Abstract
Aphids are a major family of plant insect pests. Medicago truncatula and Acyrthosiphon pisum (pea aphid, PA) are model species with a suite of resources available to help dissect the mechanism underlying plant–aphid interactions. A previous study focused on monogenic and relatively strong resistance in M. truncatula to PA and other aphid species. In this study a moderate resistance to PA was characterized in detail in the M. truncatula line A17 and compared with the highly susceptible line A20 and the more resistant line Jester. The results show that PA resistance in A17 involves both antibiosis and tolerance, and that resistance is phloem based. Quantitative trait locus (QTL) analysis using a recombinant inbred line (RIL) population (n=114) from a cross between A17 and A20 revealed that one locus, which co-segregated with AIN (Acyrthosiphon-induced necrosis) on chromosome 3, is responsible for the reduction of aphid biomass (indicator of antibiosis) for both PA and bluegreen aphid (BGA, A. kondoi), albeit to a lesser degree for PA than BGA. Interestingly, two independent loci on chromosomes 5 and 3 were identified for the plant biomass reduction (indicator of plant tolerance) by PA and BGA, respectively, demonstrating that the plant’s tolerance response to these two closely related aphid species is distinct. Together with previously identified major resistant (R) genes, the QTLs identified in this study are powerful tools to understand fully the spectrum of plant defence against sap-sucking insects and provide opportunities for breeders to generate effective and sustainable strategies for aphid control.
Keywords: Antibiosis, antixenosis, EPG, herbivory, hypersensitive response, necrosis, phloem, sap-sucking insect
Introduction
Aphids and other sap-sucking insects cause significant yield losses in agriculture globally by ingesting phloem sap and transmitting viruses, and damage may also be caused by the plant’s response to various elicitors injected by feeding aphids. Therefore, a better understanding of the physiological and genetic basis of plant–aphid interactions is of importance for effective control of aphids. Resistance to aphids and other sap feeders is frequently controlled by major dominant or semi-dominant genetic loci that, in three specific cases, are known to be homologues of classical resistance (R) genes conditioning pathogen resistance (Rossi et al., 1998; Dogimont et al., 2007; Wroblewski et al., 2007). It is hypothesized that R gene products enable the plant host to recognize specific, aphid-derived molecules (effectors) and mount a defence response against the pathogens. Aphids have evolved highly specific mouthparts termed stylets to navigate through the cuticle, epidermis, and mesophyll to establish a feeding site in the phloem sieve elements (Tjallingii, 2006; Will et al., 2009). The aphid saliva that is injected by stylets into the phloem or surrounding host tissues is likely to contain the effectors that are recognized by R proteins (Hogenhout and Bos, 2011).
To date, two aphid resistance genes have been isolated and characterized: Mi-1 from tomato conferring resistance to the potato aphid (Macrosiphum euphorbiae; Rossi et al., 1998) and Vat from melon to the cotton-melon aphid (Aphis gossypii; Dogimont et al., 2007). Both resistance genes belong to the coiled-coil nucleotide-binding site-leucine-rich repeat (CC-NBS-LRR) class of R genes. The Ra gene in lettuce, which confers resistance to the lettuce root aphid (Pemphigus bursarius L.), has been shown through reverse genetics to be a member of this same family, contained somewhere within a large R gene cluster (Wroblewski et al., 2007). Furthermore, aphid resistance has been mapped to R gene clusters in cereals (Liu et al., 2005; Sotelo et al., 2009), legumes (Githiri et al., 1996; Yang et al., 2004; Klingler et al., 2005, 2007; Kim et al., 2010), apple (Cevik and King, 2002), and lettuce (Eenink et al., 1982). R gene-mediated aphid resistance often includes multiple resistance mechanisms, including relatively better plant growth and reproduction despite aphid infestation (tolerance), deterrence of aphid settlement (antixenosis), or the complete or partial repression of aphid growth and reproduction (antibiosis; Smith, 2005). However, R gene-mediated resistance to aphids (often used in monocultured crops) can be overcome by newly evolved aphid biotypes as seen in lettuce, melon, soybean, and wheat (McCreight, 2008; Chen et al., 2009; Randolph et al., 2009; Dogimont et al., 2010; Michel et al., 2010). The use of a combination of R genes and/or quantitative basal resistance loci could provide a practical and sustainable strategy to solve the problem of resistance breakdown by new biotypes (Palloix et al., 2009).
Quantitative trait loci (QTLs) associated with aphid resistance have been identified in a limited number of crops, where distinct QTLs control specific aspects of plant resistance to aphids (e.g. antibiosis, antixenosis, or plant tolerance). In alfalfa, multiple QTLs that originated from distinct donor plants have been identified with additive effects on pea aphid (Acyrthosiphon pisum; PA) tolerance (Julier et al., 2004). In barley, three QTLs collectively explain 59% of the phenotypic variation for Russian wheat aphid (Diuraphis noxia) antibiosis (Mittal et al., 2008). Furthermore, in sorghum, distinct QTLs effecting either antibiosis or tolerance to greenbug (Schizaphis graminum) biotypes I and K have been identified (Agrama et al., 2002; Wu and Huang, 2008). Apart from the major resistance gene Vat in melon, four additive QTLs had a major effect on cotton-melon aphid resistance. These included separate QTLs affecting antixenosis and antibiosis (Boissot et al., 2010). In soybean, two QTLs controlling soybean aphid (Aphis glycines) antibiosis were detected both under glasshouse conditions and in field trials (Zhang et al., 2009).
The model legume Medicago truncatula is a host to multiple aphid species including the model aphid PA, and has emerged as an effective model system for the study of the genetic and molecular basis of aphid resistance. Single dominant loci controlling resistance to bluegreen aphid (BGA; Acyrthosiphon kondoi), spotted alfalfa aphid (SAA; Therioaphis trifolii); and PA in M. truncatula cv. Jester have been mapped to CC-NBS-LRR-rich regions on chromosome 3 of M. truncatula (Klingler et al., 2005, 2007; Gao et al., 2008). Resistance to BGA in Jester is conferred by AKR (Acyrthosiphon kondoi resistance), which conditions antibiosis, antixenosis, and tolerance, with the resistance being phloem based (Klingler et al., 2005). In the case of PA, the situation in M. truncatula appears more complex, stemming from the fact that PA is a genetically diverse species with several different biotypes with different preferential legume plant affiliations and distinct levels of damage on host plants (Birkle and Douglas, 1999; Bournoville et al., 2000). PA resistance in M. truncatula to an Australian biotype is mediated by a resistance gene independent from AKR termed APR (Acyrthosiphon pisum resistance) (Guo et al., 2009). The resistance to PA shares similarities with BGA resistance in that resistance involves a combination of antibiosis and antixenosis and is also phloem mediated (Gao et al., 2008; Guo et al., 2009). Furthermore, a single dominant gene for resistance against a European PA biotype, termed RAP1 (resistance to Acyrthosiphon pisum 1), has also been identified in the M. truncatula cv. Jemalong A17 (Stewart et al., 2009).
In addition to the monogenic and relatively pronounced aphid resistance described above, there are other forms of aphid resistance found in M. truncatula accessions. While M. truncatula cv. A17 contains the RAP1 locus against a European PA biotype, it also confers moderate resistance against BGA and the Australian PA biotype (Klingler et al., 2009; Stewart et al., 2009). A hallmark of A17’s response to the feeding of PA and BGA is the presence of necrotic flecks surrounding the aphid’s feeding site, conferred by AIL (aphid-induced lesions) for the European biotype (Stewart et al., 2009) and AIN (Acyrthosiphon-induced necrosis) for both BGA and the Australian PA biotype (Klingler et al., 2009). In previous studies, AIL did not provide resistance to the European PA biotype (Stewart et al., 2009), while the AIN locus co-segregated with the reduction of aphid colony for BGA but not for the Australian PA biotype, based on a limited number (24) of recombinant inbred lines (RILs) tested for PA (Klingler et al., 2009).
Both M. truncatula and PA are model species, with a suite of genomic and molecular resources having been generated (http://www.medicago.org; http://genouest.org/Insect/AphidBase/IAGC; Mutti et al., 2006; Town, 2006; IAGC, 2010; Carolan et al., 2011), which provide unique opportunities to dissect resistance-associated mechanisms underlying both sides of plant–aphid interactions. In the current study, the moderate resistance to the Australian PA biotype in the M. truncatula line A17 was characterized in detail and compared with the highly susceptible M. truncatula line A20 and the more resistant M. truncatula line Jester by using both choice and no-choice assays, and the electrical penetration graph (EPG) technique. It was found that the moderate PA resistance in A17 involved both antiobiosis and tolerance and that resistance was phloem based. QTL analysis using a RIL population from a cross between A17 and A20 revealed that the AIN locus also contributes to PA antibiosis and that separate, unlinked loci are responsible for plant tolerance against either PA or BGA.
Materials and methods
Plants and aphids
Two genotypes of M. truncatula were the primary focus of this study: A17 and A20. A RIL population derived from these genotypes was generated previously (Klingler et al., 2009) and used in this study for QTL analysis to identify loci important in aphid–plant interactions. Additionally, Jester, a line closely related to A17 but more resistant to PA and BGA than A17, was used as a control to compare the degree of resistance (Klingler et al., 2005; Gao et al., 2008). Seeds were germinated and plants grown as described by Klingler et al. (2009). The aphid species used were PA and BGA collected in Western Australia (Gao et al., 2007). PA and BGA were reared on faba bean (Vicia faba) and subterranean clover (Trifolium subterraneum), respectively, as described by Gao et al. (2007).
PA performance when confined on individual plants
Plants were grown in individual 0.9 litre pots in a greenhouse under natural light, and 12 replicate plants of each accession were randomly arranged. Three weeks after sowing, the largest trifoliate leaf was infested with a cohort of 10 early-instar nymphs of PA for which the weight was recorded, and caged with a mesh bag. Four days after infestation, the number and weight of the surviving aphids on each plant were recorded. The mean relative growth rate (MRGR) was calculated using the formula described in Gao et al. (2007) and this experiment is referred to as the ‘short-term experiment’. The Tukey HSD test (JMP 7.0 software; SAS Institute Inc.) was used to evaluate differences between lines in MRGR and aphid survivorship
To evaluate the response of the plant lines to the high infestation pressure in a long-term experiment, two apterous adult aphids were confined to a 2-week-old plant and the aphids were left to grow and reproduce for 9 d or 15 d on the caged plant. The aphids were confined to a seedling by placing the plant inside a transparent plastic bottle modified with a cut-off base and two large mesh-covered ventilation holes. The plants were grown in separate 0.9 litre pots. At 9 and 15 days post-infestation (dpi), the bottles were removed, the aphids were brushed off from each plant, and the aphid colony fresh weight for each plant was recorded. For both time points, 12 replicates were used and the t-test with JMP 7.0 software was used to compare the differences between A17 and A20 for each time point.
Plant damage and PA performance when allowed to move freely among plants
To assess the performance of PA and plant damage caused by aphids, 2-week-old seedlings of M. truncatula lines A17, A20, and Jester grown in separate 0.9 litre pots were infested with two apterous adult aphids. The aphids were allowed to develop, reproduce, and move freely among plants. The experiment included three biological replicates per accession. Aphid population build-up and feeding damage on plants were assessed at 3 d intervals from the third day up to 21 dpi using a scale from 1 to 5 and 0 to 5, respectively. For the aphid population build-up, the rating scale was as follows: 1=early instar nymphs present; 2=early and late instar nymphs and adults spread on most stems; 3=aphids spread on all stems and new trifoliate leaves; 4=high density of aphids on all stems and 50–80% trifoliate leaves covered with aphids; 5=plants overwhelmed by aphids with >80% covered. For feeding damage on plants, the scale was as follows: 0=no visual damage; 1=slight leaf curl, yellow spots; 2=yellowing leaves, curling leaves; 3=some leaves dying, others yellowing; 4=approximately 50% leaves browning off or dying; 5=dead.
Host selection behaviour
A17 and A20 were tested for host selection by PA alatae in a growth chamber. The experiment was set up according to Klingler et al. (2005). Two plants of each accession were randomly placed in the cage so that one plant occupied each of the four corners. Pots were spaced so that no leaves touched other plants. Fifty PA alatae were released at the platform above the plant in the centre of the each cage. Six cages representing six replicates were set up. Settling of aphids on each plant was recorded at 3, 6, 24, 48, 72, and 96 h after release. Paired t-test (JMP 7.0 software; SAS Institute Inc.) was used to measure the preference of settled alatae at each time point.
Aphid feeding behaviour
The feeding behaviour of PA on A17 and A20 was investigated by the direct-current EPG technique (Tjallingii, 1988). When plants were 3–4 weeks old, a single apterous adult PA was placed on a new fully expanded trifoliate leaf and monitored for 8 h as described by Klingler et al. (2005). The EPG output was recorded and analysed with Probe 3.0 (Wageningen, The Netherlands, www.epgsystems.eu). Sixteen biological replicates were included for each accession. EPG waveforms were scored by waveform pattern and auto power spectra [which present the waveform frequency (Hz) versus the relative magnitude with a maximum of 1, which provide a major characteristic of waveform identity]. The acquired data were further analysed by the tool based on Microsoft Excel® Workbook developed by Sarria et al. (2009). The parameters measured in this work included: the total duration of the pathway phase (C); total duration of xylem ingestion (G); total duration of salivation (E1); total duration of phloem ingestion (E2); the derailed stylet (F); total duration of non-penetration (np); and total duration of extracellular salivation (E1e). The Mann–Whitney U-test using the software SPSS 13.0 for Windows was used for the data analysis.
Genetic analysis of plant–aphid interactions
The original F6 A17×A20 genetic map was constructed using a set of 89 simple sequence repeat (SSR) markers based on 114 RILs, using the Multipoint v1.2 software according to Kamphuis et al. (2008), for a RIL selfing population instead of an F2 population. The genotype data for the 114 RIL are provided in Supplementary Table S1 available at JXB online. In Table S1, linkage groups (LGs) 4 and 8 are grouped together due to the presence of a reciprocal translocation in A17 (Kamphuis et al., 2007). The genetic map spans a total of 445.3 cM with an average distance of 4.79 cM between markers. The LGs of the genetic maps presented are similar in length to previously published maps (Thoquet et al., 2002; Choi et al., 2004; Mun et al., 2006; Kamphuis et al., 2008), and the 89 markers appear to be evenly distributed across the eight LGs. Small quantities of the A17×A20 RIL population seed are available upon request. The phenotypes for PA and BGA resistance of the F7 RILs were investigated with a subset of 87 and 93 RILs, respectively. Twelve randomized complete blocks were used with one repeat of each RIL as well as the parents randomly assigned. Six blocks were infested with PA and six blocks were used as uninfested controls. For BGA, 12 replicates from each RIL along with 12 replicates from each parent were divided into two groups, one group for infestation and the other for the non-infested control. Within each group, the six replicate genotypes were arranged in a completely randomized design as described previously by Klinger et al. (2009).
For PA, one group of seedlings was infested with two apterous adults 15 d after sowing, including an uninfested control group. At 17 dpi, the aphids were collected and the plants were cut off at the soil level. Aphid biomass, plant biomass, and symptoms such as necrosis were recorded. For BGA, 18 d after sowing, six individuals per RIL family were infested with two adult apterous BGA and six individuals were not infested (i.e. the controls). At 19 dpi, the aphids and plants were harvested and assessed in the same manner as in the PA experiment.
The QTL analysis was performed with the software package MultiQTL v2.5 using the general interval mapping and marker restoration options for a RIL-selfing population as described by Lichtenzveig et al. (2006). Using a LOD score of 3 as the minimal threshold for further analysis, the hypotheses that a single locus or two-linked loci have an effect on resistance to aphids were tested.
Results
PA performance in no-choice and choice experiments
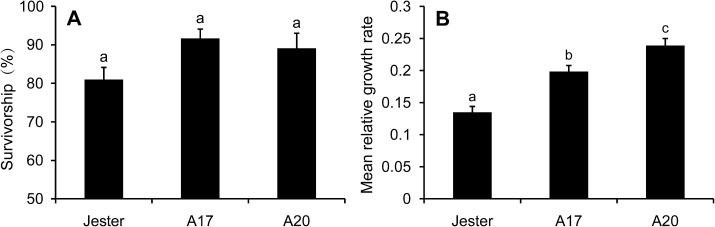
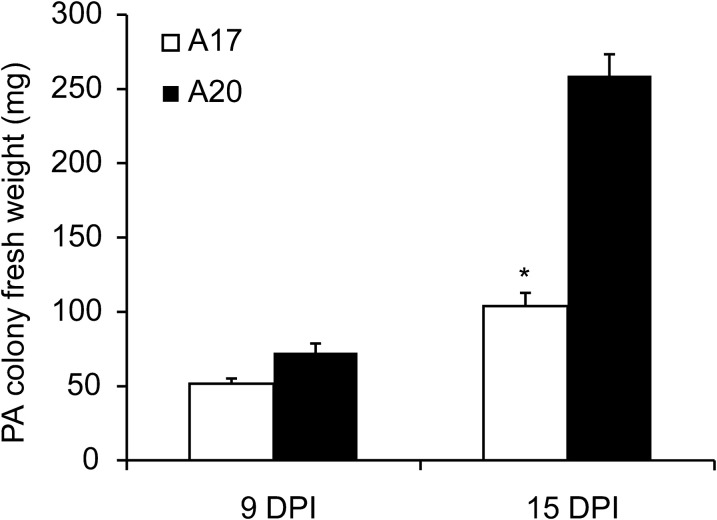
Previous studies showed evidence that A17 is relatively resistant compared with A20, but more susceptible than Jester, to the Australian PA biotype (Klingler et al., 2005, 2009; Gao et al., 2008). To characterize the PA resistance in A17 further, the MRGR and survivorship of PA on A17, A20, and Jester in a confined leaf cage were compared. In this short-term (4 d) experiment, no significant differences were found in survivorship of the nymphs infested on the different M. truncatula accessions (Fig. 1A). However, the cohorts of PA nymphs showed intermediate MRGR on A17 compared with the susceptible line A20 and the resistant line Jester (Fig. 1B). To compare the antibiotic effect of genotypes under high infestation pressure for a long period, whole-plant cages were used to confine two PA adults to individual plants. At 9 dpi the fresh weight of the PA colony was lower on A17, but not significantly different from A20, whereas by 15 dpi the PA colony fresh weight was significantly lower on A17 compared with A20 (Fig. 2).
Fig. 1.
Survivorship (A) and mean relative growth rates (B) of cohorts of PA on three different M. truncatula accessions (Jester, A17, and A20). Each value represents the mean and standard error of 12 biological replicates. Means labelled with a different letter are significantly different (Tukey HSD test, P < 0.05).
Fig. 2.
PA colony growth on A17 and A20 observed at 9 days post-infestation (dpi) and 15 dpi in a no-choice test. Values are the mean and standard error of 12 biological replicates. Means labelled with * for M. truncatula line A17 are significantly different from those for line A20 (P < 0.05).
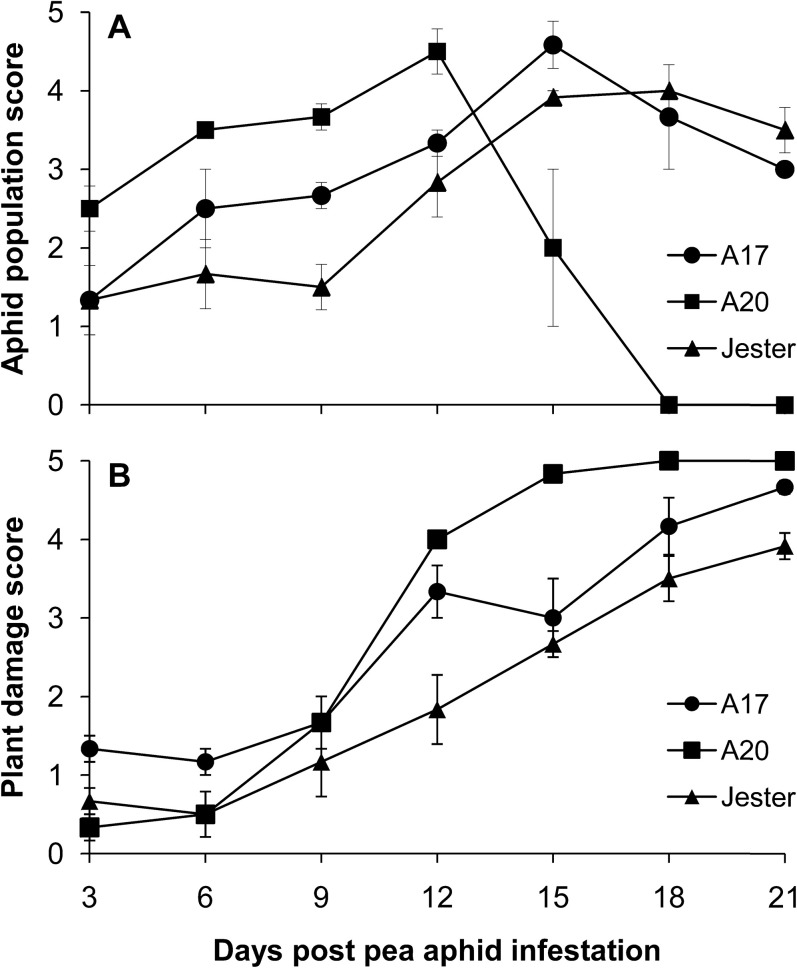
In a choice experiment, adult aphids and their progeny were allowed to move freely among different plants. Plant damage and aphid population build-up were assessed in a time-course experiment of 21 d. As shown in Fig. 3, 6 dpi with two PA adults per plant, the aphids reproduce and colonize A20 plants, building up larger aphid populations (mean score=3.5) at a higher rate than in A17 and Jester plants where the populations are smaller (mean score=2.5 and 1.7, respectively). On A20, aphid populations reached a peak (mean score=4.5) at 12 dpi. At this stage, A20 plants were starting to wilt and collapse due to the high aphid pressure. With the collapse of A20, aphids migrated to the nearby healthier plants. At 15 dpi, the aphid population on A17 reached a peak (mean score=4.6) and most A20 plants had succumbed to the PA infestation. With the death of A20 plants, and A17 plants now starting to show severe damage symptoms, aphids moved to Jester and caused the highest density of aphids on Jester at 18 dpi (mean score=3.7). At this stage, A17 showed significantly more stunting, wilting, and leaf curling compared with the resistant Jester.
Fig. 3.
Mean aphid population score (A) and mean plant damage score (B) for M. truncatula lines Jester, A17, and A20 over a 21 d period following PA infestation. Values are the mean and standard error of three biological replicates.
PA displays no host preference between A17 and A20
Antixenosis indicates the presence of plant morphological or chemical factors that adversely affect arthropod behaviour, resulting in the selection of an alternative host plant (Smith, 2005). It had previously been reported that PA showed a preference for A17 over the more resistant Jester, suggesting that antixenosis is involved in Jester’s resistance to PA (Gao et al., 2008). In order to determine whether antixenosis is involved in the moderate resistance to PA in A17 relative to the highly susceptible A20, a cohort of PA alatae were quickly dispersed from the releasing platform to allow a choice between the susceptible A20 and the resistant A17 plants. The average number of settled alatae increased on both A17 and A20 accessions up to 24 h after aphid release. Despite a trend towards more PA alatae settling on A20, there was no significant difference between the resistant A17 and susceptible A20 plants in settling behaviour over the course of the experiments, suggesting that antixenosis does not play a significant role in the resistance to PA in A17 (Supplementary Fig. S1 at JXB online).
Resistance in A17 is phloem mediated
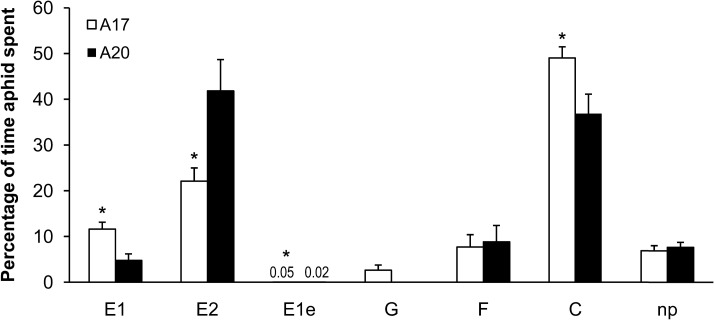
The EPG technique was used to record PA feeding behaviour on the resistant A17 and susceptible A20. EPG is a robust tool to discern activities of aphid stylets as well as locations in plant tissue in real time, including salivation into sieve elements and passive uptake of phloem sap (Tjallingii and Hogen-Esch, 1993). As shown in Fig. 4, the proportions of time that tethered apterae spent outside the plant (np, non-penetration) or contacting xylem (G) did not differ significantly between A17 and A20. On A17, aphids spent significantly more time than on A20 penetrating between cells en route to the vascular tissue (C, pathway phase) (P < 0.05) and salivating (E1) into the phloem (P < 0.05). However, the proportion of time that aphids spent on A17 ingesting phloem sap was significantly lower than on A20 (P < 0.05) (Fig. 4).
Fig. 4.
The percentage of time PA spent in various activities on A17 or A20 during an 8 h exposure to the host plants. Watery salivation in phloem (E1) indicates that aphids are injecting watery saliva into the sieve element; phloem ingestion (E2) indicates that aphids are ingesting the phloem sap; extracellular salivation (E1e) indicates watery salivation at the extracellular voltage level (note: because of the low values for Ele, the numbers were used instead of bars); xylem ingestion (G) indicates stylet penetration of tracheary elements; the derailed stylet (F) indicates penetration difficulties of stylets; pathway phase (C) indicates mostly intramural probing activities between mesophyll or parenchyma cells; non-penetration (np) indicates that stylets are outside the plant. Values are the mean and standard error of 16 biological replicates. Means labelled with * are significantly different (t-test; P < 0.05).
On A17, extracellular salivation (E1e) occurred substantially more frequently than on A20 (P < 0.05). Thirteen out of 16 E1e periods happened after E1 in A17 plants, while there were only two periods of E1e in A20 plants. Furthermore, on A17, aphids showed significantly less sustained phloem sap ingestion (sustained E2 waveform longer than 10 min) than A20 (P < 0.05).
The genetic basis of resistance to PA and BGA in M. truncatula A17
All the 47 F2:7 families that harboured an A17 allele in the AIN region displayed necrotic flecks at the site of PA infestation, confirming the previous results gained from a smaller RIL population (Klingler et al., 2009).
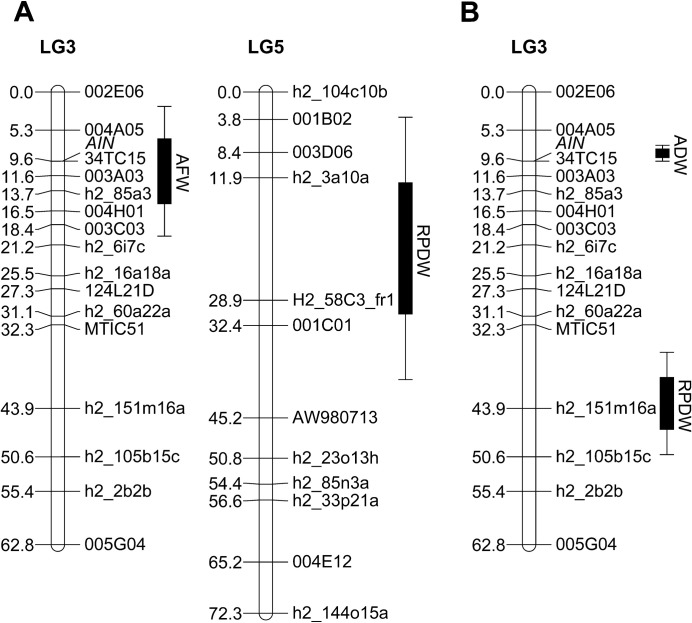
To determine the role of the AIN locus in the PA resistance, QTL analysis was performed using the aphids’ fresh weight data. This analysis identified a highly significant locus with a LOD score of 4.5 explaining 23% of aphid fresh weight (AFW; Fig. 5A; Table 1) on chromosome 3 (LG3). The AFW QTL spans across the region in which the AIN locus resides (Fig. 5A). For BGA, the aphid biomass was measured previously as aphid dry weight for 93 RILs (Klingler et al., 2009). QTL analysis on these data confirmed a major, highly significant locus with a LOD score of 35.3 explaining 87% of the phenotype for BGA dry weight (ADW) in the region containing AIN (Fig. 5B; Table 2). The AIN locus is thus the major locus controlling BGA antibiosis, consistent with previous results (Klingler et al., 2009).
Fig. 5.
Genetic map positions of QTLs involved in resistance to PA (A) and BGA (B) based on phenotype data for 87 and 93 individuals of a recombinant inbred line (RIL) population of A17×A20, respectively. Interval distances are listed in centiMorgans. The genomic locations of the QTLs are depicted on the right of a linkage group (LG), with standard deviations depicted by lines on either side. AFW, pea aphid fresh weight; RPDW, relative reduction of plant dry weight; ADW, bluegreen aphid dry weight.
Table 1.
Features of the QTLs associated with the resistance response to pea aphid (PA) in the A17×A20 M. truncatula RIL population
| Trait | LOD | LG | Position | PEVa | Response mean | Effectb |
| AFWc | 4.5 | 3 | 11.1 (9.1) | 0.23 (0.07) | 0.134 (0.003) | –0.024(0.005) |
| RPDWd | 3.9 | 5 | 21.7 (18.2) | 0.22 (0.07) | 0.513 (0.014) | –0.11(0.022) |
Proportion of explained variability.
The estimated effect; a positive value represents the effect of the A17 allele and a negative value represents the effect of the A20 allele.
Mean aphid fresh weight (g).
Relative reduction of plant dry weight caused by PA.
Table 2.
Features of the QTLs associated with the resistance response to bluegreen aphid (BGA) in the M. truncatula A17×A20 RIL population
| Trait | LOD | LG | Position | PEVa | Response mean | Effectb |
| ADWc | 35.3 | 3 | 8.5 (1.1) | 0.87 (0.04) | 0.015 (0.001) | –0.016(0.001) |
| RPDWd | 6.2 | 3 | 43.2 (7.1) | 0.33 (0.09) | 0.465 (0.012) | –0.12(0.023) |
Proportion of explained variability.
The estimated effect; a positive value represents the effect of the A17 allele and a negative value represents the effect of the A20 allele.
Mean aphid fresh weight (g).
Relative reduction of plant dry weight caused by PA.
To determine whether plant tolerance plays a role in the resistance phenotypes of A17 to PA and BGA, the mean plant dry weight of the control and aphid-infested plants was recorded to determine the relative reduction in plant dry weight caused by aphid infestation. For PA tolerance, a single QTL for relative reduction of plant dry weight (RPDW) with a LOD score of 3.9 and a proportion of explained variability (PEV) of 22% was mapped to chromosome 5 (LG5; Fig. 5A; Table 1). For BGA tolerance, a QTL for RPDW (LOD=6.2) with a PEV of 33% was identified on chromosome 3 (LG3) which was independent from that of BGA dry weight (ADW; Fig. 5B; Table 2). There are thus unlinked loci controlling plant tolerance (e.g. reduction in plant dry weight) in A17 for BGA and PA infestation and, for both aphids, these loci are also independent of the loci involved in aphid resistance (antibiosis).
Discussion
In the present study the moderate resistance to an Australian biotype of PA in the M. truncatula accession A17 was characterized and the genetic basis of the resistance to both PA and BGA in this accession was dissected. The results suggest that antibiosis is involved in A17’s resistance to the Australian biotype of PA. The antibiosis of A17 to the Australian biotype of PA is different from its response to the European PA biotype PS01 which showed a negative MRGR on A17 with lethality of aphids as early as 48 h post-infestation (Stewart et al., 2009). The Australian PA’s response to A17 appears to be distinct from that of another European biotype, LL01, in that LL01 has a much higher MRGR after 2 d compared with the Australian biotype after 4 d (Stewart et al., 2009). There thus appears to be three distinct forms of PA resistance in M. truncatula A17.
In the host selection experiments between the resistant A17 and the susceptible A20 with alatae of PA, significant differences in the settling behaviour were not observed, although the repeated experiments showed a lower number of alatae settling on A17 after 3 h and a lower aphid population on A17 than on the susceptible A20 at the early stage of PA infestation. This is in contrast to the observations seen for choice experiments between Jester and A17, where a significant difference in settling behaviour was observed 24 h post-alatae release (Gao et al., 2008). It therefore appears that antixenosis does not play a significant role in the resistance phenotype observed in A17 to the Australian PA biotype.
The EPG results showed that PA spent significantly more time salivating into the phloem sap and significantly less time ingesting phloem sap (the E2 waveform) on A17 plants than on A20 plants (Fig. 4). The longer duration of E1 salivation with shorter duration of phloem sap ingestion on A17 suggested that PA secreted more saliva into the sieve elements possibly to counter a plant defence response such as the plugging of sieve pores (Will and van Bel, 2006; Will et al., 2007, 2009). Furthermore, aphids on A17 showed significantly less sustained phloem sap ingestion (sustained E2 waveform longer than 10 min) than on A20; 10 min is a threshold often used as an indicator of phloem acceptance (Tjallingii, 2006). Deterrence compound(s) present in the phloem sap of A17 could explain the difference in sustained phloem sap ingestion between A17 and A20. Alternatively, frequent extracellular salivation (E1e) following the watery salivation (E1) in A17 generally indicates cell collapse (Tjallingii, 2006), which could be a symptom of cell death induced by recognition of effectors in the saliva of PA. This hypothesis is supported by the presence of localized necrotic flecks resembling a hypersensitive response (HR) surrounding the feeding site of PA in A17.
A17 is relatively resistant to both BGA and PA compared with A20, and A17 showed AIN lesions which resemble a HR at the site of infestation by both BGA and PA. Genetic analysis using a RIL population between A17 and A20 showed that this phenotype in response to BGA and PA was controlled by a semi-dominant locus termed AIN (Klingler et al., 2009), and the analysis of additional RILs carried out in this study further confirmed these findings.
The RIL population and associated genotyping data presented here should be a valuable tool for the wider M. truncatula community given that the parents were used by the M. truncatula genome consortium to generate the F2 reference genetic map and they segregate for various phenotypes such as pigmentation of leaves, pod shape, and the ability to establish nitrogen-fixing nodules with the ecotype-specific Sinorhizobium meliloti strains Rm41 and NRG247 (Simsek et al., 2007). Thus, this population was also used to identify QTLs involved in the resistance response to Rhizoctonia solani (J.P. Anderson, J. Lichtenzveig, and K.B. Singh, unpublished results).
Several QTLs have been identified for aphid antibiosis (Mittal et al., 2008; Stoeckli et al., 2008; Zhang et al., 2009, 2010), but only a few studies have shown distinct QTLs for antibiosis and tolerance, such as in melon to cotton-melon aphid (Boissot et al., 2010) and in sorghum to greenbug (Agrama et al., 2002; Wu and Huang, 2008).
QTL analysis on the PA biomass data revealed a significant QTL with a modest effect on aphid fresh weight (AFW) explaining 23% of the phenotype and spanning the AIN region (Fig. 5; Table 1). This modest effect could explain why previous findings using a limited set of 24 RILs of the same population by Klingler et al. (2009) concluded that AIN does not have an effect on PA biomass. In contrast the AIN locus is strongly associated with a reduction in BGA weight (Fig. 5; Table 1; Klingler et al., 2009). Interestingly, the AIL locus, which also conditions a necrotic fleck phenotype resembling a HR to a European PA biotype PS01 in A17, has been mapped to the same region on chromosome 3 (Stewart et al., 2009). The AIL locus did not have an effect on PS01 biomass, but rather the effect was mediated by two loci on chromosomes 3 and 6 termed RAP1 and RAP2, respectively. These previous experiments with the AIL or AIN locus differed from the current study in that pea aphids in the previous experiments were not allowed access to more than a single plant. It remains to be determined if the AIN phenotype to both aphid species is controlled either by the same gene or by two tightly linked genes.
The AIN locus resides in a region containing a cluster of paralogous genes of the NBS-LRR family (Klingler et al., 2009). Therefore, one or more family members may control aphid biomass by mediating antibiosis and a localized HR response following aphid attack. Under the leading models for R protein function, a protein encoded by a single gene such as AIN may ‘guard’ (monitor) a single effector target protein (or a ‘decoy’ protein) that is recognized and targeted for manipulation by effectors from different Acyrthosiphon species or biotypes (Dangl and Jones, 2001; van der Hoorn and Kamoun, 2008). Variation in the effectiveness of AIN against PA and BGA might then be explained by differences in binding affinities of the species-specific or biotype-specific effectors toward the same target protein and/or the AIN protein. Alternatively, different NBS-LRR genes in the same or adjacent clusters may recognize separate operative targets (or similar proteins acting as decoys) of distinct effectors from the different species and biotypes in the genus Acyrthosiphon. Isolation of the AIN gene will help to determine whether the same gene controls aphid-induced necrosis by BGA and PA and whether AIN plays a role in reducing the biomass of either BGA or the Australian PA biotype.
For both PA and BGA, the loci controlling relative reduction of plant biomass (RPDW) following aphid infestation were mapped to a position unlinked to AIN and antibiosis (Fig. 5). Plant biomass reduction was probably due to the nutrient loss caused by the aphids and the redirection of resources to induce a defence response as seen in other plant–aphid interactions (Edwards and Singh, 2006; Howe and Jander, 2008). Several QTLs have been identified for aphid antibiosis (Mittal et al., 2008; Stoeckli et al., 2008; Zhang et al., 2009, 2010), but only a few studies have shown distinct QTLs for antibiosis and tolerance, such as in melon to cotton-melon aphid (Boissot et al., 2010) and in sorghum to greenbug (Agrama et al., 2002; Wu and Huang, 2008). Separate loci for antibiosis and relative reduction of plant biomass suggest the independent genetic control of antibiosis and tolerance in A17. This mode of inheritance provides an opportunity to dissect two types of insect resistance simultaneously in one system.
Furthermore, the QTL termed RPDW for PA and BGA was mapped to different chromosomes for each species (Fig. 5). The different positions of QTLs mediating RPDW caused by the closely related aphids PA and BGA suggest that A17 has distinct genetic interactions with different Acyrthosiphon spp. The relative reduction of plant biomass following PA infestation is partially explained by a QTL on the short arm of chromosome 5 in A17, whereas the relative reduction of plant biomass following BGA is influenced by a QTL on the long arm of chromosome 3 in the same region where PA resistance genes RAP1 (Stewart et al., 2009) and APR (Guo et al., 2009; unpublished data) reside. This raises the possibility that the R gene cluster containing RAP1 and APR might also harbour a gene that contributes to resistance to BGA in terms of plant tolerance. This could explain how PA resistance was retained in generating the M. truncatula cv. Jester even though the breeding programme only selected for BGA and spotted alfalfa aphid resistance in Jester (Hill, 2000), since the APR locus co-segregates with the BGA RPDW QTL. The M. truncatula cv. Jester is closely related to A17 and harbours the major resistance genes AKR (for BGA resistance) and APR (for PA resistance). Further studies will help to elucidate whether the identified QTLs in A17 for BGA antibiosis and tolerance play a role in the resistance phenotype observed in Jester and whether there are interactions between AKR and these loci.
With the frequent breakdown of single dominant R genes in extensively monocultured agricultural systems, it is important to pyramid multiple resistance genes into the crops with major R genes to help inhibit the occurrence of new virulent biotypes. Plant tolerance does not affect the growth and reproduction of insects. Thus, it could not exert selection pressure on the appearance of new biotypes as antibiosis and antixenosis can, but it can help limit the damage these pests cause to crops. However, it has been a great challenge to separate tolerance from antibiosis and antixenosis and accurately quantify the effect of tolerance (Smith, 2005). In the present study, both the aphid colony biomass on each plant and the relative plant biomass reduction caused by aphid infestation were investigated in a RIL population. For both PA and BGA, separate loci for those two traits were identified. This will facilitate marker-assisted breeding of M. truncatula with tolerance to aphids and potentially other closely related Medicago species. The identification of distinct loci involved in resistance to the model aphid PA and its close relative of the same genus, BGA, in a model plant make this a powerful system to study plant defence against sap-sucking insects as well as R gene specificity and evolution.
Supplementary data
Supplementary data are available at JXB online.
Figure S1 . Settling of pea aphid alatae in a choice test conducted in a growth chamber.
Table S1 . The A17×A20 recombinant inbred line population genotype data
Acknowledgments
We would like to thank Elaine Smith, Jenny Reidy-Croft, and Julie Lawrence for technical assistance on the project. S-MG was supported by a CSIRO/China Scholarship Council fellowship. LGK was the recipient of a CSIRO/OCE Post-Doctoral Fellowship.
References
- Agrama H, Widle G, Reese J, Campbell L, Tuinstra M. Genetic mapping of QTLs associated with greenbug resistance and tolerance in Sorghum bicolor . Theoretical and Applied Genetics. 2002;104:1373–1378. doi: 10.1007/s00122-002-0923-3. [DOI] [PubMed] [Google Scholar]
- Birkle LM, Douglas AE. Low genetic diversity among pea aphid (Acyrthosiphon pisum) biotypes of different plant affiliation. Heredity. 1999;6:605–612. doi: 10.1046/j.1365-2540.1999.00509.x. [DOI] [PubMed] [Google Scholar]
- Boissot N, Thomas S, Sauvion N, Marchal C, Pavis C, Dogimont C. Mapping and validation of QTLs for resistance to aphids and whiteflies in melon. Theoretical and Applied Genetics. 2010;121:9–20. doi: 10.1007/s00122-010-1287-8. [DOI] [PubMed] [Google Scholar]
- Bournoville R, Simon JC, Badenhausser I, Girousse C, Guilloux T, André S. Clones of pea aphid, Acyrthosiphon pisum (Hemiptera: aphididae) distinguished using genetic markers, differ in their damaging effect on a resistant alfalfa cultivar. Bulletin of Entomological Research. 2000;90:33–39. [PubMed] [Google Scholar]
- Carolan JC, Caragea D, Reardon KT, et al. Predicted effector molecules in the salivary secretome of the pea aphid (Acyrthosiphon pisum): a dual transcriptomic/proteomic approach. Journal of Proteome Research. 2011;10:1505–1518. doi: 10.1021/pr100881q. [DOI] [PubMed] [Google Scholar]
- Cevik V, King GJ. High-resolution genetic analysis of the Sd-1 aphid resistance locus in Malus spp. Theoretical and Applied Genetics. 2002;105:346–354. doi: 10.1007/s00122-002-0904-6. [DOI] [PubMed] [Google Scholar]
- Chen M-S, Echegaray E, Whitworth JR, Wang H, Sloderbeck PE, Knutson A, Giles KL, Royer TA. Virulence analysis of Hessian fly populations from Texas, Oklahoma, and Kansas. Journal of Economical Entomology. 2009;102:774–780. doi: 10.1603/029.102.0239. [DOI] [PubMed] [Google Scholar]
- Choi HK, Kim D, Uhm T, et al. A sequence-based genetic map of Medicago truncatula and comparison of marker colinearity with M. sativa . Genetics. 2004;166:1463–1502. doi: 10.1534/genetics.166.3.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- Dogimont C, Bendahmane A, Burget-Bigeard E, Hagen L, Le Menn A, Pauquet J, Rouselle P, Caboche M, Chovelon V. Gene resistant to Aphis gossypii . 2007 US Patent. [Google Scholar]
- Dogimont C, Bendahmane A, Chovelon V, Boissot N. Host plant resistance to aphids in cultivated crops: genetic and molecular bases, and interactions with aphid populations. Comptes Rendus Biologies. 2010;333:566–573. doi: 10.1016/j.crvi.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Edwards O, Singh K. Resistance to insect pests: what do legumes have to offer? Euphytica. 2006;147:273–285. [Google Scholar]
- Eenink AH, Groenwold R, Dieleman FL. Resistance of lettuce (Lactuca) to the leaf aphid Nasonovia ribis nigri. 1. Transfer of resistance from L. virosa to L. sativa by interspecific crosses and selection of resistant breeding lines. Euphytica. 1982;31:291–299. [Google Scholar]
- Gao LL, Horbury R, Nair RM, Edwards OR, Singh KB. Characterization of resistance to multiple aphid species (Hemiptera: Aphididae) in Medicago truncatula . Bulletin of Entomological Research. 2007;97:41–48. doi: 10.1017/S0007485307004786. [DOI] [PubMed] [Google Scholar]
- Gao LL, Klingler JP, Anderson JP, Edwards OR, Singh KB. Characterization of pea aphid resistance in Medicago truncatula . Plant Physiology. 2008;146:996–1009. doi: 10.1104/pp.107.111971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Githiri SM, Ampong-Nyarko K, Osir EO, Kimani PM. Genetics of resistance to Aphis craccivora in cowpea. Euphytica. 1996;89:371–376. [Google Scholar]
- Guo S, Kamphuis LG, Gao L-L, Edwards OR, Singh KB. Two independent resistance genes in the Medicago truncatula cultivar Jester confer resistance to two different aphid species of the genus Acyrthosiphon . Plant Signaling and Behavior. 2009;4:328–331. doi: 10.4161/psb.4.4.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JR. Jester. Plant Varieties Journal. 2000;13:40. [Google Scholar]
- Hogenhout SA, Bos JIB. Effector proteins that modulate plant–insect interactions. Current Opinion in Plant Biology. 2011;14:422–428. doi: 10.1016/j.pbi.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Howe GA, Jander G. Plant immunity to insect herbivores. Annual Review of Plant Biology. 2008;59:41–66. doi: 10.1146/annurev.arplant.59.032607.092825. [DOI] [PubMed] [Google Scholar]
- IAGC. Genome sequence of the pea aphid Acyrthosiphon pisum . PLoS Biology. 2010;8:e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julier B, Bournoville R, Landre B, Ecalle C, Carre S. Genetic analysis of lucerne (Medicago sativa L.) seedling resistance to pea aphid (Acyrthosiphon pisum Harris) Euphytica. 2004;138:133–139. [Google Scholar]
- Kamphuis L, Lichtenzveig J, Oliver R, Ellwood S. Two alternative recessive quantitative trait loci influence resistance to spring black stem and leaf spot in Medicago truncatula . BMC Plant Biology. 2008;8:30. doi: 10.1186/1471-2229-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphuis LG, Williams A, Pfaff T, Ellwood SR, D’ Souza NK, Groves E, Singh K, Oliver RP, Lichtenzveig J. Medicago truncatula reference accession A17 has aberrant chromosomal arrangement. New Phytologist. 2007;174:299–303. doi: 10.1111/j.1469-8137.2007.02039.x. [DOI] [PubMed] [Google Scholar]
- Kim K-S, Hill C, Hartman G, Hyten D, Hudson M, Diers B. Fine mapping of the soybean aphid-resistance gene Rag2 in soybean PI 200538. Theoretical and Applied Genetics. 2010;121:599–610. doi: 10.1007/s00122-010-1333-6. [DOI] [PubMed] [Google Scholar]
- Klingler J, Creasy R, Gao L, Nair RM, Calix AS, Spafford Jacob H, Edwards OR, Singh KB. Aphid resistance in Medicago truncatula involves antixenosis and phloem-specific, inducible antibiosis, and maps to a single locus flanked by NBS-LRR resistance gene analogs. Plant Physiology. 2005;137:1445–1455. doi: 10.1104/pp.104.051243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingler JP, Edwards OR, Singh KB. Independent action and contrasting phenotypes of resistance genes against spotted alfalfa aphid and bluegreen aphid in Medicago truncatula . New Phytologist. 2007;173:630–640. doi: 10.1111/j.1469-8137.2006.01939.x. [DOI] [PubMed] [Google Scholar]
- Klingler JP, Nair RM, Edwards OR, Singh KB. A single gene, AIN, in Medicago truncatula mediates a hypersensitive response to both bluegreen aphid and pea aphid, but confers resistance only to bluegreen aphid. Journal of Experimental Botany. 2009;60:4115–4127. doi: 10.1093/jxb/erp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenzveig J, Bonfil D, Zhang H-B, Shtienberg D, Abbo S. Mapping quantitative trait loci in chickpea associated with time to flowering and resistance to Didymella rabiei the causal agent of Ascochyta blight. Theoretical and Applied Genetics. 2006;113:1357–1369. doi: 10.1007/s00122-006-0390-3. [DOI] [PubMed] [Google Scholar]
- Liu XM, Smith CM, Friebe BR, Gill BS. Molecular mapping and allelic relationships of Russian wheat aphid-resistance genes. Crop Science. 2005;45:2273–2280. [Google Scholar]
- McCreight JD. Potential sources of genetic resistance in Lactuca spp. to the lettuce aphid Nasanovia ribisnigri (Mosely) (Homoptera: Aphididae) HortScience. 2008;43:1355–1358. [Google Scholar]
- Michel AP, Mian MA, Davila-Olivas NH, Cañas LA. Detached leaf and whole plant assays for soybean aphid resistance: differential responses among resistance sources and biotypes. Journal of Economic Entomology. 2010;103:949–957. doi: 10.1603/ec09337. [DOI] [PubMed] [Google Scholar]
- Mittal S, Dahleen LS, Mornhinweg D. Barley germplasm STARS-9577B lacks a Russian wheat aphid resistance allele at a quantitative trait locus present in STARS-9301B. Crop Science. 2008;49:1999–2004. [Google Scholar]
- Mun JH, Kim DJ, Choi HK, et al. Distribution of microsatellites in the genome of Medicago truncatula: a resource of genetic markers that integrate genetic and physical maps. Genetics. 2006;172:2541–2555. doi: 10.1534/genetics.105.054791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutti NS, Park Y, Reese JC, Reeck GR. RNAi knockdown of a salivary transcript leading to lethality in the pea aphid, Acyrthosiphon pisum . Journal of Insect Science. 2006;6:38. doi: 10.1673/031.006.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palloix A, Ayme V, Moury B. Durability of plant major resistance genes to pathogens depends on the genetic background, experimental evidence and consequences for breeding strategies. New Phytologist. 2009;183:190–199. doi: 10.1111/j.1469-8137.2009.02827.x. [DOI] [PubMed] [Google Scholar]
- Randolph TL, Peairs F, Weiland A, Rudolph JB, Puterka GJ. Plant responses to seven Russian wheat aphid (Hemiptera: Aphididae) biotypes found in the United States. Journal of Economic Entomology. 2009;102:1954–1959. doi: 10.1603/029.102.0528. [DOI] [PubMed] [Google Scholar]
- Rossi M, Goggin FL, Milligan SB, Kaloshian I, Ullman DE, Williamson VM. The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proceedings of the National Academy of Sciences, USA. 1998;95:9750–9754. doi: 10.1073/pnas.95.17.9750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarria E, Cid M, Garzo E, Fereres A. Excel workbook for automatic parameter calculation of EPG data. Computers and Electronics in Agriculture. 2009;67:35–42. [Google Scholar]
- Simsek S, Ojanen-Reuhs T, Stephens SB, Reuhs BL. Strain-ecotype specificity in Sinorhizobium meliloti–Medicago truncatula symbiosis is correlated to succinoglycan oligosaccharide structure. Journal of Bacteriology. 2007;189:7733–7740. doi: 10.1128/JB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CM. Plant resistance to arthropods: molecular and conventional approaches. Dordrecht, The Netherlands: Springer; 2005. [Google Scholar]
- Sotelo P, Starkey S, Voothuluru P, Wilde GE, Smith CM. Resistance to Russian wheat aphid biotype 2 in CIMMYT synthetic hexaploid wheat lines. Journal of Economic Entomology. 2009;102:1255–1261. doi: 10.1603/029.102.0352. [DOI] [PubMed] [Google Scholar]
- Stewart SA, Hodge S, Ismail N, Mansfield JW, Feys BJ, Prosperi JM, Huguet T, Ben C, Gentzbittel L, Powell G. The RAP1 gene confers effective, race-specific resistance to the pea aphid in Medicago truncatula independent of the hypersensitive reaction. Molecular Plant-Microbe Interactions. 2009;22:1645–1655. doi: 10.1094/MPMI-22-12-1645. [DOI] [PubMed] [Google Scholar]
- Stoeckli S, Mody K, Gessler C, Patocchi A, Jermini M, Dorn S. QTL analysis for aphid resistance and growth traits in apple. Tree Genetics and Genomes. 2008;4:833–847. [Google Scholar]
- Thoquet P, Ghérardi M, Journet E, Kereszt A, Ané J, Prosperi J, Huguet T. The molecular genetic linkage map of the model legume Medicago truncatula: an essential tool for comparative legume genomics and the isolation of agronomically important genes. BMC Plant Biology. 2002;2:1. doi: 10.1186/1471-2229-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjallingii WF. Electrical recording of stylet penetration activities. In: Minks AK, Harrewijn P, editors. Aphids, their biology, natural enemies and control. Vol. 2B. Elsevier; 1988. pp. 95–108. [Google Scholar]
- Tjallingii WF. Salivary secretions by aphids interacting with proteins of phloem wound responses. Journal of Experimental Botany. 2006;57:739–745. doi: 10.1093/jxb/erj088. [DOI] [PubMed] [Google Scholar]
- Tjallingii WF, Hogen-Esch T. Fine structure of aphid stylet routes in plant tissues in correlation with EPG signals. Physiological Entomology. 1993;18:317–328. [Google Scholar]
- Town C. Annotating the genome of Medicago truncatula . Current Opinion in Plant Biology. 2006;9:122–127. doi: 10.1016/j.pbi.2006.01.004. [DOI] [PubMed] [Google Scholar]
- van der Hoorn RAL, Kamoun S. From guard to decoy: a new model for perception of plant pathogen effectors. The Plant Cell. 2008;20:2009–2017. doi: 10.1105/tpc.108.060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will T, Kornemann SR, Furch ACU, Tjallingii WF, van Bel AJE. Aphid watery saliva counteracts sieve-tube occlusion: a universal phenomenon? Journal of Experimental Biology. 2009;212:3305–3312. doi: 10.1242/jeb.028514. [DOI] [PubMed] [Google Scholar]
- Will T, Tjallingii WF, Thönnessen A, van Bel AJE. Molecular sabotage of plant defense by aphid saliva. Proceedings of the National Academy of Sciences, USA. 2007;104:10536–10541. doi: 10.1073/pnas.0703535104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will T, van Bel AJE. Physical and chemical interactions between aphids and plants. Journal of Experimental Botany. 2006;57:729–737. doi: 10.1093/jxb/erj089. [DOI] [PubMed] [Google Scholar]
- Wroblewski T, Piskurewicz U, Tomczak A, Ochoa O, Michelmore RW. Silencing of the major family of NBS-LRR-encoding genes in lettuce results in the loss of multiple resistance specificities. The Plant Journal. 2007;51:803–818. doi: 10.1111/j.1365-313X.2007.03182.x. [DOI] [PubMed] [Google Scholar]
- Wu Y, Huang Y. Molecular mapping of QTLs for resistance to the greenbug Schizaphis graminum (Rondani) in Sorghum bicolor (Moench) Theoretical and Applied Genetics. 2008;117:117–124. doi: 10.1007/s00122-008-0757-8. [DOI] [PubMed] [Google Scholar]
- Yang H, You A, Yang Z, Zhang F, He R, Zhu L, He G. High-resolution genetic mapping at the Bph15 locus for brown planthopper resistance in rice (Oryza sativa L.) Theoretical and Applied Genetics. 2004;110:182–191. doi: 10.1007/s00122-004-1844-0. [DOI] [PubMed] [Google Scholar]
- Zhang G, Gu C, Wang D. Molecular mapping of soybean aphid resistance genes in PI 567541B. Theoretical and Applied Genetics. 2009;118:473–482. doi: 10.1007/s00122-008-0914-0. [DOI] [PubMed] [Google Scholar]
- Zhang G, Gu C, Wang D. A novel locus for soybean aphid resistance. Theoretical and Applied Genetics. 2010;120:1183–1191. doi: 10.1007/s00122-009-1245-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.