Abstract
Rice OsERF922, encoding an APETELA2/ethylene response factor (AP2/ERF) type transcription factor, is rapidly and strongly induced by abscisic acid (ABA) and salt treatments, as well as by both virulent and avirulent pathovars of Magnaporthe oryzae, the causal agent of rice blast disease. OsERF922 is localized to the nucleus, binds specifically to the GCC box sequence, and acts as a transcriptional activator in plant cells. Knockdown of OsERF922 by means of RNAi enhanced resistance against M. oryzae. The elevated disease resistance of the RNAi plants was associated with increased expression of PR, PAL, and the other genes encoding phytoalexin biosynthetic enzymes and without M. oryzae infection. In contrast, OsERF922-overexpressing plants showed reduced expression of these defence-related genes and enhanced susceptibility to M. oryzae. In addition, the OsERF922-overexpressing lines exhibited decreased tolerance to salt stress with an increased Na+/K+ ratio in the shoots. The ABA levels were found increased in the overexpressing lines and decreased in the RNAi plants. Expression of the ABA biosynthesis-related genes, 9-cis-epoxycarotenoid dioxygenase (NCED) 3 and 4, was upregulated in the OsERF922-overexpressing plants, and NCED4 was downregulated in the RNAi lines. These results suggest that OsERF922 is integrated into the cross-talk between biotic and abiotic stress-signalling networks perhaps through modulation of the ABA levels.
Keywords: Abscisic acid, Magnaporthe oryzae, ERF, Oryza sativa, salt tolerance
Introduction
Plants have evolved efficient resistance mechanisms to protect themselves against pathogen infection through a combination of preformed physical and chemical barriers and induced defence systems. In plant cells, perception of conserved pathogen-associated molecular patterns (PAMPs) leads to activation of a battery of defence mechanisms, including the production of reactive oxygen species, the synthesis of antibiotic phytoalexins, deposition of callose, and expression of defence-related genes (Zhou and Chai, 2008; Clay et al., 2009). These inducible defences are regulated by complex signal transduction networks. Plant hormones have been demonstrated to play important roles in orchestrating defence responses. Salicylic acid (SA) is mostly associated with resistance to biotrophic phytopathogens, whereas jasmonic acid (JA) and ethylene (ET) are usually related to defence against necrotrophic pathogens and herbivorous insects (Rojo et al., 2003; De Vos et al., 2005; Glazebrook, 2005). However, interactions between the SA- and JA/ET-signalling pathways can be either mutually antagonistic or synergistic (Beckers and Spoel, 2006; Mur et al., 2006; Adie at al., 2007).
Recently, investigations have highlighted that abscisic acid (ABA), a global regulator of abiotic stresses, is also a key determinant in the induction of plant defence responses (Asselbergh et al., 2008a; Ton et al., 2009). Studies of mutants with ABA deficiency showed increased resistance to the necrotrophic fungus Botrytis cinerea (Audenaert et al., 2002), the bacterial pathogen Erwinia chrysanthemi (Asselbergh et al., 2008b) in tomato, and the oomycete Hyaloperonospora parasitica in Arabidopsis (Mohr and Cahill, 2003). In contrast, exogenous application of ABA enhances the susceptibility of various plants species to phytopathogens (Mohr and Cahill, 2003; Asselbergh et al., 2008b). Treatment of Arabidopsis with a virulent isolate of Pseudmonas syringae pv. tomato or ectopic expression the bacterial effector AvrPtoB induces ABA biosynthesis and modulates regulators of basal defence and callose deposition, suggesting that antagonistic interaction of ABA on SA-dependent defence responses underlies the impact of ABA as a susceptibility factor for this bacterium (de Torres-Zabala et al., 2007). Similarly, the activation-tagged Arabidopsis cds2-1D mutant with increased endogenous ABA levels accumulates JA and exhibits a complex antagonistic relationship with SA. These mutants have suppressed resistance to P. syringae and the biotrophic oomycete Hyaloperonospora arabidopsis, while the resistance to the necrotrophic fungus Alternaria brassicicola is enhanced (Fan et al., 2009). Thus, ABA has profound roles in defences mediated by cross talk with the JA- and SA-signalling pathways, depending on the specific pathosystems and infection stages (Mauch-Mani and Mauch, 2005; Asselbergh et al., 2008a; Ton et al., 2009).
Ethylene responsive factors (ERFs), composing a superfamily of transcription factors unique to plants, have been demonstrated to modulate multiple responses to abiotic and biotic stresses. For example, overexpression of OsDREB1A (Ito et al., 2006), AP37, and AP59 (Oh et al., 2009) in rice increases tolerance to drought and high salinity. Arabidopsis ERF1 (Solano et al., 1998), tomato Pti4 (Gu et al., 2002), and tobacco OPBP1 (Guo et al., 2004) bind to GCC-box-containing DNA fragments and activate expression of defence-related genes. Overexpression of tomato TSER1 in tobacco has been shown to confer resistance against Ralstonia solanacearum as a result of modulation in ABA and ET levels (Zhou et al., 2008). Similarly, transgenic tobacco plants expressing SodERF3, an ABA inducible ERF of sugarcane, display enhanced tolerance to drought and osmotic stress (Trujillo et al., 2008). Arabidopsis ORA59 is implicated the in JA- and ET-mediated defence signal pathways and has a positive impact on resistance against B. cinerea (Pré et al., 2008). Ectopic expression of TiERF1, a gene from a wild wheat relative Thinopyrum intermedium, enhances disease resistance in both wheat and tobacco through the ET-dependent pathway (Chen et al., 2008; Liang et al., 2008). Other AP2/ERF members are also reported to take part in regulation of metabolism, developmental processes, and/or defence responses in plants (Chuck et al., 1998; Fukao et al., 2006; Andriankaja et al., 2007; Oñate-Sánchez et al., 2007; Hattori et al., 2009).
Rice is not only one of the most important cereal crops in the world, but also a model monocot plant for biological research (Devos and Gale, 2000). Pathogens, salinity, and drought are among the major factors that affect the growth and productivity of the crop. Transcription factors play important roles in adaptation to these adverse environmental conditions by regulating expression of the specific responsive genes. Overexpression of transcription factor genes, such as OsWRKY45 and OsWRKY89, enhances disease resistance against Magnaporthe oryzae, the causal agent of rice blast disease, through the SA-mediated signal pathway (Shimono et al., 2007; Wang et al., 2007). On the other hand, application of exogenous ABA enhances rice susceptibility to M. oryzae, implying a negative role of ABA in defence against the pathogen (Koga et al., 2004; Bailey et al., 2009). Previous studies found that transgenic plants overexpressing OPBP1 enhanced disease resistance and salt tolerance in both tobacco and rice (Guo et al., 2004; Chen and Guo, 2008). These results promoted the study of the function of OsERF922, a close homologue of OPBP1. Expression of the OsERF922 gene was induced rapidly and strongly by inoculation with the rice blast fungus as well as by treatments with ABA and salt in rice seedlings. Knockdown of the expression of OsERF922 by means of RNA interference (RNAi) reduced the accumulation of ABA and enhanced resistance against M. oryzae.
Materials and methods
Construction of plasmids and plant transformation
The coding sequence of OsERF922 (Os01g54890) was amplified with primers E922F (5′-ACAGGATCCCCGCATGTCTCTCTCCTTG-3′) and E922R (5′-TTACTCGAGTATCACGCACTCACCGTCGCT-3′) from a rice IR72 cDNA library and ligated into a T-vector to give pUCmT-OsERF922. To construct plasmids for overexpression, the encoding region of OsERF922 was fused at its N-terminal with the sequence encoding the Flag-tag and put under the control of a maize ubiquitin promoter, generating pCo-Ubi:Flag-OsERF922. To knock down OsERF922 transcription, a fragment of OsERF922 gene (from 1 to 203 bp) was constructed in a form to generate a self-complementary hairpin RNA structure, which was driven by the cauliflower mosaic virus (CaMV35S) promoter (pCaMV35S:dsOsERF922).
For analysis of OsERF922 localization, the full-length coding sequence of OsERF922 was amplified with primers E922F and E922R and was fused in frame to the N-terminus of the green fluorescent protein (GFP) gene to generate the CaMV35S:OsERF922-GFP construct. All of the vectors constructed were verified by sequencing and introduced into rice calli of cultivar Zhonghua 17 (japonica, Zh17) by the Agrobacterium-mediated transformation method. More than 10 independent transgenic lines were obtained for each construct. The overexpressing plasmid pCo-Ubi:Flag-OsERF922 was also transformed into tobacco (Nicotiana tabacum cv. Xanthin nc) using the similar Agrobacterium-mediated method. The transgenic lines at the T2 generation were screened by planting seeds on 1/2 MS medium (containing 0.5% agar) supplemented with 40 mg l−1 hygromycin and the antibiotic resistant seedlings were used in the following experiments unless otherwise indicated.
Plant growth and treatments
Seeds of Oryza sativa Zh17 were surface sterilized, imbibed in water at 37 °C for 2 d before germination and seedlings were grown in vermiculite supplemented with mineral nutrient solution in a greenhouse around 28 °C under natural sunlight. Three-week-old rice seedlings were used for abiotic stress treatments, including spraying with 100 μM ABA (dissolved in 10 mM 4-morpholineethanosulphonic acid (MES) buffer, pH 6.0), or the submerging roots in the nutrient solution containing 150 mM NaCl.
For analysis of salt tolerance, the transgenic and wild-type (WT) plants were grown on 1/2 MS medium with 150 mM NaCl (containing 0.5% agar) under a 16/8 light/dark cycle at 28 °C for 3 weeks. These plants were then transferred to paddy pots for recovery at normal growth conditions. The plants were evaluated 8 d later.
Pathogen inoculation
Inoculation of rice blast fungi, M. oryzae, was conducted as described by Wang et al. (2007). Briefly, spores were harvested and suspended in 0.1% (w/v) gelatin to 105 conidia ml−1. Three-week-old rice plants were inoculated with the spore solution by spraying. The inoculated plants were maintained in the growth chamber at 24 °C in darkness for 24 h, then under growth conditions of a 16/8 light/dark cycle with 95% humidity. For analysis of gene expression, rice plants were inoculated with an avirulent M. oryzae strain P131 or a virulent strain P140. Leaves were sampled at designated times and used for RNA isolation.
To examine the capability of disease resistance, 3-week-old transgenic and the WT plants were inoculated with the virulent strain P140 as mentioned above. Some leaves from each line were sampled for analysis of gene expression 6 h post inoculation and disease severity was evaluated by a method of image analysis described by Fukuoka et al. (2009). The area of lesions formed 7 d after the infection was counted on the third leaves (15 cm in length) of 20 plants for each line.
Subcellular localization of OsERF922
Subcellular localization of OsERF922 in rice was examined by using the CaMV35S:OsERF922-GFP transgenic seedlings and the CaMV35S:GFP transgenic plants were used as the control. 4′-6-Diamidino-2-phenylindole (DAPI) staining was used as a nuclear marker. Fluorescence signals were detected using a confocal microscope (Eclipse TE2000, Nikon).
Western blot analysis
Total proteins were extracted from leaves of 7-d-old seedlings by grinding with small plastic pestles in the extraction buffer (100 mM HEPES, pH 7.5, 5 mM EDTA, 5 mM EGTA, 10 mM Na3VO4, 10 mM NaF, 50 mM β-glycerophosphate, 10 mM dithiothreitol, 1 mM phenylmethylsulphonyl fluoride, 5 μg ml−1 leupeptin, 5 μg ml−1 aprotinin, 5% glycerol). After centrifugation at 13,000 g for 15 min, the supernatants were transferred into clean tubes, quickly frozen in liquid nitrogen, and stored at –80 °C until use. Protein concentration was determined by using a protein assay kit (Bio-Rad) with bovine serum albumin as a standard. Protein samples (2 μg) were separated on a 10% SDS-PAGE and analysed by protein gel blot. Mouse anti-Flag antibody (Sigma-Aldrich) with 1:4000 dilution was used as the primary antibody and goat anti-mouse peroxidase-conjugated AffiniPure Goat Anti-Mouse IgG (H+L) with 1:8000 dilution was used as the secondary antibody. The membranes were visualized using a Super-Signal West Femto Trial Kit (Themor) following the manufacturer’s instructions.
Transcriptional activation assays
A reporter plasmid was constructed by replacing the full CaMV35S promoter in pBI121 using a fragment harbouring four repeats of the GCC-box and the CaMV35S minimal promoter (from –46 to +10), kindly provided by Prof. Huang (Zhang et al., 2004). Agrobacterium cells harbouring the reporter plasmid were harvested by centrifugation and resuspended in 10 mM MES, pH 5.6, 10 mM MgCl2, and 0.2 mM acetosyringone to an optical density (600 nm) of 0.7. After 4 h incubation at 28 °C, the bacteria were infiltrated into the leaves of 10-d-old pCo-Ubi:Flag-OsERF922 transgenic and WT tobacco plants using a needle-free syringe. The tissues around the infiltration sites were punched off into microcentrifuge tubes 48 h post treatment. GUS activity was determined as described using 4-methylumbelliferone as the standard (Jefferson, 1987).
Real-time quantitative RT-PCR
DNase-treated RNA (5 μg) was reverse transcribed in a total volume of 50 μl using reverse transcriptase (Takara). Real-time quantitative PCR (qPCR) was performed using 20 ng of total RNA in a 20 μl reaction volume using SYBR Green PCR MasterMix (Takara) on a StepOne Quantitative PCR system (Applied Biosystems) following the manufacturer’s protocols. Primer sequences used for the qPCR analysis are shown in Supplementary Table S1 (available at JXB online).
Determination of endogenous ABA
For estimation of endogenous ABA levels of rice seedling, samples were snap-frozen with liquid nitrogen and lyophilized. About 30 mg of dried sample was ground to powder with liquid nitrogen and extracted at 4 °C at 12 h with 1 ml of solution A (10% MeOH, 1% acetic acid) containing 50 ng of each D6-ABA (ICON Isotopes) as internal standards. The total extract was centrifuged (13,000 g, 10 min) and the pellet was re-extracted once with 1 ml of solution A. The combined supernatant was dried with N2 gas, and resultant residue was taken up in 50 μl solution A for LC-MS analysis (Fan et al., 2009).
Samples (10 μl) were then analysed by HPLC-electrospray ionization/MS-MS using an Agilent 1100 HPLC coupled to an Applied Biosystems Esquire 6000 (Bruker). Chromatographic separation was carried out on a Phenomenex Luna 3 μm C18 150 × 2.0 mm column at 35 °C. The solvent gradient used was 100% solvent A (H2O/CH3CN/CHOOH 94.9:5:0.1) to 100% solvent B (H2O/CH3CN/CHOOH 5:94.9:0.1) over 20 min. Solvent B was held at 100% for 5 min then the solvent returned to 100% A for 10 min equilibration prior to the next injection. The solvent flow rate was 200 μl min−1. Analysis of the compounds was based on appropriate multiple reaction monitoring of ion pairs for labelled and endogenous ABA.
Determination of Na+ and K+ contents
For analysis of Na+ and K+ contents, the transgenic and WT plants were grown on 1/2 MS medium with 100 mM NaCl (containing 0.5% agar) under a 16/8 light/dark cycle at 28 °C for 3 weeks. Leaves were sampled and used for analysis of Na+ and K+ contents with a flame photometer (2655-00 Digital Flame Analyzer, Cole- Parmer Instrument Company, Chicago, USA), as described by Song et al. (2005).
Results
Expression of OsERF922 was induced by Magnaporthe oryzae, ABA, and NaCl treatments
Previous studies demonstrated that tobacco OPBP1, an ERF transcription factor, is involved in disease resistance and salt tolerance (Guo et al., 2004). Multiple database searches and DNA sequence alignment analyses showed OsERF922 is one of the genes with high similarity to OPBP1 (Supplementary Fig. S1). OsERF922 has an ERF/AP2 domain, which contains the 14th Ala and 19th Asp residues, two key amino acids contributing to GCC-box binding in many ERFs (Sakuma et al., 2002). In addition to the ERF/AP2 domain, OsERF922 protein includes a relatively acidic and Ser-rich (56–81 aa) region, which might act as a transcription activation domain.
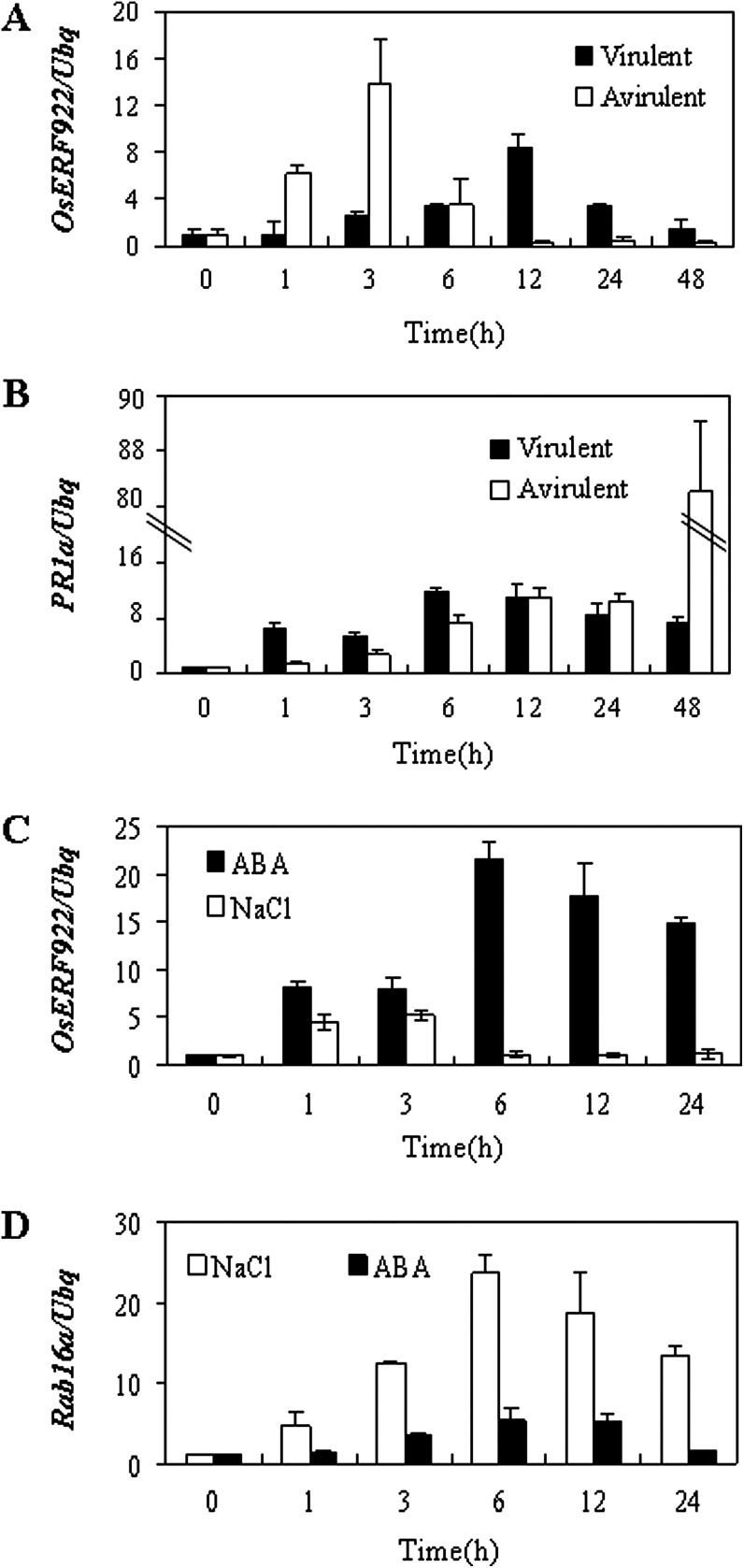
Many ERF members have been shown to take part in hormonal signal transduction in response to biotic and abiotic stresses (Zhang et al., 2004; Liang et al., 2008; Pré et al., 2008). To investigate whether OsERF922 is involved in rice defence responses, the current study used qPCR to analyse the expression patterns of OsERF922 during both the compatible and incompatible interactions between rice and rice blast fungi. Following infection with an avirulent M. oryzae strain, P131, transcription of the OsERF922 gene was induced as early as 1 h post inoculation (hpi), and its level peaked at 3 hpi with a 14-fold increase in mRNA accumulation compared with that at 0 h and decreased sharply to near basal level at 12 h (Fig. 1A). Expression of OsERF922 was also induced by inoculation with a virulent P140 strain, but the induction was slower and weaker than during the incompatible interaction (Fig. 1A). The results suggested that OsERF922 might be involved in early defence responses against pathogens. Further, this study examined the expression levels of OsERF922 in 3-week-old rice seedlings under abiotic stresses and hormone treatments. The mRNA accumulation of OsERF922 was increased remarkably after treatment with ABA (100 μM) for 1 h, reached to a maximal level after 6 h, and was maintained at a relatively high level throughout the rest of the experimental period (Fig. 1C). Similarly, expression of OsERF922 was induced about 5-fold by the NaCl treatment (Fig. 1C). Induction of PR1a (Fig. 1B) and Rab16a (Fig. 1D) revealed the efficacious pathogen and chemical treatments, respectively. However, no significant changes of OsERF922 expression were observed during methyl jasmonate or ET treatments (data not shown). The regulation of OsERF922 expression by various stresses suggests that it might play multiple roles in response to environmental stimuli.
Fig. 1.
Expression of the OsERF922 gene in response to biotic and abiotic stresses. RNA was extracted from leaves of 3-week-old seedlings treated with 100 μM abscisic acid (ABA), 100 mM NaCl, or inoculation with an avirulent Magnaporthe oryzae strain, P131, or a virulent strain, P140, for the indicated hours: (A) relative OsERF922 expression with infection; (B) relative PR1a expression with infection; (C) relative OsERF922 expression with ABA and salt treatments; (D) relative Rab16a expression with ABA and salt treatments. Transcription levels were quantified by quantitative reverse-transcription PCR and the data were normalized using the rice ubiquitin gene (Ubq) as standard. Values are mean ± SD of three separate analyses for each RNA template. The experiments were performed in duplicate using different RNA samples for the template; similar results were obtained from each duplicate.
OsERF922 protein interacts specifically with the GCC box in planta
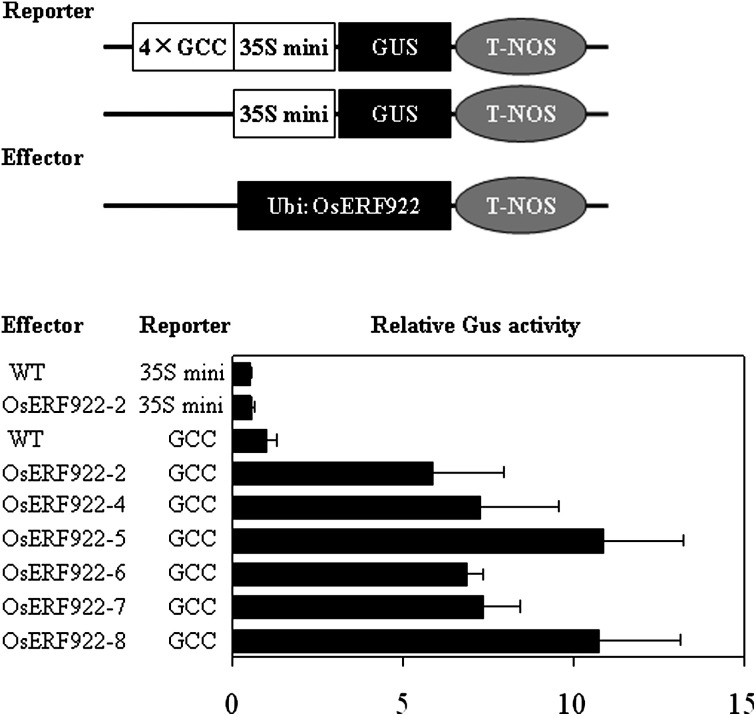
To examine the interaction of OsERF922 with the GCC box in vivo, a transient expression experiment was performed. A DNA fragment harbouring four GCC-element repeats was fused to the minimal CaMV35S promoter (–46 bp) and the GUS gene and used as a reporter plasmid (Zhang et al., 2004) (Fig. 2A). Tobacco plants overexpressing OsERF922 under the control of a maize ubiquitin promoter were used as effectors. The 4 × GCC-GUS reporter construct was introduced into the leaves of the homozygous transgenics of T3 progenies by means of Agrobacterium-mediated infiltration method (Yang et al., 2000). The GUS activity was increased about 5–10-fold in the six transgenic lines, when compared with the WT control, demonstrating that OsERF922 could bind to the GCC-box in planta and act as a transcriptional activator (Fig. 2B).
Fig. 2.
Transcriptional activation of OsERF922: the constructs of the reporter and effector genes (upper panel) and induction of GUS activities (lower panel). The GUS-reporter gene is driven by a synthetic promoter consisting of the –46 minimal CaMV 35S promoter and four copies of the GCC box, and the construct without the GCC box (35S mini) was used as the control. The effector gene was driven by a maize ubiquitin promoter and the resultant construct was transformed into tobacco. Agrobacterium cells harbouring the reporter plasmid were infiltrated into the leaves of transgenic (OsERF922) and wild-type (WT) tobacco plants using a needle-free syringe. The ratios of GUS activities were calculated from the GUS activities determined in the leaves harvested 48 h after the infiltration over those determined in the WT control. Values are means and errors calculated from triplicate samples and three independent experiments.
OsERF922 is a nuclear targeted protein
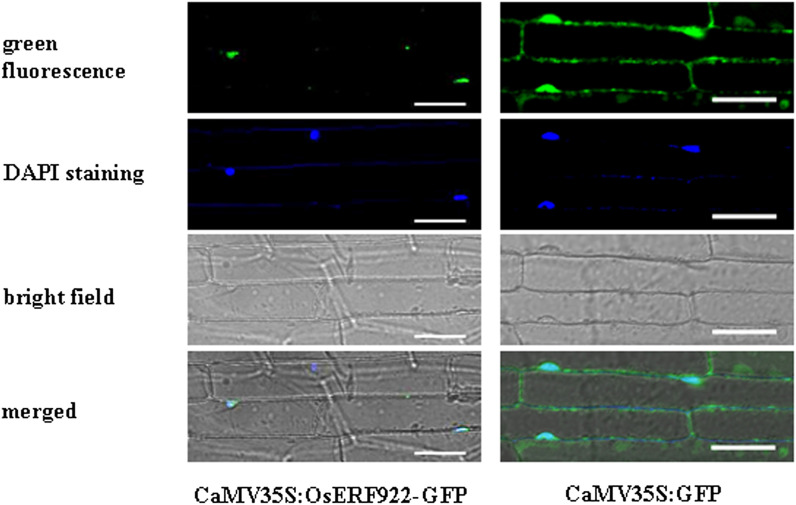
The deduced OsERF922 protein has no obvious nuclear localization signal, as predicted by PSORT analysis (Nakai and Kanehisa, 1992). To determine the subcellular localization of OsERF922 protein in rice, the construct of a CaMV35S-controlled OsERF922-GFP chimeric gene (CaMV35S:OsERF922-GFP) and the control CaMV35S:GFP vector were stably transformed into rice. Inner epidermal cells of the transgenic T1 progenies were observed by microscopy technique for fluorescence signal. Localization of the OsERF922-GFP fusion protein was visualized mainly in the nuclei (Fig. 3). These results suggest that OsERF922 could target to the nucleus.
Fig. 3.
Subcellular localization of OsERF922. The OsERF922-GFP chimeric gene was put under the control of the CaMV35S promoter and transformed into rice. The OsERF922:GFP fusion was localized exclusively to the nucleus of rice inner epidermal cells of the sheaths. GFP alone was detected in both the nucleus and the cytoplasm. Bright-field images of the inner epidermal cells are shown in the bottom half.
OsERF922 negatively regulates disease resistance in rice
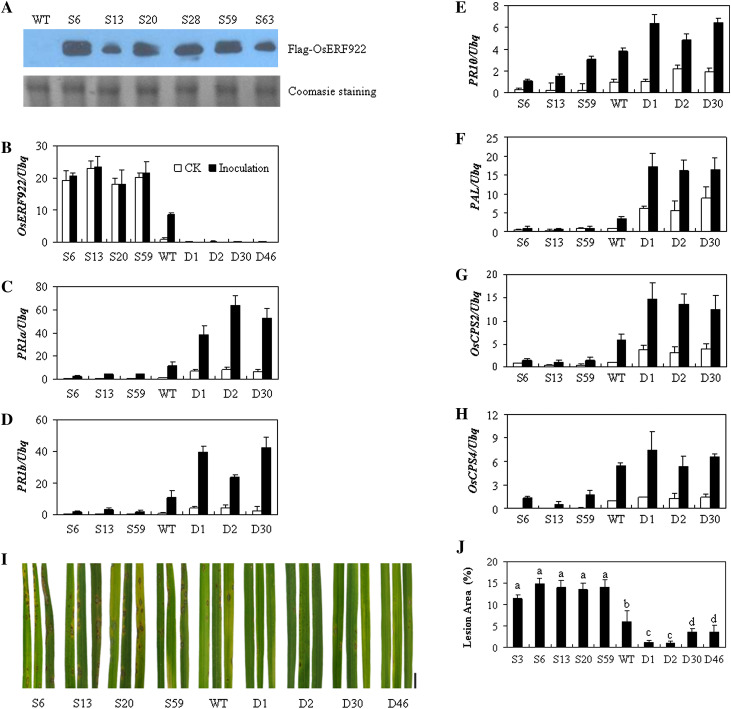
To reveal the biological function of OsERF922 in plants, full-length cDNA of OsERF922 was fused downstream of the maize ubiquitin promoter, generating a Ubi:OsERF922 construct with a Flag-tag at the N-terminus for rice transformation. For OsERF922 knockdown, a hairpin structure (CaMV35S:dsOsERF922) was used for RNA interference. More than 15 independent transgenic lines were obtained for each construct, none of which showed a significant difference in plant morphological features (such as root length, plant height, and numbers of spikelets) compared with the WT plants under normal growth conditions (data not shown). To determine the level of Flag-OsERF922 in the transgenic lines, total proteins were isolated from six randomly selected transgenic plants and analysed by Western blotting using the anti-Flag antibody. The results showed high levels of the fusion proteins in all lines tested, although the levels differ between lines (Fig. 4A).
Fig. 4.
Overexpression and knockdown of OsERF922 alter resistance to Magnaporthe oryzae and the expression of defence-related genes. (A–H) Analysis of OsERF922 expression in OsERF922 overexpression (S6, S13, S20, S28, S59, and S63) and knockdown (D1, D2, D30, D46) rice lines by Western blot (A) and quantitative reverse-transcription PCR (B, OsERF922; C, PR1a; D, PR1b; E, PR10; F, PAL; G, OsCPS2; H, OsCPS4), as described in Fig. 1. Total proteins were extracted from leaves of 7-d-old seedlings of the transgenic and wild-type plants, separated on a 10% SDS-PAGE, and detected by the mouse anti-Flag antibody. Three-week-old transgenic and the wild-type plants were inoculated with the virulent M. oryzae P140 isolate and several leaves from each line were sampled for analysis of gene expression 6 h post inoculation. Data are representative of two independent biological replicates. (I) Transgenic plants of OsERF922 were tested for resistance to M. oryzae. Leaves of 3-week-old rice plants were inoculated with conidial suspensions of M. oryzae (105 conidia ml−1) isolate P140. Leaves were detached from inoculated plants 7 d post the infection and photographed. Experiments were repeated three times with similar results. Bar, 1 cm. (J) Histograms showing the area of lesions formed on the third leaves (15 cm in length) of 20 plants for each line. Values marked with different letters are significantly different (P < 0.01, Fieldman Rank Sums test).
To examine whether the OsERF922 gene is involved in disease resistance, the transgenic plants were inoculated with the virulent fungal pathogen M. oryzae P140. As illustrated in Fig. 4I, lesions formed by pathogen infection were remarkably increased in the OsERF922-overexpressing plants of the T2 progeny compared to the WT plants, whereas the OsERF922 RNAi plants displayed a significant decrease of lesion formation. The differences were further evaluated by quantification of the lesion areas (Fig. 4J), which again indicates that knockdown of OsERF922 expression enhanced disease resistance against the rice blast pathogen.
OsERF922 downregulates the expression of defence-related genes
To examine whether the altered disease resistance phenotypes of the overexpressing lines and the RNAi lines are associated with altered defence gene expression, this study analysed transcript levels of several defence-related genes with or without pathogen inoculation using the qPCR method. The level of the OsERF922 transcript was increased more than 18-fold in the overexpressing plants and decreased 4-fold or more in the RNAi plants compared with the WT plants (Fig. 4B). As expected, expression of OsERF922 was induced in WT plants 6 h post M. oryzae inoculation, whereas its level did not change significantly in the OsERF922-overexpressing linesm which is most likely due to the high level of constitutive expression of the transgene. The transcript level remained low in the knockdown plants following pathogen infection, suggesting efficient suppression of OsERF922 in the RNAi lines. Among the defence-related genes, expression of the PR genes, including PR1a, PR1b, and PR10 were reduced in the overexpressing plants but enhanced in the RNAi plants in comparison with these in WT plants (Fig. 4C–E). Increases in expression of these genes were observed in all of the plants after inoculation with the rice blast pathogen. However, the transcript levels of the PR1a, PR1b, and PR10 genes were increased much more in the knockdown lines than those in the control plants. In contrast, the induction of these genes by the pathogen infection was compromised in the OsERF922-overexpressing lines.
Phenylalanine ammonia lyase (PAL) catalyses the first committed step of phenolic biosynthesis in plants, including SA. Analysis of PAL expression revealed slight suppression in the overexpressing lines and an increase of 5-fold or more in the RNAi lines compared with the control plants (Fig. 4F). Stronger induction of PAL expression was observed in the RNAi plants during blast fungal infection, whereas its transcript level was not increased significantly in the overexpressing plants. Diterpene cyclases, ent-copalyl diphosphate (ent-CDP) synthase (OsCPS2/OsCyc2), and syn-CDP synthase (OsCPS4/OsCyc1) catalyse the conversion of geranylgeranyl diphosphate (GGDP) to ent-CDP and syn-CDP, respectively, and ent-CDP leads to synthesis of phytocassanes A to E and oryzalexins A to F and syn-CDP leads to synthesis momilactones A and B and oryzalexin S (Kanno et al., 2006). These antimicrobial compounds, known as phytoalexins or allelochemicals, are synthesized in the rice leaf in response to infection with the blast pathogen or exposure to UV irradiation (VanEtten et al., 1994; Kato-Noguchi and Ino, 2003). Similarly to the expression pattern of the PAL gene, accumulation of OsCPS2 mRNA was increased in the RNAi lines and enhanced with the inoculation with blast fungi (Fig. 4G). However, expression of the OsCPS4 gene was suppressed more than 20-fold in the overexpressing progenies and the level was much lower than the control plants even upon the infection with M. oryzae (Fig. 4H). These results suggest that OsERF922 is involved in disease resistance as a negative regulator by suppressing defence-related genes.
OsERF922 negatively regulates salt tolerance in rice
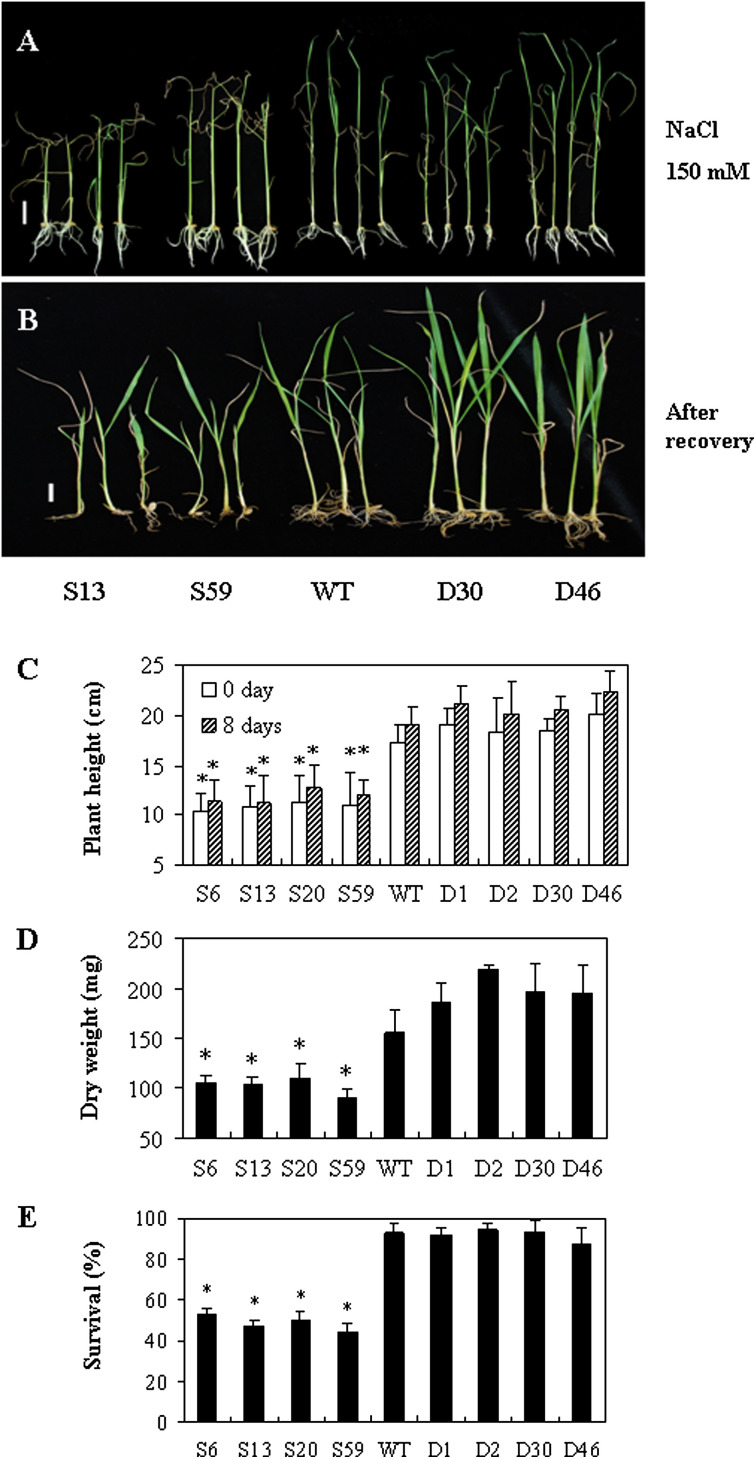
Induction of OsERF922 expression by NaCl and ABA treatments prompted this study to test the tolerance of the transgenic rice plants to NaCl. The transgenic seeds of T2 progeny and WT plants were germinated on MS medium and the seedlings were then transplanted on MS medium containing 150 mM NaCl. After growing on the NaCl medium for 3 weeks, leaves of the OsERF922-overexpressing lines became scorched particularly at the top of the leaves (Fig. 5A). The symptom was less severe in the RNAi and WT plants. After these plants were transferred to paddy pots for recovery (Fig. 5B), nearly half of the plants overexpressing OsERF922 died during the 8-d recovery period (Fig. 5E). Those plants that survived also showed significant retardation of growth and less biomass than that of the WT plants (Fig. 5C and D).
Fig. 5.
Overexpression of OsERF922 enhances salt sensitivity. (A) Transgenic and wild-type plants grown on 1/2 MS medium containing 150 mM NaCl under a 16/8 light/dark cycle at 28 °C for 3 weeks. (B) Plants after 8 d recovery in paddy pots at normal growth conditions. (C–E) Histograms showing: plant heights on the MS medium supplemented with 150 mM NaCl and after 8 d recovery (C), dry weight (D), and the percentage of survived plants (E) post the recovery. Values are means ± SE of three replicates with 10 seedlings per replicate. Asterisks indicate significant differences between the control and the overexpressing lines (P < 0.05, Fieldman Rank Sums test). Bars, 1 cm.
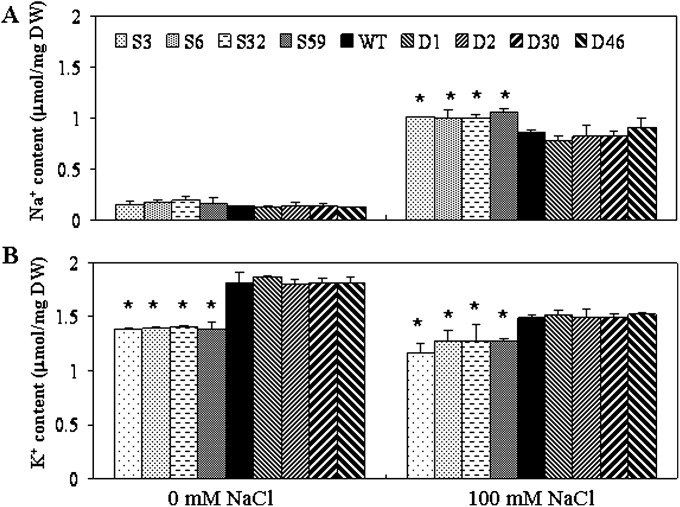
Salt tolerance in plants is the consequence of several physiological processes, such Na+ uptake, Na+/K+ balance, and ion compartmentalization. Damage of leaves caused by salinity is attributed to accumulation of Na+ in the shoot by transport of Na+ from the roots experiencing high external salt concentration (Lin et al., 2004). Thus, the current study measured the accumulation of Na+ and K+ in the rice shoots. The levels of K+ were lower in OsERF922-overexpressing lines than those in the WT and RNAi plants on 1/2 MS media, whereas the difference of Na+ accumulation was not obvious among the plants tested (Fig. 6A). During NaCl treatment, an increase of Na+ content was observed, with significantly higher accumulation in OsERF922-overexpressing lines than the WT and RNAi plants. On the other hand, the content of K+ was suppressed during the salinity treatment (Fig. 6B), in agreement with competition between Na+ and K+ uptake in the shoots (Lin et al., 2004). However, the level of K+ in the overexpressing plants was even lower than those in the WT and OsERF922 RNAi lines (Fig. 6B). The results indicate that the enhanced salt sensitivity is possibly the consequence of inhibition in K+ accumulation in OsERF922-overexpressing plants.
Fig. 6.
Na+ (A) and K+ (B) contents in the shoots of WT and the transgenic plants under non-salt-stress and 100 mM NaCl treatment for 21 d (n = 3). Asterisks indicate significant differences between the control and the overexpressing lines (P < 0.05, Fieldman Rank Sums test).
OsERF922 regulates ABA accumulation
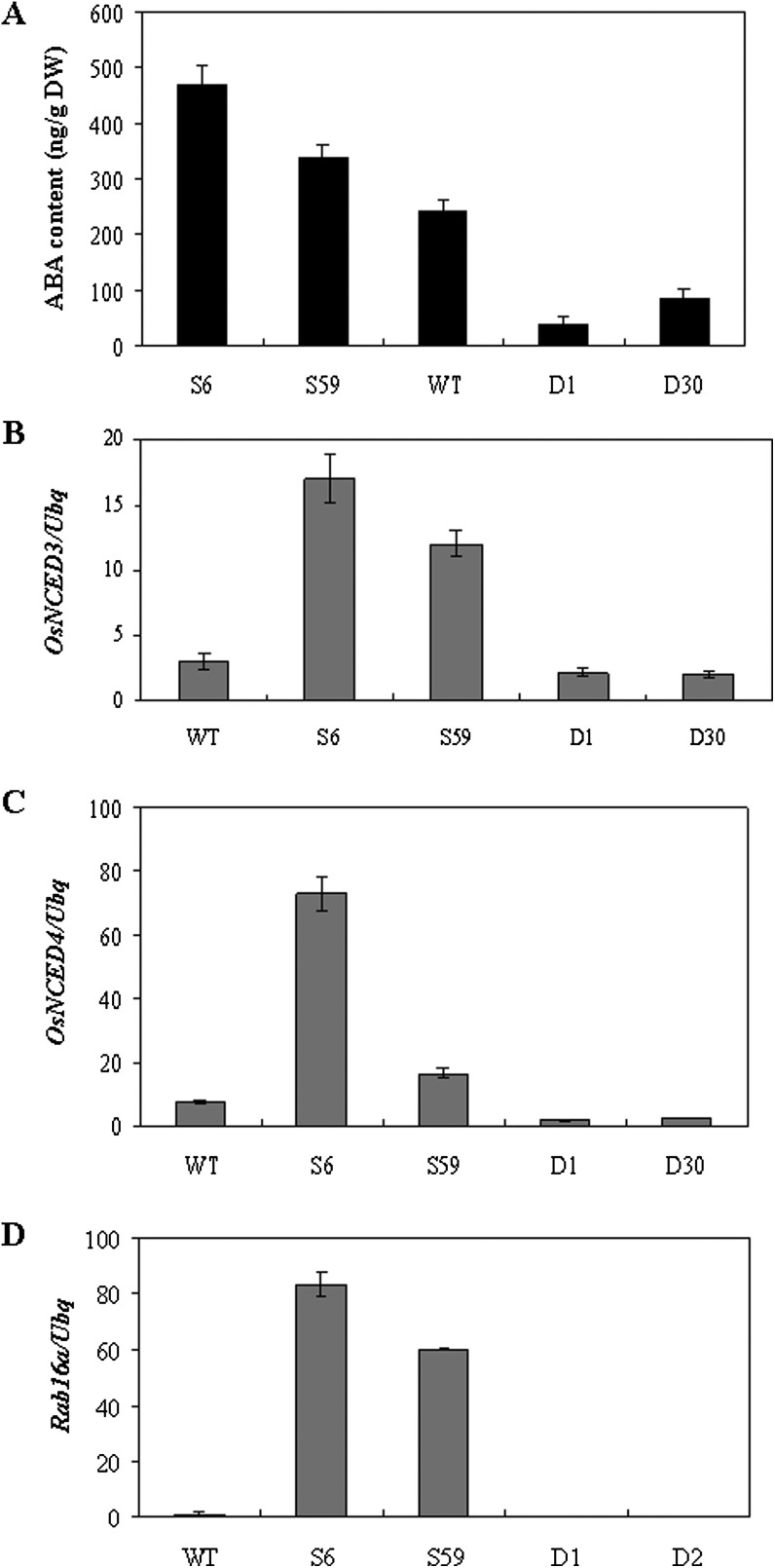
In addition to its well-established role in response to abiotic stress, ABA is also involved in defence responses. Therefore, this study measured ABA levels and expression of ABA biosynthesis-related genes. The amount of ABA was about 2-fold more in OsERF922-overexpressing line S6 and 1.4-fold in line S59 than that in the WT plant (Fig. 7A). A decrease in ABA accumulation was observed in the OsERF922 RNAi lines: in the D1 line, the ABA level in the D1 and D30 lines was about one-sixth and one-third of that in WT, respectively (Fig. 7A). 9-cis-Epoxycarotenoid dioxygenase (NCED) catalyses the conversion of 9-cis-epoxycartenoids to xanthoxin, a rate-limiting step in ABA biosynthesis (Schwartz et al., 2003). The expression of OsNCED genes was examined in the transgenic and control plants by qPCR. OsNCED3 and 4 genes were both upregulated in the OsERF922-overexpressing plants and OsNCED4 was suppressed in the RNAi plants, in comparison with the WT plants (Fig. 7B and C). Further, this study examined the expression of Rab16a, an ABA-responsive marker gene (Yamaguchi-Shinozaki et al., 1990). A much higher level of Rab16a transcripts was observed in the OsERF922-overexpressing plants as compared to the WT, whereas the expression of Rab16a was decreased in the RNAi lines (Fig. 7D). The increase of ABA accumulation was consistent with the enhanced expression of the ABA-biosynthesis and -responsive genes in the overexpressing plants.
Fig. 7.
OsERF922 alters the expression of ABA biosynthesis genes as well as ABA accumulation. (A) Endogenous levels of ABA in 3-week-old leaves were quantified by LC-MS/MS. Values are mean and SE of three independent samples. (B–D) RNA was isolated from 3-week-old leaves of the OsERF922 transgenic and the wild-type plants for the OsNCED3 (B), OsNCED4 (C) and Rab16a (D) genes and quantified as described in Fig. 1. Experiments were repeated in triplicate and similar results were obtained.
Discussion
This study reports characterization of OsERF922, which was classified into phylogenetic group IX of AP2/ERF superfamily by Nakano et al. (2006). Genes in this group have been shown to play crucial roles in biotic and abiotic stress responses. For example, overexpression of AtERF1, tomato Pti4/5, or tobacco OPBP1 enhances disease resistance (Berrocal-Lobo et al., 2002; Gu et al., 2002; Guo et al., 2004). Further, ectopic expression of OsBIERF3, a member of rice ERFs in group IX, increases disease resistance against infection by tomato mosaic virus and the bacterial wild fire pathogen, P. syringae pv. tabaci, as well as tolerance to salt stress in tobacco (Cao et al., 2006). The data indicate that these ERFs are positive regulators of defence- and/or stress-related transcription. OsERF922 is induced rapidly and strongly in response to salt, ABA, and M. oryzae pathogen (Fig. 1). The OsERF922 RNAi plants showed increased resistance against M. oryzea (Fig. 4). OsERF922 also regulates Na+/K+ homeostasis, as revealed from the increased Na+ and decreased K+ accumulation in the OsERF922-overexpressing plants. DNA binding and reporter gene activation assays, suggest that OsERF922 is a transcriptional activator with a high affinity for DNA molecules containing the GCC-box sequences (Fig. 2). In combination with the result of nuclear localization (Fig. 3), the data suggest that OsERF922 directly regulates gene expression and plays a significant role in mediating plant responses to environmental stimuli.
Through analysis of the responses to pathogen infection, OsERF922-overexpressing lines were more susceptible to rice blast fungus than that of the knockdown plants (Fig. 3), indicating OsERF922 function as a negative regulator of plant disease resistance. The question is why plants promote expression of a negative regulator in response to pathogen attack that would cause them more susceptible to the invading pathogen. One possibility is that OsEFR922 might be activated by pathogens to suppress host defence as a pathogen’s counter-defence mechanism. In the case of bacterial pathogens, a number of studies have demonstrated that plant-pathogenic effectors modulate diverse host processes to enhance virulence. AvrPtoB of P. syringae effector modifies the host ABA-signalling pathway to suppress defence responses (de Torres-Zabala et al., 2007). AvrPto functions as a kinase inhibitor, which directly interacts with receptor kinases, such as Arabidopsis and tomato FLS2, to block PAMP-triggered immunity and leads to host susceptibility (Xiang et al., 2007). Transcription activator-like effectors of Xanthomonas oryzae pv. oryzae, a causal agent of bacterial blight of rice, strongly induced rice Os8N3, OsTFX1, and OsTFIIAγ1 genes to compromise the defence mechanism and promote a state of disease susceptibility (Yang et al., 2006; Sugio et al., 2007). Further, Arabidopsis WRKY48 has also been characterized as a transcriptional activator that represses plant basal resistance (Xing et al., 2008). Induction of WRKY48 expression is not enhanced during infection by the PstDC3000hrcC mutant strain defective in the type III secretion system, suggesting that certain virulent factors from the pathogen are required to promote the expression of the negative regulator. Further experiments will be necessary to determine whether any effectors or secreted proteins from rice blast fungal pathogen are involved in expression of negative regulatory genes such as OsERF922 to attenuate host defence responses.
A large number of genes related to plant defence are induced in compatible and incompatible interactions, although the kinetics and amplitude of response are attenuated during compatible interactions (Tao et al., 2003; Vergne et al., 2007). OsERF922 expression also followed predicted patterns, showing induction by avirulent M. oryzae P131 strain was as early as 1 hpi and reached a peak expression level in 3 hpi, but it was elevated at 3 hpi and reached to the peak at 12 hpi during the compatible interaction (Fig. 1A). In the period of 3 hpi, spores attached on the surface of rice leaf are in the stages of germination and appressorium development (Talbot, 2003; Ribot et al., 2008). The results suggest that rice quickly recognizes the invading pathogen through spores, germ tubes, contaminated hypha, and/or other unknown secreted factors and elicits innate defence responses or enhances host susceptibility. Likely, OsERF922 is an important component of the signalling pathway. MAL10, a barley resistance protein against the powdery mildew fungus, has been shown to directly associate with HvWRKY1/2 transcription factors upon recognition of the fungal avirulence A10 effector (Shen et al., 2007). Importantly, the identified WRKY proteins are repressors of PAMP-activated basal defence, revealing MAL10 functions as a de-repressor of the immune response. Arabidopsis AtERF104, also an ERF member of group IX, is characterized as a substrate of MAPK 6 (Bethke et al., 2009), which is a positive regulator of the flg22-mediated basal defence. Interestingly, both the overexpressor and the mutant of erf104 showed enhanced susceptibility to B. cinerea and a non-adapted bacterial pathogen, P. syringae pv. phaseolicola. It has been demonstrated that ERF104 must be maintained at an optimal level and any alterations of this crucial threshold can tip the ‘signalling balance’ (Bethke et al., 2009). OsMPK5, a MAPK protein in rice, is inducible by infection of rice blast fungi as well as ABA and abiotic stresses and its role is well documented as negatively modulating broad-spectrum disease resistance and positively regulating abiotic tolerance (Xiong and Yang, 2003). It will be interesting to reveal whether OsMPK5 and OsERF922 are involved in the same pathways during biotic and abiotic stress responses.
This study indicates that OsERF922 functions as a transcriptional activator in plants (Fig. 2), although it suppresses expression of defence genes and resistance against rice blast fungal pathogen (Fig. 4). It is plausible that OsERF922 first activates some unknown negative regulators, instead of suppressing defence genes directly. In turn, the negative regulators repress defence-related genes and compromise disease resistance. OsERF922 could also play a role in preventing overactivation of defence reactions that may have an overall fitness cost. Nevertheless, identification of the available targets of OsERF922 will provide useful tools to dissect the poorly understood mechanism of transactivators acting negatively on plant immune pathways.
The phytohormone ABA is a key regulator of responses to diverse abiotic stresses as well as in plant growth and development. A high ABA content is often associated with stresses such as drought, cold, and salt. The transgenic lines of overexpressing OsERF922 showed more sensitivity to salty conditions in comparison with the WT plants. An explanation is probably due to the disturbance of Na+/K+ homeostasis (Fig. 6). Similarly, the transgenic plants of OsAP2-39, another rice ERF gene are more susceptible to drought comparing with the WT plants, although the transgenic plants contain higher amount of ABA (Yaish et al., 2010). Increasing reports have shown that ABA may also participate directly or indirectly in responses to a range of pathogens. Using ABA-deficient mutants, inhibition of ABA biosynthesis or exogenous application of ABA, several studies have shown a positive correlation between endogenous ABA levels and resistance to pathogens (Adie et al., 2007; Fan et al., 2009), while others have documented a negative role for this hormone in plant defence reactions (Audenaert et al., 2002; Mohr and Cahill, 2003; de Torres-Zabala et al., 2007, 2009; Asselbergh et al., 2008b; Jensen et al., 2008). In rice plants, ABA has been shown as an activator of resistance against rice brown spot-causing Cochliobolus miyabeanus (De Vleesschauwer et al., 2010). However, ABA appears to play a negative role in response to rice blast fungus, since leaves treated with ABA become slightly more susceptible to M. oryzae (Koga et al., 2004). The current data also indicate that ABA might enhance host susceptibility. Expression of ABA biosynthetic genes and accumulation of ABA were decreased in the OsERF922 RNAi plants, in line with enhanced resistance against M. oryzae, whereas overexpressing plants were more susceptible to the pathogen (Fig. 4). Recently, the blast fungus M. oryzae was found to produce and secrete ABA, which antagonizes the SA-signalling pathway to attenuate host defence responses (Jiang et al., 2010). These results imply that fungus- and host-derived ABA may play a critical role in the interactions between rice plant and the hemi-biotrophic fungus.
PAL is a key enzyme catalysing the first committed step of the phenylpropanoid biosynthetic pathway, leading to production of hormone SA and antimicrobial phytoalexins. ABA suppression of PAL activity is a possible mechanistic explanation for its negative impact on disease resistance. Application of ABA reduced the accumulation of SA and PAL expression in Arabidopsis, consistent with enhanced susceptibility to an avirulent stain of P. syringae pv. tomato (Mohr and Cahill, 2007) and the production of the phytoalexin glyceollin is reduced in an ABA-dependent manner in soybean upon infection with Phytophthora sojae (Mohr and Cahill, 2003). In agreement with this, the current study also found that PAL expression was elevated and further increased by inoculation with M. oryzea in OsERF922 RNAi plants coincident with the low level of ABA accumulation. In contrast, the pathogen-inducible PAL expression was suppressed in the overexpression lines (Fig. 4F). The large majority of phytoalexins isolated in rice are diterpenoids with the exception of the flavanone sakuranetin (Kodama et al., 1992). The diterpenoids have been classified into phytocassanes A to E, oryzalexins A to F, momilactones A and B, and oryzalexin S, according to their structures of diterpene hydrocarbon precursors (Kanno et al., 2006). Expression of OsCPS2 and OsCPS4, two cyclase genes leading to the synthesis of the diterpenoid phytoalexins, was suppressed significantly at the basal levels as well as under the conditions of pathogen inoculation (Figs. 4G and H). In mutants deficient in ABA biosynthesis, the synthesis of capsidiol, a sesquiterpene phytoalexin in tobacco is induced higher than WT plants when infected with B. cinerea or elicited with elicitors, while the application of exogenous ABA reversed the elevated effect (Mialoundama et al., 2009). Collectively, the data indicate that ABA is implicated in the biosynthesis of phytoalexins in challenged plants, which affects host susceptibility.
Recently, Bailey and associates (2009) have found that exogenous application of ABA drastically reduces endogenous ET levels in rice and enhances susceptibility to M. oryzea. Antisense suppression of OsEIN2, a key component of ET signal transduction, confers ET insensitivity and ABA supersensitivity and enhances rice blast susceptibility, while the resistance against C. miyabeanus is increased (De Vleesschauwer et al., 2008, 2010). Further, the OsEIN2 antisense plants are more tolerant to drought, salinity, and cold. Interestingly, rice ETHYLENE INSESITIVE3-LIKE 1, an orthologue of Arabidopsis EIN3 has been found to interact with OsMPK5, which increases ET level and resistance against M. oryzae and decreases the ABA level and tolerance to abiotic stresses in the RNAi-mediated OsMPK5 suppression plants (Xiong and Yang, 2003; Bailey et al., 2009). Based on the model of ET signal transduction in Arabidopsis (Yoo et al., 2009), EIN3 and EIN3-LIKE (EIL) proteins transduce the signal from the upstream component EIN2 to downstream components including the ERF transcription factors. These results indicate that ABA antagonistically interacts with ET-signalling networks and consequently influences disease resistance as well as abiotic stress tolerance, and also implies that OsERF922 is possibly such a candidate downstream of OsEIL1 signal transduction. Overlap between abiotic and biotic stress signal transduction pathways is well known, including some similar mechanisms, such as the generation of reactive oxygen species and nitric oxide (Pastori and Foyer 2002), and the involvement of MAPK-signalling cascades (Xiong and Yang, 2003; Fujita et al., 2006) and various transcription factors. Increased expression of transcription factors, such as a barley ERF and rice OsMyb4, lead to enhanced resistance to pathogen attack and tolerance to abiotic stresses (Vannini et al., 2006; Jung et al., 2007). This study’s data suggest that the negative regulation of OsERF922 on basal resistance and salt tolerance is mediated through the ABA-signalling pathway, suggesting that OsERF922 is integrated into the complicated cross-talk between biotic and abiotic stress-signalling networks.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Table S1 . Primers used for qRT-PCR.
Supplementary Fig. S1 . OsERF922 sequences and phylogenetic analysis.
Acknowledgments
The authors thank Prof Huang for providing the GUS reporter plasmid and Dr Withers for edited the manuscript. This work was supported by the State Basic Research and Development Plan (2011CB100702), the Natural Science Foundation of China (30900927 and 30871611), and the Program for Changjiang Scholars and Innovative Research Team in University (IRT042).
References
- Adie BAT, Perez-Perez J, Perez-Perez MM, Godoy M, Sanchez-Serrano JJ, Schmelz EA, Solano R. ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis . The Plant Cell. 2007;19:1665–1681. doi: 10.1105/tpc.106.048041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriankaja A, Boisson-Dernier A, Frances L, Sauviac L, Jauneau A, Barker DG, de Carvalho-Niebel F. AP2-ERF transcription factors mediate Nod factor dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. The Plant Cell. 2007;19:2866–2885. doi: 10.1105/tpc.107.052944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselbergh B, Achuo EA, Höfte M, Van Gijsegem F. Abscisic acid-deficiency leads to rapid activation of tomato defense responses upon infection with Erwinia chrysanthemi . Mol Plant Pathol. 2008b;9:11–24. doi: 10.1111/j.1364-3703.2007.00437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselbergh B, De Vleesschauwer D, Höfte M. Global switches and fine-tuning: ABA modulates plant pathogen defense. Molecular Plant–Microbe Interactions. 2008a;21:709–719. doi: 10.1094/MPMI-21-6-0709. [DOI] [PubMed] [Google Scholar]
- Audenaert K, De Meyer GB, Höfte M. Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiology. 2002;128:491–501. doi: 10.1104/pp.010605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TA, Zhou X, Chen J, Yang Y. Role of ethylene, abscisic acid and MAP kinase pathways in rice blast resistance. In: GL Wang, B Valent., editors. Advances in Genetics, Genomics and Control of Rice Blast Disease. Dordrecht, The Netherlands: Springer; 2009. pp. 185–190. [Google Scholar]
- Beckers GJM, Spoel SH. Fine-tuning plant defence signalling: salicylate versus jasmonate. Plant Biology. 2006;8:1–10. doi: 10.1055/s-2005-872705. [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A, Solano R. Constitutive expression of ETHYLENE RESPONSE FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. The Plant Journal. 2002;29:23–32. doi: 10.1046/j.1365-313x.2002.01191.x. [DOI] [PubMed] [Google Scholar]
- Bethke G, Unthan T, Uhrig JF, Poschl Y, Gust AA, Scheel D, Lee J. Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proceedings of the National Academy of Sciences, USA. 2009;106:8067–8072. doi: 10.1073/pnas.0810206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Wu Y, Zheng Z, Song F. Overexpression of the rice EREBP-like gene OsBIERF3 enhances disease resistance and salt tolerance in transgenic tobacco. Physiological and Molecular Plant Pathology. 2006;67:202–211. [Google Scholar]
- Chen L, Zhang ZY, Liang HX, Liu HX, Du LP, Xu HJ, Xin ZY. Overexpression of TiERF1enhances resistance to sharp eyespot in transgenic wheat. Journal of Experimental Botany. 2008;59:4195–4204. doi: 10.1093/jxb/ern259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Guo Z. Tobacco OPBP1 enhances salt tolerance and disease resistance of transgenic rice. International Journal of Molecular Sciences. 2008;9:2601–2613. doi: 10.3390/ijms9122601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Meeley RB, Hake S. The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1 . Genes and Development. 1998;12:1145–1154. doi: 10.1101/gad.12.8.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science. 2009;323:95–101. doi: 10.1126/science.1164627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres-Zabala M, Bennett MH, Truman WH, Grant MR. Antagonism between salicylic and abscisic acid reflects early host-pathogen conflict and moulds plant defence responses. The Plant Journal. 2009;59:375–386. doi: 10.1111/j.1365-313X.2009.03875.x. [DOI] [PubMed] [Google Scholar]
- de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, Rodriguez Egea P, Bögre L, Grant M. Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. The EMBO J. 2007;26:1434–1443. doi: 10.1038/sj.emboj.7601575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleesschauwer D, Djavaheri M, Bakker PAHM, Höfte M. Pseudomonas fluorescens WCS374r-induced systemic resistance in rice against Magnaporthe oryzae is based on pseudobactin-mediated priming for a salicylic acid-repressible multifaceted defense response. Plant Physiology. 2008;148:1996–2012. doi: 10.1104/pp.108.127878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleesschauwer D, Yang Y, Cruz CV, Höfte M. Abscisic acid-induced resistance against the brown spot pathogen Cochliobolus miyabeanus in rice involves MAP kinase-mediated repression of ethylene signaling. Plant Physiology. 2010;152:2036–2052. doi: 10.1104/pp.109.152702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos M, Van Oosten VR, Van Poecke RM, et al. Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Molecular Plant–Microbe Interactions. 2005;18:923–937. doi: 10.1094/MPMI-18-0923. [DOI] [PubMed] [Google Scholar]
- Devos KM, Gale MD. Genome relationships: the grass model in current research. The Plant Cell. 2000;12:637–646. doi: 10.1105/tpc.12.5.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Hill L, Crooks C, Doerner P, Lamb C. Abscisic acid has a key role in modulating diverse plant-pathogen interactions. Plant Physiology. 2009;150:1750–1761. doi: 10.1104/pp.109.137943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Current Opinion in Plant Biology. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Fukao T, Xu K, Ronald PC, Bailey-Serres J. A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. The Plant Cell. 2006;18:2021–2034. doi: 10.1105/tpc.106.043000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka S, Saka N, Koga H, et al. Loss of function of a proline-containing protein confers durable disease resistance in rice. Science. 2009;325:998–1001. doi: 10.1126/science.1175550. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annual Review of Phytopathology. 2005;43:5–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- Gu Y, Wildermuth MC, Chakravarthy S, Loh Y, Yang C, He X, Han Y, Martin GB. Tomato transcription factors Pti4, Pti5, and Pti6 activate defense responses when expressed in Arabidopsis . The Plant Cell. 2002;14:817–831. doi: 10.1105/tpc.000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Chen X, Wu X, Ling J, Xu P. Overexpression of the AP2/EREBP transcription factor OPBP1 enhances disease resistance and salt tolerance in tobacco. Plant Molecular Biology. 2004;55:607–618. doi: 10.1007/s11103-004-1521-3. [DOI] [PubMed] [Google Scholar]
- Hattori Y, Nagai K, Furukawa S, et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature. 2009;460:1026–1030. doi: 10.1038/nature08258. [DOI] [PubMed] [Google Scholar]
- Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiology. 2006;47:141–153. doi: 10.1093/pcp/pci230. [DOI] [PubMed] [Google Scholar]
- Jefferson R. Assaying chimeric genes in plants: the Gus gene fusion system. Plant Molecular Biology Rep. 1987:387–405. [Google Scholar]
- Jensen MK, Hagedorn PH, de Torres-Zabala M, Grant MR, Rung JH, Collinge DB, Lyngkjaer MF. Transcriptional regulation by an NAC (NAM-ATAF1, 2-CUC2) transcription factor attenuates ABA signalling for efficient basal defence towards Blumeria graminis f. sp hordei in Arabidopsis . The Plant Journal. 2008;56:867–880. doi: 10.1111/j.1365-313X.2008.03646.x. [DOI] [PubMed] [Google Scholar]
- Jiang C-J, Shimono M, Sugano S, Kojima M, Yazawa K, Yoshida R, Inoue H, Hayashi N, Sakakibara H, Takatsuji H. Abscisic acid interacts antagonistically with salicylic acid signaling pathway in rice-Magnaporthe grisea interaction. Molecular Plant–Microbe Interactions. 2010;23:791–798. doi: 10.1094/MPMI-23-6-0791. [DOI] [PubMed] [Google Scholar]
- Jung J, Won SY, Suh SC, Kim H, Wing R, Jeong Y, Hwang I, Kim M. The barley ERF-type transcription factor HvRAF confers enhanced pathogen resistance and salt tolerance in Arabidopsis . Planta. 2007;225:575–588. doi: 10.1007/s00425-006-0373-2. [DOI] [PubMed] [Google Scholar]
- Kanno Y, Otomo K, Kenmoku H, Mitsuhashi W, Yamane H, Oikawa H, Toshima H, Matsuoka M, Sassa T, Toyomasu T. Characterization of a rice gene family encoding type-A diterpene cyclases. Bioscience, Biotechnology, and Biochemistry. 2006;70:1702–1710. doi: 10.1271/bbb.60044. [DOI] [PubMed] [Google Scholar]
- Kato-Noguchi H, Ino T. Rice seedlings release momilactone B into the environment. Phytochemistry. 2003;63:551–554. doi: 10.1016/s0031-9422(03)00194-8. [DOI] [PubMed] [Google Scholar]
- Kodama O, Miyakawa J, Akatsuka T, Kiyosawa S. Sakuranetin, a flavanone phytoalexin from ultraviolet-irradiated rice leaves. Phytochemistry. 1992;31:3807–3809. [Google Scholar]
- Koga H, Dohi K, Mori M. Abscisic acid and low temperatures suppress the whole plant-specific resistance reaction of rice plants to the infection of Magnaporthe grisea . Physiological and Molecular Plant Pathology. 2004;65:3–9. [Google Scholar]
- Liang H, Lu Y, Liu H, Wang F, Xin Z, Zhang Z. A novel activator-type ERF of Thinopyrum intermedium, TiERF1, positively regulates defence responses. Journal of Experimental Botany. 2008;59:3111–3120. doi: 10.1093/jxb/ern165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HX, Zhu MZ, Yano M, Gao JP, Liang ZW, Su WA, Hu XH, Ren ZH, Chao DY. QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theoretical and Applied Genetics. 2004;108:253–260. doi: 10.1007/s00122-003-1421-y. [DOI] [PubMed] [Google Scholar]
- Mauch-Mani B, Mauch F. The role of abscisic acid in plant–pathogen interactions. Current Opinion in Plant Biology. 2005;8:409–414. doi: 10.1016/j.pbi.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Mialoundama AS, Heintz D, Debayle D, Rahier A, Camara B, Bouvier F. Abscisic acid negatively regulates elicitor-induced synthesis of capsidiol in wild tobacco. Plant Physiology. 2009;150:1556–1566. doi: 10.1104/pp.109.138420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr PG, Cahill DM. Abscisic acid influences the susceptibility of Arabidopsis thaliana to Pseudomonas syringae pv. tomato and Peronospora parasitica . Functional Plant Biology. 2003;30:461–469. doi: 10.1071/FP02231. [DOI] [PubMed] [Google Scholar]
- Mohr PG, Cahill DM. Suppression by ABA of salicylic acid and lignin accumulation and the expression of multiple genes, in Arabidopsis infected with Pseudomonas syringae pv. tomato . Functional Integrated Genomics. 2007;7:181–191. doi: 10.1007/s10142-006-0041-4. [DOI] [PubMed] [Google Scholar]
- Mur LAJ, Kenton P, Atzorn R, Miersch O, Wasternack C. The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiology. 2006;140:249–262. doi: 10.1104/pp.105.072348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiology. 2006;140:411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S-J, Kim YS, Kwon C-W, Park HK, Jeong JS, Kim J-K. Overexpression of the transcription factor AP37 in rice improves grain yield under drought condition. Plant Physiology. 2009;150:1368–1379. doi: 10.1104/pp.109.137554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñate-Sánchez L, Anderson JP, Young J, Singh KB. AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defence. Plant Physiology. 2007;143:400–409. doi: 10.1104/pp.106.086637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastori GM, Foyer CH. Common components, networks, and pathways of cross-tolerance to stress. The central role of ‘redox’ and abscisic acid-mediated controls. Plant Physiology. 2002;129:460–468. doi: 10.1104/pp.011021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pré M, Atallah M, Champion A, De Vos M, Pieterse CJ, Memelink J. The AP2/ERF-domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiology. 2008;147:1347–1357. doi: 10.1104/pp.108.117523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribot C, Hirsch J, Balzergue S, Tharreau D, Notteghem JL, Lebrun MH, Morel JB. Susceptibility of rice to the blast fungus, Magnaporthe grisea . Journal of Plant Physiology. 2008;165:114–124. doi: 10.1016/j.jplph.2007.06.013. [DOI] [PubMed] [Google Scholar]
- Rojo E, Solano R, Sanchez-Serrano JJ. Interactions between signaling compounds involved in plant defense. Journal of Plant Growth Regulation. 2003;22:82–98. [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochemical and Biophysical Research Communications. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Qin X, Zeevaart JAD. Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiology. 2003;131:1591–1601. doi: 10.1104/pp.102.017921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P. Nuclear activity of MLA immune receptors links isolate-specific and basal disease resistance responses. Science. 2007;315:1098–1103. doi: 10.1126/science.1136372. [DOI] [PubMed] [Google Scholar]
- Shimono M, Sugano S, Jiang CJ, Ono K, Toki S, Takatsuji H. Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. The Plant Cell. 2007;19:2064–2076. doi: 10.1105/tpc.106.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR. Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSEFACTOR1. Genes and Development. 1998;12:3703–3714. doi: 10.1101/gad.12.23.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Feng G, Tian C, Zhang F. Strategies for adaptation of Suaeda physophora, Haloxylon ammodendron and Haloxylon persicum to a saline environment during seed-germination stage. Annals of Botany. 2005;96:399–405. doi: 10.1093/aob/mci196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio A, Yang B, Zhu T, White FF. Two type III effector genes of Xanthomonas oryzae pv. oryzae control the induction of the host genes OsTFIIAγ1 and OsTFX1 during bacterial blight of rice. Proceedings of the National Academy of Sciences, USA. 2007;104:10720–10725. doi: 10.1073/pnas.0701742104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot NJ. On the trail of a cereal killer: exploring the biology of Magnaporthe grisea . Annual Review of Microbiology. 2003;57:177–202. doi: 10.1146/annurev.micro.57.030502.090957. [DOI] [PubMed] [Google Scholar]
- Tao Y, Xie Z, Chen W, Glazebrook J, Chang H-S, Han B, Zhu T, Zou G, Katagiri F. Quantitative nature of arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae . The Plant Cell. 2003;15:317–330. doi: 10.1105/tpc.007591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton J, Flors V, Mauch-Mani B. The multifaceted role of ABA in disease resistance. Trends in Plant Science. 2009;14:310–317. doi: 10.1016/j.tplants.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Trujillo LE, Sotolongo M, Menendez C, Ochogavia ME, Coll Y, Hernandez I, Borras-Hidalgo O, Thomma BP, Vera P, Hernandez L. SodERF3, a novel sugarcane ethylene responsive factor (ERF), enhances salt and drought tolerance when overexpressed in tobacco plants. Plant Cell Physiology. 2008;49:512–525. doi: 10.1093/pcp/pcn025. [DOI] [PubMed] [Google Scholar]
- VanEtten HD, Mansfield JW, Bailey JA, Farmer EE. Two classes of plant antibiotics: phytoalexins versus ‘phytoanticipins’. The Plant Cell. 1994;6:1191–1192. doi: 10.1105/tpc.6.9.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini C, Iriti M, Bracale M, Locatelli F, Faoro F, Croce P, Pirona R, Di Maro A, Coraggio I, Genga A. The ectopic expression of the rice Osmyb4 gene in Arabidopsis increases tolerance to abiotic, environmental and biotic stresses. Physiological and Molecular Plant Pathology. 2006;69:26–42. [Google Scholar]
- Vergne E, Ballini E, Marques S, et al. Early and specific gene expression triggered by rice resistance gene Pi33 in response to infection by ACE1 avirulent blast fungus. New Phytologist. 2007;174:159–171. doi: 10.1111/j.1469-8137.2007.01971.x. [DOI] [PubMed] [Google Scholar]
- Wang H, Hao J, Chen X, Hao Z, Wang X, Lou Y, Peng Y, Guo Z. Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Molecular Biology. 2007;65:799–815. doi: 10.1007/s11103-007-9244-x. [DOI] [PubMed] [Google Scholar]
- Xiang T, Zong N, Zou Y, et al. Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Current Biology. 2007;18:74–80. doi: 10.1016/j.cub.2007.12.020. [DOI] [PubMed] [Google Scholar]
- Xing DH, Lai ZB, Zheng ZY, Vinod KM, Fan BF, Chen ZX. Stress and pathogen-induced Arabidopsis WRKY48 is a transcriptional activator that represses plant basal defense. Molecular Plant. 2008;1:459–470. doi: 10.1093/mp/ssn020. [DOI] [PubMed] [Google Scholar]
- Xiong LZ, Yang YN. Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen activated protein kinase. The Plant Cell. 2003;15:745–759. doi: 10.1105/tpc.008714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaish MW, El-Kereamy A, Zhu T, Beatty PH, Good AG, Bi Y-M, Rothstein SJ. The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genetics. 2010 doi: 10.1371/journal.pgen.1001098. 6, pii, e1001098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Mundy J, Chua N-H. Four tightly linked rab genes are differentially expressed in rice. Plant Molecular Biology. 1990;14:29–39. doi: 10.1007/BF00015652. [DOI] [PubMed] [Google Scholar]
- Yang B, Sugio A, White FF. Os8N3 is a host disease susceptibility gene for bacterial blight of rice. Proceedings of the National Academy of Sciences, USA. 2006;103:10503–10508. doi: 10.1073/pnas.0604088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Li R, Qi M. In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. The Plant Journal. 2000;22:543–551. doi: 10.1046/j.1365-313x.2000.00760.x. [DOI] [PubMed] [Google Scholar]
- Yoo S-D, Cho Y, Sheen J. Emerging connections in the ethylene signaling network. Trends in Plant Science. 2009;14:270–279. doi: 10.1016/j.tplants.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang D, Chen J, Yang Y, Huang Z, Huang D, Wang XC, Huang R. Tomato stress-responsive factor TSRF1 interacts with ethylene responsive element GCC box and regulates pathogen resistance to Ralstonia solanacearum . Plant Molecular Biology. 2004;55:825–834. doi: 10.1007/s11103-004-2140-8. [DOI] [PubMed] [Google Scholar]
- Zhou J, Zhang H, Yang Y, Zhang Z, Zhang H, Hu X, Chen J, Wang X-C, Huang R. Abscisic acid regulates TSRF1-mediated resistance to Ralstonia solanacearum by modifying the expression of GCC box-containing genes in tobacco. Journal of Experimental Botany. 2008;59:645–652. doi: 10.1093/jxb/erm353. [DOI] [PubMed] [Google Scholar]
- Zhou J-M, Chai J. Plant pathogenic bacterial type III effectors subdue host responses. Current Opinion in Microbiology. 2008;11:179–185. doi: 10.1016/j.mib.2008.02.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.