell Biol 12: 505–516 [DOI] [PubMed] [Google Scholar]
- Lorković ZJ, Herrmann RG, Oelmüller R. (1997) PRH75, a new nucleus-localized member of the DEAD-box protein family from higher plants. Mol Cell Biol 17: 2257–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Peng L, Guo J, Lu Q, Lu C, Zhang L. (2007) LPA2 is required for efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 19: 1980–1993 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Majeran W, Cai Y, Sun Q, van Wijk KJ. (2005) Functional differentiation of bundle sheath and mesophyll maize chloroplasts determined by comparative proteomics. Plant Cell 17: 3111–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, Friso G, Asakura Y, Qu X, Huang M, Ponnala L, Watkins KP, Barkan A, van Wijk KJ. (2012) Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: a new conceptual framework for nucleoid functions. Plant Physiol 158: 156–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, Friso G, Ponnala L, Connolly B, Huang M, Reidel E, Zhang C, Asakura Y, Bhuiyan NH, Sun Q, et al. (2010) Structural and metabolic transitions of C4 leaf development and differentiation defined by microscopy and quantitative proteomics in maize. Plant Cell 22: 3509–3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthes A, Schmidt-Gattung S, Köhler D, Forner J, Wildum S, Raabe M, Urlaub H, Binder S. (2007) Two DEAD-box proteins may be part of RNA-dependent high-molecular-mass protein complexes in Arabidopsis mitochondria. Plant Physiol 145: 1637–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingam A, Toffano-Nioche C, Brunaud V, Boudet N, Kreis M, Lecharny A. (2004) DEAD-box RNA helicases in Arabidopsis thaliana: establishing a link between quantitative expression, gene structure and evolution of a family of genes. Plant Biotechnol J 2: 401–415 [DOI] [PubMed] [Google Scholar]
- Nishimura K, Ashida H, Ogawa T, Yokota A. (2010) A DEAD box protein is required for formation of a hidden break in Arabidopsis chloroplast 23S rRNA. Plant J 63: 766–777 [DOI] [PubMed] [Google Scholar]
- Ohnishi S, Pääkkönen K, Koshiba S, Tochio N, Sato M, Kobayashi N, Harada T, Watanabe S, Muto Y, Güntert P, et al. (2009) Solution structure of the GUCT domain from human RNA helicase II/Gu beta reveals the RRM fold, but implausible RNA interactions. Proteins 74: 133–144 [DOI] [PubMed] [Google Scholar]
- Olinares PD, Kim J, Davis JI, van Wijk KJ. (2011) Subunit stoichiometry, evolution, and functional implications of an asymmetric plant plastid ClpP/R protease complex in Arabidopsis. Plant Cell 23: 2348–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olinares PD, Ponnala L, van Wijk KJ. (2010) Megadalton complexes in the chloroplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry, and hierarchical clustering. Mol Cell Proteomics 9: 1594–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier JB, Cai Y, Sun Q, Zabrouskov V, Giacomelli L, Rudella A, Ytterberg AJ, Rutschow H, van Wijk KJ. (2006) The oligomeric stromal proteome of Arabidopsis thaliana chloroplasts. Mol Cell Proteomics 5: 114–133 [DOI] [PubMed] [Google Scholar]
- Prikryl J, Watkins KP, Friso G, van Wijk KJ, Barkan A. (2008) A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res 36: 5152–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudella A, Friso G, Alonso JM, Ecker JR, van Wijk KJ. (2006) Downregulation of ClpR2 leads to reduced accumulation of the ClpPRS protease complex and defects in chloroplast biogenesis in Arabidopsis. Plant Cell 18: 1704–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutschow H, Ytterberg AJ, Friso G, Nilsson R, van Wijk KJ. (2008) Quantitative proteomics of a chloroplast SRP54 sorting mutant and its genetic interactions with CLPC1 in Arabidopsis. Plant Physiol 148: 156–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Williams-Carrier R, Barkan A. (2005) RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell 17: 2791–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šimková K, Kim C, Gacek K, Baruah A, Laloi C, Apel K. (2012) The chloroplast division mutant caa33 of Arabidopsis thaliana reveals the crucial impact of chloroplast homeostasis on stress acclimation and retrograde plastid-to-nucleus signaling. Plant J 69: 701–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. (1985) Measurement of protein using bicinchoninic acid. Anal Biochem 150: 76–85 [DOI] [PubMed] [Google Scholar]
- Stern DB, Goldschmidt-Clermont M, Hanson MR. (2010) Chloroplast RNA metabolism. Annu Rev Plant Biol 61: 125–155 [DOI] [PubMed] [Google Scholar]
- Sun X, Feng P, Xu X, Guo H, Ma J, Chi W, Lin R, Lu C, Zhang L. (2011) A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat Commun 2: 1–10 [DOI] [PubMed] [Google Scholar]
- Till B, Schmitz-Linneweber C, Williams-Carrier R, Barkan A. (2001) CRS1 is a novel group II intron splicing factor that was derived from a domain of ancient origin. RNA 7: 1227–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, et al. (2004) Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135: 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker R, Barkan A. (1995) Nuclear genes required for post-translational steps in the biogenesis of the chloroplast cytochrome b6f complex in maize. Mol Gen Genet 249: 507–514 [DOI] [PubMed] [Google Scholar]
- Wang Y, Duby G, Purnelle B, Boutry M. (2000) Tobacco VDL gene encodes a plastid DEAD box RNA helicase and is involved in chloroplast differentiation and plant morphogenesis. Plant Cell 12: 2129–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins KP, Rojas M, Friso G, van Wijk KJ, Meurer J, Barkan A. (2011) APO1 promotes the splicing of chloroplast group II introns and harbors a plant-specific zinc-dependent RNA binding domain. Plant Cell 23: 1082–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B, Friso G, Kim J, Rudella A, Rodríguez VR, Asakura Y, Sun Q, van Wijk KJ. (2009) Large scale comparative proteomics of a chloroplast Clp protease mutant reveals folding stress, altered protein homeostasis, and feedback regulation of metabolism. Mol Cell Proteomics 8: 1789–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ. (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 3: e1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.