Abstract
Pds5 and Wpl1 act as anti-establishment factors preventing sister-chromatid cohesion until counteracted in S-phase by the cohesin acetyl-transferase Eso1. However, Pds5 is also required to maintain sister-chromatid cohesion in G2. Here, we show that Pds5 is essential for cohesin acetylation by Eso1 and ensures the maintenance of cohesion by promoting a stable cohesin interaction with replicated chromosomes. The latter requires Eso1 only in the presence of Wapl, indicating that cohesin stabilization relies on Eso1 only to neutralize the anti-establishment activity. We suggest that Eso1 requires Pds5 to counteract anti-establishment. This allows both cohesion establishment and Pds5-dependent stable cohesin binding to chromosomes.
Keywords: cohesion, Pds5, Eso1, Psm3, Wapl
Introduction
Sister-chromatid cohesion is mediated by cohesin, a tripartite ring-shaped complex made of a Smc1–Smc3 heterodimer associated with Scc1, which recruits Scc3, Pds5 and Wapl subunits (Psm1, Psm3, Rad21, Psc3, Pds5 and Wpl1, respectively, in fission yeast) [1–4]. How sister-chromatid cohesion is made and maintained is of fundamental importance but still incompletely understood. Cohesin is deposited on chromatin by the loading complex (Mis4/Ssl3 in fission yeast), and unless in the event of a DNA double-strand break, cohesin is converted into cohesive structures exclusively during S-phase [5]. In fission yeast and mammals, cohesion establishment correlates with a change in the mode of cohesin interaction with chromosomes. In G1 cells, cohesin cycles chromatin on and off as a result of repeated events of cohesin loading and unloading. By contrast, after S-phase a fraction of cohesin is stably bound to chromosomes and ensures long-term cohesion [6–8]. This change in cohesin dynamics may involve the inactivation of unloading activities, activation of stabilization factors or both. Cohesin unloading is stimulated by Wapl. The deletion of the wpl1 gene in fission yeast or Wapl depletion in human cells slows down cohesin removal from chromosomes [7, 9]. Add-back experiments in Xenopus mitotic extracts have shown that both Pds5 and Wapl are required for cohesin removal from chromatin [10]. Pds5 and Wapl are also described as anti-establishment factors. Cohesion establishment during S-phase requires the neutralization of an anti-establishment activity by a conserved acetyl-transferase (Eso1 in fission yeast) that catalyses acetylation of a pair of conserved lysine residues within the core cohesin subunit Smc3. Acetyl-mimicking forms of Smc3 or mutations affecting Wapl, Scc3 or Pds5 can bypass the requirement for the acetyl-transferase, indicating that these factors are all part of the anti-establishment pathway [4]. Among these, Pds5 and Wapl form a stable subcomplex that interacts with cohesin and promotes its removal from chromatin [9, 11], suggesting that anti-establishment and releasing activity might be synonymous, as recently suggested [4]. Accordingly, the stable mode of cohesin’s interaction with fission yeast chromosomes is Eso1-dependent, but not when the wpl1 gene is deleted [6, 7]. The deletion of pds5 also bypasses Eso1 requirement [12]. It is clear, however, that the inactivation of Pds5 or Wapl leads to different outcomes. In budding yeast, deletion of WPL1 generates moderate cohesion defects [11, 13, 14], while Pds5 is essential for cell viability and its inactivation in postreplicative cells leads to a loss of sister-chromatid cohesion [15–17]. Wapl and Pds5 functions are even more clearly separable in fission yeast. The deletion of wpl1 has no detrimental effect on sister-chromatid cohesion establishment and maintenance [7]. Strains deleted for pds5 are viable and proficient for sister-chromatid cohesion in actively dividing cycling cells, showing that Pds5 is dispensable for the establishment of sister-chromatid cohesion [12, 18]. However, cohesion is progressively eroded during the G2-phase and is eventually lost when the duration of G2 is extended [12]. We report here two new key findings related to Pds5 function. First, we show that Pds5 authorizes cohesion establishment by allowing Eso1-mediated neutralization of the anti-establishment. Pds5 is therefore required both for the anti-establishment activity and to bring about the mechanism that counteracts it. Second, we show that Pds5 is essential for the stable mode of cohesin interaction with replicated chromosomes, giving a straightforward explanation for its role in cohesion maintenance. We suggest a model in which Pds5 couples DNA replication with sister-chromatid cohesion establishment and maintenance.
Results and discussion
Pds5 is required for Psm3 acetylation by Eso1
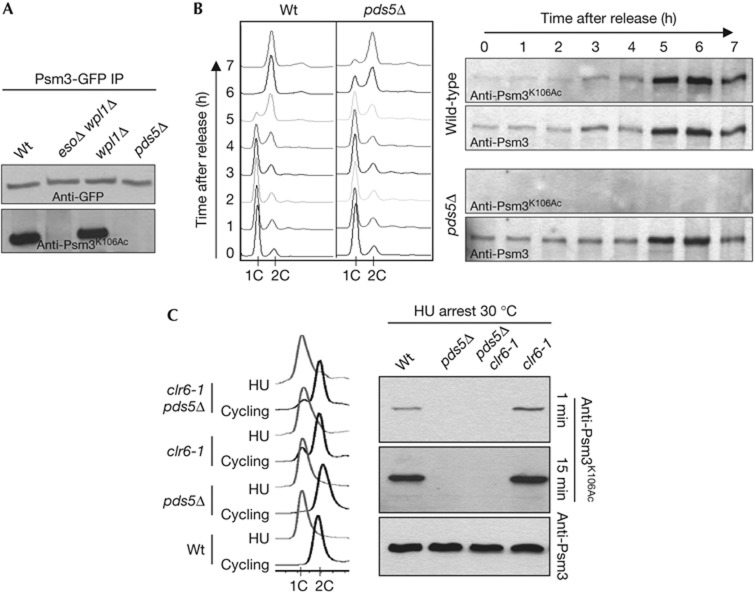
In yeasts and mammals, Smc3 is acetylated on two adjacent lysine residues, which correspond to K105 and K106 in fission yeast Psm3. The acetylated form rises during S-phase and is maintained throughout G2 [7, 19]. Fig 1A shows that Psm3-GFP immunopurified from cycling Schizosaccharomyces pombe cells (∼80% G2) reacts strongly with anti-Psm3K106Ac antibodies, but not when purified from pds5Δ cells. To address whether the lack of acetylation is due to defective acetylation during S-phase, as opposed to a failure to maintain acetylation during G2, we analysed synchronous cultures progressing through S-phase and cells arrested in early S-phase using hydroxyurea (Fig 1B,C). Again, Psm3 from pds5Δ cells did not react with the anti-Psm3K106Ac antibody. To observe whether de-acetylation would be responsible for the absence of acetylated Psm3, the experiment was repeated in a thermosensitive mutant background for Clr6 that was recently reported as the S. pombe Psm3 de-acetylase [19]. Psm3 acetylation was slightly elevated in hydroxyurea-arrested clr6-1 cells at 30 °C, as reported [19], but remained undetectable in pds5Δ clr6-1 (Fig 1C). These data show that Pds5 is essential for Psm3K106Ac, and are consistent with Pds5 being required for de novo Psm3 acetylation. As Pds5 and Eso1 interact together [12], loss of Pds5 may prevent Eso1’s access to cohesin.
Figure 1.
Pds5 is required for Psm3K106 acetylation. (A) GFP-tagged Psm3 was immunopurified from cycling cells and probed with anti-Psm3K106Ac antibodies. (B) Cells were arrested in G1 by nitrogen starvation and released into rich medium. Progression into S-phase was monitored by measuring DNA content by flow cytometry. Total protein extracts were probed with anti-Psm3K106Ac antibodies and the total amount of Psm3 with anti-Psm3 antibodies. (C) DNA content analysis of cycling cells at 30 °C and after 4-h incubation with hydroxyurea. Total protein extracts were probed with Psm3K106Ac and Psm3 antibodies. The short exposure time shows a slight increase in Psm3 acetylation in the clr6-1 mutant, as reported [19]. The longer exposure (15 min) shows that Psm3 acetylation remains undetectable in a pds5Δ background. GFP, green fluorescent protein; Hu, hydroxyurea; IP, immunoprecipitation; Wt, wild type.
Pds5 is essential for the stable mode of cohesin binding
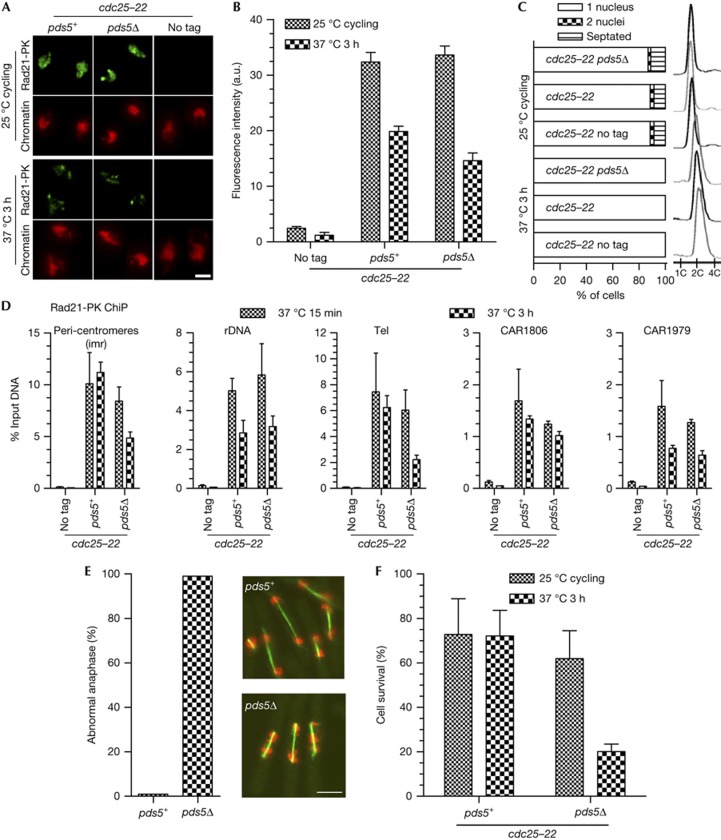
Fission yeast cells lacking the pds5 gene are viable [12, 18], indicating that sister-chromatid cohesion can be achieved without Pds5. However, cells progressively lose cohesion during the G2-phase when its duration is extended by the use of the thermosensitive cdc25-22 mutation, which prevents entry into mitosis at 37 °C. As expected from the severe loss of cohesion during G2, cells experienced massive chromosome segregation defects during their first mitosis upon release from the cdc25-22 block [12]. We repeated this experiment to assess Rad21 association with chromosomes during the G2 arrest, globally by nuclear spreads and at specific loci by chromatin immunoprecipitation (ChiP). We looked at the three major sites of cohesin binding in the fission yeast genome: the peri-centromeric regions, the non-transcribed spacer sequences within the ribosomal DNA gene cluster and telomeres [7], and two cohesin-associated regions (CAR1806 and CAR1979 [6]) in euchromatin. Before the temperature shift (cycling cells, ∼80% in G2, Fig 2C), Rad21 was bound to chromatin in pds5+ as well as in pds5Δ cells (Fig 2A,B). In agreement with the progressive loss of sister-chromatid cohesion shown by Tanaka et al [12], pds5Δ cells were unable to properly segregate their chromosomes upon release from the G2 arrest, as shown by massive chromosome segregation defects (Fig 2E) and poor cell survival (Fig 2F). Nuclear spreads and ChiP indicated that pds5Δ cells retained a sizable amount of chromatin-bound Rad21 after the 3-h time period at 37 °C (Fig 2A,B), suggesting that although they are chromatin-bound, those cohesin complexes may not ensure functional cohesion.
Figure 2.
Examination of Rad21 binding to chromosomes in pds5Δ cells upon prolonged G2 arrest. Cycling cells were shifted to 37 °C. The cdc25-22 mutation blocks entry into mitosis and keep cells in G2. (A) Images of Rad21-PK immunofluorescence on nuclear spreads from cells collected before and after 3 h at 37 °C. Chromatin was counterstained with DAPI. Bar=2 μm. (B) Quantification of Rad21 bound to chromosomes. Fluorescence intensity was measured for 50–100 nuclei per sample. Error bar=95% confidence interval of the mean with α=0.05. (C) Cell cycle staging showing that cells were predominantly in G2 before the temperature shift (one nucleus and 2C DNA content) and remained in G2 throughout the experiment. (D) ChiP assay showing the amount of Rad21-PK bound at the indicated loci, shortly (15 min) and 3 h after the 37 °C temperature shift. Error bar=s.d. from four ChiPs. (E) Cytological analysis of cells undergoing their first mitosis upon release from the cdc25-22 arrest. After 3 h at 37 °C, cells were shifted back to 25 °C and fixed 1 h later when most cells were in anaphase. The mitotic spindles were stained by indirect immunofluorescence against tubulin (green) and DNA was stained with DAPI (red, pseudocolour). Bar=5 μm. The frequency of abnormal anaphases was determined from the examination of 80–100 anaphases per sample. Abnormal anaphases were defined as cells with a spindle length>5 μm displaying DAPI-stained material that had not reached the spindle poles. (F) Cell survival was determined by plating cells back to 25 °C and scoring the number of colony-forming units. Error bar=s.d. from four (control) and five (pds5Δ) experiments. ChIP, chromatin immunoprecipitation; DAPI, 4,6-diamidino-2-phenylindole.
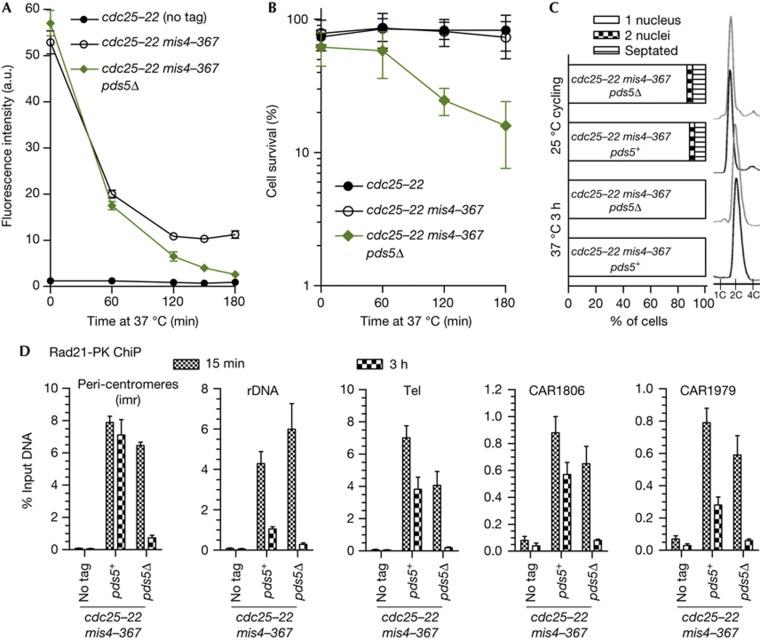
In fission yeast and mammals, a fraction of cohesin (∼20–30%) is stably bound to chromosomes after S-phase and ensures functional cohesion, while the bulk of cohesin remains labile [6–8]. We hypothesized that pds5Δ cells may be unable to generate the stable cohesin fraction. To test this, pds5Δ cells were arrested in G2 as before by cdc25-22 and further cohesin deposition was prevented by the use of the thermosensitive mis4-367 mutation in the cohesin loader Mis4 [6, 20]. Chromatin-bound cohesin was monitored by nuclear spreads and ChiP against Rad21. In an otherwise wild-type background, chromatin-bound Rad21 decreased with time to reach a plateau after ∼120 min (Fig 3A; [7]). Sister-chromatid cohesion was not affected by the 3-h time period at 37 °C (supplementary Fig S1 online), and cell viability remained high upon release from the G2 block (Fig 3B), indicating that cohesin that remains bound to chromatin (the stable fraction) provides functional cohesion, as reported [7]. By contrast, in pds5Δ cells, chromatin-bound Rad21 decreased to background levels (Fig 3A), sister-chromatid cohesion was severely compromised (supplementary Fig S1 online) and cell survival was low upon release from the cdc25-22 arrest (Fig 3B). ChiP analyses confirmed that the stable cohesin fraction was severely affected by the lack of Pds5 (Fig 3D). The effect is particularly dramatic within the peri-centromeric domains (imr). In an otherwise wild-type background, cohesin binds mostly in the stable mode at that site, as the amount of bound Rad21 does not decrease with time (Fig 3D and [7]). By contrast, in pds5Δ, Rad21 dissociated from these regions during the 3-h period of G2 arrest. At rDNA and telomeres, ∼20% and 50% of Rad21 is bound in the stable mode in pds5+cells, respectively [7]. In pds5Δ, however, Rad21 dissociates close to background levels (Fig 3D). The two euchromatic sites analysed behaved similarly. At CAR1806 and CAR1979, ∼60% and ∼35% of Rad21 was stably bound in pds5+, respectively [7], but Rad21 dropped to background levels in pds5Δ. We conclude that the stable cohesin fraction is not made in pds5Δ cells and this causes a failure to maintain sister-chromatid cohesion. We propose that cohesion establishment does occur in pds5Δ, but these cohesive cohesin complexes are not stable enough on chromatin and cohesion is lost when the cohesive pool is exhausted.
Figure 3.
Pds5 is required for the stable mode of cohesin interaction with chromosomes. (A) Kinetics of Rad21 dissociation from G2 chromosomes upon inactivation of the cohesin loader. Cycling cells were shifted to 37 °C to shut off cohesin loading (mis4-367) while keeping cells in G2 (cdc25-22). Chromatin-bound Rad21 was measured at the indicated time points by nuclear spreads (50–100 nuclei per sample). Error bar=95% confidence interval of the mean with α=0.05. (B) Cell survival over time. Error bar=s.d. from three experiments. (C) Cell cycle staging showing that cells were predominantly in G2 before the temperature shift and remained in G2 throughout the experiment. (D) ChiP assay showing the amount of Rad21-PK bound at the indicated loci, shortly (15 min) and 3 h after the 37 °C temperature shift. Error bar=s.d. from three ChiPs. ChIP, chromatin immunoprecipitation.
In budding yeast, Pds5 seems to be continuously required after S-phase, as its inactivation in postreplicative cells leads to a loss of sister-chromatid cohesion [15–17]. We found that Pds5 co-localized with the subpopulation of Rad21 that binds chromatin in the stable mode, suggesting that cohesin stabilization may require a continuous interaction of Pds5 with cohesin (supplementary Fig S2 online).
Pds5 can stabilize cohesin independently from Eso1-Wpl1
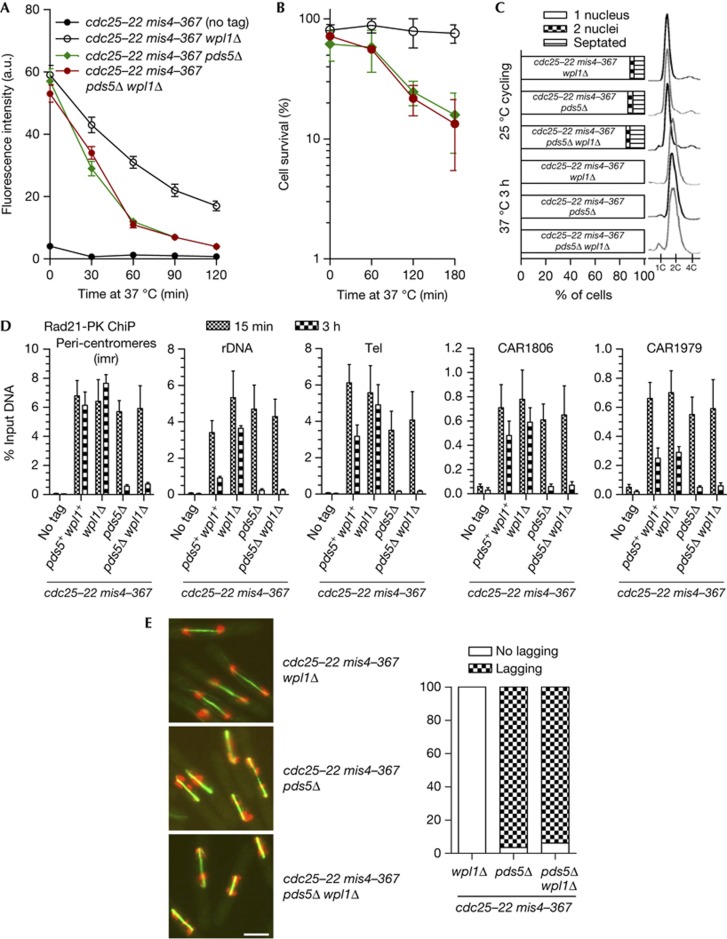
The establishment of cohesion and the formation of the stable cohesin fraction require Eso1, whose main function is counteracting Wpl1 [6, 7, 21]. As Pds5 is required for Eso1 function (Fig 1), pds5Δ cells may be unable to stabilize cohesin binding to chromosomes because cohesin remains sensitive to Wpl1. To test this, we asked whether the deletion of wpl1 would restore stable cohesin binding in a pds5Δ background. Examination of chromatin-bound Rad21 by nuclear spreads (Fig 4A) indicates that the kinetics of Rad21 release from chromatin is very similar in pds5Δ and pds5Δ wpl1Δ, showing that Wpl1 is not responsible for cohesin removal in pds5Δ. Nuclear spreads and ChiP (Fig 4A,D) showed that the stable cohesin fraction is not restored in pds5Δ wpl1Δ. Consistently, cells experienced massive chromosome segregation defects upon release from the cdc25-22 arrest (Fig 4E), and cell survival was low (Fig 4B). Therefore, deletion of wpl1 does not rescue the pds5Δ defect, ruling out the possibility of a residual Wpl1 activity causing a failure to generate the stable cohesin fraction. This experiment also confirms that wpl1Δ cells are proficient for cohesion maintenance. The deletion of wpl1 does not affect sustained Rad21 and Pds5 binding to chromosomes (Fig 4A,D; supplementary Fig S3 online); chromosome segregation occurred normally and cells remained viable upon release from the 3-h time of G2 arrest (Fig 4B,E). Wpl1 is therefore not required for the maintenance of sister-chromatid cohesion, and cohesion maintenance in wpl1Δ cells is Pds5-dependent.
Figure 4.
The absence of a stable cohesin fraction in pds5Δ is not rescued by wpl1 deletion. Cycling cells were shifted to 37 °C to shut off cohesin loading (mis4-367) while keeping cells in G2 (cdc25-22). (A) Kinetics of Rad21 dissociation from chromatin by nuclear spreads. Fluorescence intensity was measured for 50–100 nuclei per sample. Error bar=95% confidence interval of the mean with α=0.05. (B) Cell survival over time. Error bar=s.d. from three experiments. (C) Cell cycle staging before and after the temperature shift. (D) ChiP assay showing the amount of Rad21-PK bound at the indicated loci, shortly (15 min) and 3 h after the 37 °C temperature shift. Error bar=s.d. from four ChiPs. (E) Cytological analysis of cells undergoing their first mitosis upon release from the cdc25-22 arrest. Green: tubulin, red: DAPI. Bar=5 μm. The frequency of abnormal anaphases was determined as in Fig 2 from the examination of 80–100 anaphases per sample. ChIP, chromatin immunoprecipitation; DAPI, 4,6-diamidino-2-phenylindole.
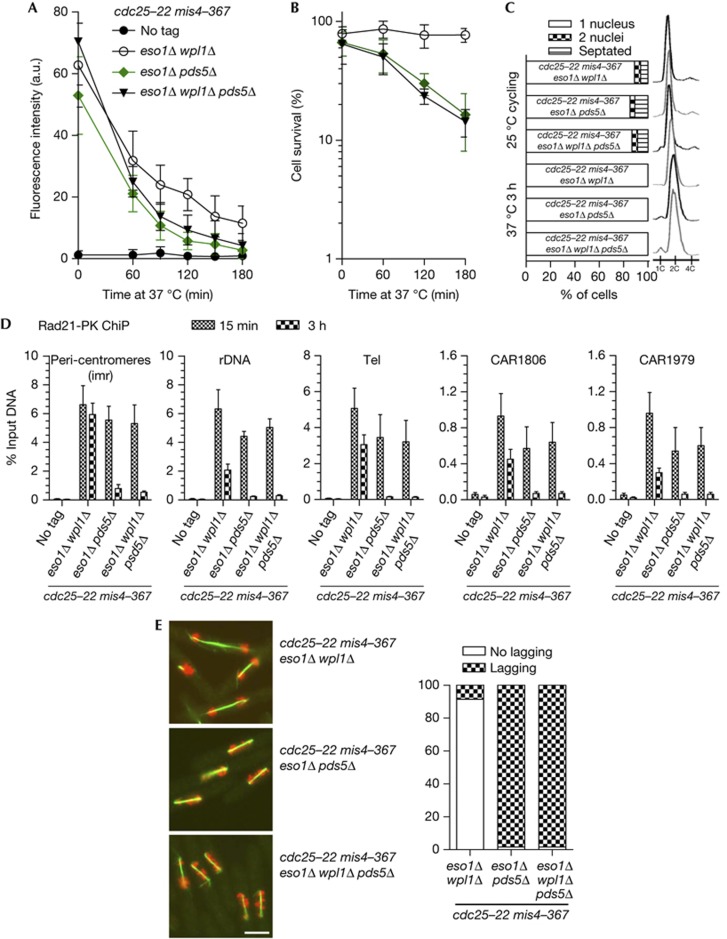
Pds5 is required for Psm3 acetylation (Fig 1), suggesting that the inability of pds5Δ cells to generate a stable cohesin interaction with chromatin may stem from the lack of Psm3 acetylation. However, we previously showed that the stable cohesin fraction is obtained in eso1Δ wpl1Δ, and sister-chromatid cohesion is robust even after prolonged G2 or metaphase arrest, showing that Psm3 acetylation is dispensable for cohesion maintenance [7]. Accordingly, the amount of chromatin-bound Pds5 was very similar in wild-type and eso1Δ wpl1Δ (supplementary Fig S3). Crucially, deletion of pds5 in this context abolished the formation of the stable cohesin fraction (Fig 5A,D). Cell survival dropped with time upon prolonged G2 arrest (Fig 5B), and abnormal chromosome segregation was observed when cells were released into mitosis (Fig 5E). These experiments demonstrate that cells deleted for wpl1 or both eso1 and wpl1 are proficient for cohesion maintenance in a Pds5-dependent manner. Therefore, neither the Eso1 acetyl-transferase nor Psm3 acetylation per se are part of the basic mechanism by which Pds5 promotes a stable cohesin interaction with chromosomes. Eso1 is fully dispensable for cohesion establishment and maintenance in a wpl1Δ background, but is absolutely required for cohesion establishment in wild type [7, 21]. We conclude that the sole but essential function of Eso1 is to neutralize the anti-establishment during S-phase as a prerequisite for cohesion establishment and maintenance.
Figure 5.
Pds5’s function in cohesion maintenance remains essential in the absence of both Eso1 and Wpl1. (A) Kinetics of Rad21 dissociation from G2 chromosomes upon inactivation of the cohesin loader. Cycling cells were shifted to 37 °C to shut off cohesin loading (mis4-367) while keeping cells in G2 (cdc25-22). Chromatin-bound Rad21 was measured by nuclear spreads (50–100 nuclei per sample). Error bar=95% confidence interval of the mean with α=0.05. (B) Cell survival over time. Error bar=s.d. from three experiments. (C) Cell cycle staging showing that cells were predominantly in G2 before the temperature shift and remained in G2 throughout the experiment. (D) ChiP assay showing the amount of Rad21-PK bound at the indicated loci, shortly (15 min) and 3 h after the 37 °C temperature shift. Error bar=s.d. from four ChiPs. (E) Cytological analysis of cells undergoing their first mitosis upon release from the cdc25-22 arrest. Green: tubulin, red: DAPI. Bar=5 μm. The frequency of abnormal anaphases was determined as in Fig 2 from the examination of 80–100 anaphases per sample. ChIP, chromatin immunoprecipitation; DAPI, 4,6-diamidino-2-phenylindole.
The data presented here highlight Pds5 as a major regulator of cohesin. Pds5 and Wpl1 are components of the anti-establishment activity (pds5Δ and wpl1Δ are eso1Δ suppressors) and act together in G1 to promote cohesin removal ([6] and supplementary Fig S4 online). In S-phase, Eso1 neutralizes the anti-establishment activity, authorizing cohesion establishment. Eso1 function is Pds5-dependent, as all pds5Δ phenotypes are unmodified by the deletion of eso1 (Figs 4, 5 and [12]) and Psm3 acetylation is abolished in pds5Δ (Fig 1). As the cohesin acetyl-transferase may associate with replication forks [22–24], the dependency of Eso1 on Pds5 provides a robust system to ensure that the anti-establishment is counteracted when the replisome encounters cohesin and cohesion must be made. Pds5 is subsequently required for sustained sister-chromatid cohesion during the G2-phase, and we have shown here that Pds5 ensures this function by promoting the stable mode of cohesin interaction with chromosomes. The mechanism by which cohesin becomes stably bound remains enigmatic. Eso1-mediated neutralization of the anti-establishment activity in S-phase is a prerequisite, but is unlikely to be the sole event. In wpl1Δ cells, the anti-establishment is abolished and the rate of cohesin removal is reduced but not abolished. This can be observed in G1 ([6] and supplementary Fig S4 online) and G2 for the labile cohesin pool [7]. Likewise, in mammalian cells, Wapl depletion increases the residence time of the labile cohesin fraction on G2 chromatin (from ∼8 to ∼16 min) but does not lead to a full stabilization [9]. In other words, the sole neutralization of the anti-establishment is not sufficient to generate the stable binding mode. Altogether, these data suggest that the stable mode of cohesin interaction with chromosomes requires the neutralization of the anti-establishment, the Pds5 protein, but also passage through S-phase. This may involve a conformational change in cohesin driven by the replisome, other S-phase-specific factors of both. Clarifying these issues will be of crucial importance to deepen our understanding of sister-chromatid cohesion establishment and maintenance.
Methods
The strains used are listed in supplementary Table S1 online. Media and genetic techniques were as described [25]. Synthetic medium EMM2 was used unless otherwise stated. EMM2 lacking a nitrogen source was used to arrest cells in G1, and YPD to release cells into the cell cycle. Cell cycle arrest in early S-phase was induced by addition of 12 mM hydroxyurea to exponentially growing cells at 30 °C. Cell cycle block in G2 by cdc25-22 was verified by 4,6-diamidino-2-phenylindole and calcofluor staining of ethanol-fixed cells (n=200) to score the rise in mononucleate cells at the expense of binucleated and septated cells. DNA content was measured by flow cytometry after Sytox Green staining of ethanol-fixed cells. Tubulin immunofluorescence and spindle length measurements were done as described [26]. Nuclear spreads, ChiP, immunoprecipitation and western blotting were done as described [7].
Supplementary Material
Acknowledgments
This work was supported by the Centre National de la Recherche Scientifique, l’Université Victor Segalen, la Région Aquitaine, l’Association pour la Recherche sur le Cancer and l’Agence Nationale de la Recherche. A.F. was supported by a fellowship from the Ministère de la Recherche et de l’Enseignement Supérieur.
Authors contributions: J.P.J. and S.V. designed the study. All authors participated in the collection of the data. A.F., S.V. and J.P.J. analysed the data. J.P.J. and S.V. drafted the manuscript with input from A.F.
Footnotes
The authors declare that they have no conflict of interest.
References
- Peters JM, Tedeschi A, Schmitz J (2008) The cohesin complex and its roles in chromosome biology. Genes Dev 22: 3089–3114 [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH (2009) Cohesin: its roles and mechanisms. Annu Rev Genet 43: 525–558 [DOI] [PubMed] [Google Scholar]
- Skibbens RV (2009) Establishment of sister chromatid cohesion. Curr Biol 19: R1126–R1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K (2011) Cohesin: a catenase with separate entry and exit gates? Nat Cell Biol 13: 1170–1177 [DOI] [PubMed] [Google Scholar]
- Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE (2008) Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol 24: 105–129 [DOI] [PubMed] [Google Scholar]
- Bernard P, Schmidt CK, Vaur S, Dheur S, Drogat J, Genier S, Ekwall K, Uhlmann F, Javerzat JP (2008) Cell-cycle regulation of cohesin stability along fission yeast chromosomes. EMBO J 27: 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feytout A, Vaur S, Genier S, Vazquez S, Javerzat JP (2011) Psm3 acetylation on conserved lysine residues is dispensable for viability in fission yeast but contributes to Eso1-mediated sister chromatid cohesion by antagonizing Wpl1. Mol Cell Biol 31: 1771–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich D, Koch B, Dupeux F, Peters JM, Ellenberg J (2006) Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr Biol 16: 1571–1578 [DOI] [PubMed] [Google Scholar]
- Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters JM (2006) Wapl controls the dynamic association of cohesin with chromatin. Cell 127: 955–967 [DOI] [PubMed] [Google Scholar]
- Shintomi K, Hirano T (2009) Releasing cohesin from chromosome arms in early mitosis: opposing actions of Wapl-Pds5 and Sgo1. Genes Dev 23: 2224–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland BD et al. (2009) Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol Cell 33: 763–774 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Hao Z, Kai M, Okayama H (2001) Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. EMBO J 20: 5779–5790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shahar TR, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F (2008) Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science 321: 563–566 [DOI] [PubMed] [Google Scholar]
- Sutani T, Kawaguchi T, Kanno R, Itoh T, Shirahige K (2009) Budding yeast Wpl1(Rad61)-Pds5 complex counteracts sister chromatid cohesion-establishing reaction. Curr Biol 19: 492–497 [DOI] [PubMed] [Google Scholar]
- Hartman T, Stead K, Koshland D, Guacci V (2000) Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J Cell Biol 151: 613–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizza S, Tanaka T, Hochwagen A, Eisenhaber F, Nasmyth K (2000) Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr Biol 10: 1557–1564 [DOI] [PubMed] [Google Scholar]
- Stead K, Aguilar C, Hartman T, Drexel M, Meluh P, Guacci V (2003) Pds5p regulates the maintenance of sister chromatid cohesion and is sumoylated to promote the dissolution of cohesion. J Cell Biol 163: 729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SW, Read RL, Norbury CJ (2002) Fission yeast Pds5 is required for accurate chromosome segregation and for survival after DNA damage or metaphase arrest. J Cell Sci 115: 587–598 [DOI] [PubMed] [Google Scholar]
- Kagami A, Sakuno T, Yamagishi Y, Ishiguro T, Tsukahara T, Shirahige K, Tanaka K, Watanabe Y (2011) Acetylation regulates monopolar attachment at multiple levels during meiosis I in fission yeast. EMBO Rep 12: 1189–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Drogat J, Maure JF, Dheur S, Vaur S, Genier S, Javerzat JP (2006) A screen for cohesion mutants uncovers Ssl3, the fission yeast counterpart of the cohesin loading factor Scc4. Curr Biol 16: 875–881 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Yonekawa T, Kawasaki Y, Kai M, Furuya K, Iwasaki M, Murakami H, Yanagida M, Okayama H (2000) Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol Cell Biol 20: 3459–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna MA, Skibbens RV (2003) Mechanical link between cohesion establishment and DNA replication: Ctf7p/Eco1p, a cohesion establishment factor, associates with three different replication factor C complexes. Mol Cell Biol 23: 2999–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S (2006) PCNA controls establishment of sister chromatid cohesion during S phase. Mol Cell 23: 723–732 [DOI] [PubMed] [Google Scholar]
- Skibbens RV (2004) Chl1p, a DNA helicase-like protein in budding yeast, functions in sister-chromatid cohesion. Genetics 166: 33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Bernard P, Hardwick K, Javerzat JP (1998) Fission yeast bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis. J Cell Biol 143: 1775–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.