Abstract
The phloem plays a crucial role in assimilate and nutrient transport, pathogen response, and plant growth and development. Yet, few species have yielded pure phloem exudate and, if proteins need to be analysed, those species may not have sequenced genomes, making identification difficult. The enrichment of Arabidopsis thaliana phloem exudate in amounts large enough to allow for metabolite and protein analysis is described. Using this method, it was possible to identify 65 proteins present in the Arabidopsis phloem exudate. The majority of these proteins could be grouped by response to pathogens, stress, or hormones, carbon metabolism, protein interaction, modification, and turnover, and transcription factors. It was also possible to detect 11 proteins that play a role in lipid/fatty acid metabolism (aspartic protease, putative 3-β-hydroxysteroid dehydrogenase, UDP-sulphoquinovose synthase/SQD1, lipase, PIG-P-like protein: phosphatidylinositol-N-acetylglucosaminyltransferase), storage (glycine-rich protein), binding (annexin, lipid-associated family protein, GRP17/oleosin), and/or signalling (annexin, putative lipase, PIG-P-like protein). Along with putative lipid-binding proteins, several lipids and fatty acids could be identified. Only a few examples exist of lipids (jasmonic acid, oxylipins) or lipid-binding proteins (DIR1, acyl-CoA-binding protein) in the phloem. Finding hydrophobic compounds in an aqueous environment is not without precedence in biological systems: human blood contains a variety of lipids, many of which play a significant role in human health. In blood, lipids are transported while bound to proteins. The present findings of lipids and lipid-binding proteins in phloem exudates suggest that a similar long-distance lipid signalling exists in plants and may play an important role in plant growth and development.
Keywords: Fatty acids, lipid-binding proteins, lipids, long-distance signalling, long-distance transport, phloem, phloem exudates, proteome
Introduction
Plants have developed two long-distance transport systems: the xylem for water and mineral distribution from the roots to the remainder of the plant; and the phloem, which was originally thought to be responsible simply for the transport of assimilates. The latter has evolved to optimize longitudinal flow in the sieve elements by removing any obstacles in the form of organelles and by increasing the porosity of the cell walls at the longitudinal ends (van Bel and Knoblauch, 2000). While it is possible that sieve elements may still contain nuclei, vacuoles, and organelles during their early development, those cell components disintegrate, leaving the sieve elements with only the plasma membrane and a thin cytoplasm containing endoplasmic reticulum, phloem-specific plastids, and a few, often dilated, mitochondria (Shah and Jacob, 1969; Cronshaw, 1981; Behnke and Schulz, 1983; Behnke, 1991; van Bel and Knoblauch, 2000). No functional protein synthesis machinery is thought to exist. As a result, sieve tube proteins are assumed to be synthesized in the companion cells and transported into the sieve elements via plasmodesmata (Raven, 1991; Hayashi et al., 2000). However, recent findings by Lin et al. (2009) suggest that at least some components of the protein synthesis machinery are present in pumpkin phloem sap, raising the possibility that limited translation may occur.
The view of the phloem function has evolved from that of simple assimilate transport to a trafficking system for pathogen response and developmental regulators (Citovsky and Zambryski, 2000; Ding et al., 2003; Wu et al., 2003; Haywood et al., 2005). The phloem is essential for the transport of mineral nutrients, plant viruses, virus-induced silencing, defence and resistance against pathogen infection, and signalling of environmental conditions (Lucas et al., 1995; Ryabov et al., 1999; Burton et al., 2000; Seo et al., 2001; Yoo et al., 2004; Suárez-López, 2005; Kehr, 2006; Lough and Lucas, 2006). It contains a multitude of compounds: small molecules (Chen et al., 2001; Corbesier et al., 2003), peptides and proteins (Fisher et al., 1992; Sakuth et al., 1993; Schobert et al., 1995; Kühn et al., 1997; Marentes and Grusak, 1998; Kehr et al., 1999; Xoconostle-Cazares et al., 1999; Haebel and Kehr, 2001; Hoffmann-Benning et al., 2002; Giavalisco et al., 2006; Lin et al., 2009), and nucleic acids (Kühn et al., 1997; Ruiz-Medrano et al., 1999; Yoo et al., 2004; Haywood et al., 2005). Moreover, interactions between several proteins or RNA and proteins to facilitate or regulate phloem transport appear more and more likely (Xoconostle-Cazares et al., 1999; Aoki et al., 2002, 2005; Yoo et al., 2004; Lee et al., 2005; Kehr and Butz, 2008; Ham et al., 2009). The contents of the phloem are now known to be so complex that phloem transport has been called the ‘information superhighway’ of plants (Jorgensen et al., 1998). It is important for the signalling of environmental conditions as well as in the pathogen response of plants.
Many plant viruses have hijacked the phloem transport mechanisms to spread throughout the infected plant (Waigmann et al., 2004; Gopinath and Kao, 2007). On the other hand, plants are able to show a systemic response to infection, which makes long-distance communication within the plant essential (Durrant and Dong, 2004; Truman et al., 2007). Plant hormones such as systemin and jasmonic acid (JA), which elicit a pathogen response, and several pathogenesis-related proteins have been shown to be transported in the phloem in response to pathogen infection (Schillmiller and Howe, 2005; Truman et al., 2007; Thorpe et al., 2007; summarized in Lough and Lucas, 2006).
Similarly, the floral stimulus is formed in the leaves in response to an inductive photoperiod and translocated through the phloem to the apical meristem (Lang, 1965; Zeevaart, 1976). It was shown that the interaction between CONSTANS (CO) and FLOWERING LOCUS T (FT) in the plant is essential for the induction of flowering (An et al., 2004; Ayre and Turgeon 2004) and that the FT protein in Arabidopsis and cucurbits and its analogue Hd3a in rice (Kojima et al., 2002; Corbesier et al., 2007; Lin et al., 2007; Tamaki et al., 2007) are the long-distance signal for flowering.
The difficulty in working with plant phloem lies in its capacity to seal itself upon wounding. Only in few plant species is it possible to obtain reasonable amounts of phloem exudate through cuts of the petiole (cucurbits; Balachandran et al., 1997; Haebel and Kehr, 2001), shallow cuts or punctures in the stem or petiole (lupine; Marentes and Grusak, 1998; Hoffmann-Benning et al., 2002; Brassica napus; Giavalisco et al., 2006), or by using aphids (Fisher et al., 1992; Doering-Saad et al., 2002; Walz et al., 2002; Barnes et al., 2004). The first two of the approaches mentioned above are not feasible due to the fragile nature of Arabidopsis inflorescence stems and petioles. Aphids introduce several proteins into the phloem with their saliva (Kehr, 2006; Will and van Bel, 2006). This leads to a limited transcriptional reprogramming (Thompson and Goggin, 2006) and the reduction in normal callose deposition and P-protein accumulation, and has the potential to change the phloem composition. For this reason, it was decided to use a method employing EDTA, which had been successfully used in other plants (King and Zeevaart, 1974; Hoffmann-Benning et al., 2002). Phloem sap obtained in this way can be used to analyse proteins, small molecules, lipids, and RNAs (Guelette et al., 2007; Deeken et al., 2008).
With the analytical methodology becoming increasingly more sensitive, the complexity of the phloem exudates becomes even more apparent. The next logical step in the study of the function of phloem macromolecules is not only the establishment of comprehensive databases of vascular genes and phloem-mobile transcripts, proteins, and metabolites, but to understand the functions of the phloem macromolecules. This will help to better understand previously unknown long-distance transport and signalling pathways. The present study represents a further step towards the characterization of phloem-mobile proteins and metabolites. A successful attempt to collect Arabidopsis phloem exudate in large enough amounts to allow for the analysis of metabolites, proteins, and, to a lesser extent, RNA, is described. While the role of RNA, viruses, and many proteins is extensively studied, little is known about fatty acids and lipids and their transport throughout the phloem (Madey et al., 2002; Guelette et al., 2007; Behmer et al., 2010). The present findings suggest that lipids and lipid-binding proteins are indeed present in the phloem sap. Since the phloem is an aqueous environment, lipids are not necessarily expected. Yet, other aqueous biological systems such as human blood contain a variety of lipids. Typically these are bound to proteins. Some lipid–protein complexes are transported to other tissues for storage, modification, or degradation; others transport essential vitamins or play a role in signalling (Blaner, 1989; Glatz et al., 1995; Charbonneau et al., 2009). In several cases, these lipid–protein complexes serve as messengers and affect transcription factor activity (Tontonoz et al., 1994; Nagy and Szanto, 2005). Here it is hypothesized that plant phloem lipids and lipid-binding proteins have regulatory and transport functions in plants similar to those in animals and that they may play an important role in stress-related and developmental signalling beyond what is already known about hormones such as JA.
Materials and methods
Growth of plant material
Arabidopsis thaliana var. Columbia seedlings were surface sterilized in 20% bleach/1% Triton X-100 for 20 min, washed several times with water, and plated on a Petri dish containing half-strength Murashige and Skoog Basal medium (Sigma-Aldrich). They were incubated for 2 d in darkness in the refrigerator followed by a 5 d incubation at 22 °C. Seedlings were planted in soil and grown at 22 °C during the day and at 18 °C during the 8 h night (LD).
Harvest of phloem exudate
Phloem exudate harvest was started 2–3 h after the onset of light. Arabidopsis rosette leaves from 6-week-old plants were cut and re-cut in 20 mM K2-EDTA, pH 7.0, placed in groups of 10 in a microtube containing 1.2 ml of the same solution, and incubated for 1 h in a dark and humid atmosphere. After 1 h, the solution was discarded. The petioles were washed thoroughly with water to remove all residual EDTA and placed in a new microtube containing 1.2 ml of deionized water. Exudates were collected for 6–8 h in a dark, humid atmosphere, immediately frozen in liquid nitrogen, lyophilized, and stored at –80 °C. For preparation of phloem RNA, 100 U of RNase inhibitor (Roche) was added per ml of deionized water into which the phloem exudate was collected.
Analysis of proteins in the phloem exudates
Phloem exudate was analysed using an Ultrafast Microprotein Analyzer system with a peptide microtrap and a C18 reverse-phase column (Michrom BioResources, Auburn, CA, USA). Samples were resuspended in 100 μl of deionized water and loaded onto the peptide microtrap. The gradient went from 5% acetonitrile (ACN)/0.1% trifluoroacetic acid (TFA) in water to 65% ACN/0.1% TFA in water in 42 min. The flow rate was 500 μl min−1. Fractions of interest were further analysed using matrix-assisted laser desorption ionization-time of flight-mass spectrometry (MALDI-TOF-MS) or Edman sequencing. Edman sequencing was performed by the MSU- Research Technology Support Facility.
Alternatively, phloem exudates from 20 leaves per replicate (three replicates in total) were subjected to 15% SDS–PAGE. Protein bands were excised and digested with trypsin according to Shevchenko et al. (1996). The digested peptides were extracted into 60% ACN/1% TFA and the appearance of tryptic fragments was monitored using MALDI-TOF-MS. For liquid chromatography–tandem mass spectrometry (LC-MS/MS), samples were dried and resuspended into 2% ACN/0.1% formic acid. LC-MS was performed on a Capillary LC system (Waters Corp., Milford, MA, USA) coupled to an LCQ DECA ion trap mass spectrometer (Thermofinnigan, San Jose, CA, USA) equipped with a nanospray ionization source. The sample was trapped onto a Peptide Cap Trap (Michrom BioResources) and flushed onto a 5 cm×75 μm ID picofrit column packed with 5 μm ProteoPep C18 material (New Objective, Woburn, MA, USA). Peptides were eluted with a gradient of 2–95% ACN in 0.1% formic acid at a flow rate of 200 nl min−1 for 60 min. Peptides/proteins were identified using the programs SEQUEST, MASCOT, or gpm.org. Carbamidomethyl cysteine was set as the fixed modification and oxidation of methionine was allowed. Up to two missed tryptic sites were permitted. Peptide tolerance was set to 2.5 Da, and MS/MS tolerance was set to 0.8 Da. Positive identifications were confirmed by individually comparing MS/MS spectra. Positive identification required at least two unique peptides per proteins counting only peptides with significant scores (95% confidence per peptide; >2.5 for SEQUEST). Gupta and Pevzner (2009) have suggested that inclusion of single-hit proteins does not necessarily increase the false discovery rate. Four additional proteins with a single peptide identification were included due to the fact that their mass corresponded to the predicted size from the gel and they were seen in at least two independent preparations. One protein was submitted to Edman sequencing (Ed in Supplementary Table S1 available at JXB online) at the Michigan State University Research Technology Support Facility. Database searches using individual tryptic fragments were performed using the BLAST searches at NCBI (http://www.ncbi.nlm.nih.gov/blast). Predicted function/localization of proteins is based on the NCBI, GO (http://www.ebi.ac.uk/QuickGO/), and/or Bar (http://bar.utoronto.ca/welcome.htm) databases as well as the cited literature.
Analysis of metabolites in the phloem exudates (modified from Roessner et al., 2000)
Phloem exudate from 20 Arabidopsis leaves was resuspended, pooled, and phase partitioned with chloroform:methanol (1:1). Both phases were dried individually and derivatized using methoxyamine in pyridine and BSTFA [N,O-bis-(trimethyl silyltrifluoroacetamide)] with 1% trimethylchlorosilane. The metabolites were then separated by gas chromatography–mass spectrometry (GC-MS) using a JOEL XMS-AX505H system with a DB17/MS column (J&W Scientific, 30 m×0.32 mm ID×0.25 μm film; temperature program: 70-5, 5/min-320-5; data in Fig. 1) or an Agilent 5890N GC system coupled to a 5973 MSD using a DB 5MS column (J&W Scientific, 30 m×0.25 mm ID×0.25 μm film; temperature program: 50-2, 10/min-320-5; data in Supplementary Fig. S3 at JXB online). Identification was carried out using the NIST library. Only compounds with a score >80 that passed additional individual comparison were included in the list. Data sets from four independent preparations were compared. Metabolites included in Table 1 had to be present in at least three of those preparations and absent in the controls. Two sets of controls were used for the phloem metabolite analysis: one ‘blank’ sample applying the complete GC-MS preparation and analysis to 1.2 ml of deionized water (only reagent peaks were visible; not shown) and one minus-EDTA sample where leaves/petioles had never been submerged in EDTA but collection occurred directly into deionized water (see Fig. 1). Reagent peaks were visible only in the first 10 min of each GC-MS. To confirm the presence, identity, and relative ratios of sucrose:glucose and sucrose:fructose, standard solutions of these sugars were derivatized both individually and as a 1:1:1 mixture, and analysed using GC-MS as described above. Analysis of the peak areas from the chromatogram of the mixture confirmed the maintenance of the 1:1:1 ratio between those sugars and thus no impact of BSTFA (not shown).
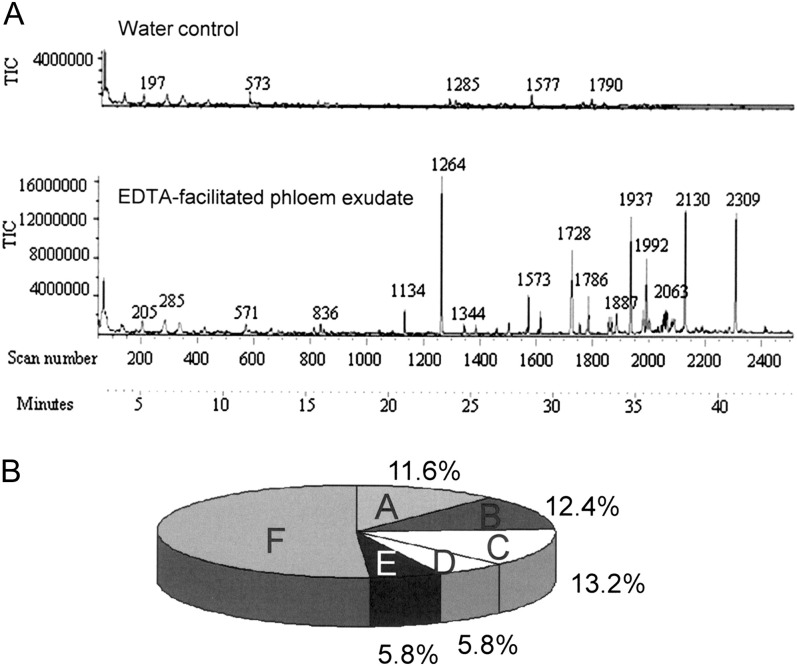
Fig. 1.
(A) Gas chromatogram showing the separation of metabolites from the chloroform phase of Arabidopsis thaliana phloem exudates. The bottom panel displays metabolites present in the exudates of petioles incubated first in EDTA to prevent the sealing of the phloem. The top panel demonstrates the absence of any major metabolite peaks if the petioles were incubated in water (water control). The water control was fitted to the same scale as the EDTA extract. Both panels are a typical representative of three biological replicates. (B) Distribution of metabolites detected in Arabidopsis phloem exudate. They were grouped into amino acids (A), sugars and sugar derivatives (B), fatty acids/-esters (C), organic acids and phosphates (D), others (E), and unknowns (F).
Table 1.
Metabolites identified in Arabidopsis thaliana phloem exudate
| Amino acids | Sugar/-alcohols phosphates | Fatty acids/-esters | Organic acids | Others |
| Alanine | Ribose | Nonanoic acid* | Propanoic acid (1) | Uridine |
| Valine | Fructose | Dodecanoic acid* | Ethyl-phosphoryc acid | Bis-ethyl-ethane amine |
| Leucine | Glucose | Octadecanoic acid butyl ester | Butanoic acid | Phytosterol-like cpd (13) |
| Glycine | Galactitol | Tetradecanoic acid* | Malic acid (4) | Urea |
| Isoleucine | Galactonic acid | Pentadecanoic acid | 2-Methoxy-pentanedioic acid | Cinnamic acid |
| Serine | Myo-inositol (9) | Octadecanedioic acid | Acetic acid | Dehydroabietic acid (12) |
| Threonine | 5 Hexoses (7) | Palmitelaidic acid | Maleic acid (3) | Cyclopentanapth azepinone |
| Aspartic acid | 2-O-Glycerol-galactopyranoside | Palmitic acid (8) | Succinic acid | Phosphate |
| Glutamine | Myo-inositol-P | Octadecanoic acid methyl ester | Propanedicarboxylic acid (6) | |
| Proline (5) | Sucrose | 11-trans octadecanoic acid | ||
| Phenylalanine | Melibiose (14) | Octadecanoic acid (11) | ||
| Asparagine | Eicosanoic acid | |||
| Tyrosine | 9,12,2-OH Octadecanoic acid methyl ester | |||
| Docosanoic acid methyl ester* | ||||
| Oleic acid (10) |
Lipids marked with an asterisk are related to those found by Madey et al. (2002). Numbers correspond to peak # in Supplementary Figure S3 at JXB online.
Analysis of phloem exudate for the presence of mRNA
RNA was prepared using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA), digested with DNase for 30 min at 37 °C, and repurified. RNA concentration and quality were determined with a NanoDrop ND-1000 UV-Vis spectrophotometer (NanoDrop Technologies). The 260/280 ratios of the mRNA were in the range of 1.68–2.16. A 200 ng aliquot of mRNA was reverse transcribed using the Omniscript RT Kit (Qiagen). Semi-quantitative RT-PCR to ensure phloem sap purity was performed monitoring 18S (F, TCAACTTTCGATGGTAGGATAGTG; R, CCGTGTCAGGATTGGGTAATTT; product size, 144 bp), Rubisco small subunit (SSU; F, CATCCCCGGCTCAGGCC; R, AAGGAATCCACTTGTTGCGG; product size, 230 bp), Rubisco large subunit (LSU; F, GCTACCACATCGAGCCCG; R, CGGCACAAAATAAGAAACGG; product size, 372 bp), pathogen and circadian controlled (PCC1; At3g22231; F, TCCTCACTCCTCAGCTCCTC; R, GTTTGGGCAACGACTTCTGT; product size, 363 bp), and ubiquitin-conjugating enzyme (UBC9; At4g27960; F, TGGCTTCGAAAAGGATCTTG; R, TCGATATGGTGAGTGCAGGA; product size, 290 bp). The annealing temperature for all reactions was 53 °C/40 cycles using a PCR master mix (Promega; Taq DNA polymerase). The identity of the PCR products was confirmed based on the expected product size (see figures in parentheses above) and sequencing of representative bands: PCR products were cloned into pGEM T-Easy (Promega), transformed into Escherichia coli strain DH5α using heat shock treatment, and inserts sequenced following a plasmid preparation (Wizard plus SV Miniprep; Promega).
Analysis of lipids in phloem exudates
Phloem exudates were phase partitioned against chloroform:methanol (1:1, v/v), concentrated under N2, and submitted to either thin-layer chromatography (TLC; acetone:methanol:water; 90:30:7) or LC-MS. For the latter, the hydrophobic components of the phloem exudates were separated using a LC-20AD Ultrafast liquid chromatograph (Shimadzu, Kyoto, Japan) on a C18 column (5 cm×2.1 mm×2.7 μm; Supelco, Bellefon, PA, USA) with a gradient from 30% solvent A (10 mM ammonium acetate) to 100% solvent B (100% isopropanol/10 mM ammonium acetate), and identified using electrospray ionization (ESI)-MS (3200 QTRAP; AB/MDS). Initial identification of lipids was based on their m/z plus the presence of at least two predicted MS/MS fragments using the LipidView software by AB/MDS. To improve lipid identification, subsequent lipid analysis was performed using a Waters LCT Premier mass spectrometer (LC-TOF-MS) with multiplexed CID (collision-induced dissociation, at 20, 35, 50, 65, and 80 V) and a gradient from 10% A (10 mM ammonium formate) to 99% B (acetonitrile:isopropanol; 1:2) on the column described above using both positive and negative ion mode. Data are representatives of three biological replicates.
Results and Discussion
Extraction of phloem exudate without cellular cross-contamination
Collection and analysis of the phloem sap are complicated by the fact that, in most plants, the phloem seals itself upon wounding, and no exudates are secreted. To prevent this, Arabidopsis petioles were incubated for 1 h in EDTA. This initial exudate was discarded. The petioles were washed, followed by collection of the exudate into water using the method previously described for Perilla (King and Zeevaart 1974; Hoffmann-Benning et al., 2002) and modified for Arabidopsis (Maeda et al., 2006; Guelette et al., 2007; Deeken et al., 2008; see Supplementary Fig. S1 at JXB online). These authors had thoroughly tested the effect of EDTA on the phloem exudation in Perilla crispa. They had shown that exudation was maximal during the first 4 h and continued at a slightly reduced rate after that. They also determined that EDTA stimulates the exudation of assimilates and leads to less callose accumulation in the sieve pores. The conclusion was that EDTA chelates Ca2+ ions, which would otherwise participate in reactions that seal the phloem (King and Zeevaart, 1974). Indeed, it was shown later that, as a result of plasma membrane leakage, injury, or turgor disruption, P-protein crystalloids present in legume sieve tubes disperse and block the sieve pores. This process is facilitated by Ca2+ and inhibited by EDTA (Knoblauch et al., 2001). At the same time, no effect of EDTA on the cell ultrastructure was observed, indicating that it can be safely used for phloem exudate collection. A similar method used to analyse glucosinolates and tocopherols, respectively, in the phloem also showed no adverse effect on phloem loading and transport (Chen et al., 2001; Maeda et al., 2006). To summarize, phloem exudate collection methods using a range of EDTA concentrations from 10 mM to 20 mM have been published and have shown no adverse effects. To reduce interference of EDTA with chromatography and gel electrophoresis, the plants were moved into water after 1 h and only the later part of the exudate was used (Supplementary Fig. S1 at JXB online; Hoffmann-Benning et al., 2002). Thus, rather than exudation into EDTA, phloem is exudated into water (EDTA-facilitated exudation).
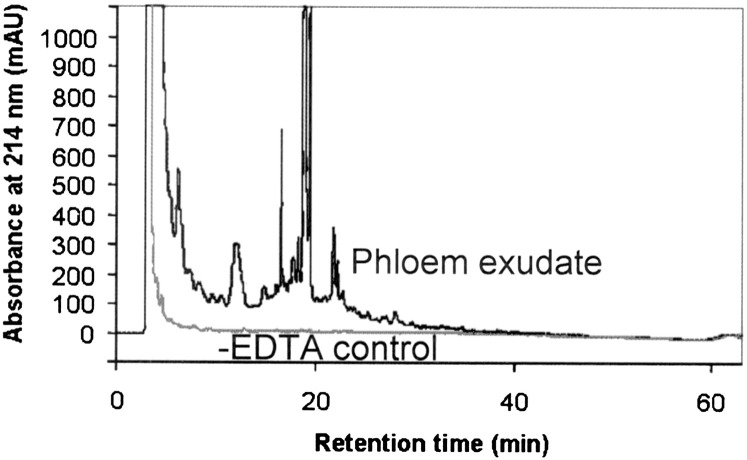
Collection of exudates for different lengths of time showed the same protein pattern after 1, 5, and 9 h of exudation. Particularly during the first 5 h the density of all protein bands increases with the increase in exudation time, confirming that as the collection time increases, more protein is accumulated and that exudation has not ceased during longer collection times. Except for a band at ∼50 kDa, the density of bands relative to each other appears the same and no new bands appear, suggesting that cell damage is unlikely and that collection for 5–9 h is optimal (Supplementary Fig. S2 at JXB online). To account for proteins or metabolites that would leak from cut cells, the same number of leaves was incubated for the first hour in water instead of EDTA and then the exudate was collected into water. Exudation into water without prior EDTA treatment does not yield metabolites or proteins detectable by GC-MS (Fig. 1) or high-performance liquid chromatography (HPLC; Fig. 2). On the other hand, collection into water after a short incubation with EDTA leads to the detection of a large number of proteins and metabolites (Figs 1–7).
Fig. 2.
HPLC chromatogram of Arabidopsis phloem exudates extracted into water after previous incubation with EDTA (black) or without EDTA (water control; grey).
Fig. 7.
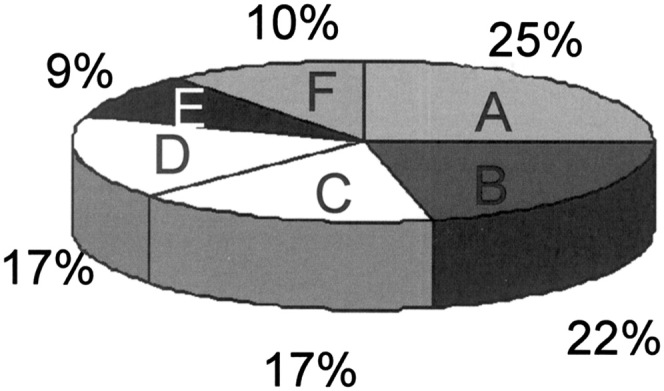
Distribution of proteins detected in Arabidopsis phloem exudate. Proteins were grouped by function into pathogen/stress response (A), carbohydrate metabolism (B), protein binding/modification/metabolism (C), lipid/fatty acid binding/metabolism/signalling/storage (D), DNA/RNA/nucleotide binding (E), and others/unknown function (F).
Extraction of mRNA from leaves, petioles, and phloem exudate followed by RT-PCR shows the presence of Rubisco small and large subunit mRNA in leaves and petioles but not in the phloem exudate (Fig. 3). On the other hand, mRNAs for UBC9 (At4g27960) and PCC1 (At3g22231) were found to be present in the phloem exudates (Fig. 3). Both had previously been described in phloem exudates (Deeken et al., 2008). The absence of Rubisco mRNAs indicates that compounds detected and identified are indeed largely derived from the phloem sap and are not due to intracellular contamination leaked through the cut surface or membranes. The amounts of fructose and glucose relative to sucrose were determined using GC-MS after exudation for 1 h. Identification of fructose, sucrose, and glucose was based on the retention time of individual standards as well as a mixture, and on identification through the NIST library. It revealed that sucrose was the predominant sugar/metabolite in the exudates, with a peak area ratio of 1:7.6 and 1:8 for glucose:sucrose and fructose:sucrose, respectively (Supplementary Fig. S3 at JXB online). This matches the findings of Deeken et al. (2008) whose ratios of glucose to sucrose (96 nmol g FW−1) in phloem exudates were between 1:4 and 1:8. Peaks eluting in the first 10 min were identified as reagent peaks and are marked with an asterisk. True metabolite peaks are indicated with a number and cross-referenced in Table 1.
Fig. 3.
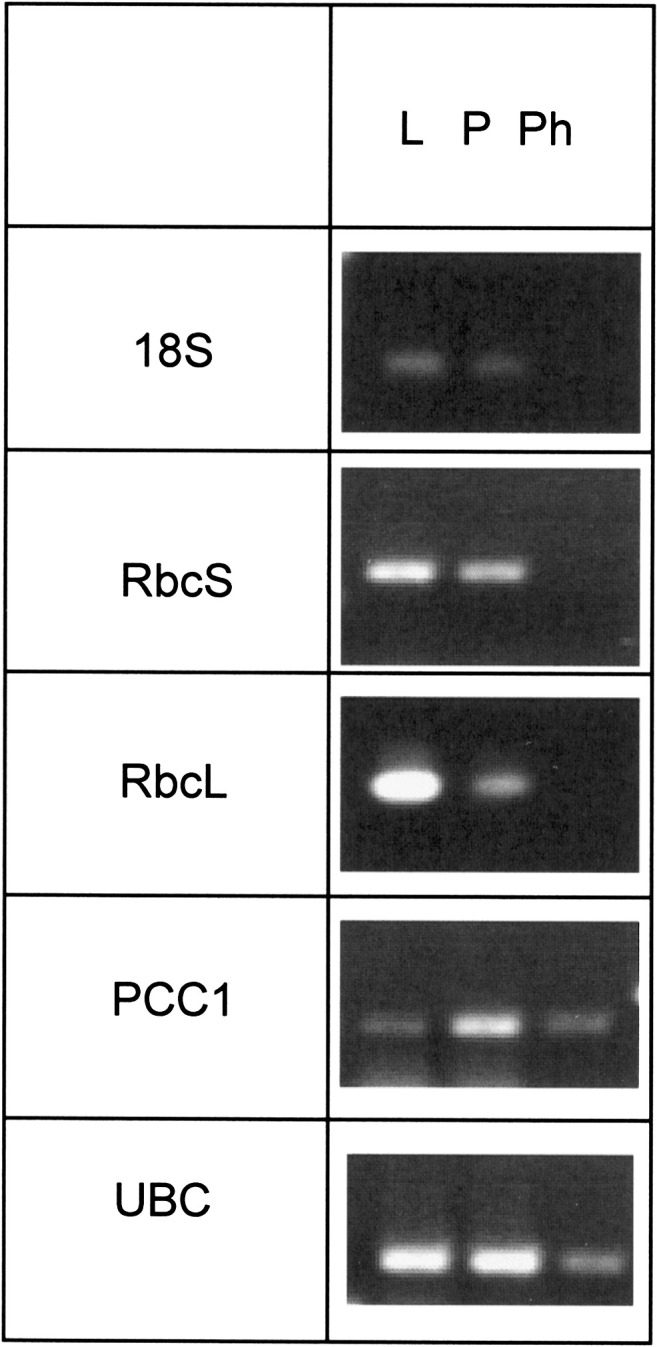
Presence of mRNAs for 18S, Rubisco small and large subunit (RbcS and RbcL), ubiquitin-conjugating enzyme (UBC), and PCC1 (pathogen and circadian controlled) in samples from leaf (L), petiole (P), and phloem exudates (Ph) as determined by RT-PCR. The picture shows a representative of three independent biological replicates.
Identification of metabolites in Arabidopsis phloem exudate
Metabolites were analysed using total phloem exudates as well as separated hydrophobic and hydrophilic fractions. GC-MS revealed the presence of at least 121 distinct metabolite spectra in the phloem exudates. Of these, ∼50% gave rise to spectra of known identity (Table 1; Fig. 1A, B: hydrophobic fraction). Eleven percent of the metabolite spectra belonged to amino acids, 12% to sugars and sugar derivatives, 13% to fatty acids and their derivatives, and 6% to organic acids and phosphates. Amino acids and sugars (monosaccharides and disaccharides) are assimilates expected to be present in the phloem. As expected, sucrose was the most abundant metabolite present (Supplementary Fig. S3 at JXB online).
The presence of myo-inositol and myo-inositol phosphate in the phloem has interesting implications. Myo-inositol phosphate is largely known as a precursor of phytic acid, a storage compound for phosphate in seeds (Hegeman and Grabau, 2001). It also plays a central role in plant metabolism, signal transduction, and cell wall biosynthesis (Loewus and Murthy, 2000). Since both molecules can be readily converted into each other by highly regulated enzymes (myo-inositol kinase; myo-inositol phosphatase), myo-inositol phosphate may also be a source for myo-inositol. This, in turn, is rate limiting for the synthesis of phosphatidylinositol bisphosphate and can thus either directly or indirectly play a role in signalling in the phloem (Ananieva and Gillaspy, 2009; Munnik and Vermeer, 2010).
It was possible to detect several fatty acids and their derivatives (Table 1). The presence of lipids/fatty acids in the phloem has been reported before, when small amounts of lipids and large amounts of free fatty acids were identified in the phloem of canola (Madey et al., 2002). Recently, Behmer et al. (2010) suggested the presence of phytosteroids and cholesterol in the phloem sap. Like in Madey et al. (2002), fatty acids were detected in this study. Several of these were short and medium length and medium length, saturated fatty acids (non-anoic acid, dodecanoic acid, tetradecanoic acid, and pentadecanoic acid), including odd-chain length fatty acids (propanoic acid, nonanoic acid, and pentadecanoic acid). Propanoic acid and butanoic acid are grouped with organic acids, although they are considered to be short-chain fatty acids (Oxford Dictonary of Chemistry; www.cyberlipid.org). Saturated, even-numbered fatty acids make up 10–40% of the total fatty acids in most naturally occurring lipids, with C14–C18 fatty acids being the most abundant (http://lipidlibrary.aocs.org). Shorter chain and odd-numbered fatty acids are not typically present in plant membranes and, thus, appear to be specific to the plant phloem. Yet their function remains unknown. They could simply be transported in the phloem; however, a role in pathogen response or signalling has been discussed in the literature (Sanz et al., 1998; Hamberg et al., 1999).
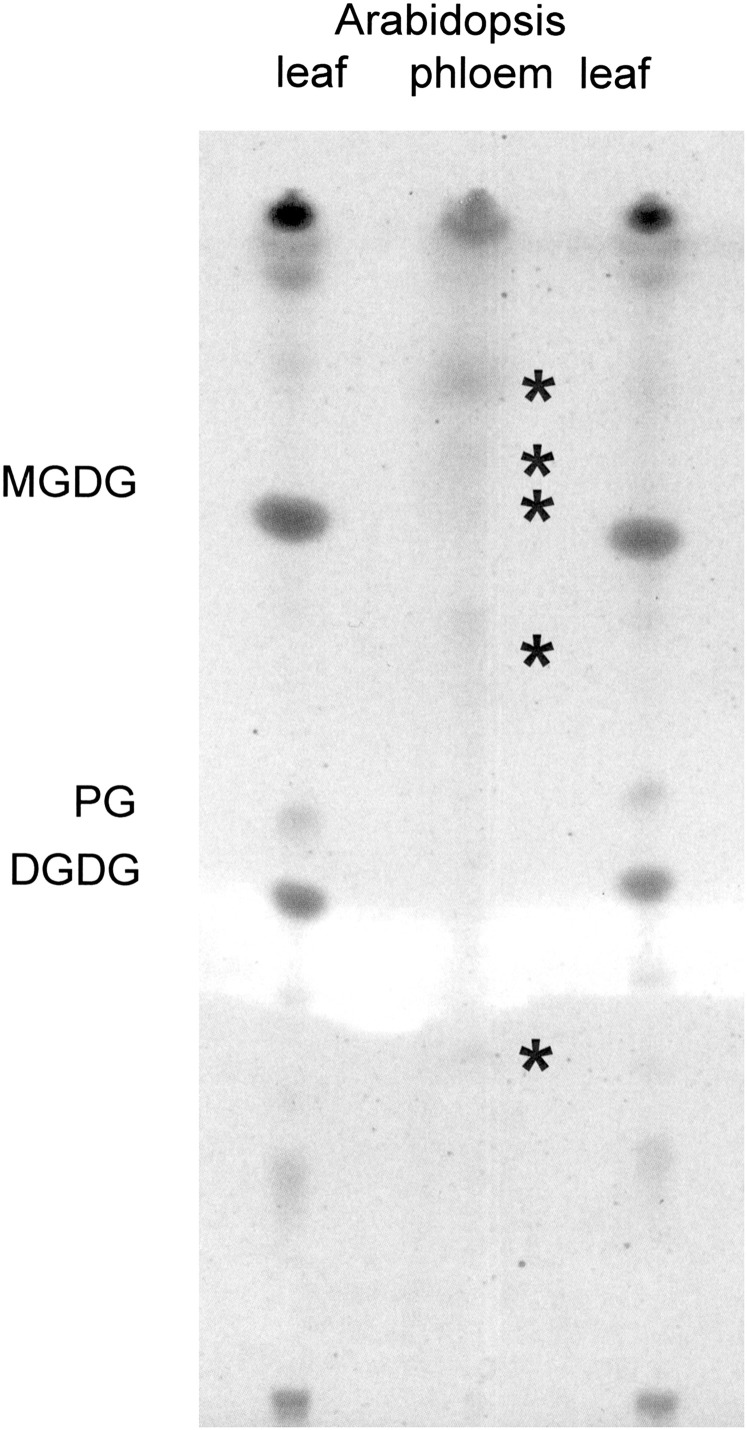
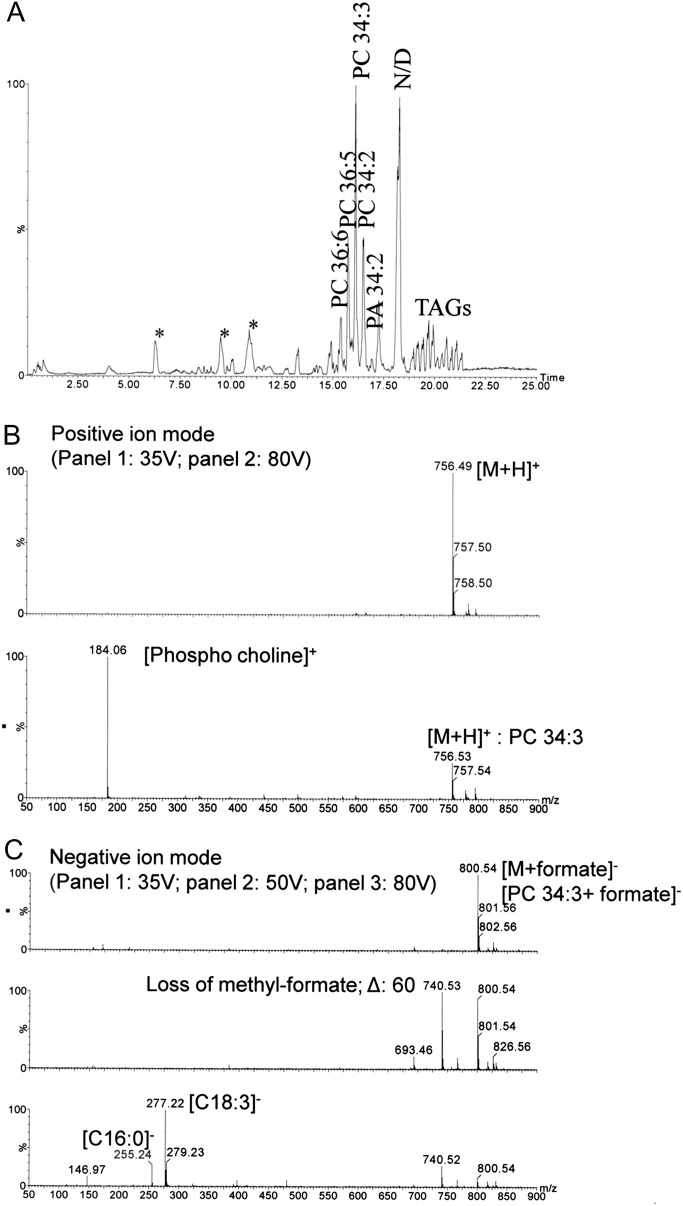
The presence of fatty acids raises the possibility of more complex lipids in the phloem sap. TLC confirmed the presence of a small amount of lipids in the phloem exudate (Fig. 4). Five of the lipids have retention times different from those present in leaves. This suggests that they are not contaminants but may instead perform separate functions in the phloem. LC-MS confirmed the occurrence of lipids and other hydrophobic compounds in exudates (Fig. 5A ). Mass over charge-based characterization of ions in conjunction with detection of at least two predicted MS/MS fragments using lipid profiler as well as manual interpretation (Fig. 5B) suggests the presence of phosphatidic acid (PA), lysophosphatidic acid (LPA), phosphatidylglycerol (PG), phosphtidylinositol (PI) and its phosphates (PIPs), di- and triacylglycerols (TAGs), and phosphatidylcholine (PC). An example is given for the lipid at the retention time of 16.1 min [PC 34:3 (16:0, 18:3); Fig. 5B]. PA and PIPs are known second messengers in plants (Wang, 2004). However, little is known about their long-distance transport. These findings raise the possibility that they could also play a role in long-distance signalling.
Fig. 4.
Thin-layer chromatogram comparing lipids from Arabidopsis leaves and phloem exudates. The asterisks indicate lipids specific for phloem exudates. PG, phosphatidylglycerol; MGDG, monogalactosyldiacyglycerol; DGDG, digalactosyldiacylglycerol.
Fig. 5.
(A) Lipid profile of phloem exudates of Arabidopsis (chloroform phase) using LC-MS/positive ion mode. Mass spectra of the lipid at a retention time of 16.1 min were generated at aperture 1 voltages of 35, 50, and 80 V in both positive (B) and negative (C) ion mode to show lipid fragmentation. PC, phosphatidylcholine; PA, phosphatidic acid; TAG, triacyl glycerol; *, detergent; N/D, not determined due to multiple peaks in the spectra. Chromatograms are representatives of three biological replicates for each sample.
Identification of proteins present in Arabidopsis phloem exudate
In the past, several labs have isolated phloem exudates and separated the proteins therein. Some of these experiments were to follow-up specific proteins or look at specific functions (Schobert et al., 1995; Kühn et al., 1997; Balachandran et al., 1997; Xoconostle-Cazares et al., 1999; Hoffmann-Benning et al., 2002; Lee et al., 2005); others attempted to obtain a more comprehensive protein profile (Fisher et al., 1992; Sakuth et al., 1993; Marentes and Grusak 1998; Kehr et al., 1999; Haebel and Kehr, 2001; Giavalisco et al., 2006). Over 1000 proteins have been detected in those exudates (Lough and Lucas, 2006; Lin et al., 2009). Many of those, however, have just been characterized as spots on a gel with no clear identification (Kehr, 2006). In addition, many of those spots on the gel correspond to identical proteins with modifications such as phosphorylation and glycosylation (Nakamura et al., 1995; Taoka et al., 2007). A previous very comprehensive proteomics study of phloem proteins was done in a close relative of Arabidopsis, Brassica napus, and yielded 101 distinct genes out of 140 gel spots (Giavalisco et al., 2006). This shows that large numbers of phloem proteins can be identified if the species has its genome sequenced or is closely related to one that does. The fact that many of the protein spots identified from their two-dimensional gels correspond to identical proteins (22% based on identical accession numbers) suggests that a large number of post-translational modifications are present in phloem proteins.
Using a combination of HPLC and Edman sequencing or SDS–PAGE (Fig. 6) followed by in-gel digests and LC-MS/MS, it was possible to distinguish 65 proteins present in Arabidopsis phloem exudate (Supplementary Table S1 at JXB online; Figs 6, 7). Many (23%) of those appear to be identical to the proteins described by Giavalisco et al. (2006) for Brassica (indicated on Supplementary Table S1). The identified proteins could be grouped into the following categories based on their predicted or known function according to Fig. 7.
Fig. 6.
SDS–PAGE of proteins from Arabidopsis phloem exudate.
Carbohydrate metabolism
This group included 14 proteins, most of which had been found in the phloem of other plant species (Walz et al., 2002; Giavalisco et al., 2006). It was discussed that all enzymes of the glycolytic pathway are present in the phloem but are repressed due to the low oxygen levels (Geigenberger, 2003). Fructose-2,6-bisphosphatase and triosephosphate isomerase have now been added to this list.
Stress, pathogen, or hormone response proteins (16)
Several disease-resistance proteins (RPP1-WsB and RPP8-like protein), components of the myrosinase system (myrosinase and myrosinase-binding protein), as well as several other pathogen response proteins (a thaumatin-like protein, major latex protein, and Bet v1 allergen-like protein) were detected. All had been described previously in the phloem (Giavalisco et al., 2006; Lin et al., 2009; Dafoe et al., 2010). The Bet v1 family protein belongs to the group of PR-10 pathogenesis-related proteins. Experiments have shown that it has the chemical ability to bind fatty acids and cytokinins (Mogensen et al., 2002) as well as brassinosteroids, and may play a role in plant growth and development (Markovic-Housley et al., 2003), but probably not in brassinosteroid trafficking (Symons and Reid, 2004). Some proteins in this group have been shown to be induced by auxin (actin 7/2, Zn-binding dehydrogenase, and glutathione S-transferase), abscisic acid (uncharacterized phloem protein UPP3), JA (myrosinase-binding protein), or to play a role during oxidative (superoxide dismutase CSD 1 and 2; Walz et al., 2002) or anaerobic stress (alanine aminotransferase; Good and Muench, 1992), or in cold, drought, and salt tolerance (glyoxylase I, monodehydroascorbate reductase, and carbonic anhydrase). The function and presence in the phloem of most of these proteins has been described by Giavalisco et al. (2006) and Lin et al. (2009). This is the first description of the putative Zn-binding dehydrogenase, alanine aminotransferase, and glyoxylase 1 in the phloem.
Nucleotide-, DNA-, or RNA binding proteins (six)
These include a possible transcription factor (unidentified phloem protein UPP6) as well as RNA-binding proteins (GRP7 and UPP5). In addition, several of the stress response proteins listed above contain nucleotide-binding sites (RPP8, ethylene-responsive RNA-helicase, and 14-3-3 like proteins—two subunits). Most could be interesting in light of the fact that mRNA appears to be transported through the phloem and acts as a signalling molecule, a process that is facilitated by RNA-binding proteins (Heintzen et al., 1997; Xoconostle-Cazares et al., 1999; Yoo et al., 2004; Aoki et al., 2005; Deeken et al., 2008; Ham et al., 2009; Lin et al., 2009).
‘Plastidal’ proteins (11)
As in Giavalisco et al. (2006) in B. napus, a small number of proteins which have previously been described as associated with chloroplasts or mitochondria were found. In general, most of the proteins present in phloem exudates are assumed to be synthesized in the companion cell and transported into the sieve elements (Fisher et al., 1992; Sakuth et al., 1993; Dannenhofer et al., 1997; Ruiz-Medrano et al., 2001) though a new discussion about protein translation in phloem sap has been initiated (Lin et al., 2009). Eleven proteins were found that are known to be located predominantly in the chloroplast. Among these are the two Rubisco subunits which have been previously described in phloem exudates (Marentes and Grusak, 1998; Giavalisco et al., 2006; Lin et al., 2009; Rodriguez-Medina et al., 2011). Since the ‘plastidal’ proteins detected are mostly nuclear encoded, it is possible that they also have been actively transported into the sieve element from companion cells through plasmodesmata and serve a related function here. This transport has been shown to be a highly regulated process needing the presence of chaperones (Aoki et al., 2002, 2005) or kinases (Lee et al., 2005). A series of green fluorescent protein (GFP) transport and other experiments has shown that the size exclusion limit (SEL) for proteins trafficking through plasmodesmata companion cells to the sieve element can be up to 67 kDa, though some labs report a SEL of only 25 kDa (Oparka et al., 1999; Fisher and Cash-Clark, 2000; Kim et al., 2002; Stadler et al., 2005; Martens et al., 2006). This would be sufficient to explain the presence of the vast majority of proteins (plastidal and others) detected in the phloem. Another explanation for their presence lies in the fact that during development, sieve elements have gradually lost their nucleus and most of their organelles (van Bel, 2003; Lough and Lucas, 2006). However, angiosperm sieve elements still contain P- and S-type plastids and few dilated mitochondria, though the functionality of the latter is still debated (van Bel, 2003). The plastidal proteins detected here could be components of those organelles.
Protein modification, folding, turnover, and stability, and protein–protein interaction (11 proteins)
The presence and the role of components of the translational apparatus, the ubiquitin–proteasome pathway, as well as that of kinases, proteases, and chaperones had been described previously and will not be discussed here (Aoki et al., 2005; Nakamura et al., 1995; Schobert et al., 1995; Giavalisco et al., 2006; Lin et al., 2009). One protein, annotated as a phosphoglycerate mutase/putative acid phosphatase-like protein, is interesting in the context of phosphate homeostasis or signalling. It contains a histidine phosphatase domain which is found in histidine acid phosphatases and phytases (myo-inositol hexakisphosphate phosphohydrolases). Proteins with this domain function in metabolism, signalling, and regulation (http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid=209303). Though not (yet) annotated as such, one possible function, rather than cleaving a phosphate of a protein, could be to liberate the phosphate from the myo-inositol backbone (Mullaney and Ullah, 1998). It could thus play a major role not just in phosphate metabolism, but also in regulating the relative ratios of myo-inositol phosphate and myo-inositol available for osmoprotection and signalling. This is of particular significance since its potential substrates have also been detected in the phloem exudate (see above).
14-3-3 proteins are small conserved proteins, which bind an array of regulatory proteins such as kinases, phosphatases, ATPases, and transmembrane receptors. They play an important role in regulatory processes and signal transduction. In plants they activate transcription by binding to activators in the nucleus (Ferl, 1996; Fu et al., 2000; Emi et al., 2001). They have been shown to act as intracellular receptors for rice Hd3a and, thus, could play a role in flowering (Taoka et al., 2011). Interestingly, free Raf kinases appear to be bound to 14-3-3 proteins in animal systems; however, once they bind to liposomes/membranes, the 14-3-3 protein dissociates (Hekman et al., 2006). This suggests that 14-3-3 proteins may modulate binding of proteins to lipids.
Lipid/fatty acid-binding proteins
Eleven proteins with a role in lipid/fatty acid metabolism (aspartic protease, 3-β-hydroxy dehydrogenase, SQD1, and class 3 lipase) and storage (glycine-rich protein), binding (annexin, lipid-associated family protein, and GRP17), and signalling (annexin, putative GDSL lipase, MLP/Bet vI-like protein, and PIG-P; Table 2) were detected. Lipids and fatty acids are not typically expected in the phloem exudates. As the phloem exudate is an aqueous environment, it is not too unexpected that the lipids described above may be bound to proteins. Annexin and a lipid transfer protein (LTP) had previously been reported in the phloem exudate. LTP/DIR1 was shown to play a role in systemic acquired resistance (Maldonado et al., 2002; Mitton et al., 2009). However, no connection was made to phloem lipids. Annexin, one of the lipid-binding proteins, can be cytoplasmic but has also been shown to bind phospholipids/membranes and is involved in Ca2+ signalling as well as callose formation (Andrawis et al., 1993; Mortimer et al., 2008). Under stress conditions, Medicago annexin AnxMs2 localizes in the nucleolus even though it contains no typical nuclear localization signal. This suggests that it plays a role in intracellular membrane trafficking and signal transduction (Kovács et al., 1998).GRP17 has previously been identified in Arabidopsis pollen (Mayfield et al., 2001). It contains an oleosin domain. Seed-localized oleosin prevent lipid aggregation and control oil-body size (Mayfield and Preuss, 2000). It is conceivable that it plays a similar role in the phloem sap.
Table 2.
Putative lipid-binding proteins identified from phloem exudates
| Protein identification | Accession number | Mol. wt (kDa) |
| Lipase class 3 family protein | At4g16820 | 58 |
| Aspartic protease | At4g04460 | 56 |
| GRP17 (oleosin) | At5g07530 | 53 |
| Putative PIG-P | At2g39435 | 50 |
| Sqd1 | At4g33030 | 44 |
| Putative lipase/hydrolase | At1g29660 | 40 |
| Annexin | At1g35720 | 36 |
| Putative 3-β-hydroxysteroid dehydratase/isomerase | At2g37660 | 35 |
| Lipid-associated family protein | At4g39730 | 20 |
| Bet vI allergen | At1g23130 | 18 |
| GRP7 | At2g21660 | 17 |
Two proteins that are listed above as pathogen response proteins are the major latex protein-like protein and the Bet vI allergen-like protein. They belong to the Bet vI-like superfamily, which contains proteins with low sequence similarity but a common hydrophobic fold. Members of this superfamily include ceramide transfer proteins, phosphatidylcholine transfer proteins, phosphatidyinositol transfer proteins, acetyl-CoA hydrolases, and HD-Zip transcription factors from plants (Radauer et al., 2008). The discovery of representatives of the proteins in phloem exudates suggests that they may play a role not just in transmembrane lipid transport but also in long-distance lipid transport and signalling.
Three other proteins could have a role in long-distance lipid signalling: two putative lipases and a putative PIG-P protein. Lipases have been implicated in salicylic acid signalling (Feys et al., 2001) and wound response (Guan and Nothnagel, 2004). The PIG-P protein is subunit P of a phosphatidylinositol N-acetylglucosaminyltransferase. The enzyme catalyses the transfer of N-acetylglucosamine to phosphatidylinositol in the first step of glycosylphosphatidylinositol (GPI) anchor synthesis. GPI anchors are glycolipids which anchor proteins to membranes (Watanabe et al., 2000). Interestingly, myo-inositol, which is not only necessary for the synthesis of phosphatidyl bisphosphate (PIP2), a signalling compound, but can also be a part of the GPI anchor, was also found. In animals, GPI anchors may directly interact with receptor proteins and signal neural and brain development (Peles et al., 1997; Ferrando-Miguel et al., 2004). Only few GPI-anchored proteins have been found in plants. One group, the arabinogalactan proteins (AGPs), have been shown to contain a classical C-terminal GPI anchor signal sequence (Schultz et al., 1998): their synthesis is thought to occur in the endoplasmic reticulum followed by secretion into the apoplast. There the AGP could be cleaved by phospholipases and act as signal molecules. It is possible that GPI-anchored proteins participate in cell-to-cell or even long-distance interaction. Similarly, inositol-3-phosphate and PIP2 are important second messengers in animals as well as in plants (Ananieva and Gillaspy; 2009; Munnik and Vermeer, 2010); in plants, PA and LPA also appear to be relevant (Katagiri et al., 2005; Munnik and Testerink, 2009). PA has been proposed to play a role in ABA-, wound-, and pathogen-related signalling (Katagiri et al., 2005; Testerink and Munnik, 2005). Representatives of all three lipid groups (PIP, PA, and LPA) were identified in the samples and confirmed by MS/MS.
Throughout this study of phloem exudates, several fatty acids and lipids, some of which are known factors in intracellular signaling (PA and PIP) have been detected. In addition, proteins with the predicted, or in some cases known, ability to bind and transport these lipids and again a role in intracellular signalling were found (i.e. annexin; Andrawis et al., 1993). Together, this suggests that these lipids may not only be involved in intracellular signalling but may also play an important role in long-distance lipid signalling and that the lipid-binding proteins function as ‘chaperones’ which move lipids into and/or throughout the phloem. Therefore, lipids and lipid-binding proteins appear to have a larger and more complex function in systemic and developmental signalling throughout the plant than previously thought—a novel concept that could open up an entirely new field of study.
Supplementary data
Suppplementary data are available at JXB online.
Figure S1. Flow chart of phloem extraction and experimental set-up.
Figure S2. Time course of the appearance of proteins collected into water.
Figure S3. GC-MS profile of total (non-phase-partitioned) phloem exudates after 1 h of exudation into water (after EDTA treatment).
Table S1. Proteins identified in Arabidopsis thaliana phloem exudate.
Supplementary Material
Acknowledgments
The authors wish to thank Dr J.A.D. Zeevaart for help in getting this project started. Metabolite and protein analysis was performed by BSG, UFB, and SHB at the MSU-Mass Spectrometry Facility: we thank Beverly Chamberlin and Dr A.D. Jones for their support during the metabolite and lipid analysis. The project was funded in part by MSU intramural grant # 05-IRGP-313 to SHB and by the USDA National Institute of Food and Agriculture, Hatch project number MICL02233.
References
- An HL, Roussot C, Suárez-López P, et al. CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development. 2004;131:3615–3626. doi: 10.1242/dev.01231. [DOI] [PubMed] [Google Scholar]
- Ananieva EA, Gillaspy GE. Switches in nutrient and inositol signaling. Plant Signaling and Behavior. 2009;4:304–306. doi: 10.4161/psb.4.4.8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrawis A, Solomon M, Delmer DP. Cotton fiber annexins—a potential role in the regulation of callose synthase. The Plant Journal. 1993;3:763–772. doi: 10.1111/j.1365-313x.1993.00763.x. [DOI] [PubMed] [Google Scholar]
- Aoki K, Kragler F, Xoconostle-Cázares B, Lucas WJ. A subclass of plant heat shock cognate 70 chaperones carries a motif that facilitates trafficking through plasmodesmata. Proceedings of the National Academy of Sciences, USA. 2002;99:16342–16347. doi: 10.1073/pnas.252427999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Yamaya T, Sakakibara H. CmPP16-1-interacting phloem sap proteins are involved in the regulation of CmPP16-1 long-distance movement. Plant and Cell Physiology. 2005;46:S151. [Google Scholar]
- Ayre BG, Turgeon R. Graft transmission of a floral stimulant derived from CONSTANS. Plant Physiology. 2004;135:2271–2278. doi: 10.1104/pp.104.040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran S, Xiang Y, Schobert C, Thompson GA, Lucas WJ. Phloem sap proteins from Cucurbita maxima and Ricinus communis have the capacity to traffic cell to cell through plasmodesmata. Proceedings of the National Academy of Sciences, USA. 1997;94:14150–14155. doi: 10.1073/pnas.94.25.14150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes A, Bale J, Constantinidou C, Aston P, Jones A, Pritchard J. Determining protein identity from sieve element sap in Ricinus communis L. by quadrupole time of flight (Q-TOF) mass spectrometry. Journal of Experimental Botany. 2004;55:1473–1481. doi: 10.1093/jxb/erh161. [DOI] [PubMed] [Google Scholar]
- Blaner WS. Retinol-binding protein: the serum transport protein for vitamin A. Endocrine Reviews. 1989;10:308–316. doi: 10.1210/edrv-10-3-308. [DOI] [PubMed] [Google Scholar]
- Behmer ST, Grebenok RJ, Douglas AE. Plant sterols and host plant suitability for a phloem feeding insect. Functional Ecology. 2010;25:484–491. [Google Scholar]
- Behnke H-D. Distribution and evolution of forms and types of sieve-element plastids in the dicotyledons. ALISO. 1991;3:167–182. [Google Scholar]
- Behnke H-D, Schulz A. The development of specific sieve-element plastids in would phloem of Coleus blumei (S-type) and Pisum sativum (P-type), regenerated from amyloplast-containing parenchyma cells. Protoplasma. 1983;114:125–132. [Google Scholar]
- Burton RA, Gibeaut DM, Bacic A, Findlay K, Hamilton A, Boulcombe DC, Fincher GB. Virus-induced silencing of a plant cellulose synthase. The Plant Cell. 2000;12:691–706. doi: 10.1105/tpc.12.5.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonneau D, Beauregard M, Tajmir-Riahi H- A. Structural analysis of human serum albumin complexes with cationic lipids. Journal of Physical Chemistry B. 2009;113:1777–1781. doi: 10.1021/jp8092012. [DOI] [PubMed] [Google Scholar]
- Chen S, Petersen BL, Olsen CE, Schulz A, Halkier BA. Long-distance phloem transport of glucosinolates in Arabidopsis. Plant Physiology. 2001;127:194–201. doi: 10.1104/pp.127.1.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V, Zambryski P. Systemic transport of RNA in plants. Trends in Plant Science. 2000;5:52–54. doi: 10.1016/s1360-1385(99)01540-x. [DOI] [PubMed] [Google Scholar]
- Corbesier L, Coral V, Seonghoe J, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis . Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- Corbesier L, Prinsen E, Jacqmard A, Lejeune P, Van Onckelen H, Perilleux C, Bernier G. Cytokinin levels in leaves, leaf exudate and shoot apical meristem of Arabidopsis thaliana during floral transition. Journal of Experimental Botany. 2003;54:2511–2517. doi: 10.1093/jxb/erg276. [DOI] [PubMed] [Google Scholar]
- Cronshaw J. Phloem structure and function. Annual Review of Plant Physiology. 1981;32:465–484. [Google Scholar]
- Dafoe NJ, Gowen BE, Constabel CP. Thaumatin-like proteins are differentially expressed and localized in phloem tissues of hybrid poplar. BMC Plant Biology. 2010;10:191. doi: 10.1186/1471-2229-10-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenhoffer JM, Schulz A, Skaggs MI, Bostwick DE, Thompson GA. Expression of the phloem lectin is developmentally linked to vascular differentiation in cucurbits. Planta. 1997;201:405–414. [Google Scholar]
- Deeken R, Ache P, Kajahn I, Klinkenberg J, Bringmann G, Hedrich R. Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem-based transcripts hidden in complex datasets of microarray experiments. The Plant Journal. 2008;55:746–759. doi: 10.1111/j.1365-313X.2008.03555.x. [DOI] [PubMed] [Google Scholar]
- Ding B, Itaya A, Qi YJ. Symplasmic protein and RNA traffic: regulatory points and regulatory factors. Current Opinion in Plant Biology. 2003;6:596–602. doi: 10.1016/j.pbi.2003.09.010. [DOI] [PubMed] [Google Scholar]
- Doering-Saad C, Newbury HJ, Bale JS, Pritchard J. Use of aphid stylectomy and RT-PCR for the detection of transporter mRNAs in sieve elements. Journal of Experimental Botany. 2002;53:631–637. doi: 10.1093/jexbot/53.369.631. [DOI] [PubMed] [Google Scholar]
- Doering-Saad C, Newbury HJ, Couldridge CE, Bale JS, Pritchard J. A phloem-enriched cDNA library from Ricinus: insights into phloem function. Journal of Experimental Botany. 2006;57:3183–3193. doi: 10.1093/jxb/erl082. [DOI] [PubMed] [Google Scholar]
- Durrant WE, Dong X. Systemic acquired resistance. Annual Review of Phytopathology. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- Emi T, Kinoshita T, Shimazaki K. Specific binding of vf14-3-3a isoform to the plasma membrane H+-ATPase in response to blue light and fusicoccin in guard cells of broad bean. Plant Physiology. 2001;125:1115–1125. doi: 10.1104/pp.125.2.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferl RJ. 14-3-3 proteins and signal transduction. Annual Review of Plant Physiology and Plant Molecular Biology. 1996;47:49–73. doi: 10.1146/annurev.arplant.47.1.49. [DOI] [PubMed] [Google Scholar]
- Ferrando-Miguel R, Cheon MS, Lubec G. Protein levels of genes encoded on chromosome 21 in fetal Down Syndrome brain (Part V): overexpression of phosphatidyl-inositol-glycan class P protein (DSCR5) Amino Acids. 2004;26:255–261. doi: 10.1007/s00726-004-0065-9. [DOI] [PubMed] [Google Scholar]
- Feys BJ, Moisan LJ, Newman MA, Parker JE. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO Journal. 2001;20:5400–5411. doi: 10.1093/emboj/20.19.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher DB, Cash-Clark CE. Sieve tube unloading and post-phloem transport of fluorescent tracers and proteins injected into sieve tubes via severed aphid stylets. Plant Physiology. 2000;123:125–137. doi: 10.1104/pp.123.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher DB, Wu Y, Ku MSB. Turnover of soluble-proteins in the wheat sieve tube. Plant Physiology. 1992;100:1433–1441. doi: 10.1104/pp.100.3.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu HA, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annual Review of Pharmacology and Toxicology. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- Geigenberger P. Response of plant metabolism to too little oxygen. Current Opinion in Plant Biology. 2003;6:247–256. doi: 10.1016/s1369-5266(03)00038-4. [DOI] [PubMed] [Google Scholar]
- Giavalisco P, Kapitza K, Kolasa A, Buhtz A, Kehr J. Towards the proteome of Brassica napus phloem sap. Proteomics. 2006;6:896–909. doi: 10.1002/pmic.200500155. [DOI] [PubMed] [Google Scholar]
- Glatz JFC, Boerchers T, Spener F, van der Vusse GJ. Fatty acids in cell signalling: modulation by lipid binding proteins. Prostaglandins, Leukotrienes and Essential Fatty Acids. 1995;52:121–127. doi: 10.1016/0952-3278(95)90010-1. [DOI] [PubMed] [Google Scholar]
- Good AG, Muench DG. Purification and characterization of an anaerobically induced alanine aminotransferase from barley roots. Plant Physiology. 1992;99:1520–1525. doi: 10.1104/pp.99.4.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath K, Kao CC. Replication-independent long-distance trafficking by viral RNAs in Nicotiana benthamiana . The Plant Cell. 2007;19:1179–1191. doi: 10.1105/tpc.107.050088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Nothnagel EA. Binding of arabinogalactan proteins by Yariv phenylglycoside triggers wound-like responses in Arabidopsis cell cultures. Plant Physiology. 2004;135:1346–1366. doi: 10.1104/pp.104.039370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelette BS, Chamberlin B, Benning UF, Hoffmann-Benning S. Indications of lipids/lipid signaling in the phloem exudates of Arabidopsis thaliana and Perilla ocymoides . In: Benning C, Ohlrogge J, editors. Proceedings of the 17th International Symposium of Plant Lipids. Michigan State University, E. Lansing; 2007. pp. 92–95. [Google Scholar]
- Gupta N, Pevzner PA. False discovery rates of protein identifications: a strike against the two-peptide rule. Journal of Proteome Research. 2009;8:4173–4181. doi: 10.1021/pr9004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haebel S, Kehr J. Matrix-assisted laser desorption/ionization time of flight mass spectrometry peptide mass fingerprints and post source decay: a tool for the identification and analysis of phloem proteins from Cucurbita maxima Duch. separated by two-dimensional polyacrylamide gel electrophoresis. Planta. 2001;215:586–593. doi: 10.1007/s004250100523. [DOI] [PubMed] [Google Scholar]
- Ham BK, Brandom JL, Xoconostle-Cázares B, Ringgold V, Lough TJ, Lucas WJ. Polypyrimidine tract binding protein, CmRBP50, forms the basis of a pumpkin phloem ribonucleoprotein complex. The Plant Cell. 2009;21:197–215. doi: 10.1105/tpc.108.061317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberg M, Sanz A, Castresana C. Alpha-oxidation of fatty acids in higher plants. Identification of a pathogen-inducible oxygenase (PIOX) as an alpha-dioxygenase and biosynthesis of 2-hydroperoxylinolenic acid. Journal of Biological Chemistry. 1999;274:24503–24513. doi: 10.1074/jbc.274.35.24503. [DOI] [PubMed] [Google Scholar]
- Hayashi H, Fukuda A, Suzui N, Fujimaki S. Proteins in the sieve element–companion cell complexes: their detection, localization and possible functions. Australian Journal of Plant Physiology. 2000;27:489–496. [Google Scholar]
- Haywood V, Yu TS, Huang NC, Lucas WJ. Phloem long-distance trafficking of gibberellic acid-insensitive RNA regulates leaf development. The Plant Journal. 2005;42:49–68. doi: 10.1111/j.1365-313X.2005.02351.x. [DOI] [PubMed] [Google Scholar]
- Hekman M, Albert S, Galmiche A, Rennefahrt UE, Fueller J, Fischer A, Puehringer D, Wiese S, Rapp UR. Reversible membrane interaction of BAD requires two C-terminal lipid binding domains in conjunction with 14-3-3 protein binding. Journal of Biological Chemistry. 2006;281:17321–17336. doi: 10.1074/jbc.M600292200. [DOI] [PubMed] [Google Scholar]
- Hegeman CE, Grabau EA. A novel phytase with sequence similarity to purple acid phosphatases is expressed in cotyledons of germinating soybean seedlings. Plant Physiology. 2001;12:1598–1608. doi: 10.1104/pp.126.4.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzen C, Nater M, Apel K, Staiger D. AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA. 1997;94:8515–8520. doi: 10.1073/pnas.94.16.8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann-Benning S, Gage DA, McIntosh L, Kende H, Zeevaart JAD. Comparison of peptides in the phloem sap of flowering and non-flowering Perilla and lupine plants using microbore HPLC followed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Planta. 2002;216:140–147. doi: 10.1007/s00425-002-0916-0. [DOI] [PubMed] [Google Scholar]
- Jorgensen RA, Atkinson RG, Forster RLS, Lucas WJ. An RNA-based information superhighway in plants. Science. 1998;279:1486–1487. doi: 10.1126/science.279.5356.1486. [DOI] [PubMed] [Google Scholar]
- Katagiri T, Ishiyama K, Kato T, Tabata S, Kobayashi M, Shinozaki K. An important role of phosphatidic acid in ABA signaling during germination in Arabidopsis thaliana. The Plant Journal. 2005;43:107–117. doi: 10.1111/j.1365-313X.2005.02431.x. [DOI] [PubMed] [Google Scholar]
- Kehr J. Phloem sap proteins: their identities and potential roles in the interaction between plants and phloem-feeding insects. Journal of Experimental Botany. 2006;57:767–774. doi: 10.1093/jxb/erj087. [DOI] [PubMed] [Google Scholar]
- Kehr J, Butz A. Long distance transport and movement of RNA through the phloem. Journal of Experimental Botany. 2008;59:85–92. doi: 10.1093/jxb/erm176. [DOI] [PubMed] [Google Scholar]
- Kehr J, Haebel S, Blechschmidt-Schneider S, Willmitzer L, Steup M, Fisahn J. Analysis of phloem protein patterns from different organs of Cucurbita maxima Duch. by matrix-assisted laser desorption/ionization time of flight mass spectroscopy combined with sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Planta. 1999;207:612–619. doi: 10.1007/s004250050525. [DOI] [PubMed] [Google Scholar]
- Kim JY, Yuan Z, Cilia M, Khalfan-Jagani Z, Jackson D. Intercellular trafficking of a Knotted 1 green fluorescent protein fusion in the leaf and shoot meristem of Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2002;99:4103–4108. doi: 10.1073/pnas.052484099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Zeevaart JAD. Enhancement of phloem exudation from cut petioles by chelating-agents. Plant Physiology. 1974;53:96–103. doi: 10.1104/pp.53.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblauch M, Peters WS, Ehlers K, van Bel AJE. Reversible calcium-regulated stopcocks in legume sieve tubes. The Plant Cell. 2001;13:1221–1230. doi: 10.1105/tpc.13.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant and Cell Physiology. 2002;43:1096–1105. doi: 10.1093/pcp/pcf156. [DOI] [PubMed] [Google Scholar]
- Kovács I, Ayaydin F, Oberschall A, Ipacs I, Bottka S, Pongor S, Dudits D, Toth EC. Immunolocalization of a novel annexin-like protein encoded by a stress and abscisic acid responsive gene in alfalfa. The Plant Journal. 1998;15:185–197. doi: 10.1046/j.1365-313x.1998.00194.x. [DOI] [PubMed] [Google Scholar]
- Kühn C, Franceschi VR, Schulz A, Lemoine R, Frommer WB. Macromolecular trafficking indicated by localization and turnover of sucrose transporters in enucleate sieve elements. Science. 1997;275:1298–1300. doi: 10.1126/science.275.5304.1298. [DOI] [PubMed] [Google Scholar]
- Lang A. Physiology of flower formation. In: Ruhland W, editor. Handbuch der Pflanzenphysiologie XV/1. Berlin: Springer; 1965. pp. 1380–1536. [Google Scholar]
- Lee J-Y, Taoka K-I, Yoo B-C, Ben-Nissan G, Kim D-J, Lucas WJ. Plasmodesmal-associated protein kinase in tobacco and arabidopsis recognizes a subset of non-cell-autonomous proteins. The Plant Cell. 2005;17:2817–2831. doi: 10.1105/tpc.105.034330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M-K, Belanger H, Lee Y-J, et al. FT protein may act as the long-distance florigenic signal in the cucurbits. The Plant Cell. 2007;19:1488–1506. doi: 10.1105/tpc.107.051920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M-K, Lee Y-J, Lough TJ, Phinney B, Lucas WJ. Analysis of the pumpkin phloem proteome provides functional insights into angiosperm sieve tube function. Molecular and Cellular Proteomics. 2009;8:343–356. doi: 10.1074/mcp.M800420-MCP200. [DOI] [PubMed] [Google Scholar]
- Loewus FA, Murthy PPN. Myo-inositol metabolism in plants. Plant Science. 2000;150:1–19. [Google Scholar]
- Lough TJ, Lucas WJ. Integrative plant biology: role of phloem long-distance macromolecular trafficking. Annual Review of Plant Physiology. 2006;57:203–232. doi: 10.1146/annurev.arplant.56.032604.144145. [DOI] [PubMed] [Google Scholar]
- Lucas WJ, Bouche-Pillon S, Jackson DP, Nguyen L, Baker L, Ding B, Hake S. Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science. 1995;270:1980–1983. doi: 10.1126/science.270.5244.1980. [DOI] [PubMed] [Google Scholar]
- Madey E, Nowack LM, Thompson JE. Isolation and characterization of lipid in phloem sap of canola. Planta. 2002;214:625–634. doi: 10.1007/s004250100649. [DOI] [PubMed] [Google Scholar]
- Maeda H, Song W, SageTL, DellaPenna D. Tocopherols play a crucial role in low-temperature adaptation and phloem loading in Arabidopsis. The Plant Cell. 2006;18:2710–2732. doi: 10.1105/tpc.105.039404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado AM, Doerner P, Dixon RA, Lamb CJ, Cameron RK. A putative lipid transfer protein involved in systemic resistance signaling in Arabidopsis . Nature. 2002;419:399–403. doi: 10.1038/nature00962. [DOI] [PubMed] [Google Scholar]
- Marentes E, Grusak MA. Mass determination of low-molecular-weight proteins in phloem sap using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Journal of Experimental Botany. 1998;49:903–911. [Google Scholar]
- Markovic-Housley Z, Degano M, Lamba D, Roepenack-Lahaye E, Clemens S, Susani M, Ferreira F, Scheiner O, Breiteneder H. Crystal structure of a hypoallergenic isoform of the major birch pollen allergen Bet v 1 and its likely biological function as a plant steroid carrier. Journal of Molecular Biology. 2003;325:123–133. doi: 10.1016/s0022-2836(02)01197-x. [DOI] [PubMed] [Google Scholar]
- Martens HJ, Roberts AG, Oparka KJ, Schulz A. Quantification of plasmodesmatal endoplasmatic reticulum coupling between sieve elements and companion cells using fluorescence redistribution after photobleaching. Plant Physiology. 2006;142:471–480. doi: 10.1104/pp.106.085803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield JA, Fiebig A, Johnstone SE, Preuss D. Gene families from the Arabidopsis thaliana pollen coat proteome. Science. 2001;292:2482–2485. doi: 10.1126/science.1060972. [DOI] [PubMed] [Google Scholar]
- Mayfield JA, Preuss D. Rapid initiation of Arabidopsis pollination requires the oleosin-domain protein GRP17. Nature Cell Biology. 2000;2:128–130. doi: 10.1038/35000084. [DOI] [PubMed] [Google Scholar]
- Mitton FM, Pinedo ML, de la Canal L. Phloem sap of tomato plants contains a DIR1 putative ortholog. Journal of Plant Physiology. 2009;166:543–547. doi: 10.1016/j.jplph.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Mogensen JE, Wimmer R, Larsen J N, Spangfort MD, Otzen DE. The major birch allergen, Bet v 1, shows affinity for a broad spectrum of physiological ligands. Journal of Biological Chemistry. 2002;277:23684–23692. doi: 10.1074/jbc.M202065200. [DOI] [PubMed] [Google Scholar]
- Mortimer JC, Laohavisit A, Macpherson N, Webb A, Brownlee C, Battey NH, Davies JM. Annexins: multifunctional components of growth and adaptation. Journal of Experimental Botany. 2008;59:533–544. doi: 10.1093/jxb/erm344. [DOI] [PubMed] [Google Scholar]
- Mullaney EJ, Ullah AHJ. Identification of a histidine acid phosphatase (phyA)-like gene in Arabidopsis thaliana . Biochemical and Biophysical Research Communications. 1998;251:252–255. doi: 10.1006/bbrc.1998.9452. [DOI] [PubMed] [Google Scholar]
- Munnik T, Testerink C. Plant phospholipid signaling: ‘in a nutshell’. Journal of Lipid Research. 2009;50:S260–S265. doi: 10.1194/jlr.R800098-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MunnikT, Vermeer JEM. Osmotic stress-induced phosphoinositde and inositol signaling in plants. Plant, Cell and Environment. 2010;33:655–669. doi: 10.1111/j.1365-3040.2009.02097.x. [DOI] [PubMed] [Google Scholar]
- Nagy L, Szanto A. Roles for lipid activated transcription factors. Molecular Nutrition and Food Research. 2005;49:1072–1074. doi: 10.1002/mnfr.200500097. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Hayashi H, Mori S, Chino M. Detection and characterization of protein-kinases in rice phloem sap. Plant and Cell Physiology. 1995;36:19–27. [Google Scholar]
- Oparka KJ, Roberts AG, Boevink P, Cruz SS, Roberts I, Pradel KS, Imlau A, Kotlizky G, Sauer N, Epel B. Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell. 1999;97:743–754. doi: 10.1016/s0092-8674(00)80786-2. [DOI] [PubMed] [Google Scholar]
- Peles E, Nativ M, Lustig M, Grumet M, Schilling J, Martinez R, Plowman GD, Schlessinger J. Identification of a novel contactin-associated transmembrane receptor with multiple domains implicated in protein–protein interactions. EMBO Journal. 1997;16:978–988. doi: 10.1093/emboj/16.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radauer C, Lackner P, Breiteneder H. The Bet v 1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evolutionary Biology. 2008;8:286–304. doi: 10.1186/1471-2148-8-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA. Long-term functioning of enucleate sieve elements: possible mechanisms of damage avoidance and damage repair. Plant, Cell and Environment. 1991;14:139–146. [Google Scholar]
- Rodriguez-Medina C, Atkins CA, Mann AJ, Jordan ME, Smith PMC. Macromolecular composition of phloem exudate from white lupin (Lupinus albus L.) BMC Plant Biology. 2011;11:36. doi: 10.1186/1471-2229-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L. Simultaneous analysis of metabolites in potato tuber by gas chromatography–mass spectrometry. The Plant Journal. 2000;23:131–142. doi: 10.1046/j.1365-313x.2000.00774.x. [DOI] [PubMed] [Google Scholar]
- Ruiz-Medrano R, Xoconostle-Cázares B, Lucas WJ. Phloem long-distance transport of CmNACP mRNA: implications for supracellular regulation in plants. Development. 1999;126:4405–4419. doi: 10.1242/dev.126.20.4405. [DOI] [PubMed] [Google Scholar]
- Ruiz-Medrano R, Xoconostle-Cázares B, Lucas WJ. The phloem as conduit for inter-organ communication. Current Opinion in Plant Biology. 2001;4:202–209. doi: 10.1016/s1369-5266(00)00162-x. [DOI] [PubMed] [Google Scholar]
- Ryabov EV, Robinson DJ, Taliansky ME. A plant virus-encoded protein facilitates long-distance movement of heterologous viral RNA. Proceedings of the National Academy of Sciences, USA. 1999;96:1212–1217. doi: 10.1073/pnas.96.4.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuth T, Schobert C, Pecsvaradi A, Eichholz A, Komor E, Orlich G. Specific proteins in the sieve-tube exudate of Ricinus-communis L seedlings—separation, characterization and in-vivo labeling. Planta. 1993;191:207–213. [Google Scholar]
- Sanz A, Moreno JI, Castresana C. PIOX, a new pathogen-induced oxygenase with homology to animal cyclooxygenase. The Plant Cell. 1998;10:1523–1537. doi: 10.1105/tpc.10.9.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Howe GA. Systemic signaling in the wound response. Current Opinion in Plant Biology. 2005;8:369–377. doi: 10.1016/j.pbi.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Schobert C, Grossmann P, Gottschalk M, Komor E, Pecsvaradi A, Zurnieden U. Sieve-tube exudate from Ricinus-communis L seedlings contains ubiquitin and chaperones. Planta. 1995;196:205–210. [Google Scholar]
- Schultz C, Gilson P, Oxley D, Youl J, Bacic A. GPI-anchors on arabinogalactan-proteins: implication for signalling in plants. Trends in Plant Science. 1998;3:426–431. [Google Scholar]
- Seo HS, Song JT, Cheong J-J, Lee Y-H, Lee Y-W, Hwang I, Lee JS, Choi YD. Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proceedings of the National Academy of Sciences, USA. 2001;98:4788–4793. doi: 10.1073/pnas.081557298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah JJ, Jacob R. Development and structure of phloem in the petiole of Langenaria siceraria (Mol.) Standl. and Momordica charantia L. Annals of Botany. 1969;33:855–863. [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Analytical Chemistry. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Stadler R, Wright KM, Lauterbach C, Amon G, Gahrtz M, Feuerstein A, Oparka KJ, Sauer N. Expression of GFP-fusions in arabidopsis companion cells reveals non-specific protein trafficking into sieve elements and identifies a novel post-phloem domain in roots. The Plant Journal. 2005;41:319–331. doi: 10.1111/j.1365-313X.2004.02298.x. [DOI] [PubMed] [Google Scholar]
- Suárez-López P. Long-range signalling in plant reproductive development. International Journal of Developmental Biology. 2005;49:761–771. doi: 10.1387/ijdb.052002ps. [DOI] [PubMed] [Google Scholar]
- Symons GM, Reid JB. Brassinosteroids do not undergo long-distance transport in pea. Implications for the regulation of endogenous brassinosteroid levels. Plant Physiology. 2004;135:2196–2206. doi: 10.1104/pp.104.043034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- Taoka K-I, Ham B-K, Xoconostle-Cázares B, Rojas MR, Lucas WJ. Reciprocal phosphorylation and glycosylation recognition motifs control NSAAP1 interaction with pumpkin phloem proteins and their cell-to-cell movement. The Plant Cell. 2007;19:1866–1884. doi: 10.1105/tpc.107.052522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka K, Ohki I, Tsuji H, F, et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature. 2011;476:332–335. doi: 10.1038/nature10272. [DOI] [PubMed] [Google Scholar]
- Testerink C, Munnik T. Phosphatidic acid: a multifunctional stress signaling lipid in plants. Trends in Plant Sciences. 2005;10:368–375. doi: 10.1016/j.tplants.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Thompson GA, Goggin FL. Transcriptomics and functional genomics of plant defence induction by phloem-feeding insects. Journal of Experimental Botany. 2006;57:755–766. doi: 10.1093/jxb/erj135. [DOI] [PubMed] [Google Scholar]
- Thorpe MR, Ferrieri AP, Herth MM, Ferrieri RA. (11)C-imaging: methyl jasmonate moves in both phloem and xylem, promotes transport of jasmonate, and of photoassimilate even after proton transport is decoupled. Planta. 2007;226:541–551. doi: 10.1007/s00425-007-0503-5. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPARg2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Truman W, Bennett MH, Kubigsteltig I, Turnbull C, Grant M. Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proceedings of the National Academy of Sciences, USA. 2007;104:1075–1080. doi: 10.1073/pnas.0605423104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bel AJE. The phloem, a miracle of ingenuity. Plant, Cell and Environment. 2003;26:125–149. [Google Scholar]
- van Bel AJE, Knoblauch M. Sieve element and companion cell: the story of the comatose patient and the hyperactive nurse. Australian Journal of Plant Physiology. 2000;27:477–487. [Google Scholar]
- Waigmann E, Ueki S, Trutnyeva K, Citovsky V. The ins and outs of nondestructive cell-to-cell and systemic movement of plant viruses. Critical Reviews in Plant Sciencse. 2004;23:195–250. [Google Scholar]
- Walz C, Juenger M, Schad M, Kehr J. Evidence for the presence and activity of a complete antioxidant defence system in mature sieve tubes. The Plant Journal. 2002;31:189–197. doi: 10.1046/j.1365-313x.2002.01348.x. [DOI] [PubMed] [Google Scholar]
- Wang X. Lipid signaling. Current Opinion in Plant Biology. 2004;7:329–336. doi: 10.1016/j.pbi.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Watanabe R, Murakami Y, Marmor MD, Inoue N, Maeda Y, Hino J, Kangawa K, Julius M, Kinoshita T. Initial enzyme for glycosylphosphatidylinositol biosynthesis requires PIG-P and is regulated by DPM2. EMBO Journal. 2000;19:4402–4411. doi: 10.1093/emboj/19.16.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will T, van Bel AJE. Physical and chemical interactions between aphids and plants. Journal of Experimental Botany. 2006;57:729–737. doi: 10.1093/jxb/erj089. [DOI] [PubMed] [Google Scholar]
- Wu X, Dinneny JR, Crawford KM, Rhee Y, Citovsky V, Zambryski PC, Weigel D. Modes of intercellular transcription factor movement in the Arabidopsis apex. Development. 2003;130:3735–3745. doi: 10.1242/dev.00577. [DOI] [PubMed] [Google Scholar]
- Xoconostle-Cazares B, Yu X, Ruiz-Medrano R, Wang HL, Monzer J, Yoo BC, McFarland KC, Franceschi VR, Lucas WJ. Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science. 1999;283:94–98. doi: 10.1126/science.283.5398.94. [DOI] [PubMed] [Google Scholar]
- Yoo BC, Kragler F, Varkonyi-Gasic E, Haywood V, Archer-Evans S, Lee Y M, Lough TJ, Lucas WJ. A systemic small RNA signaling system in plants. The Plant Cell. 2004;16:1979–2000. doi: 10.1105/tpc.104.023614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD. Physiology of flower formation. Annual Review of Plant Physiology and Plant Molecular Biology. 1976;27:321–348. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.