Abstract
Methylation of histone H3 at lysine 4 (H3K4) is a conserved feature of active chromatin catalyzed by methyltransferases of the SET1-family (SET1A, SET1B, MLL1, MLL2, MLL3 and MLL4 in humans). These enzymes participate in diverse gene regulatory networks with a multitude of known biological functions, including direct involvement in several human disease states. Unlike most lysine methyltransferases, SET1-family enzymes are only fully active in the context of a multi-subunit complex, which includes a protein module comprised of WDR5, RbBP5, ASH2L and DPY-30 (WRAD). These proteins bind in close proximity to the catalytic SET domain of SET1-family enzymes and stimulate H3K4 methyltransferase activity. The mechanism by which WRAD promotes catalysis involves elements of allosteric control and possibly the utilization of a second H3K4 methyltransferase active site present within WRAD itself. WRAD components also engage in physical interactions that recruit SET1-family proteins to target sites on chromatin. Here, the known molecular mechanisms through which WRAD enables the function of SET1-related enzymes will be reviewed.
Keywords: SET1, MLL, WDR5, RbBP5, ASH2L, DPY-30
INTRODUCION
Methylation of the epsilon amino group of lysine residues is a major regulatory mechanism influencing chromatin structure and function, catalyzed by the S-adenosylmethionine (SAM)-dependent lysine methyltransferase superfamily of enzymes [1]. Among the substrates of lysine methylation identified to date, core histones H3 and H4 are the most well studied, where specific sites of methylation can modulate local protein:protein interactions occurring on the chromatin fiber. In most instances, regulation by lysine methylation is mediated by a class of proteins possessing methyl-lysine recognition domains (e.g. chromodomains and PHD domains), also known as chromatin readers, which associate with lysine residues preferentially in their methylated or unmmethylated states [2]. Individual lysine methyltransferases are highly substrate specific and product specific (in performing mono-, di- and/or tri-methylation), which affords diverse regulatory functions within this class of enzymes. Indeed, lysine methyltransferases regulate a multitude of nuclear processes, including heterochromatin formation, transcription, DNA replication, and DNA repair [1].
Methylation of histone H3 at lysine 4 (H3K4) is well-studied modification linked to chromatin in its active or poised state [3, 4]. An early study from the Allis laboratory found H3K4 methylation to be highly enriched within the transcriptionally active macronucleus relative to the transcriptionally inert micronucleus of the ciliated protozoan Tetrahymena [5]. This early observation laid the groundwork for a large body of subsequent studies across diverse species, correlating the presence of H3K4 methylation with transcriptional activity [e.g. 6, 7]. Studies mapping histone lysine methylation across eukaryotic genomes have universally identified H3K4me3 as strongly enriched near the transcriptional start site of active genes, whereas H3K4me2 and H3K4me1 are found more broadly across active genes and at enhancer elements [6–9]. Several chromatin readers are known to translate the presence of H3K4 methylation into positive effects on transcriptional activity, including CHD1, BPTF and TAF3 [10–12].
The SET1 gene in the budding yeast Saccharomyces cerevisiae was discovered by several laboratories as encoding the first known H3K4 methyltransferase, a catalytic function performed through its conserved SET domain [13–16]. Since Set1 is the only H3K4 methyltransferase in this organism, a SET1-null yeast strain completely lacks all H3K4me1, me2 and me3 in bulk chromatin, resulting in a host of pleoiotropic phenotypes, including defects in growth and defects in telomeric chromatin structure [13, 14, 16]. Yeast Set1, like its orthologs in higher eukaryotes (SET1A/B in humans), is recruited to chromatin by the serine 5-phosphorylated C-terminal domain of RNA polymerase II (pol II) [8, 17–20]. This interaction with the initiating form of pol II results in the occupancy of Set1 near the 5′-end of active genes, which correlates closely with the peak of H3K4me3 [8, 17–20]. In metazoans, SET1A/B proteins seem to function analogously to yeast Set1 in performing the majority of transcription-coupled H3K4 methylation found at active genes, presumably through co-transcriptional recruitment [18–21]. Unlike their yeast counterpart, human SET1A/B proteins are also recruited to chromatin via an interaction with non-methylated CpG island sequences, mediated by the SET1A/B-associated protein CFP1 [22, 23].
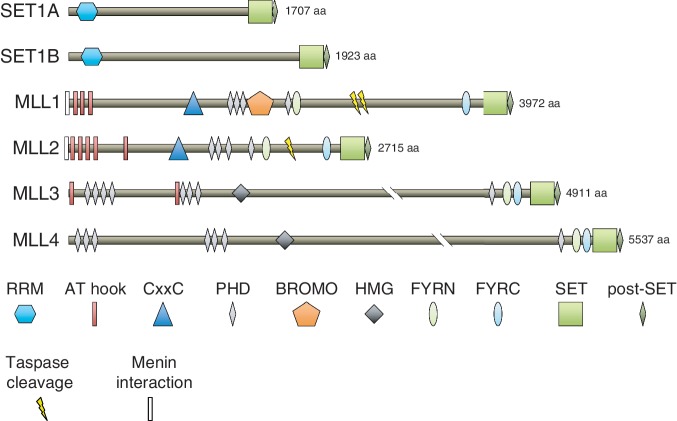
In addition to the global function of SET1A/B, metazoans possess additional homologs that perform H3K4 methylation in a more gene-specific fashion, enzymes known as MLL1 to 4 in humans or Trx/Trr in Drosophila [24]. Trithorax (Trx) was discovered genetically in flies as a homeotic mutant, with Trx alleles displaying body segment specification defects resulting from Hox gene mis-expression [25, 26]. Notably, one of the hypomorphic alleles of Trx (Z11) identified in a forward genetic screen has been mapped to its SET domain and disrupts H3K4 methyltransferase activity [27, 28]. The Trx orthologs in mammals, MLL1 and MLL2, also catalyze H3K4 methylation via their SET domain, with a conserved role in positively regulating HOX gene expression to specify segmental identity [29–32]. Trr in Drosophila and its human orthologs, MLL3 and MLL4, function in a specialized manner to co-activate nuclear hormone receptors via catalysis of H3K4 methylation [33–35]. MLL3 and MLL4 have also been implicated in immunoglobulin class-switching [36]. While all members of the SET1-family possess a similar H3K4 methyltransferase domain at their C-terminus, SET1A/B proteins are likely to maintain the majority of global H3K4 methylation [20, 21], with MLL1 to 4 catalyzing H3K4 methylation at only specific genomic intervals [29, 37–39]. One exception to this is the oocyte, where MLL2 appears to maintain bulk H3K4me3 [40]. The unique functions among each SET1-family member is mediated by the divergent domain architecture outside of the catalytic SET domain (Figure 1). It should be noted that the nomenclature of MLL2 and MLL4 genes is often confused in the literature. Here, we refer to MLL4 (also known as ALR, encoded on human chromosome 12q13) as the closer homolog of MLL3, whereas we refer to MLL2 (also known as TRX2 and Wbp7, encoded on human chromosome 19q13) as the closer homolog of MLL1.
Figure 1:
The human SET1-family of H3K4 methyltransferases. Domain architecture is indicated. The SET and post-SET domains participate in methylation catalysis.
Many of the SET1-family proteins have been causally implicated in pathogenic processes such as cancer. MLL1 was first cloned based on its is involvement in chromosomal translocations found in Mixed Lineage Leukemia, a cancer of the hematopoietic system often seen in infants where malignant cells express markers of both myeloid and lymphoid lineages [41, 42]. Chromosomal translocations associated with this disease rearrange the MLL1 gene and lead to the expression of leukemogenic fusion proteins with deregulated functions. Several excellent reviews discuss the mechanisms of epigenetic alteration by MLL1-fusion proteins, which is outside of the scope of this review [43, 44]. In addition, inactivating somatic mutations of MLL3 and MLL4 have been identified in a wide spectrum of different cancer types, suggesting a tumor suppressor function for these family members [45–48]. Moreover, Kabuki syndrome, which is associated with multiple congenital abnormalities, has recently been attributed to germline inactivating mutations of MLL4 [49]. Importantly, the disease-related functions of MLL3/4 in cancer and the role of MLL4 in Kabuki syndrome have yet to be characterized mechanistically.
WRAD ASSOCIATES WITH SET1-FAMILY H3K4 METHYLTRANSFERASES TO STIMULATE CATALYTIC ACTIVITY AND PRODUCT SPECIFICITY
Purification of yeast Set1 complexes, also known as COMPASS, by several independent laboratories revealed several copurifying subunits, including Swd3, Bre2/Spp1, Swd1 and Sdc1 proteins, which encode orthologs of the mammalian proteins WDR5, ASH2L, RbBP5 and DPY-30 (WRAD), respectively [15, 16, 50] (Figure 2A). Later purifications of SET1-related complexes from fly and human cells also identified an association with WRAD proteins, indicating these factors are a highly conserved repertoire of core-subunits for all SET1-family complexes [20, 21, 33, 35, 51, 52]. Deletions of any WRAD-encoding gene in yeast leads to phenotypes and gene expression alterations that resemble those observed with SET1 mutants, suggesting a functionally relevant interaction among these proteins in vivo [16, 50]. Furthermore, WRAD components are also required to maintain global levels of H3K4 methylation in cells [13, 14, 16]. In studies performed in both yeast and in mammalian cells, WDR5 and RbBP5 have been found to be essential to maintain H3K4me1, H3K4me2 and H3K4me3, whereas ASH2L and DPY-30 are principally required for H3K4me3 and to a lesser extent H3K4me2 [53–56]. WDR5 and RbBP5 proteins are also essential for the stability of the Set1 complex in yeast, as Set1 levels become undetectable in cell extracts when either subunit is depleted [54]. In contrast, ASH2L and DPY-30 orthologs appear to be dispensable for yeast Set1 complex integrity; however, a purified COMPASS complex lacking these factors displays reduced H3K4 methylation activity in vitro [53]. The requirement for WRAD in mammalian cells to maintain global H3K4me3 is likely to reflect the interaction with SET1A/B, as opposed to MLL1-4 [21, 29]. Nonetheless, WRAD represents a conserved set of proteins that associate with SET1-related H3K4 methyltransferases to promote their stability and/or catalytic activity.
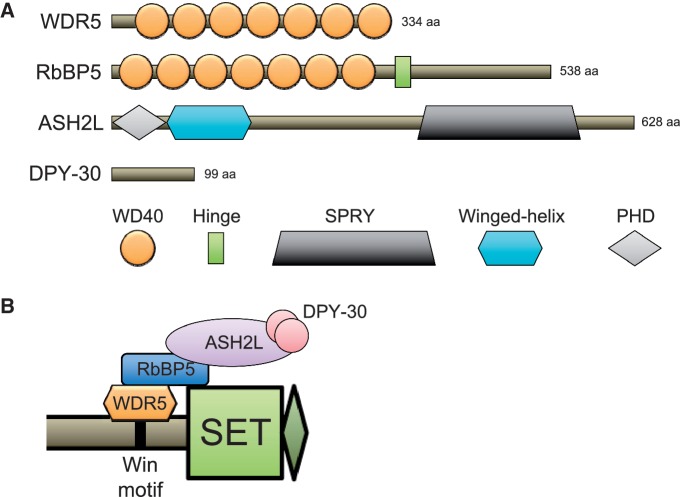
Figure 2:
The WRAD module. (A) The domain architecture of WDR5, RbBP5, ASH2L and DPY-30. (B) The current model of WRAD assembly with the Win-motif and SET domain portions of SET1-family proteins.
A wealth of insights into the biochemical mechanism of WRAD has been obtained from studying a reconstituted complex of purified subunits in association with the SET domain of MLL1. Through examination of the pairwise interactions of MLL1 with individual WRAD subunits purified from insect cells or bacteria, it has been demonstrated that each subunit assembles in a hierarchical interaction arrangement (MLL1 ⇔ WDR5 ⇔ RbBP5 ⇔ ASH2L ⇔ DPY-30) [55, 57] (Figure 2B). The beta-propeller WD40 domains of WDR5 engage in a strong 1:1 interaction with an unstructured region of MLL1 called the Win motif located approximately 139 amino acids N-terminal to the SET domain catalytic site [58, 59]. The Win motif of MLL1 is crucial under most assay conditions for the association with WRAD [58]. The opposing face of the WDR5 beta-propeller domain can simultaneously engage in direct interactions with a region of RbBP5 adjacent to its hinge region [60, 61]. In addition to interacting with WDR5, RbBP5 directly binds to the SPRY domain of ASH2L and engages in a weak direct physical interaction with MLL1 [55, 62]. ASH2L also binds directly to a homodimer of DPY-30 and is essential for its association with MLL1 [57]. Based on these interactions, WDR5 and RbBP5 are thought to function as a scaffold for the subsequent association of ASH2L and DPY-30 (Figure 2B).
In the absence of any interacting proteins, purified MLL1 is a relatively weak H3K4 methyltransferase with product specificity limited to mono-methylation [55, 57, 62]. By assembling individual components with purified MLL1, it has been found that a stoichiometric association with WDR5 and RbBP5 only leads to a modest increase in H3K4 methylation activity, which, like MLL1 alone, is limited in product specificity to mono-methylation [57]. The addition of stoichiometric amounts of ASH2L to the MLL1-WDR5-RbBP5 complex results in a profound increase in H3K4 methyltransferase activity (a >300-fold increase in the rate constant) [55, 57]. Moreover, the inclusion of ASH2L shifts the product specificity of the complex to that of a H3K4 di-methyltransferase, with a minimal activity of the complex to catalyze trimethylation [57]. The addition of DPY-30 to the complex results in a further increase in activity without shifting product specificity [56, 57]. Hence, these studies indicate that WRAD stimulates the intrinsic H3K4 methyltransferase activity of MLL1 and influences its capacity to perform multiple lysine methylation transfers to its substrate.
One issue that has yet to be addressed is whether all SET1-family enzymes utilize WRAD in an identical manner to what has been determined with reconstituted MLL1 complexes. This issue is particularly relevant since the SET domain of MLL1 is dispensable for many of its essential functions in vivo [37]; hence, the methyltransferase domains of the other five SET1-family members might be more active and/or relevant in vivo. Recent evidence suggests that WDR5-RbBP5-ASH2L associates with a Win motif of all six human SET1-family members, resulting in the stimulation of their H3K4 methyltransferase activity in a reconstituted system; however, effects on product specificity were not examined [63]. Interestingly, MLL3 displays a stronger intrinsic H3K4 methyltransferase activity than the other family members in the absence of associated factors, suggesting that it might be uniquely capable of methylating chromatin in a WRAD-independent manner [63]. A purified yeast Set1 protein also completely lacks H3K4 methyltransferase activity in vitro but is dramatically stimulated by its association with purified WRAD orthologs [64]. Unlike MLL1-WRAD, a reconstituted Set1-WRAD complex can carry out mono-, di- and tri-methylation of H3K4, consistent with in vivo observations in yeast and metazoan cells [20, 21, 64]. At present, a major unresolved question is why WRAD can stimulate H3K4 tri-methylation by Set1 but can only promote H3K4 di-methylation activity for MLL1. In addition, the product specificity of MLL2, MLL3 and MLL4, alone or in association with WRAD, also is yet to be determined.
One explanation for the poor intrinsic catalytic activity of SET1-family enzymes is suggested through comparison of the crystal structure of the MLL1 SET domain with other SET domain containing lysine methyltransferases which display greater intrinsic activity [65]. Most of the active SET domains are known to form a closed hydrophobic channel that connects the substrate lysine and cofactor SAM on opposite faces of the protein [reviewed in 66]. Importantly, the closed channel formed by the SET-I, SET-C and postSET subregions of the SET domain optimally align the methyl leaving group of SAM with the episilon-amine-receiving group of lysine. In addition, a carbonyl cage present in the closed channel provides an optimal chemical environment for the nucleophilic substitution mechanism. In the MLL1 SET domain, however, the SET-I and postSET regions fail to interact to form a closed channel, resulting in the substrate being bound in an open cleft between the two lobes [65]. This open arrangement allows mobility of the substrate, hence resulting in an ineffective conformation for methyl transfer. This open SET domain configuration of MLL1 is likely to impose, at least in part, the WRAD-requirement to facilitate methylation [65]. One possibility is that the WRAD interaction allosterically closes the catalytic channel into an optimal configuration for catalysis. As will be discussed below, a non-mutually exclusive alternative is that the open channel allows access to a second H3K4 methyltransferase active site present in WRAD itself to perform additional methyl transfers [57, 62, 67]. It will be important in future studies to determine whether the open SET domain conformation is present in other SET1-family enzymes.
Another feature that restricts the product specificity of isolated SET1-family enzymes relates to the presence of a tyrosine in the ‘switch position’ of the SET domain [68, 69]. It is known that the differing product specificity among members of the SET domain family is related to the available volume of the active site being able to accommodate pre-methylated substrates. A major determinant of product specificity is the presence of phenylalanine or a tyrosine in the critical ‘switch position’ of the SET domain. SET-domain containing enzymes, such as Set7/9, possess a tyrosine in the switch position resulting in a smaller active site that can only catalyze mono-methylation. In contrast, Dim5 and G9a enzymes possess phenylalanine in the switch position, resulting in a larger active site that can accommodate rotation of pre-methylated substrates, thus allowing processive methylation [66]. Interestingly, all of the SET1-related enzymes possess a tyrosine in the switch position, which is predicted to render their enzymatic activity limited to mono-methylation, as is seen with purified MLL1 in vitro. Accordingly, substituting the switch position tyrosine with alanine (MLL1Y3942F) converts MLL1 into an effective tri-methyltransferase enzyme without any requirement for WRAD binding when using histone H3 tail peptides as substrates [57]. When using nucleosomes as a substrate, however, purified MLL1Y3942F is only capable of mono-methylating H3K4 [67]. The addition of WRAD to MLL1Y3942F restores the di- and tri-methylation activity of this mutant, suggesting that WRAD regulates how the MLL1 SET domain engages nucleosomal substrates [67]. A similar tyrosine to alanine substitution in yeast Set1 also elevates its tri-methyltransferase activity in vitro and can bypass the requirement for one of the ASH2L-like subunits (Spp1) to allow H3K4 trimethylation in vivo [70]. Based on these studies, it can be inferred that the presence of tyrosine in the switch position of SET1-family proteins limits their intrinsic activity to mono-methylation, however upon binding to WRAD this limitation can be overcome to promote additional degrees of methylation.
EVIDENCE FOR AN INTRINSIC H3K4 METHYLTRANSFERASE ACTIVITY PRESENT IN WRAD
Recombinant WRAD proteins in solution readily form an independent complex without an associated SET domain, which can also be observed in crude nuclear extracts [54, 55, 57]. An interesting study found that WRAD, in the absence of an associated SET domain, harbors an intrinsic H3K4 mono-methyltransferase activity [57]. This observation was remarkable, as none of the WRAD subunits possess homology to known lysine methyltransferases. The methyltransferase activity of WRAD is detectable, albeit extremely weak, when using a histone H3 tail peptide as a substrate; however, WRAD is incapable of methylating H3K4 in the context of a nucleosome [67]. Like the activity of other known lysine methyltransferases, catalytic activity of WRAD is zinc dependent, inhibited by S-adenosylhomocysteine, and displays Michaelis–Menton kinetics [67]. In this study, it was found that the catalytic activity could be minimally reconstituted with a subcomplex of WDR5, RbBP5 and ASH2L and cannot be observed with any individual purified subunit [67]. An independent study has also found a weak H3K4 methyltransferase activity that can be detected within a heterodimer of ASH2L and RbBP5 [62]. The authors used UV crosslinking to show that ASH2L, like MLL1, binds directly to the methyl-donor SAM [62]. Unlike MLL1, ASH2L is only capable of SAM binding when in a complex with both RbBP5 and MLL1. Collectively, these independent observations support WRAD possessing an intrinsic H3K4 methyltransferase activity, but only in the context of the fully assembled complex.
What role could a second H3K4 methyltransferase catalytic center have in the context of the MLL1 core complex? Several interesting models have emerged by examining total H3K4 methyltransferase activity of various MLL1 SET domain mutations assembled in the context of WRAD [62, 67]. Substitution of asparagine 3906 of the MLL1 SET domain with alanine disrupts a key hydrogen bond with SAM, thereby abolishing its intrinsic H3K4 methyltransferase activity [67]. Interesting MLL1N3906A assembled with WRAD can still methylate an H3 peptide with pre-existing H3K4me0 or H3K4me1, whereas WRAD alone can only mono-methylate an H3K4me0 substrate [67]. This result suggests that the product specificity of WRAD is expanded to perform dimethylation when physically associated with MLL1. Furthermore, an MLL1 Y3874A/K3878A double mutant is similarly compromised in its intrinsic H3K4 methyltransferase activity, but when assembled with RbBP5/ASH2L, this trimeric complex regains largely normal activity [62]. Taken together, these studies support the possibility that total H3K4 methylation activity might be accomplished through dual catalytic sites present in MLL1 and WRAD, which function sequentially to perform two methylation reactions on a single lysine side chain.
While these studies are provocative, the weak overall catalytic activity of WRAD, as compared to other known methyltransferases, necessitates further investigation to validate the ‘dual active site’ model of SET1-family mediated H3K4 methylation. Structural studies to visualize substrate bound to the WRAD catalytic site will be essential to prove this mechanism in fact exists.
ROLES FOR WRAD IN RECRUITMENT OF SET1-FAMILY PROTEINS TO CHROMATIN
Beyond the regulatory roles in catalysis outlined above, WRAD components can also engage in protein:protein, protein:RNA and protein:DNA interactions that serve to recruit and/or stabilize SET1-family complexes at their target sites in the genome. ASH2L has been shown to interact with sequence-specific transcription factors, such as Mef2d and Ap2delta, to promote recruitment of SET1-family proteins as coactivators, resulting in H3K4 methylation at occupied cis-elements [71, 72]. Recently, crystal structures of the N-terminal PHD-like domain of ASH2L unexpectedly revealed a winged-helix fold, which is a known DNA-binding domain found in Forkhead-family transcription factors [73, 74]. Indeed, the winged-helix domain of ASH2L can bind to DNA, albeit 1000 times weaker than the forkhead transcription factor FoxO1 [73, 74]. While this domain has rather limited sequence specificity, it appears to be important for stable binding of ASH2L to chromatin in vivo [73, 74].
WDR5 has also been shown to engage in several direct interactions that influence complex recruitment. WDR5 can bind to the transcription factor Oct4, thus recruiting SET1-family enzymes to co-activate gene regulatory networks that sustain self-renewal of embryonic stem cells [75]. WDR5 can also interact with histone H3 via its WD40 beta propeller domain, which is also the same domain that interacts with the Win motif of SET1-family proteins [76]. Some studies have found that WDR5 preferentially binds to H3 when methylated at H3K4 [76, 77], whereas another study failed to observe methylation-specific binding [78]. A recent report indicates that WDR5 preferentially binds to H3 when it is symmetrically dimethylated at H3R2 (H3R2me2s), which was also identified by the authors as a novel histone mark found at active gene promoters in a mammalian cells [79]. Interestingly, asymmetric H3R2 methylation specifically impedes the interaction with WDR5 [79]. The global colocalization of H3K4me3 and H3KR2me2s at a genome-wide scale supports the idea that WDR5 might bind this modification to promote SET1-family recruitment [79]. One factor in considering histone binding by WDR5 as a mechanism for complex recruitment is that it cannot occur simultaneously with its interaction with the Win-motif. Hence, it seems unlikely that this interaction alone could tether SET1-family proteins to their target sites in the genome; however, it could still promote substrate presentation or recruitment if the association/disassociation events occurred in a dynamic fashion with other stabilizing interactions [55].
Finally, WDR5 has also been found to associate with the long intergenic noncoding RNA (lincRNA) called HOTTIP, which is transcribed from the 5′-end of the HOXA locus [80]. In this study, it was found that knocking down HOTTIP with siRNA resulted in diminished H3K4me3 and MLL1/WDR5 recruitment to the HOXA locus [80]. The investigators showed that recombinant WDR5 protein, and not other components of the MLL1 core complex, can directly interact with HOTTIP in vitro. While the molecular details of this physical interaction are yet to be determined, this study suggests that SET1-family proteins might be tethered to specific genomic sites in cis by an association with lincRNAs. An additional study also identified another lincRNA, called Mistral, transcribed from the HoxA locus which also mediates MLL1 recruitment, although it is not known if this is mediated by a direct WDR5 interaction [81]. It will be interesting to determine the generality of this mechanism to other lincRNAs and SET1-family members.
SUMMARY
The pattern of H3K4 mono-, di-, and tri-methylation across eukaryotic genomes exists in a highly organized arrangement to influence diverse gene-regulatory networks in a temporally dynamic manner. This global chromatin state is regulated through an elaborate multi-component system comprised of six SET1-related H3K4 methyltransferases, their associated WRAD module, other regulators that influence complex recruitment and/or activity (e.g. CFP1), and the opposing activity of H3K4 demethylases. It can be speculated that a reliance of SET1-family proteins on WRAD for catalysis exists to enable ‘tuning’ of methylation levels at particular genomic sites for enhanced regulatory potential. An important aspect of H3K4 methylation that currently defies explanation is regarding its functional importance in gene regulation. Using in vitro systems, it has been observed that H3K4 methylation has only a minor effect on transcriptional activity [82]. Reducing the level of DPY-30 in embryonic stem cells results in a global reduction of H3K4me3, yet self-renewal and proliferation of these cells were unaffected [56]. However, DPY-30-deficient ES cells display a specific defect in undergoing neural differentiation [56]. Thus, despite H3K4me3 being a general feature of active chromatin, its non-redundant function in transcriptional regulation is apparently highly dependent on gene- and cell type-context. Based on this, it will be a worthwhile endeavor in future studies to explore the biological significance of H3K4 methylation in vivo to identify additional roles in normal physiology and in disease processes.
Key points.
The SET1-family of enzymes is involved in maintenance of H3K4 methylation, a chromatin mark that is associated with transcriptional activation.
Catalysis of H3K4 methylation by SET1-family of enzymes is dependent on association with a protein module comprised of WDR5, RbBP5, ASH2L and DPY-30 (WRAD).
WRAD has been found to catalyze H3K4 methylation independently of an associated SET1-protein, implicating a dual active site mechanism for methylation.
WRAD can interact with RNA, DNA, and other proteins to recruit SET1-family proteins to sites along the chromatin fiber.
Biographies
Patricia Ernst, PhD, is an associate professor in the Department of Genetics, and Department of Microbiology and Immunology, Dartmouth Medical School. Her lab's research focuses on the mechanisms that control the balance between self-renewal and differentiation in hematopoiesis and leukemogenesis, particularly as influenced by MLL1.
Christopher Vakoc, MD/PhD, is an assistant professor at Cold Spring Harbor Laboratory. His research focuses on mechanisms of chromatin regulation as it relates to the pathogenesis of cancer. His laboratory uses MLL-fusion acute myeloid leukemia as a model to study the role of epigenetic regulators in this disease.
FUNDING
Christopher Vakoc is supported by funds from the Don Monti Memorial Research Foundation, Laurie Strauss Leukemia Foundation, Sass Foundation, Edward P. Evans Foundation, and F.M. Kirby Foundation. Patricia Ernst is supported by grants from the American Cancer Society (RSG-10-242-LIB), Gabrielle’s Angel Foundation, Lauri Strauss Foundation and the National Institute of Health (HL090036), as well as funds from the Norris Cotton Cancer Center (CA023108).
References
- 1.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–69. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 2.Taverna SD, Li H, Ruthenburg AJ, et al. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–40. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20:341–8. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 5.Strahl BD, Ohba R, Cook RG, et al. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc Natl Acad Sci USA. 1999;96:14967–72. doi: 10.1073/pnas.96.26.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernstein BE, Kamal M, Lindblad-Toh K, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–81. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Santos-Rosa H, Schneider R, Bannister AJ, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–11. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 9.Heintzman ND, Stuart RK, Hon G, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 10.Wysocka J, Swigut T, Xiao H, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 11.Sims RJ, 3rd, Chen CF, Santos-Rosa H, et al. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 2005;280:41789–92. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vermeulen M, Mulder KW, Denissov S, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 13.Krogan NJ, Dover J, Khorrami S, et al. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem. 2002;277:10753–5. doi: 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- 14.Briggs SD, Bryk M, Strahl BD, et al. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286–95. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roguev A, Schaft D, Shevchenko A, et al. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20:7137–48. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagy PL, Griesenbeck J, Kornberg RD, et al. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci USA. 2002;99:90–4. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng HH, Robert F, Young RA, et al. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–19. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 18.Lee JH, Skalnik DG. Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A Histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol Cell Biol. 2008;28:609–18. doi: 10.1128/MCB.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JH, Tate CM, You JS, et al. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J Biol Chem. 2007;282:13419–28. doi: 10.1074/jbc.M609809200. [DOI] [PubMed] [Google Scholar]
- 20.Ardehali MB, Mei A, Zobeck KL, et al. Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. EMBO J. 2011;30:2817–28. doi: 10.1038/emboj.2011.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu M, Wang PF, Lee JS, et al. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol. 2008;28:7337–44. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomson JP, Skene PJ, Selfridge J, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–6. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tate CM, Lee JH, Skalnik DG. CXXC finger protein 1 restricts the Setd1A histone H3K4 methyltransferase complex to euchromatin. FEBS J. 2010;277:210–23. doi: 10.1111/j.1742-4658.2009.07475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuettengruber B, Martinez AM, Iovino N, et al. Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol. 2011;12:799–814. doi: 10.1038/nrm3230. [DOI] [PubMed] [Google Scholar]
- 25.Ingham PW. trithorax and the regulation of homeotic gene expression in Drosophila: a historical perspective. Int J Dev Biol. 1998;42:423–9. [PubMed] [Google Scholar]
- 26.Ingham PW. A clonal analysis of the requirement for the trithorax gene in the diversification of segments in Drosophila. J Embryol Exp Morphol. 1985;89:349–65. [PubMed] [Google Scholar]
- 27.Katsani KR, Arredondo JJ, Kal AJ, et al. A homeotic mutation in the trithorax SET domain impedes histone binding. Genes Dev. 2001;15:2197–202. doi: 10.1101/gad.201901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith ST, Petruk S, Sedkov Y, et al. Modulation of heat shock gene expression by the TAC1 chromatin-modifying complex. Nat Cell Biol. 2004;6:162–7. doi: 10.1038/ncb1088. [DOI] [PubMed] [Google Scholar]
- 29.Milne TA, Briggs SD, Brock HW, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–17. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura T, Mori T, Tada S, et al. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119–28. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 31.Yu BD, Hess JL, Horning SE, et al. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–8. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 32.Glaser S, Schaft J, Lubitz S, et al. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development. 2006;133:1423–32. doi: 10.1242/dev.02302. [DOI] [PubMed] [Google Scholar]
- 33.Goo YH, Sohn YC, Kim DH, et al. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol Cell Biol. 2003;23:140–9. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sedkov Y, Cho E, Petruk S, et al. Methylation at lysine 4 of histone H3 in ecdysone-dependent development of Drosophila. Nature. 2003;426:78–83. doi: 10.1038/nature02080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Issaeva I, Zonis Y, Rozovskaia T, et al. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol. 2007;27:1889–903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daniel JA, Santos MA, Wang Z, et al. PTIP promotes chromatin changes critical for immunoglobulin class switch recombination. Science. 2010;329:917–23. doi: 10.1126/science.1187942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terranova R, Agherbi H, Boned A, et al. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc Natl Acad Sci USA. 2006;103:6629–34. doi: 10.1073/pnas.0507425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lubitz S, Glaser S, Schaft J, et al. Increased apoptosis and skewed differentiation in mouse embryonic stem cells lacking the histone methyltransferase Mll2. Mol Biol Cell. 2007;18:2356–66. doi: 10.1091/mbc.E06-11-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee S, Lee DK, Dou Y, et al. Coactivator as a target gene specificity determinant for histone H3 lysine 4 methyltransferases. Proc Natl Acad Sci USA. 2006;103:15392–7. doi: 10.1073/pnas.0607313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andreu-Vieyra CV, Chen R, Agno JE, et al. MLL2 is required in oocytes for bulk histone 3 lysine 4 trimethylation and transcriptional silencing. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 42.Gu Y, Nakamura T, Alder H, et al. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell. 1992;71:701–08. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 43.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–33. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 44.Mohan M, Lin C, Guest E, et al. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat Rev Cancer. 2010;10:721–8. doi: 10.1038/nrc2915. [DOI] [PubMed] [Google Scholar]
- 45.Parsons DW, Li M, Zhang X, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–9. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watanabe Y, Castoro RJ, Kim HS, et al. Frequent alteration of MLL3 frameshift mutations in microsatellite deficient colorectal cancer. PLoS One. 2011;6:e23320. doi: 10.1371/journal.pone.0023320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gui Y, Guo G, Huang Y, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43:875–78. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morin RD, Mendez-Lago M, Mungall AJ, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ng SB, Bigham AW, Buckingham KJ, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42:790–3. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller T, Krogan NJ, Dover J, et al. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci USA. 2001;98:12902–7. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mohan M, Herz HM, Smith ER, et al. The COMPASS family of H3K4 methylases in Drosophila. Mol Cell Biol. 2011;31:4310–8. doi: 10.1128/MCB.06092-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yokoyama A, Wang Z, Wysocka J, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–49. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schneider J, Wood A, Lee JS, et al. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol Cell. 2005;19:849–56. doi: 10.1016/j.molcel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 54.Steward MM, Lee JS, O'Donovan A, et al. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol. 2006;13:852–4. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- 55.Dou Y, Milne TA, Ruthenburg AJ, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–9. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 56.Jiang H, Shukla A, Wang X, et al. Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell. 2011;144:513–25. doi: 10.1016/j.cell.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patel A, Dharmarajan V, Vought VE, et al. On the mechanism of multiple lysine methylation by the human mixed lineage leukemia protein-1 (MLL1) core complex. J Biol Chem. 2009;284:24242–56. doi: 10.1074/jbc.M109.014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patel A, Vought VE, Dharmarajan V, et al. A conserved arginine-containing motif crucial for the assembly and enzymatic activity of the mixed lineage leukemia protein-1 core complex. J Biol Chem. 2008;283:32162–75. doi: 10.1074/jbc.M806317200. [DOI] [PubMed] [Google Scholar]
- 59.Song JJ, Kingston RE. WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J Biol Chem. 2008;283:35258–64. doi: 10.1074/jbc.M806900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Odho Z, Southall SM, Wilson JR. Characterization of a novel WDR5-binding site that recruits RbBP5 through a conserved motif to enhance methylation of histone H3 lysine 4 by mixed lineage leukemia protein-1. J Biol Chem. 2010;285:32967–76. doi: 10.1074/jbc.M110.159921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Avdic V, Zhang P, Lanouette S, et al. Structural and biochemical insights into MLL1 core complex assembly. Structure. 2011;19:101–8. doi: 10.1016/j.str.2010.09.022. [DOI] [PubMed] [Google Scholar]
- 62.Cao F, Chen Y, Cierpicki T, et al. An Ash2L/RbBP5 heterodimer stimulates the MLL1 methyltransferase activity through coordinated substrate interactions with the MLL1 SET domain. PLoS One. 2011;5:e14102. doi: 10.1371/journal.pone.0014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang P, Lee H, Brunzelle JS, et al. The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases. Nucleic Acids Res. 2012 doi: 10.1093/nar/gkr1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takahashi YH, Westfield GH, Oleskie AN, et al. Structural analysis of the core COMPASS family of histone H3K4 methylases from yeast to human. Proc Natl Acad Sci USA. 2011;108:20526–31. doi: 10.1073/pnas.1109360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Southall SM, Wong PS, Odho Z, et al. Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks. Mol Cell. 2009;33:181–91. doi: 10.1016/j.molcel.2008.12.029. [DOI] [PubMed] [Google Scholar]
- 66.Dillon SC, Zhang X, Trievel RC, et al. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Patel A, Vought VE, Dharmarajan V, et al. A novel non-SET domain multi-subunit methyltransferase required for sequential nucleosomal histone H3 methylation by the mixed lineage leukemia protein-1 (MLL1) core complex. J Biol Chem. 2011;286:3359–69. doi: 10.1074/jbc.M110.174524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xiao B, Jing C, Wilson JR, et al. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003;421:652–6. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- 69.Zhang X, Yang Z, Khan SI, et al. Structural basis for the product specificity of histone lysine methyltransferases. Mol Cell. 2003;12:177–85. doi: 10.1016/s1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takahashi YH, Lee JS, Swanson SK, et al. Regulation of H3K4 trimethylation via Cps40 (Spp1) of COMPASS is monoubiquitination independent: implication for a Phe/Tyr switch by the catalytic domain of Set1. Mol Cell Biol. 2009;29:3478–86. doi: 10.1128/MCB.00013-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tan CC, Sindhu KV, Li S, et al. Transcription factor Ap2delta associates with Ash2l and ALR, a trithorax family histone methyltransferase, to activate Hoxc8 transcription. Proc Natl Acad Sci USA. 2008;105:7472–7. doi: 10.1073/pnas.0711896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rampalli S, Li L, Mak E, et al. p38 MAPK signaling regulates recruitment of Ash2L-containing methyltransferase complexes to specific genes during differentiation. Nat Struct Mol Biol. 2007;14:1150–6. doi: 10.1038/nsmb1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen Y, Wan B, Wang KC, et al. Crystal structure of the N-terminal region of human Ash2L shows a winged-helix motif involved in DNA binding. EMBO Rep. 2011;12:797–803. doi: 10.1038/embor.2011.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sarvan S, Avdic V, Tremblay V, et al. Crystal structure of the trithorax group protein ASH2L reveals a forkhead-like DNA binding domain. Nat Struct Mol Biol. 2011;18:857–9. doi: 10.1038/nsmb.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ang YS, Tsai SY, Lee DF, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–97. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wysocka J, Swigut T, Milne TA, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–72. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 77.Han Z, Guo L, Wang H, et al. Structural basis for the specific recognition of methylated histone H3 lysine 4 by the WD-40 protein WDR5. Mol Cell. 2006;22:137–44. doi: 10.1016/j.molcel.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 78.Couture JF, Collazo E, Trievel RC. Molecular recognition of histone H3 by the WD40 protein WDR5. Nat Struct Mol Biol. 2006;13:698–703. doi: 10.1038/nsmb1116. [DOI] [PubMed] [Google Scholar]
- 79.Migliori V, Muller J, Phalke S, et al. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat Struct Mol Biol. 2012;19:136–44. doi: 10.1038/nsmb.2209. [DOI] [PubMed] [Google Scholar]
- 80.Wang KC, Yang YW, Liu B, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bertani S, Sauer S, Bolotin E, et al. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell. 2011;43:1040–6. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 82.Pavri R, Zhu B, Li G, et al. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–17. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]